Abstract
The adenovirus E4 open reading frame 4 (E4orf4) protein is a multifunctional viral regulator that is involved in the temporal regulation of viral gene expression by modulating cellular and viral genes at the transcription and translation levels and by controlling alternative splicing of adenoviral late mRNAs. When expressed individually, E4orf4 induces apoptosis in transformed cells. Using oligonucleotide microarray analysis, validated by quantitative real time PCR, we found that MYC (also known as c-Myc) is downregulated early after the induction of E4orf4 expression. As a result, Myc protein levels are reduced in E4orf4-expressing cells. MYC downregulation is observed both when E4orf4 is expressed individually and within the context of viral infection. E4orf4 reduces MYC transcription but does not affect transcriptional elongation or RNA stability. An interaction with the PP2A-B55 subunit is required for the downregulation of MYC by E4orf4. Since Myc overexpression was previously shown to inhibit adenovirus replication, the downregulation of Myc by E4orf4 would contribute to efficient virus infection.
Efficient virus replication requires the reprogramming of the host cell during viral infection. One important mechanism by which viruses rewire the cell is the modulation of cellular transcription. A number of reports have shown that adenovirus infection leads to alterations in the transcription of several hundreds, if not thousands, of genes. The differentially expressed genes belong to several Gene Ontology groups and are altered during various stages of viral infection (33, 47). One of the genes that are downregulated during adenovirus infection is MYC (27, 47).
The MYC family of genes encodes multifunctional regulators of cellular mechanisms, which play a role in most major cellular processes including cell proliferation, cell growth, differentiation, genomic stability, cell motility, and cell adhesion. Myc proteins can also induce apoptosis when overexpressed in the presence of limiting survival signals or cell stress and can induce cellular senescence in human fibroblasts (reviewed in reference 1). Myc is a transcription factor, which can both activate and repress the transcription of its target genes. Myc activates transcription when bound to its DNA consensus sequence (the E box) but represses transcription when tethered to promoters by other proteins (1). It has been estimated that Myc can bind ∼25,000 sites in the human genome and regulate up to 10 to 15% of human genes (9, 25), underscoring the importance of Myc as a fundamental cellular regulator. It is not surprising, therefore, that Myc is targeted during the infection of several viruses, including adenovirus. Using various virus mutants, it was previously reported that the adenovirus E1B-55-kDa and E4orf6 proteins are partially responsible for reducing MYC steady-state RNA levels and that the deletion of E4orf3 enhances the downregulation of MYC RNA. Since Myc overexpression was reported to inhibit adenovirus replication, a reduction in MYC levels would counteract the detrimental effect of Myc on viral infection (27). However, the mechanisms underlying the negative regulation of MYC RNA and additional viral proteins participating in this mode of regulation have not previously been identified.
The adenovirus E4orf4 protein has been shown to downregulate the expression of the cellular AP-1 transcription factor, consisting of c-fos and JunB, as well as the expression of early viral genes (6, 19, 29, 36). The timely downregulation of these genes contributes to the temporal control of the progression of adenovirus infection. The effect of E4orf4 on gene expression was generated by both transcriptional and translational mechanisms (36), and E4orf4 was further shown to affect protein translation through an interaction with the mTOR pathway (38). In addition, E4orf4 was reported to induce the hypophosphorylation of various viral and cellular proteins (16, 36), to regulate the alternative splicing of adenovirus mRNAs (16), and to induce p53-independent apoptosis in transformed cells (22, 31, 40). All E4orf4 functions known to date are mediated by an interaction between E4orf4 and cellular protein phosphatase 2A (PP2A) (17, 38).
PP2A is usually composed of three subunits, a catalytic C subunit, a scaffolding A subunit, and one of several regulatory B subunits (Bα-δ/B55, B′α-ɛ/B56, B" [PR72, PR130, PR59, and PR48], and B‴) encoded by unrelated gene families. The B subunits were previously reported to dictate the cellular localization and substrate specificity of the phosphatase (reviewed in reference 43). We previously showed that E4orf4 interacts with PP2A through a direct association with the phosphatase Bα/B55 regulatory subunit (19) or the B′/B56 subunit (42). Although the interaction of E4orf4 with an active PP2A enzyme is required for all E4orf4 functions known to date, different functions may be mediated by interactions with different PP2A complexes. For example, PP2A holoenzymes containing the Bα/B55 subunit, but not those containing the B′/B56 subunit, mediate E4orf4-induced apoptosis (42).
In this work, we show that E4orf4 is one of the adenovirus proteins that downregulate Myc expression by collaborating with a PP2A complex containing the Bα/B55 subunit to decrease MYC transcription.
MATERIALS AND METHODS
Cell lines, plasmids, and viral mutants.
Cell lines containing tetracycline-inducible E4orf4 (clone 13) or an empty vector (T-REX) were prepared using the T-Rex tetracycline-regulated mammalian expression system (Invitrogen) in HEK293 (10) and H1299 (34) cells. These cell lines were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum guaranteed to be tetracycline free (BD Bioscience), 5 μg/ml blasticidine (Invitrogen), and 200 μg/ml zeocin (Invitrogen). Cell lines expressing a PP2A-B55 short hairpin RNA (shRNA) (IR-B55) or containing an empty vector (NTO) were derived from 293 cells by cotransfection of pSuper-Bα (44) or the empty vector, together with pBabePuro (35), followed by a 2-week selection in medium containing 1 μg/ml puromycin.
Additional plasmids used in this work include pcDNA4/TO, pcDNA6/TR (Invitrogen), pcDNA4/TO-E4orf4, pCMV (13), pCMV-E4orf4 (19), pCMV-PP2A-B55-HA (41), pLS153/6 (A. Krumm), MYC Del-1 luciferase (11), pRL-0 Renilla luciferase (Promega), and pEGFP-C1 (BD Bioscience). A PP2A-B55 mutant resistant to the PP2A-B55 shRNA was generated using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's protocols. The primers used were forward primer GTCTGTGCAAGTGGCAAGCGTAAAAAGGATGAAATAAGTGTTGACAGCC and reverse primer GGCTGTCAACACTTATTTCATCCTTTTTACGCTTGCCACTTGCACAGAC.
Adenoviral mutants dl366*, lacking the complete E4 region, and dl366*+E4orf4, lacking all E4 open reading frames (ORFs) except E4orf4, were previously described (14). Infections were carried out at 10 PFU/cell.
RNA preparation, microarray analysis, quantitative real time PCR, and semiquantitative reverse transcription (RT)-PCR.
Total RNA was prepared from control and doxycycline-induced cells at various times postinduction (two independent plates per sample) using Trizol reagent (Invitrogen). The RNA was analyzed by an Affymetrix Human Genome Focus array (see protocols at http://www.affymetrix.com/support/technical/manual/expression_manual.affx). The 8,757 probe sets contained in the Affymetrix Human Focus oligonucleotide array were filtered using the MAS 5 algorithm. A list of 4,994 “valid” probe sets was compiled, representing probe sets with signals higher than 20, and detected as being present (P) in at least one sample. Treated and control samples were compared in the various time points. The comparison generated a list of “active genes” representing probe sets changed by at least twofold (as calculated from the MAS 5 log ratio values of ≥1 or ≤−1), and these were detected as being “increased” or “decreased” (P value of 0.0025) or as being “marginally increased” or “marginally decreased” (P value 0.003) in treated samples compared to control samples in at least one time point. This list excluded upregulated genes in all treated samples with signals lower than 20 or detected as being absent and also excluded downregulated genes with baseline signals lower than 20 or detected as being absent in the control samples.
For quantitative real time PCR experiments, 2 μg RNA was reverse transcribed using random hexamers (Promega) and SuperScript II (Invitrogen). Fifty nanograms of the cDNA was used for quantitative PCR with specific primers, fluorescence-labeled nucleotides, and a specific probe (Assay on Demand; Applied Biosystems). The fluorescent PCR products were quantified using ABI Prism 7000, and the results were analyzed using ABI Prism 7000 software. Results for the relevant genes were normalized to results for the housekeeping gene GAPDH, which served as a control.
For semiquantitative RT-PCR, RNA was reverse transcribed and amplified using the Reverse-iT kit (ABgene, Epsom, United Kingdom). The primers used included MYC exon 1 forward primer GGGATCGCGCTGAGTATAAAA, MYC exon 1 reverse primer GAAGCCCCCTATTCGCTCC, MYC exon 3 forward primer TCGGAAGGACTATCCTGCTG, MYC exon 3 reverse primer GTGTGTTCGCCTCTTGACATT, GAPDH forward primer ACCCCTTCATTGACCTCAACT, and GAPDH reverse primer ATGCCAGTGAGCTTCCCGTT. Samples were taken during various amplification cycles, and DNA was chromatographed on agarose gels.
Transfections, Western blot analysis, and antibodies.
Cells were plated in 60-mm culture dishes, and transfections were carried out using the JetPie reagent (Polyplus-Transfection Inc., New York, NY). Cells were harvested 24 or 48 h later, and cell extracts were prepared in lysis buffer (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.5% Nonidet P-40, and a 1/10 volume of Complete protease inhibitor cocktail [Roche]). Proteins were analyzed by Western blot analysis and chemiluminescence using antibodies against the following proteins or protein tags: E4orf4 (40), PP2A-B55 (41), c-Myc (9E10 or C33; Santa Cruz), α-tubulin (Sigma), and hemagglutinin (HA) (Covance). Blots were scanned and images were processed using Adobe Photoshop.
Nuclear run-on experiments.
Extraction of nuclei, transcription reactions, and the hybridization procedure were carried out as described previously (18) but have been modified in several ways. T-REX and clone 13 cells were induced with 1 μg/ml doxycycline for 6 h. Cells were harvested (2 × 107 cells per sample), and nuclei were prepared in 1 ml NP-40 lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% [vol/vol] NP-40), pelleted by centrifugation at 500 × g for 5 min, gently resuspended in 100 μl of nucleus storage buffer (50 mM Tris-HCl [pH 8.5], 5 mM MgCl2, 0.1 mM EDTA, 40% [vol/vol] glycerol), immediately frozen in liquid nitrogen, and stored at −80°C for later use. Nuclear DNA was transcribed in vitro by adding an equal volume of 2× reaction mix (50 mM HEPES [pH 7.5]; 5 mM MgCl2; 5 mM dithiothreitol; 300 mM KCl; 10% glycerol; 1 mM concentration each of ATP, GTP, and CTP; and 100 μCi of an 800 Ci/mmol [α-32P]UTP stock [GE Healthcare]) to the thawed nuclei followed by 30 min of incubation at room temperature. DNase I was added to a final concentration of 10 μg/ml, and the incubation was continued for 10 min at 37°C. Radioactively labeled RNA was purified with Trizol reagent according to the manufacturer's instructions, precipitated with ethanol, and resuspended in diethylpyrocarbonate-treated water containing 50% formamide. The labeled RNA was boiled for 5 min, chilled on ice, and hybridized to DNA immobilized on a nitrocellulose membrane (Schleicher & Schuell) in 3 ml of hybridization buffer (50% formamide, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10× Denhardt's solution, 0.2% sodium dodecyl sulfate) for 72 h at 42°C. After hybridization, the filters were washed twice at room temperature in 2× SSC for 5 min, twice at 60°C in 2× SSC for 30 min, once at 37°C in 2× SSC containing 10 μg/ml RNase A for 30 min, and twice at room temperature in 2× SSC-0.1% sodium dodecyl sulfate for 15 min. The filters were exposed to a phosphoscreen, and signals were scanned using a phosphorimager. TINA software was used to quantify the results.
RESULTS
Microarray analysis identified MYC as being an early downregulated target of E4orf4.
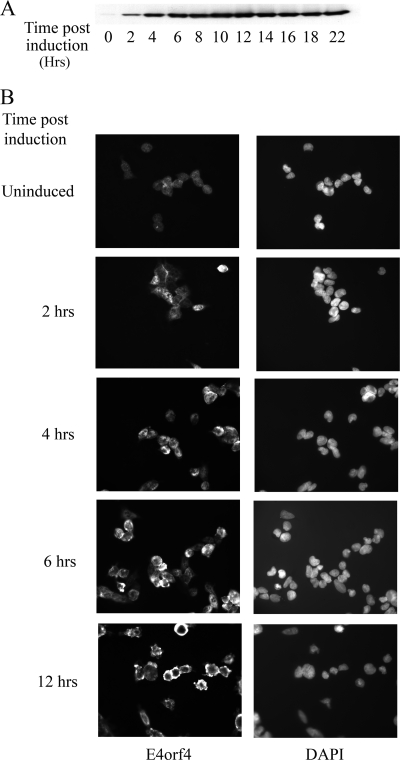
HEK293-derived cell lines, either expressing E4orf4 from a tetracycline-inducible promoter (clone 13) or containing an empty vector (T-REX), were constructed. E4orf4 expression was induced rapidly in clone 13 cells following the addition of doxycycline (Fig. 1A), and membrane blebbing, an early manifestation of E4orf4-induced apoptosis, was detected starting 6 h after the addition of doxycycline (Fig. 1B). To find early transcription events that may be induced by E4orf4, an Affymetrix microarray was used to examine differentially expressed genes at early times after the induction of E4orf4. RNA was prepared from clone 13 cells treated with doxycycline for 4, 6, or 10 h; from clone 13 cells treated with ethanol (used to dissolve doxycycline) (uninduced) for 10 h; or from the control cell line treated with either ethanol or doxycycline for 10 h. The six RNA samples were subjected to microarray analysis, and the Affymetrix MAS 5 algorithm was used to analyze the results. The expression of 4,994 genes out of the 8,757 human sequences present on the Affymetrix Human Genome Focus array could be detected in at least one of the tested samples. The expression of each gene was normalized to its expression in the uninduced clone 13. We chose genes with a greater-than-twofold change between induced and uninduced clone 13 cells and discarded those that manifested a twofold or higher difference between induced and uninduced control cells or whose expression was significantly different in uninduced clone 13 and control cell lines. Of the genes that were affected at the earliest time point, the expression of the MYC gene (also known as c-Myc) was found to be affected the most, showing a 12.7-fold decrease at 4 h and remaining low, with an 8.1-fold reduction at 6 h and a 15.7-fold reduction at 10 h.
FIG. 1.
Induction of E4orf4 expression in clone 13 cells. E4orf4 expression was induced in clone 13 cells by 1 μg/ml doxycycline. At the indicated times postinduction, cells were either harvested and subjected to Western blot analysis with E4orf4-specific antibodies (A) or fixed and stained with E4orf4-specific antibodies and with DAPI (4′,6′-diamidino-2-phenylindole) (B).
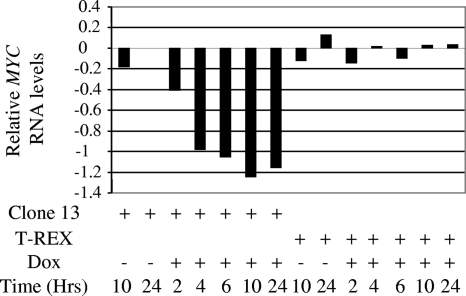
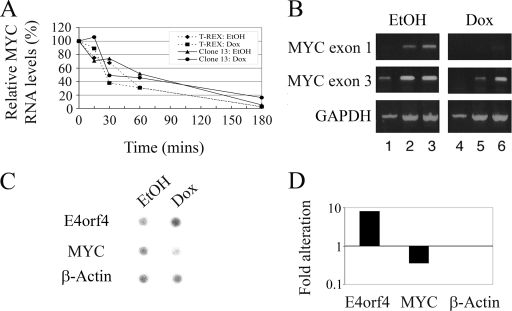
To validate the microarray results, quantitative real time PCR was performed in triplicates, and results for MYC expression were normalized to GAPDH levels. The results were further normalized to those of clone 13 cells treated with ethanol for 24 h. As seen in Fig. 2, MYC RNA levels started to decrease at 2 h post-E4orf4 induction (2.6-fold reduction), with a rapid decrease between 2 and 4 h (down to 9.5-fold reduction), which paralleled a significant increase in cellular E4orf4 levels but which preceded early manifestations of apoptosis (Fig. 1). MYC levels remained low at least up to 24 h postinduction. No significant changes in MYC expression were observed in the control cell line, which was subjected to similar induction, or in uninduced clone 13 cells.
FIG. 2.
Quantitative real time PCR analysis of MYC RNA levels following induction of E4orf4 expression. Clone 13 and control T-REX cells were treated with 1 μg/ml doxycycline (Dox) or with the ethanol vehicle. Two plates of cells were harvested at each of the indicated times postinduction, and RNA was prepared. The various RNA samples were subjected to quantitative real time PCR analysis using primers for MYC and GAPDH. MYC RNA levels were normalized to those of the housekeeping GAPDH RNAs, and relative quantification was obtained by further normalizing these results to MYC levels in clone 13 cells treated with ethanol for 24 h. Relative MYC RNA levels representing log10(relative quantification) are shown.
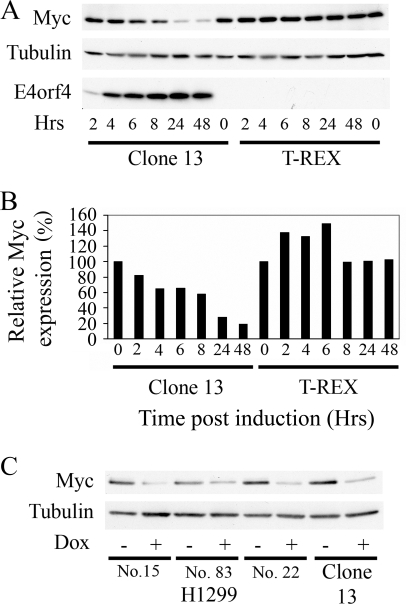
To determine whether Myc protein levels were altered following the induction of E4orf4 expression, Western blot analysis was performed using protein extracts from clone 13 and the control cell line, and the signals were quantified by densitometry. As shown in Fig. 3A and B, Myc protein levels gradually decreased following the induction of E4orf4 in clone 13 cells. They were reduced 1.5-fold at 4 h postinduction and 1.7-fold at 8 h and reached minimal levels at 24 h (3.5-fold) and 48 h (5.3-fold). Myc levels in the control cell line were not reduced and were even increased early after induction with doxycycline. No significant changes in the levels of α-tubulin were observed in both cell lines. To find whether Myc levels were reduced in other cells following the induction of E4orf4 expression, three H1299-derived cell lines expressing E4orf4 from the tetracycline-inducible promoter were examined. As shown in Fig. 3C, Myc levels decreased in the H1299-derived cell lines 24 h after E4orf4 induction, similarly to their decrease in clone 13 cells.
FIG. 3.
Downregulation of Myc protein upon induction of E4orf4 expression. (A) Clone 13 and T-REX cells were subjected to treatment with 1 μg/ml doxycycline (Dox) for the indicated times in hours (Hrs). Cells were then harvested, and proteins were subjected to Western blot analysis with antibodies to Myc, α-tubulin, and E4orf4. (B) Results of the Western blot stained with Myc-specific antibodies were quantified by densitometry. Values at time zero of induction in both clone 13 and T-REX cells were defined as 100%, and relative Myc protein levels are plotted for each time point. (C) E4orf4 expression was induced by a 24-h treatment of various cell clones derived from H1299 cells and the 293-derived clone 13 cells with 1 μg/ml doxycycline (Dox). Proteins were then extracted and subjected to Western blot analysis with Myc- and α-tubulin-specific antibodies.
Study of the contribution of E4orf4 to MYC downregulation within the context of virus infection.
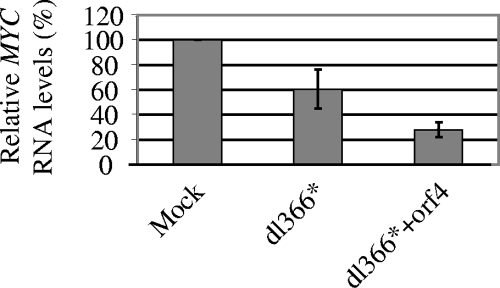
It was previously reported that an adenovirus infection leads to the downregulation of MYC RNA levels, thus possibly counteracting the detrimental stabilization of the Myc protein by the adenovirus E1A protein (27). Deletions of E1B-55-kDa and E4orf6 proteins partially inhibited MYC downregulation but were not sufficient for fully preventing it (27). In contrast, another report showed that individually expressed E4orf6 contributes to the accumulation of cytoplasmic MYC RNAs (12). To determine whether E4orf4 contributes to MYC downregulation in the context of a viral infection, we used viral mutants lacking the E4 region, thus eliminating the effect of other E4 ORFs. H1299 cells were either mock infected or infected with an adenovirus lacking the E4 region (dl366*) or with an adenovirus lacking all E4 ORFs except E4orf4 (dl366*+orf4). RNA was harvested from cells 6 h postinfection, quantitative real time PCR was carried out with primers to MYC and to GAPDH, and MYC levels were normalized to GAPDH levels. In two independent experiments, MYC levels were reduced 1.5- to 2.2-fold following infection with dl366* virus compared to mock infection, and the addition of E4orf4 (dl366*+orf4) reduced these levels by a further 2.1- to 2.2-fold (Fig. 4). These results indicate that E4orf4 contributes to the adenovirus-induced downregulation of MYC, although other viral products, such as the E1B-55-kDa protein, may also be involved.
FIG. 4.
E4orf4-induced downregulation of MYC RNA levels in the context of virus infection. H1299 cells were mock infected or infected with dl366* and dl366*+orf4 viruses at 10 PFU/cell. RNAs were prepared from these cells 6 h postinfection and subjected to quantitative real time PCR with primers to MYC and to GAPDH. MYC RNA levels were normalized to GAPDH levels, and the resulting relative quantification is shown as a percentage of MYC levels in mock-infected cells. The graph shows the average of two independent experiments, and error bars represent standard deviations.
Investigation of mechanisms underlying E4orf4-induced MYC downregulation.
We have used several approaches to determine the mechanisms underlying the E4orf4-induced downregulation of MYC expression. First, we tested whether E4orf4 affected MYC RNA stability. Actinomycin D was added to control cells (T-REX) and to E4orf4-expressing clone 13 cells 5 h after the induction of E4orf4. RNA was extracted from the cells at various times after the addition of the drug, and analysis of MYC RNA levels was carried out by quantitative real time PCR using GAPDH RNA as a control. MYC levels present in each class of treated cells prior to the addition of actinomycin D were defined as 100%. As shown in Fig. 5A, E4orf4 did not decrease MYC RNA stability.
FIG. 5.
Investigation of the mechanisms underlying E4orf4-induced MYC downregulation. (A) T-REX and clone 13 cells were treated with 1 μg/ml doxycycline (Dox) or with ethanol (EtOH) vehicle for 5 h. Actinomycin D was then added (5 μg/ml), and cells from two plates per each point were harvested at the indicated times after the addition of the drug. RNA was prepared and subjected to quantitative real time PCR analysis with primers to MYC and the housekeeping GAPDH gene. Relative quantification was obtained by normalizing MYC results to GAPDH levels. The values at the time of addition of actinomycin D (time zero) were defined as 100% for each group of treated cells. Broken line with diamonds, T-REX with EtOH treatment; broken line with squares, T-REX with doxycycline treatment; solid line with triangles, clone 13 with EtOH treatment; solid line with circles, clone 13 with doxycycline treatment. (B) RNAs were extracted from clone 13 cells treated with ethanol or 1 μg/ml doxycycline for 6 h and were subjected to semiquantitative RT-PCR analysis with primers to MYC exon 1 or 3 and to GAPDH. Amplified cDNAs were taken for agarose gel analysis after 21 (lanes 1 and 4), 24 (lanes 2 and 5), and 27 (lanes 3 and 6) amplification cycles for the MYC reactions and after 17 (lanes 1 and 4), 22 (lanes 2 and 5), and 25 (lanes 3 and 6) amplification cycles for the GAPDH reaction. (C) Clone 13 cells were treated with ethanol or 1 μg/ml doxycycline for 6 h, and nuclei were prepared and subjected to nuclear run-on analysis. Hybridization of the 32P-labeled RNA products to a filter carrying DNA sequences of E4orf4, MYC, and β-ACTIN was visualized by use of a PhosphorImager. (D) Results of C were quantified using TINA software. The alteration shown here using a logarithmic scale is the ratio between intensities of hybridization for doxycycline- and EtOH-treated cells normalized to the ratio for the control β-ACTIN gene.
It was previously reported that MYC RNA synthesis can be regulated at the level of transcriptional elongation. Thus, sequences found in the intron between the first and second exons can cause premature transcription termination (28, 39). Since we examined MYC RNA levels using primers from the second and third exons in previous experiments, it was possible that the observed alterations reflected an E4orf4 effect on transcription elongation and that sequences of the first exon would be equally represented in control and E4orf4-expressing cells. However, performing semiquantitative RT-PCRs with primers from both exon 1 and exon 3 revealed that the expression of both exons was similarly reduced in E4orf4-expressing cells (Fig. 5B). These results suggest that E4orf4 does not inhibit MYC transcriptional elongation through sequences in intron 1.
Next, we used a gene reporter assay to test whether E4orf4 affected MYC transcription. The reporter plasmid contained a 2.5-kb genomic fragment encompassing the MYC promoter and sequences including close to 300 bp downstream of the major P2 transcription start site. This promoter element was inserted upstream of a luciferase reporter gene (11). The reporter plasmid was introduced into 293 cells in the presence or absence of E4orf4 or into clone 13 cells, induced or uninduced by doxycycline. A plasmid expressing a Renilla luciferase was cotransfected into the same cells and served as a transfection control. However, no E4orf4-induced changes in luciferase expression were observed in any of the systems tested (results not shown).
It is possible that E4orf4 affects MYC transcription, but the E4orf4-responsive element lies further upstream of the transcription start site. Alternatively, the response to E4orf4 may require changes in chromatin that cannot be reproduced by the transient expression of reporter plasmids. To address the first possibility, we cloned a longer 7-kb upstream MYC promoter sequence next to the luciferase reporter described above. However, similarly to the shorter promoter sequence, this segment did not respond to E4orf4 expression in transient assays (results not shown). It remains to be seen whether further upstream sequences are required to mediate E4orf4-induced MYC downregulation. To address the second possibility, we generated stable cell lines containing a luciferase reporter linked to the 2.5-kb or the 7-kb MYC promoter element. However, the introduction of E4orf4 into these cells did not reduce luciferase activity (results not shown). Thus, we could not reconstitute the E4orf4-induced reduction of MYC expression with exogenous MYC promoter sequences.
To examine in vivo whether MYC transcription was downregulated by E4orf4, nuclear run-on assays were performed. Clone 13 cells were induced by doxycycline or with ethanol vehicle as a control, and nuclei were harvested 6 h later and subjected to run-on analysis as described previously (18). The intensity of hybridization of the 32P-labeled RNA products to a filter carrying DNA sequences of E4orf4, MYC, β-ACTIN, and an empty plasmid was quantified using a PhosphorImager. For each treatment (ethanol or doxycycline), the hybridization intensity of the empty plasmid was subtracted from all points, and results were normalized to β-ACTIN, whose transcription changed very little upon doxycycline induction (reduced by 20%). Figure 5C and D demonstrates that whereas E4orf4 transcription was enhanced 8-fold following induction by doxycycline, MYC transcription was reduced 2.9-fold. Thus, E4orf4-induced downregulation of MYC expression is carried out, at least in part, on the transcriptional level.
The PP2A-B55 subunit is required for the E4orf4-induced downregulation of MYC expression.
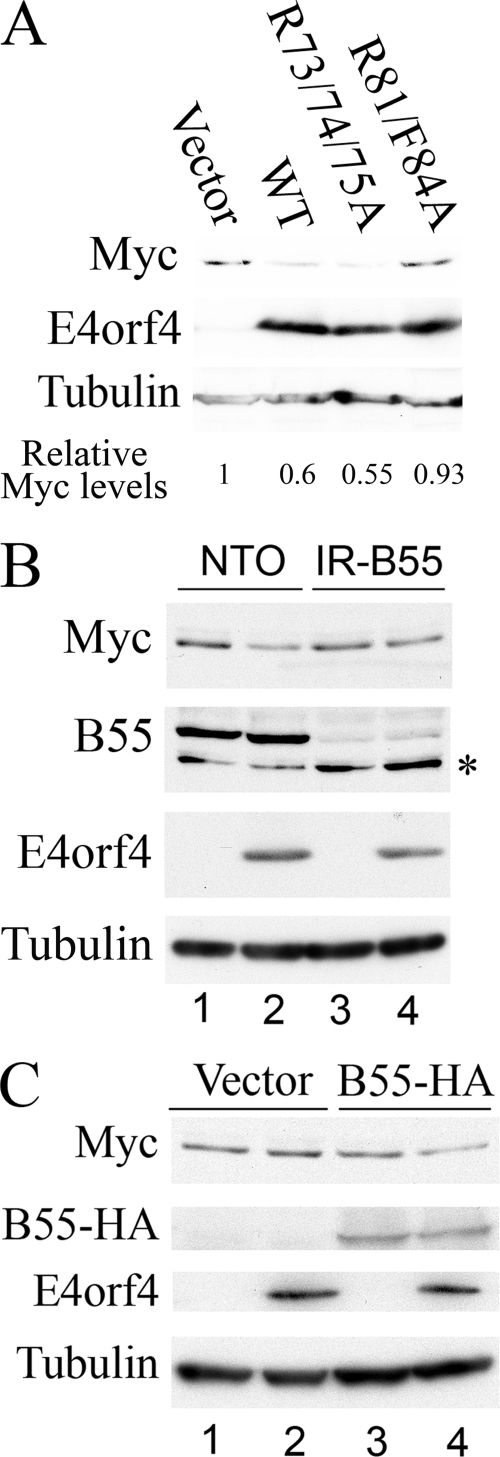
Since it was previously shown that PP2A participates in all known functions of E4orf4, we investigated whether PP2A is involved in the E4orf4-induced downregulation of MYC expression. Constructs expressing wild-type (WT) E4orf4 and E4orf4 mutants unable to bind either PP2A (R81/F84A) (30) or Src (the R4 mutant [R73/74/75A]) (7, 41) were transfected into 293 cells at a high efficiency, and Myc protein levels were analyzed by Western blots 2 days posttransfection. As shown in Fig. 6A, WT E4orf4 and the R4 mutant (R73/74/75A), which binds PP2A but not Src (7, 41), reduced Myc protein levels efficiently, whereas a mutant that does not bind PP2A (R81/F84A) (30) did not reduce Myc levels. These results suggested that PP2A binding may be required for the downregulation of Myc by E4orf4. To further establish the involvement of PP2A in the E4orf4-induced downregulation of Myc expression, we generated a 293-derived cell line in which the expression of the PP2A-B55 subunit was knocked down using an appropriate shRNA construct (IR-B55) and a negative control cell line expressing wild-type levels of the B55 subunit (NTO). Since the transfection of IR-B55 cells was consistently less efficient than the transfection of NTO cells, the two cell lines were transfected with E4orf4 and a green fluorescent protein transfection control, and only green fluorescent protein-transfected cells were collected for analysis using the BD FACSAria cell sorting system. As shown in Fig. 6B, E4orf4 reduced the levels of the Myc protein in the control cell line expressing PP2A-B55 but not in the cell line in which PP2A-B55 was knocked down. To verify that the different effects on Myc expression in NTO and IR-B55 cells can be attributed to the presence or absence of the PP2A-B55 subunit, we prepared a PP2A-B55 mutant cDNA in which silent mutations caused resistance to knockdown by the PP2A-B55 shRNA construct. When this mutant was cotransfected into IR-B55 cells with E4orf4, E4orf4-induced Myc downregulation was restored (Fig. 6C). Thus, we conclude that an interaction with the B55-containing PP2A heterotrimer is required for the downregulation of Myc by E4orf4.
FIG. 6.
The PP2A-B55 subunit is required for E4orf4-induced downregulation of Myc expression. (A) 293 cells were transfected with an empty vector, WT E4orf4, or the E4orf4 R4 (R73/74/75A) or R81/F84A mutant. Cells were harvested 24 h later, and proteins were subjected to Western blot analysis sequentially stained with antibodies to Myc, E4orf4, and α-tubulin. Densitometry was used to quantify Myc and α-tubulin levels, and Myc levels were normalized to α-tubulin levels. Myc expression in vector-transfected cells was defined as 1, and relative Myc levels are shown below the blot. (B) NTO (lanes 1 and 2) and IR-B55 (lanes 3 and 4) cells were transfected with WT E4orf4 (lanes 2 and 4) or an empty vector (lanes 1 and 3) and with a plasmid expressing green fluorescent protein. Cells were harvested 24 h later, and green fluorescent protein-transfected cells were collected using a FACSAria cell sorter. Proteins were extracted and subjected to West-ern blot analysis and sequentially stained with antibodies to Myc, PP2A-B55, E4orf4, and α-tubulin. The asterisk marks a nonspecific protein band. (C) IR-B55 cells were transfected with plasmids expressing a PP2A-B55-HA mutant resistant to the B55 shRNA (lanes 3 and 4), E4orf4 (lanes 2 and 4), or empty vectors (lane 1). Cells were harvested 24 h later, and proteins were subjected to Western blot analysis and sequentially stained with antibodies to Myc, the ΗΑ tag, Ε4orf4, and α-tubulin.
DISCUSSION
We have shown here that the adenovirus E4orf4 protein downregulates MYC at the transcriptional level (Fig. 5) and that an interaction with a PP2A holoenzyme containing the B55 subunit is required for this process (Fig. 6). MYC downregulation is observed both when E4orf4 is expressed individually (Fig. 2 and 3) and within the context of viral infection (Fig. 4).
The cellular regulation of MYC is highly intricate and involves several layers of complexity, including the regulation of transcription initiation and elongation, RNA processing, RNA stability, translation, and protein degradation (reviewed in reference 8). This complexity facilitates the response of the MYC gene to numerous incoming signals, allowing it to serve as a hub of cellular regulation. Each combination of physiological conditions may result in different Myc levels leading to a different cellular outcome.
The downregulation of MYC by E4orf4 is carried out at the transcriptional level (Fig. 5) and not through changes in RNA stability or as a result of changes in premature RNA termination. Interestingly, however, the effect of E4orf4 on MYC transcription could not be reconstituted by promoter-driven reporter assays. It was indeed previously reported that several features of MYC expression could not be recapitulated using exogenous MYC promoter sequences, whether transiently or stably transfected, integrated, or episomal, as well as transgenes passaged through the germ line in mice (26). Thus, it is possible that E4orf4 affects MYC transcription through sequences that lie far upstream of the MYC transcription start site or that E4orf4 affects a unique chromatin conformation typical of the MYC locus, which cannot be reconstituted by short MYC sequences inserted elsewhere in the genome.
The kinetics of the downregulation of MYC RNA and Myc protein appeared to differ. RNA levels were reduced 2.6-fold at 2 h after induction and 9.5-fold at 4 h after induction of E4orf4 (Fig. 2). This significant decrease in MYC RNA levels precedes early manifestations of E4orf4-induced apoptosis and is thus likely to be a direct E4orf4 effect and not an indirect effect of cell death. The reduction in Myc protein levels was slower and reached 1.7-fold at 8 h, 3.5-fold at 24 h, and 5.3-fold at 48 h (Fig. 3). The slower decrease in Myc protein levels could be partially explained by an increased stability of the Myc protein in clone 13 cells derived from the E1A-expressing 293 cell line. E1A was previously shown to stabilize the Myc protein (27), thus delaying the reduction in protein levels. In addition, it was shown that E4orf4 itself could stabilize the ectopically expressed Myc protein, possibly through its interaction with a PP2A complex containing the B′/B56 subunit (2). Thus, E4orf4 appears to have opposing effects on Myc levels by interacting with two different PP2A complexes: the interaction with a B55-containing PP2A holoenzyme mediates the downregulation of MYC transcription as shown here, whereas the interaction with a B56-containing PP2A complex may mediate the stabilization of the Myc protein (2). Opposing effects generated by viral regulators were described previously and are not unique to E4orf4. For example, E1A can both stabilize the p53 tumor suppressor and repress p53 transcriptional activity (reviewed in reference 4). E4orf4 itself was shown to inhibit the activity of the APC/C ubiquitin ligase while simultaneously activating its activator, Cdc28 (21). Such opposing effects may facilitate the fine-tuning of the cellular response to viral infection, modulated by various sets of physiological conditions.
Since Myc is a key cellular regulator, it is not surprising that it serves as a major target for numerous viruses. Examples include viruses that alter Myc expression levels such as adenovirus (this work; 27), polyomavirus (20, 48), parvoviruses (23), cytomegalovirus (5), and human immunodeficiency virus (45); viruses encoding proteins that alter Myc protein activity or stability (3, 15, 46); or viral proteins that affect MYC mRNA transport to the cytoplasm and its stabilization (12). Similarly to the participation of PP2A in the virus-induced downregulation of MYC shown here (Fig. 6), other viruses were also shown to affect MYC expression through interactions with PP2A. Examples include the upregulation of MYC RNA levels by polyomavirus small T antigen (20) and Myc protein stabilization by simian virus 40 small T antigen (46).
Adenovirus has been shown to encode different proteins that target Myc, including E1A, which stabilizes the Myc protein (27); E4orf6, which, when expressed individually, enhances the export of AV-rich element-containing mRNAs including MYC mRNA to the cytoplasm and stabilizes these mRNAs (12); and E4orf4 and possibly other adenoviral proteins, which significantly reduce MYC RNA levels in adenovirus-infected cells (this work; 27). The finding that several proteins encoded by the same virus target MYC at different levels, as exemplified by adenovirus, underscores the potential importance of MYC to virus replication and is reminiscent of the multilevel targeting of p53 during viral infection (reviewed in reference 4). It has indeed been shown that overexpressed Myc inhibits adenovirus replication, interferes with the proper formation of viral replication centers, and decreases virus yield (27), indicating that the downregulation of Myc by adenovirus proteins, including E4orf4, is beneficial for viral multiplication. On the other hand, it was previously shown that E4orf4 inhibits viral DNA synthesis in a certain genetic background, whereas E4orf3 and E4orf6 antagonize this effect (32). E4orf3 also appears to antagonize the downregulation of MYC RNA levels, since an E4orf3 mutant (E4inorf3) caused a further dramatic reduction in MYC levels compared with those of WT adenovirus (27). E4orf6, however, was reported to contribute to the downregulation of MYC expression within the context of viral infection (27). Thus, Myc is probably not the only target of the E4 ORFs affecting adenoviral DNA replication.
It has been shown that Myc has a biphasic effect on the adenovirus major late promoter (MLP) in vitro. At lower concentrations, Myc upregulates the MLP through E-box Myc binding sites; however, at higher concentrations, Myc represses transcription by a mechanism dependent on initiator elements in the MLP (24). E4orf4 was reported to stabilize the Myc protein (2), and E4orf6 possibly increases cytoplasmic MYC RNA levels under some conditions (12). Therefore, the downregulation of MYC RNA levels by E4orf4 may counteract excessive virally induced Myc overexpression and increase late protein expression, thus contributing to the progression of viral infection. It has indeed been shown that the deletion of E4orf4 from the adenoviral genome (dl359) reduces late protein expression and viral production by 50% compared to WT virus-infected cells (37). This effect may be attributed, at least in part, to the absence of an E4orf4-induced downregulation of Myc expression.
In summary, the adenovirus E4orf4 protein downregulates MYC transcription as part of the virus strategy to create an optimal cellular environment for its replication, and this is yet another E4orf4 function requiring an interaction of the viral protein with a PP2A complex containing the B55 subunit.
Acknowledgments
We are grateful to S. Lavi, Tel Aviv University, Tel Aviv, Israel; P. Hearing, Stony Brook University, Stony Brook, NY; S. Strack, University of Iowa, IowaCity, IA; A. Aronheim and M. Avital-Shacham, Technion, Haifa, Israel; A. Krumm, University of Washington, Seattle, WA; and B. Vogelstein and K. W. Kinzler, Johns Hopkins Oncology Center, Baltimore, MD, for their generous gifts of cell lines, viruses, and plasmids. We thank N. Amariglio and J. Jacob-Hirsch for their advice and assistance.
The work was supported (in part) by the Israel Science Foundation (grant 319/04) and by the Israel Cancer Association.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6635-645. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, H. K., and R. C. Sears. 2006. Protein phosphatase 2A regulatory subunit B56α associates with c-Myc and negatively regulates c-Myc accumulation. Mol. Cell. Biol. 262832-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awasthi, S., A. Sharma, K. Wong, J. Zhang, E. F. Matlock, L. Rogers, P. Motloch, S. Takemoto, H. Taguchi, M. D. Cole, B. Luscher, O. Dittrich, H. Tagami, Y. Nakatani, M. McGee, A. M. Girard, L. Gaughan, C. N. Robson, R. J. Monnat, Jr., and R. Harrod. 2005. A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol. Cell. Biol. 256178-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Israel, H., and T. Kleinberger. 2002. Adenovirus and cell cycle control. Front. Biosci. 7d1369-d1395. [DOI] [PubMed] [Google Scholar]
- 5.Boldogh, I., S. AbuBakar, and T. Albrecht. 1990. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science 247561-564. [DOI] [PubMed] [Google Scholar]
- 6.Bondesson, M., K. Ohman, M. Mannervik, S. Fan, and G. Akusjarvi. 1996. Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J. Virol. 703844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champagne, C., M. C. Landry, M. C. Gingras, and J. N. Lavoie. 2004. Activation of adenovirus type 2 early region 4 ORF4 cytoplasmic death function by direct binding to Src kinase domain. J. Biol. Chem. 27925905-25915. [DOI] [PubMed] [Google Scholar]
- 8.Chung, H. J., and D. Levens. 2005. c-myc expression: keep the noise down! Mol. Cells 20157-166. [PubMed] [Google Scholar]
- 9.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 171115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, F. L., J. Smiley, W. C. Russel, and R. Nairn. 1977. Characteristics of a human cell line transformed by human adenovirus type 5. J. Gen. Virol. 3659-72. [DOI] [PubMed] [Google Scholar]
- 11.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 2811509-1512. [DOI] [PubMed] [Google Scholar]
- 12.Higashino, F., M. Aoyagi, A. Takahashi, M. Ishino, M. Taoka, T. Isobe, M. Kobayashi, Y. Totsuka, T. Kohgo, and M. Shindoh. 2005. Adenovirus E4orf6 targets pp32/LANP to control the fate of ARE-containing mRNAs by perturbing the CRM1-dependent mechanism. J. Cell Biol. 17015-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinds, P., C. A. Finlay, R. S. Quartin, S. J. Baker, E. R. Fearon, B. Vogelstein, and A. J. Levine. 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1571-580. [PubMed] [Google Scholar]
- 14.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 632605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalra, N., and V. Kumar. 2006. The X protein of hepatitis B virus binds to the F box protein Skp2 and inhibits the ubiquitination and proteasomal degradation of c-Myc. FEBS Lett. 580431-436. [DOI] [PubMed] [Google Scholar]
- 16.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393185-187. [DOI] [PubMed] [Google Scholar]
- 17.Kleinberger, T. 2004. Induction of transformed cell-specific apoptosis by the adenovirus E4orf4 protein. Prog. Mol. Subcell. Biol. 36245-267. [DOI] [PubMed] [Google Scholar]
- 18.Kleinberger, T., Y. B. Flint, M. Blank, S. Etkin, and S. Lavi. 1988. Carcinogen-induced trans activation of gene expression. Mol. Cell. Biol. 81366-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinberger, T., and T. Shenk. 1993. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J. Virol. 677556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klucky, B., B. Koch, M. Radolf, P. Steinlein, and E. Wintersberger. 2004. Polyomavirus tumorantigens have a profound effect on gene expression in mouse fibroblasts. Oncogene 234707-4721. [DOI] [PubMed] [Google Scholar]
- 21.Kornitzer, D., R. Sharf, and T. Kleinberger. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in S. cerevisiae and interacts with the anaphase promoting complex/cyclosome. J. Cell Biol. 154331-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavoie, J. N., M. Nguyen, R. C. Marcellus, P. E. Branton, and G. C. Shore. 1998. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J. Cell Biol. 140637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., E. Werner, M. Hergenhahn, R. Poirey, Z. Luo, J. Rommelaere, and J. C. Jauniaux. 2005. Expression profiling of human hepatoma cells reveals global repression of genes involved in cell proliferation, growth, and apoptosis upon infection with parvovirus H-1. J. Virol. 792274-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, L. H., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 134070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 1008164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, J., and D. Levens. 2006. Making myc. Curr. Top. Microbiol. Immunol. 3021-32. [DOI] [PubMed] [Google Scholar]
- 27.Lohr, K., O. Hartmann, H. Schafer, and M. Dobbelstein. 2003. Mutual interference of adenovirus infection and myc expression. J. Virol. 777936-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.London, L., R. G. Keene, and R. Landick. 1991. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol. Cell. Biol. 114599-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannervik, M., S. Fan, A. C. Strom, K. Helin, and G. Akusjarvi. 1999. Adenovirus E4 open reading frame 4-induced dephosphorylation inhibits E1A activation of the E2 promoter and E2F-1-mediated transactivation independently of the retinoblastoma tumor suppressor protein. Virology 256313-321. [DOI] [PubMed] [Google Scholar]
- 30.Marcellus, R. C., H. Chan, D. Paquette, S. Thirlwell, D. Boivin, and P. E. Branton. 2000. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Bα subunit of protein phosphatase 2A. J. Virol. 747869-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcellus, R. C., J. N. Lavoie, D. Boivin, G. C. Shore, G. Ketner, and P. E. Branton. 1998. The early region 4 orf4 protein of human adenovirus type 5 induces p53-independent cell death by apoptosis. J. Virol. 727144-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medghalchi, S., R. Padmanabhan, and G. Ketner. 1997. Early region 4 modulates adenovirus DNA replication by two genetically separable mechanisms. Virology 2368-17. [DOI] [PubMed] [Google Scholar]
- 33.Miller, D. L., C. L. Myers, B. Rickards, H. A. Coller, and S. J. Flint. 2007. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 8R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, and A. F. Gazdar. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7171-180. [PubMed] [Google Scholar]
- 35.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller, U., T. Kleinberger, and T. Shenk. 1992. Adenovirus E4orf4 protein reduces phosphorylation of c-Fos and E1A proteins while simultaneously reducing the level of AP-1. J. Virol. 665867-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Shea, C., K. Klupsch, S. Choi, B. Bagus, C. Soria, J. Shen, F. McCormick, and D. Stokoe. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 241211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Shea, C. C., S. Choi, F. McCormick, and D. Stokoe. 2005. Adenovirus overrides cellular checkpoints for protein translation. Cell Cycle 4883-888. [DOI] [PubMed] [Google Scholar]
- 39.Pan, Q., and R. U. Simpson. 1999. c-myc intron element-binding proteins are required for 1,25-dihydroxyvitamin D3 regulation of c-myc during HL-60 cell differentiation and the involvement of HOXB4. J. Biol. Chem. 2748437-8444. [DOI] [PubMed] [Google Scholar]
- 40.Shtrichman, R., and T. Kleinberger. 1998. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J. Virol. 722975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shtrichman, R., R. Sharf, H. Barr, T. Dobner, and T. Kleinberger. 1999. Induction of apoptosis by adenovirus E4orf4 protein is specific to transformed cells and requires an interaction with protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 9610080-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shtrichman, R., R. Sharf, and T. Kleinberger. 2000. Adenovirus E4orf4 protein interacts with both Bα and B′ subunits of protein phosphatase 2A, but E4orf4-induced apoptosis is mediated only by the interaction with Bα. Oncogene 193757-3765. [DOI] [PubMed] [Google Scholar]
- 43.Sontag, E. 2001. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell. Signal. 137-16. [DOI] [PubMed] [Google Scholar]
- 44.Van Kanegan, M. J., D. G. Adams, B. E. Wadzinski, and S. Strack. 2005. Distinct protein phosphatase 2A heterotrimers modulate growth factor signaling to extracellular signal-regulated kinases and Akt. J. Biol. Chem. 28036029-36036. [DOI] [PubMed] [Google Scholar]
- 45.Wen, W., S. Chen, Y. Cao, Y. Zhu, and Y. Yamamoto. 2005. HIV-1 infection initiates changes in the expression of a wide array of genes in U937 promonocytes and HUT78 T cells. Virus Res. 11326-35. [DOI] [PubMed] [Google Scholar]
- 46.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6308-318. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, H., F. Granberg, and U. Pettersson. 2007. How adenovirus strives to control cellular gene expression. Virology 363357-375. [DOI] [PubMed] [Google Scholar]
- 48.Zullo, J., C. D. Stiles, and R. L. Garcea. 1987. Regulation of c-myc and c-fos mRNA levels by polyomavirus: distinct roles for the capsid protein VP1 and the viral early proteins. Proc. Natl. Acad. Sci. USA 841210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]