Abstract
Cytomegalovirus replication depends upon a betaherpesvirus-conserved 150-kDa tegument phosphoprotein (pp150; encoded by UL32) that supports the final steps in virion maturation at cytoplasmic assembly compartments. Amino acid substitutions were introduced into conserved region 1 (CR1) and CR2 of pp150, affecting a region that may interact with nucleocapsids. Two independent CR2 point mutants (N201A and G207A) failed to support viral replication in evaluations by a transient complementation assay or after reconstruction into recombinant viruses. An assembly compartment-like cytoplasmic inclusion developed in UL32 mutant virus-infected cells that was similar to that of wild-type virus-infected cells. The cellular localization of the trans-Golgi marker Golgin-97 suggested differences in the organization of the assembly compartment compared to that of wild-type virus-infected cells. Replication-defective CR2 point mutants exhibited the same phenotype as that of a virus carrying a complete deletion of the UL32 open reading frame in these assays. Electron micrographs of fibroblasts at 3 or 5 days postinfection with a deletion mutant (ΔUL32) grown on UL32-complementing cells showed a similar number and morphology of capsids in the nucleus, but the cytoplasmic region associated with virion assembly appeared highly vesiculated and contained few recognizable nucleocapsids or complete virus particles. These data demonstrate that the principle role of pp150 is to retain nucleocapsid organization through secondary envelopment at the assembly compartment.
Human cytomegalovirus (HCMV) DNA replication, capsid assembly, and encapsidation of viral DNA all occur within nuclear replication compartments (27, 29). Nucleocapsid egress from the nucleus (30, 36) and translocation to the cytoplasm precede the final maturation steps (6). While many properties are established and are common to all herpesviruses, the final steps of maturation (tegumentation, final envelopment, and the release of mature progeny) remain poorly understood (6). HCMV maturation most likely requires the primary envelopment of nucleocapsids at the inner nuclear membrane followed by deenvelopment at the outer nuclear membrane (27, 29), as proposed for other herpesviruses (25). Evidence has accumulated to support HCMV tegumentation and envelopment at cytoplasmic vesicles derived from endosomes (42). Cellular vesicle transport machinery carries mature virions to the plasma membrane for release into the extracellular space and spread to adjacent cells. Maturation appears to have common (28) as well as distinguishing (6) characteristics in the different herpesvirus subfamilies (25). For HCMV, the cytoplasmic maturation steps occur in a clearly distinguishable assembly compartment (AC) that includes early endosomes as well as the Golgi apparatus. The AC or cytoplasmic inclusion, together with a kidney-shaped nucleus and nuclear inclusion, gives HCMV-infected cells a distinct morphology (39) resembling an owl's eye. The components of the cellular secretory apparatus become reoriented within the AC in conjunction with virus maturation and release (12). The formation of the AC is associated with the accumulation of late viral proteins and is blocked when infection is carried out in the presence of inhibitors of either viral DNA replication or encapsidation. Virion tegument proteins (pp65 and pp28) (34), nucleocapsids containing viral DNA (2), and virion envelope glycoproteins (gB, gH, and gM) (22, 23, 34) all localize to the AC. This specialized adaptation appears to condense cellular areas involved in protein synthesis and vesicle transport machinery (12). The endoplasmic reticulum becomes located toward the periphery of the AC, with endoplasmic reticulum-to-Golgi-intermediate, trans-Golgi network, and endosomal compartments becoming arranged concentrically toward the center (12). Although distinct ACs are observed only in betaherpesviruses, this reorganization of cellular compartments has long been associated with the final steps in HCMV maturation (27, 29).
Genetic evidence has shown that several herpesvirus core (UL48, UL71, UL76, UL77, UL93, UL94, UL95, and UL99) and two betaherpesvirus conserved (UL32 and UL96) tegument protein genes play critical roles during HCMV replication in human fibroblasts (HFs) (14, 29, 46). The contribution of the myristoylated and palmitoylated tegument protein pp28 (encoded by UL99) has been extensively investigated to define its role in secondary (or final) envelopment at the AC (8, 20, 40). Amino-terminal myristoylation, phosphorylation, and acidic character all contribute to pp28 function (20, 38), and although maturation continues even when pp28 is deleted, evidence suggests that localization to the AC is one critical aspect for function (37). Phosphoprotein 150 (pp150), a major tegument constituent (15, 27) encoded by UL32, is absolutely essential for cytoplasmic maturation steps that occur within the AC (2). As a betaherpesvirus-conserved protein, pp150 may contribute to the unique features common to this subfamily of herpesviruses, such as the formation of the AC. In addition to pp28 and pp150, phosphorylation by the viral protein kinase (encoded by UL97), acting together with cell cycle kinases (18), contributes to the formation of the AC (3) even though this herpesvirus core enzyme has been associated primarily with nuclear maturation events (21, 45). In the absence of UL97, aggregates of viral structural proteins accumulate in both the nucleus and cytoplasm (3, 31). The most abundant HCMV tegument protein, pp65 (encoded by UL83), is completely dispensable for virus replication and maturation, although this protein contributes to dense body production (35) and appears to play an immunomodulatory role early during infection (1). Other tegument proteins that act early in infection, such as UL82 (5) and UL69 (16), also may contribute to maturation. In addition to tegument proteins, the virion envelope glycoprotein (g) complex gM:gN is required for replication and also may contribute to maturation steps within the AC (22), whereas other essential viral envelope glycoproteins (gB and gH:gL) appear to be involved in entry (11).
pp150, which is encoded by UL32, is an abundant tegument protein associated with nucleocapsid-containing enveloped particles (virions and noninfectious particles) released by HCMV-infected cells (19), although it is only a minor component of dense bodies (32). Studies primarily focused on simian CMV have implicated pp150 in direct interactions with nucleocapsids (4), and this potentially occurs via capsid triplexes based on ultrastructural studies (10, 43). A bacmid-derived deletion mutant (14) lacking the UL32 open reading frame (ORF) (ΔUL32) fails to release infectious virus particles, although cells apparently release pp65-rich particles (2). This is consistent with a role of pp150 in virion maturation but not in dense-body formation. The amino-terminal one-third of pp150 contains two relatively short domains, conserved region 1 (CR1) and CR2, that are required for function (2). These same regions are required for interaction with purified nucleocapsids (4), suggesting an important role in virion maturation (4). The colocalization of fully functional enhanced green fluorescent protein (eGFP)-tagged pp150 with the major capsid protein (MCP) at nuclear replication compartments has been taken as evidence that pp150 is incorporated into nascent HCMV particles simultaneously with or immediately following capsid assembly (33), which is consistent with an earlier localization study (17). pp150-eGFP-expressing virus confirmed that pp150 associates with nucleocapsids in the nucleus and follows virus maturation through primary envelopment and deenvelopment steps (33).
When the replication block in ΔUL32 virus-infected HFs was investigated, viral structural proteins and viral DNA were shown to accumulate in the cytoplasm within the AC, indicating a role for pp150 in final maturation steps rather than in nuclear maturation events (2). This observation seemed to run counter to the suspected role of pp150-nucleocapsid interaction at an earlier, nuclear tegumentation step and suggested that the localization of pp150 to the nucleus early in HCMV infection (33) simply identifies the point where this tegument protein is added to the nucleocapsid and does not identify the point in replication where pp150 functions. The phenotype of UL32 mutant virus as well as complementation studies carried out with pp150 expression constructs (2) all pointed to a critical cytoplasmic role for this protein within the AC (34). Thus, the characterization of pp150 and various mutant pp150 forms by the complementation of ΔUL32 virus replication revealed the role of pp150 in the final cytoplasmic maturation steps rather than in the initial tegumentation, primary envelopment, or egress of nucleocapsids from the nucleus (2). All three kinetic classes of HCMV genes are expressed in ΔUL32-infected cells. MCP and gB, as well as newly synthesized viral DNA, localized to the cytoplasm in the absence of pp150, suggesting that nucleocapsid accumulation occurs independently of pp150 but that secondary envelopment requires pp150 (2). As with the earlier characterization of deletion mutants (4), CR1 and CR2 were shown to be essential for pp150 function, presumably because these regions are important for interaction with the nucleocapsid. Neither O-linked glycosylation nor an intact carboxyl terminus was required for pp150 function. The simian CMV, but not the murine CMV, pp150 homolog complemented UL32 mutant virus. This work suggested that pp150 was functionally required for envelopment at the AC, suggesting that a critical interaction between pp150 and nucleocapsids occurs within the AC.
In this study, point mutants affecting the amino-terminal CR1 and CR2 of pp150 were screened for their ability to complement the UL32 mutant virus in a transient assay. Two mutants carrying mutations in CR2 complemented poorly. These point mutants and additional mutations were reconstructed into Towne-BAC to evaluate mutant virus growth properties by immunofluorescence assay and electron microscopy (EM). Our investigation shows a role for pp150 in virion maturation, the stabilization of nucleocapsids, and, potentially, the reorganization of the AC.
MATERIALS AND METHODS
Cells.
Primary foreskin-derived HFs were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FetalClone III (HyClone, Logan, UT) at 37°C with 5% CO2. HFs between passages 5 and 15 were used for transfections. Medium was changed every other day in transfected cell cultures. A pp150-DsRed-expressing cell line (UL32DsR-HF) was produced by the cotransfection of pON5001 (described below) along with plasmids LTRVSVG, JK3, and CMVtat into 293T cells as described previously (2). pp150-DsRed-expressing retrovirus isolated from the supernatants of transfected 293T cells was used to infect low-passage HFs. Stable UL32DsR-HF cells were selected with 400 μg/ml Geneticin (Gibco/Invitrogen, Carlsbad, CA).
DNA constructs, mutagenesis, and secondary spread assays.
The wild-type (WT) UL32 ORF cloned in the pLNCX vector (pON2780) has been reported previously (2). DsRed fluorescent protein was PCR amplified from the vector pDsRed-Monomer-N1 (Clontech Inc., Mountain View, CA) and cloned into ClaI/PmeI-digested pON2780 to make the C-terminal DsRed-monomer-tagged pp150 expression plasmid (pON5001). The entire UL32 ORF was cloned into pUC19 utilizing BglII/ClaI restriction sites, producing pON5002, a template for QuickChange mutagenesis (Qiagen Inc., Valencia, CA). Inserted mutations were confirmed by the diagnosis of new restriction sites that were introduced along with the desired mutations. UL32 ORF mutants were transferred to pLNCX (26) using BglII/ClaI restriction sites and sequenced in this region using UL32 primers to confirm the insertion of the desired mutations without undesirable changes. Secondary spread assays were performed as described previously (2). Briefly, primary HFs in either 24- or 6-well tissue culture dishes were cotransfected 24 h postseeding with ΔUL32-BAC, the pp71 expression plasmid, and one of the UL32 LNCX constructs. Medium was changed every other day until day 10 posttransfection, when cells were fixed in 3.7% formaldehyde-phosphate-buffered saline (PBS) for 10 min prior to evaluation by epifluorescent microscopy. Viral spread was defined as three or more adjacent eGFP-positive cells.
BAC mutagenesis and recombinant viruses.
pSIM6 plasmid (13), encoding lambda-red functions (a generous gift from Tim Barnett, Emory Children's Center), was introduced into Escherichia coli carrying Towne-BAC (14) by electroporation. Bacterial artificial chromosome (BAC) recombineering protocols (41) were followed to mutate regions of UL32. Briefly, E. coli cells carrying Towne-BAC and pSIM6 were made electroporation competent by being grown to an optical density at 600 nm of 0.4 to 0.6 and being washed twice with ice-cold nanopure water. A PCR amplicon carrying an overhang of 50 bp of homology to both flanks of the desired deletion region from Towne-BAC, in addition to the full kanamycin resistance (Kanr) and Bacillus subtilis levansucrase (SacB) cassette, was then introduced into these competent cells by electroporation. Kanr colonies were selected on Luria-Bertani-kanamycin plates and were screened for sucrose sensitivity. Sucrose-sensitive colonies were further screened by PCR for the proper orientation and location of the Kanr-SacB cassette. Cultures (5 ml) of PCR-positive colonies were grown overnight, and BAC DNA was extracted for digestion with HindIII to confirm the expected restriction pattern. BACs were reconstituted with a PCR fragment containing the WT or mutant UL32 and screened for the loss of Kanr as well as sucrose sensitivity. The UL32 region was sequenced to confirm the presence of the desired mutation, as well as the absence of any undesired modifications. Supernatants from transfections of replication-competent BACs were harvested 9 days posttransfection for the recovery of recombinant viruses. Replication-incompetent ΔUL32 virus (14) was grown on HFs stably expressing pp150-DsRed protein. Single-step growth curves were performed by infecting confluent HFs at a multiplicity of infection (MOI) of 5.0 in triplicate and removing supernatants at days 1, 3, 5, and 6 postinfection. These supernatants were diluted 1,000- to 100,000-fold for titration by plaque assay on HFs, which were counted 6 days postinfection by eGFP fluorescence.
Antibodies.
c-Myc-labeled pp150 was detected by mouse monoclonal anti-c-myc antibody (Zymed Inc., San Francisco, CA). Native or nontagged pp150 was detected using monoclonal antibody (MAb) 36-14 (34), gM:gN complexes were detected using MAb 14-16A (7), and MCP was detected using MAb 28-4 (9) (gifts from William Britt, University of Alabama, Birmingham). Anti-pp28 MAb was purchased from Virusys Corporation, Sykesville, MD. Hoechst 33258 (AnaSpec Corp., San Jose, CA) staining was used to identify nuclei. Anti-Golgin-97 monoclonal antibody (Molecular Probes-Invitrogen Inc., Carlsbad, CA) was used to detect the trans-Golgi network. The secondary antibodies Texas red anti-mouse immunoglobulin G (H+L) or Texas red anti-rabbit immunoglobulin G (H+L) were purchased from Vector Laboratories, Inc. (Burlingame, CA).
Microscopy.
Samples were prepared using established protocols for immunofluorescence assay and confocal fluorescence microscopy. Briefly, cells (HFs) were grown on coverslip inserts in 24-well tissue culture dishes. At the end point, cells were fixed in either 3.7% formaldehyde for 10 min or 4% paraformaldehyde for 20 min and were incubated in 50 mM NH4Cl in PBS for 10 min to reduce autofluorescence. This followed a wash, incubation in 0.5% Triton X-100 for 20 min to permeabilize the cells, and finally a wash and incubation with primary and secondary antibodies. Coverslips were retrieved from the wells and were mounted on glass slides with a drop of mounting medium (2.5% DABCO in Fluoromont G) and dried overnight before imaging. Images were acquired on a Carl-Zeiss LSM 510 META confocal fluorescent microscope or Zeiss Axio Imager A1 epifluorescent microscope. Samples for EM were prepared by infecting HFs with Towne or ΔUL32 virus (MOI of 1). Monolayer cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 2 h at 4°C. Cells then were washed with the same buffer and postfixed with buffered 1.0% osmium tetroxide at room temperature for 1 h. Following several washes with 0.1 M cacodylate buffer, cells were dehydrated with ethanol, infiltrated, and embedded in Eponate 12 resin (Ted Pella Inc., Redding, CA). Ultrathin sections (60 to 70 nm) of monolayer cells were cut and counterstained using uranyl acetate and lead citrate. The examination of ultrathin sections was carried out on a Hitachi H-7500 transmission electron microscope.
RESULTS
Point mutations in conserved regions disrupt pp150 function.
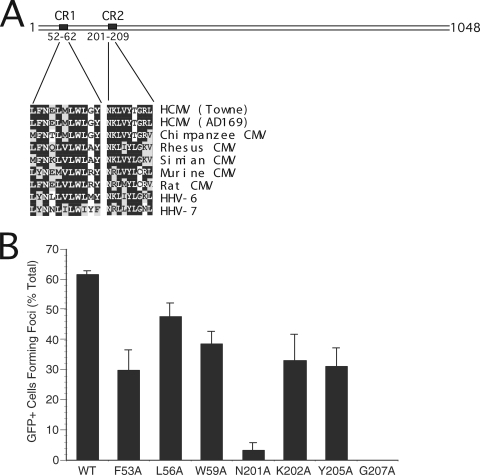
Based on the observations that clustered mutations within CR1 or CR2 disrupt HCMV pp150 function and that the simian CMV pp150 homolog complements ΔUL32 virus (2), we aligned CR1 and CR2 from sequenced betaherpesvirus pp150 homologs (Fig. 1A) to identify conserved amino acids. We hypothesized that if CR1 and CR2 form a critical binding site, individual amino acid mutations in these regions would significantly disrupt pp150 function. Based on conservation in primate betaherpesviruses, three amino acids in CR1 (F53, L56, and W59) and four amino acids in CR2 (N201, K202, Y205, and G207) were mutated individually to alanine. Mutant protein expression constructs were tested for their ability to complement ΔUL32 replication in secondary spread assays (Fig. 1B). CR1 point mutants complemented ΔUL32, although all three (F53A, L56A, and W59A) exhibited reduced activity compared to that of WT pp150 (Fig. 1B). While CR2 point mutants K202A and Y205A complemented ΔUL32, though with reduced activity compared to that of the WT (Fig. 1B), CR2 point mutants N201A and G207A failed to complement or complemented very poorly. In an earlier study from Gibson's laboratory (4), the amino-terminal region of simian CMV pp150 (amino acids [aa] 1 to 194), which contains CR1 but not CR2, failed to bind to isolated capsids in an in vitro assay, whereas a fragment containing both CR1 and CR2 (aa 1 to 275), but not the C-terminal region, was as efficient as full-length pp150 in binding to simian CMV nucleocapsids. Our functional analysis supports a role for this amino-terminal domain, particularly CR2, in HCMV pp150 function. Although individual CR1 mutants did not destroy function, we confirmed the importance of two adjacent conserved amino acids (L58I and W59D) previously shown to be critical in complementation assays (2). This mutant was not investigated further, as we focused on point mutants within CR2 that play a critical role in pp150 function. Critical amino acids within either CR1 (L58 and W59 together) or CR2 (N201 or G207) may contribute to pp150 function by controlling the interaction of this tegument protein with important partners such as nucleocapsids.
FIG. 1.
(A) Line diagram of pp150 (1,048 aa) illustrating amino-terminal CR1 (L52 to Y62) and CR2 (N201 to L209). The alignment of amino acid sequences of CR1 and CR2 from different betaherpesviruses is shown in the expanded region. The Towne and AD169 strains of HCMV are aligned with chimpanzee, rhesus, simian, mouse, and rat CMV species and also with human herpesviruses-6 and -7. (B) Secondary spread assay evaluation of plasmids expressing WT or mutant forms of UL32 to complement ΔUL32-BAC. Individual point mutations in CR1 (F53A, L56A, and W59A) and CR2 (N201A, K202A, Y205A, and G207A) were tested by cotransfection with ΔUL32-BAC DNA in HFs.
Replication defects of recombinant UL32 mutant viruses.
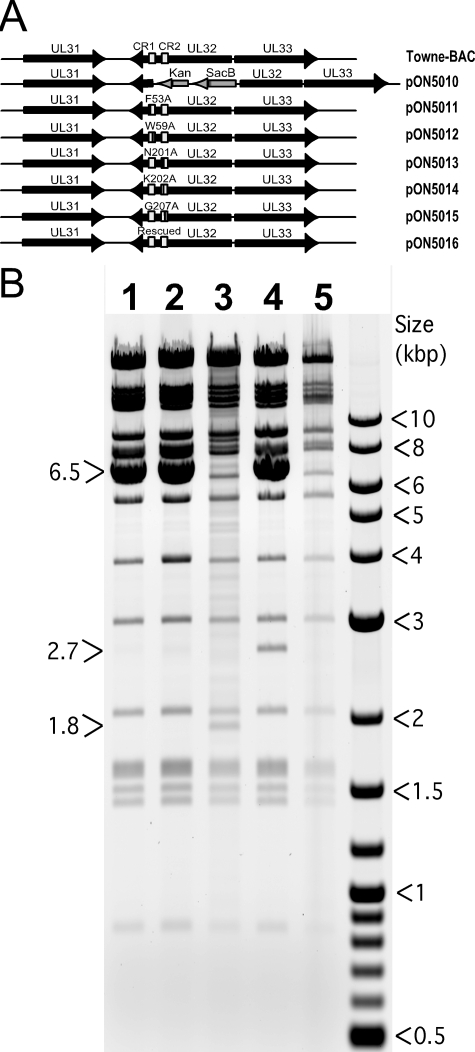
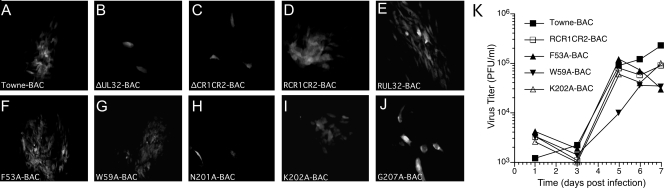
Both of the critical CR2 point mutants (N201A and G207A), as well as three additional mutants (F53A, W59A, and K202A) that partially complemented ΔUL32 replication, were introduced into Towne-BAC (Fig. 2) by lambda-red-mediated recombination. The introduction of the CR2 point mutations that failed to complement yielded replication-incompetent bacmids (N201A-BAC and G207A-BAC). These generated single eGFP-positive cells following transfection, like ΔUL32 or a bacmid that carried deletions in both CR1 and CR2 (ΔCR1ΔCR2-BAC) (Fig. 3). Bacmids carrying mutations that did not eliminate function in secondary spread assays (F53A-BAC, W59A-BAC, or K202A-BAC) were replication competent and generated eGFP-positive plaques similar in size and morphology to those of Towne-BAC (Fig. 3). Single-step growth curves for viruses that were recovered without complementation revealed some variability in replication, but overall the final yields were within a 10-fold range (Fig. 3K). W59A-BAC mutant virus, in particular, appeared to replicate slower and to reduced levels; however, on repeated evaluation, plaque size and other infection parameters were not readily distinguishable from either parental Towne-BAC or viruses carrying other UL32 mutations that did not compromise viral replication (K202A-BAC or F53A-BAC). Although N201A-BAC and G207A-BAC were replication incompetent on HFs, these bacmids produced eGFP-positive plaques when complemented by transfection into pp150-DsRed-expressing HFs (data not shown), confirming that mutant virus growth defects were due to nonfunctional pp150 and not to other unintended alterations in the viral genome. These two mutants also were complemented by the WT pp150 expression plasmid (pON2780) when cotransfected into HFs. This pattern was previously shown for a mutant virus with a complete deletion of the UL32 ORF (2). Thus, the two CR2 point mutants that were nonfunctional in the secondary spread assay also failed to function when reconstructed into recombinant virus. The consistency in growth phenotype when pp150 mutations were evaluated within mutant viruses and in secondary spread assays supports the utility of transient complementation as a screen that can be used in advance of constructing recombinant viruses. Our observations on the behavior of pp150 CR2 mutants suggests that previously characterized clustered and deletion mutations in both CR1 and CR2 (2) also would yield replication-defective viruses upon reconstruction. Here, the behavior of individual CR2 mutants illustrates the critical role that pp150 plays in HCMV replication and provides a set of mutants to compare the replication defect during ΔUL32 infection, when pp150 is not made, to the replication defect in which a nonfunctional protein is made during infection.
FIG. 2.
(A) Schematic representation of UL32 mutations introduced into the Towne-BAC genome using lambda-red-mediated recombination. The portion of UL32 containing CR1 and CR2 was replaced with a Kanr-SacB cassette (pON5010) that was used to derive pON5011-pON5016 bacmids as shown. (B) Electrophoretic separation of HindIII-digested BAC DNAs on agarose gel (0.8%) as a diagnostic for the correct insertion of the Kanr-SacB cassette and the integrity of BAC genome. Digests of G207A-BAC (lane 1), N201A-BAC (lane 2), ΔUL32-BAC (lane 3), ΔUL32-Kanr-SacB-BAC (lane 4), and Towne-BAC (lane 5) are shown. The presence of the pSIM6 plasmid (6.5 kb) confers lambda red functions to these BACs. This plasmid was lost during the growth of bacterial culture in the absence of ampicillin (lanes 3 and 5). The 2.7-kb band results from the insertion of the Kanr-SacB cassette, and the 1.8-kb band results from insertion of the Kanr cassette in the Towne-BAC genome.
FIG. 3.
Replication properties of BAC-reconstituted replication-competent viruses. (A to J) Fluorescent images of eGFP-positive cells at day 10 posttransfection of HFs with Towne-BAC, ΔUL32-BAC, ΔCR1ΔCR2-BAC, rescued (R) CR1CR2-BAC, RUL32-BAC, F53A-BAC, W59A-BAC, N201A-BAC, K202A-BAC, and G207A-BAC. (K) Single-step growth curves (MOI of 5.0) of Towne-BAC and replication-competent UL32 mutant BAC-derived recombinant viruses. Virus titers were determined by plaque assays of supernatants collected daily. The standard deviation from the mean of each titer is within the symbols.
Localization of viral structural proteins within the AC.
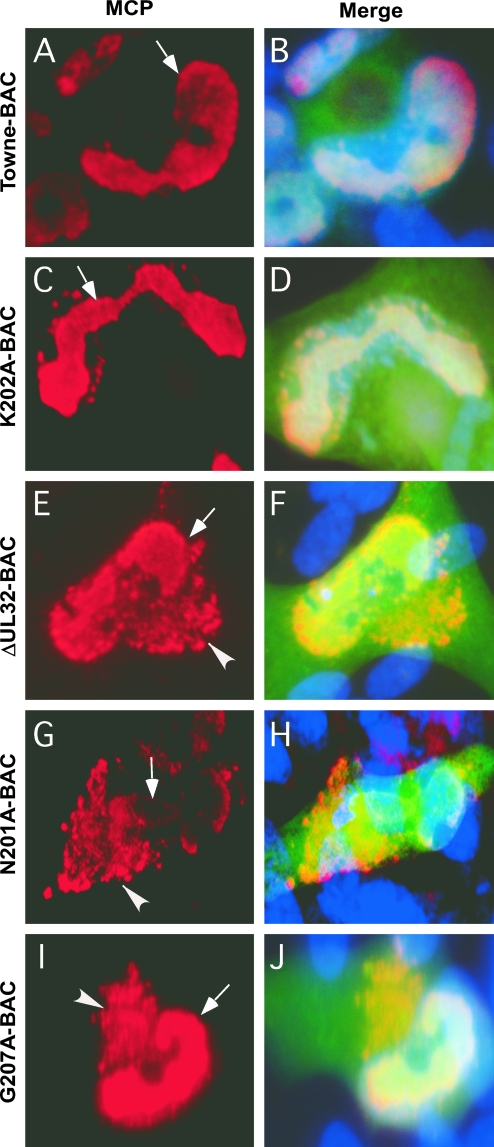
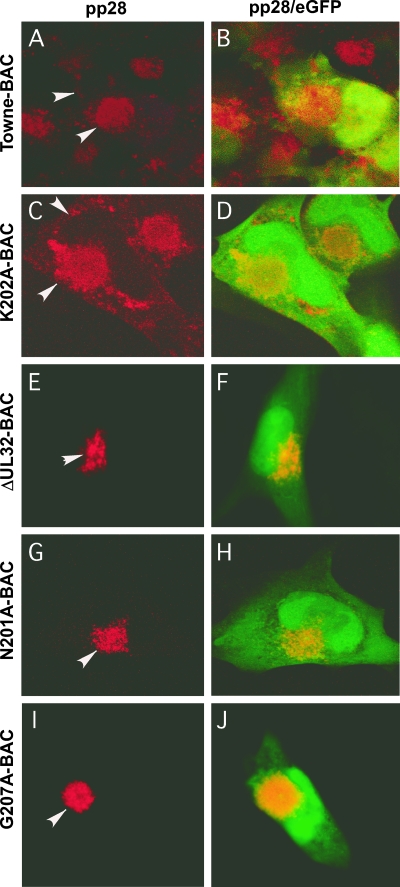
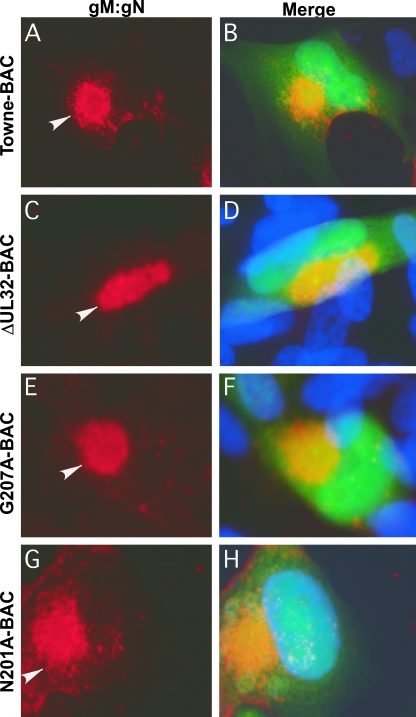
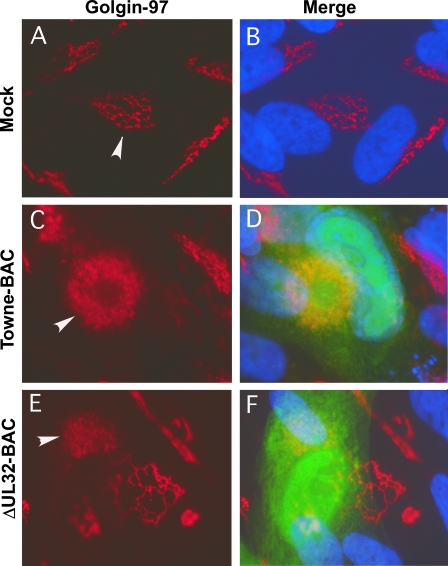
We sought to evaluate the block in virus maturation during infection with ΔUL32 or UL32 point mutant bacmids compared to that with Towne-BAC. The primary localization of either WT or mutant pp150 was cytoplasmic, with a concentration in the AC (2). Functional or nonfunctional single or clustered point mutant proteins localized similarly to WT protein in infected cells (2 and data not shown). MCP was much more prominent in the cytoplasm of ΔUL32 virus-infected cells than in WT virus-infected cells (2). This localization pattern was interpreted as representing an accumulation of nucleocapsids blocked at a maturation step based on the movement of bromodeoxyuridine (BrdU)-labeled DNA from the nuclear site of synthesis to the AC in Towne-BAC and ΔUL32 virus-infected cells (2). MCP localized with a similar distribution to nuclear and cytoplasmic sites in ΔUL32-BAC-, N201A-BAC-, and G207A-BAC-infected HFs (Fig. 4), in contrast to the more exclusive nuclear localization pattern observed with replication-competent K202A-BAC or Towne-BAC (Fig. 4). The increased distribution of cytoplasmic MCP in mutant virus-infected cells in which maturation did not occur demonstrates that HCMV reaches a similar point in maturation regardless of whether pp150 is absent or is expressed as a nonfunctional protein. In either setting, the pattern is consistent with a block in maturation at the AC when pp150 is absent or compromised. We further characterized the impact of pp150 on AC development in virus-infected cells by evaluating the localization of tegument protein pp28, a tegument protein that accumulates within the AC in Towne-BAC-infected cells and where evidence from several laboratories implicates its acquisition during final envelopment (8, 20, 40). Mutant ΔUL32-BAC-, N201A-BAC-, or G207A-BAC-infected cells (2) exhibited pp28 staining that was almost exclusively within the AC, whereas pp28 localized to the AC as well as in a general punctate pattern throughout the cytoplasm in Towne-BAC-infected cells or replication-competent K202A-BAC-infected cells (Fig. 5). Given that the envelopment and release of virus is compromised in ΔUL32-BAC-, N201A-BAC-, and G207A-BAC-infected cells, this difference in cytoplasmic distribution of pp28 correlated with the maturation of virions. The final virion protein we evaluated was the mature, assembled gM:gN envelope glycoprotein complex, detected using monoclonal antibody 14-16A (24). gM:gN localized within the same AC-associated pattern during infection when either replication-defective mutant virus or Towne-BAC virus infections were evaluated (Fig. 6). A similar AC localization pattern also was observed when additional replication-competent or replication-defective UL32 mutants were examined (data not shown). These results suggest that gM:gN complex formation and localization are not influenced by pp150 function. Based on cellular marker Golgin-97, trans-Golgi compartments were dramatically increased in size, shape, and intensity during infection, with formation of an AC in either WT or UL32 mutant virus infection. Golgin-97 appeared less organized within the AC in ΔUL32-BAC-infected cells than in the reported cylindrical arrangement (12) of trans-Golgi compartments in Towne-BAC infection (Fig. 7). The staining patterns for HCMV pp28 and MCP together with the Golgin-97 localization pattern and prior observations on ΔUL32 (2) suggest a pp150-dependent virion maturation function in which nucleocapsids are present in the AC but fail to proceed further to vesicle transport-associated release. Assembled envelope gM:gN (Fig. 6) and gB (2), markers of membranes that identify envelopment sites, appeared to localize within the AC whether pp150 was functional or not. While an AC forms in WT as well as UL32 mutant virus-infected cells, MCP and pp28 tegument protein provided the most compelling evidence that there is a difference in the organization of AC when pp150 is absent (or nonfunctional) compared to when it is functional and maturation proceeds. This defect was explored further using EM to evaluate the structure and contents of the AC at different times in Towne-BAC and ΔUL32-BAC virus-infected cells.
FIG. 4.
Immunofluorescence localization of MCP in HFs transfected with Towne-BAC (A, B), K202A-BAC (C, D), ΔUL32-BAC (E, F), N201A-BAC (G, H), or G207A-BAC (I, J). HFs were fixed 9 days posttransfection and stained with mouse monoclonal anti-MCP primary antibody and anti-mouse Texas red secondary antibody (A, C, E, G, and I) before immunofluorescence imaging at ×1,000 magnification. Hoechst 33258 staining was used to identify nuclei, in merged images that include contributions from Texas red and virus-encoded eGFP (B, D, F, H, and J). Arrows point to nuclear MCP in all panels, and arrowheads point to the cytoplasmic MCP detected in ΔUL32-BAC-, N201A-BAC-, or G207A-BAC-transfected cells in which pp150 is nonfunctional.
FIG. 5.
Immunofluorescent localization of pp28 to the AC in HFs transfected with Towne-BAC (A, B), K202A-BAC (C, D), ΔUL32-BAC (E, F), N201A-BAC (G, H), or G207A-BAC (I, J). HFs were fixed 9 days posttransfection and stained with mouse monoclonal anti-pp28 primary antibody and anti-mouse Texas red secondary antibody (A, C, E, G, and I) before immunofluorescence imaging at ×1,000 magnification. Arrowheads point to pp28 within the AC and cytoplasms of transfected cells. Merged images include contributions from Texas red and eGFP (B, D, F, H, and J).
FIG. 6.
Immunofluorescent localization of gM:gN in HFs transfected with Towne-BAC (A, B), ΔUL32-BAC (C, D), G207A-BAC (E, F), or N201A-BAC (G, H). HFs were fixed 9 days posttransfection and stained with mouse monoclonal 14-16A primary antibody against gM:gN complex and anti-mouse Texas red secondary antibody (A, C, E, and G) before immunofluorescence imaging at ×1,000 magnification. Arrowheads point to gM:gN within the AC detected in Towne-BAC and K202A-BAC infections in which pp150 is functional and also in ΔUL32-BAC-, N201A-BAC-, or G207A-BAC-transfected cells in which pp150 is nonfunctional. Hoechst 33258 staining was used to identify nuclei, which are depicted as merged images that include contributions from Texas red and eGFP (B, D, F, and H).
FIG. 7.
Immunofluorescence localization of the trans-Golgi network in HFs. Either mock-transfected (Mock) (A, B), Towne-BAC-transfected (C, D), or ΔUL32-BAC-transfected (E, F) HFs were fixed 9 days posttransfection and stained with mouse monoclonal anti-Golgin-97 primary antibody and anti-mouse Texas red secondary antibody before imaging at ×1,000 magnification. Arrowheads mark the trans-Golgi network within individual cells.
Localization of nucleocapsids and virus particles during UL32 mutant virus infection.
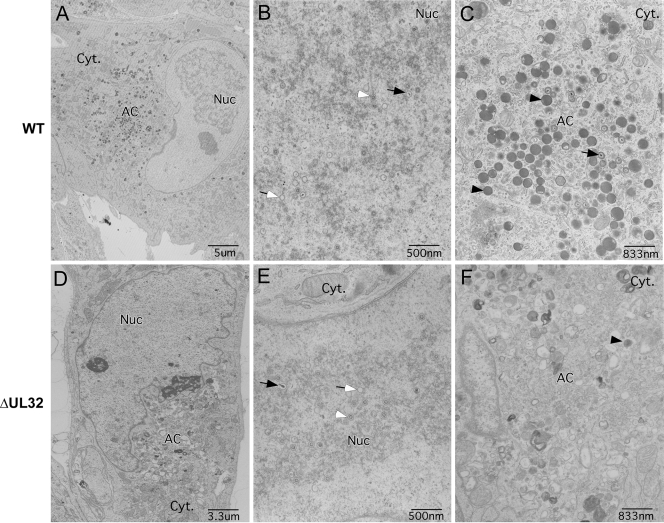
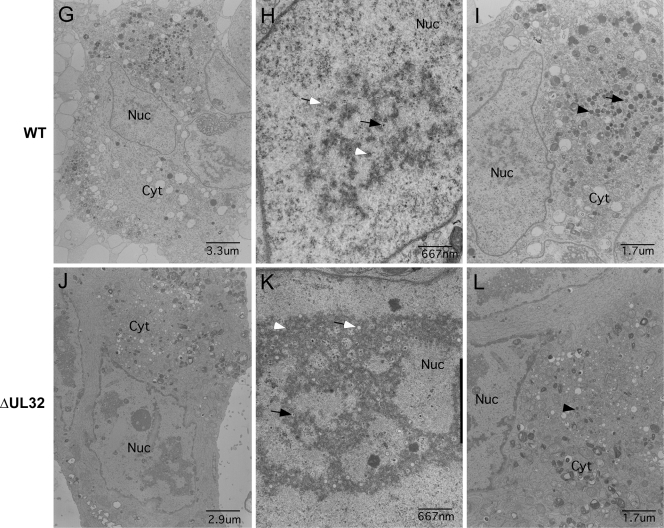
HFs infected with Towne-BAC and ΔUL32-BAC viruses were analyzed by transmission EM for differences in cytoplasmic structure and the distribution of virus particles. Mutant virus stocks were prepared on UL32DsR-HFs. At day 3 postinfection, virus-infected cultures were prepared for transmission EM as described in Materials and Methods, and images were collected for comparison. We used ΔUL32 for these studies, because point mutants recombined with resident pp150 when propagated on UL32DsR-HFs, whereas the large deletion mutant did not. Virus particles containing DNA or scaffold, as well as many that remained empty, were observed at comparable levels in the nuclei of Towne-BAC as well as ΔUL32-BAC virus-infected HFs (Fig. 8B and E and Table 1). Whereas Towne-BAC virus-infected cells had mature virus particles with distinct double rings and an electron-dense core representing viral DNA, as well as immature virus particles that either contained scaffold or were empty and dense bodies within the AC by 3 days postinfection (Fig. 8C), virus particles were rarely observed in the cytoplasm of ΔUL32-infected cells at this time (Fig. 8F). Instead, the AC region within ΔUL32-infected cells contained many vesicles with no consistent size or symmetry. The smaller vesicles (less than 50 nm) appeared similar in size and number to the vesicles that also could be identified in the AC of Towne-BAC-infected cells (Fig. 8C), although bigger vesicles (50 to 300 nm) were present predominantly in ΔUL32-infected cells at this time (Fig. 8F). At 5 days postinfection, abundant virus particles and dense bodies were associated with the AC and other parts of the cytoplasm of Towne-BAC-infected cells (Fig. 8I). In ΔUL32-BAC-infected cells, however, few virus particles or dense bodies were detected, although the few that were identified were associated with the AC-like region. This region remained highly vesicular and had a range of forms that could be associated with incomplete, unstable, or misformed virus particles. Most appeared similar to the vesicles that were detected in the AC of ΔUL32-BAC-infected cells at day 3 postinfection, although some vesicles at day 5 more resembled virus particles with a dense core but no electron-dense tegument layer. Some vesicles were asymmetrical, and many were more electron transparent or much larger than nucleocapsids. The possibility exists that these are either improperly formed or degraded nucleocapsids. The abundance and similar distribution of capsids present in the nuclei in mutant and WT virus infection (Fig. 8H and K; Table 1) suggest degradation in the cytoplasm. The few intact nucleocapsids present in the cytoplasm of mutant virus-infected cells was in contrast to the fluorescent localization of MCP in the AC of ΔUL32-BAC-infected cells, as well as BrdU pulse-chase data suggesting nucleocapsid egress to the cytoplasm (2). This led us to suspect that maturing nucleocapsids are unstable following egress from the nucleus in the absence of pp150.
FIG. 8.
Transmission electron micrographs of HFs illustrating differences between Towne-BAC and ΔUL32 virus infections at day 3 (A through F) and day 5 (G through L) postinfection. Nuc, nucleus; Cyt, cytoplasm. Black arrows, C capsids; black arrowheads, dense bodies; white arrows, empty capsids; white arrowheads, B capsids.
TABLE 1.
Nuclear distribution of capsid types in Towne-BAC or ΔUL32 virus-infected HFsa
| Capsid type | % of capsids with indicated morphologyb at 3 days postinfection
|
% of capsids with indicated morphologyb at 5 days postinfection
|
||
|---|---|---|---|---|
| Towne-BAC | ΔUL32-BAC | Towne-BAC | ΔUL32-BAC | |
| A | 10 | 8 | 6 | 10 |
| B | 56 | 62 | 69 | 57 |
| C | 17 | 12 | 4 | 14 |
| Nontypeable | 17 | 18 | 21 | 19 |
Data shown are from counts of two different EM fields for each day and infection. The capsids with hard-to-distinguish features were categorized as nontypeable.
See reference 29.
In order to determine whether MCP detected in the cytoplasm of ΔUL32-infected cells was free or nucleocapsid associated, transfected cells were cultured in the presence of 20 μM 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidizole (BDCRB), an inhibitor of viral DNA encapsidation (44), and evaluated for the distribution of MCP. MCP was not detected in the cytoplasm of either ΔUL32-infected or Towne-BAC-infected cells when maintained in the presence of BDCRB but was detected in the nucleus in both cases. Both MCP and viral DNA (2) present in the cytoplasm of ΔUL32-infected cells likely originated in the nucleus based on the evidence from BrdU pulse-chase experiments (2) and BDCRB experiments. These data together strongly suggest that nucleocapsids form in ΔUL32-infected nuclei; however, these nucleocapsids remain either incompletely formed or unstable based on the rare appearance of virus particles in the cytoplasm by EM. This lack of cytoplasmic virus particles correlates with the restricted localization of pp28 to the AC in the UL32 mutant virus infections of HFs in fluorescent assays.
Based on these results, the assembly of virus particles in the nuclei of ΔUL32-infected cells appeared normal, but subsequent events involving nuclear egress and cytoplasmic maturation appeared compromised. The major differences in the cytoplasmic accumulation of recognizable virions could result either from defects during egress from the nucleus or entry into the cytoplasm. The mechanism underlying such nucleocapsid instability may be resolved with a greater understanding of the host cell processes involved in these steps.
DISCUSSION
HCMV pp150 is necessary for virion maturation, apparently as nucleocapsids egress from the nucleus and translocate to the cytoplasmic AC. On the basis of existing data, this tegument protein is not needed for the formation of nucleocapsids, encapsidation, or nuclear egress. CR1 and CR2 are necessary (2), and small clustered mutations made within either of these CR compromise pp150 function. Here, single-point mutations introduced into conserved amino acids within CR2 were shown to disrupt pp150 function. The phenotype of these mutants was similar to the phenotype of a virus carrying a complete deletion of the ORF (ΔUL32), whether point mutants were surveyed by transient complementation of ΔUL32 or following reconstruction into recombinant bacmids. Single-point mutants in conserved CR2 (N201A and G207A) as well as clustered point mutations in either CR1 or CR2 (2) prevent this protein from functioning, in agreement with a long-suspected role for this region in interactions with nucleocapsids (4).
We gained further insights into the role of pp150 during virion maturation by studying the patterns of viral structural protein and DNA accumulation in the absence of pp150, as well as during infections in which nonfunctional point mutants were made. In either case, we observed identical defects in virus particle assembly and maturation. Important findings were as follows. (i) Point mutations in CR2 amino acids N201 and G207 individually disrupted pp150 function. (ii) Nonfunctional pp150 resulted in a cytoplasmic block late in viral replication. Virus particles failed to exit the AC even though gM:gN and gB complexes localized properly. (iii) The organization of the AC was altered in the absence of pp150 based on the localization of trans-Golgi network and EM studies comparing Towne-BAC and UL32 mutant virus infections of HFs. (iv) Fewer virus particles accumulated in the cytoplasm of infected cells in the absence of pp150, although nuclear steps in assembly remained unaffected. These data implicate pp150 exclusively in cytoplasmic maturation events and suggest that this tegument protein controls the fate of nucleocapsids as they associate with assembly sites within the AC. Alternatively, pp150 may either stabilize nucleocapsid structure and prevent a time-dependent decay or contribute to the formation of the AC from cellular vesicle transport machinery.
The identification of two critical amino acids in CR2 reinforces the importance of interactions between pp150 and viral or cellular targets that depend on this region in support of virion maturation. The most relevant interaction is likely to be with nucleocapsids, as proposed by Baxter and Gibson, based on work with simian and human CMV (4). The apparent instability that we observed using ΔUL32 virus infection is best accommodated through a mechanism by which pp150 is responsible for stabilizing nucleocapsids that have been transported from the nuclear site of assembly to the cytoplasm, where envelopment occurs. The new twist from our data is the observation that pp150 function is necessary in the cytoplasm of infected cells, rather than in the nucleus where capsid assembly and the encapsidation of viral DNA occur. Given the common phenotype of viruses that express nonfunctional pp150 point mutants or that carry a complete deletion of the UL32 ORF, our data suggest that CR2-dependent interactions contribute to the cytoplasmic fate of preassembled nucleocapsids.
Virus maturation is blocked at the AC in the absence of pp150 function, as supported by the restricted localization of pp28 and MCP to these compartments in pp150 mutant virus-infected cells but not in Towne-BAC or functional pp150 mutant virus infections. Based on transmission EM images, cytoplasmic virus particles were infrequent in ΔUL32-BAC infections, which is surprisingly different from Towne-BAC infections, even though the nuclear assembly of nucleocapsids appeared to be similar. The localization of pp28 was consistent with a failure of envelopment at the AC, as discussed previously (2). While MCP and viral DNA were transported from the nucleus to the cytoplasm regardless of pp150 function, nucleocapsids per se were not readily detected in the cytoplasm when cells were examined following EM. These results were consistent with a pp150-independent transport of viral structural components out of the nucleus but an accumulation of structural components in the vicinity of the AC. Differences in fixation procedures are known to influence the detection of MCP in the cytoplasm, in agreement with differences in epitope exposure that are associated with the pp150 stabilization of nucleocapsids (2). Taken together with EM studies, this exposure of MCP epitope may be an indication of capsid instability in the absence of pp150. In addition, gM:gN complexes localized normally in AC regardless of pp150 function, confirming that the assembly of gM:gN complexes is independent of pp150 or virion maturation. Our observations suggest that pp150 function contributes to the unique organization of the AC (12). The area coinciding with the AC was found to be highly vesiculated in EM studies in ΔUL32 virus infections compared to a compartmentalized region of virus particles and dense bodies in Towne-BAC infections. Despite many morphological similarities in mutant and Towne-BAC virus-infected cells, Golgin 97 staining revealed a compromised orientation of cellular compartments constituting the AC.
This study advances the understanding of the function of viral tegument proteins in maturation. A number of important issues remain to be resolved, including (i) whether pp150 is required at a stage in nuclear export, (ii) the identification of pp150 interaction partners, and (iii) the position of pp150 within the virion and proximity to the capsid.
Acknowledgments
We acknowledge William J. Britt at the University of Alabama, Birmingham, for providing 14-16A, 28-4, and 36-14 antibodies. We thank Katherine Schaefer-Hales at the cell imaging and microscopy core at The Winship Cancer Institute for help with confocal microscopy and Hong Yi at the School of Medicine Microscopy Core for Electron Microscopy. We appreciate the help of William J. Kaiser in cell culture.
PHS grant (RO1 AI20211) funded the research.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 7810995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 808199-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzeh, M., A. Honigman, A. Taraboulos, A. Rouvinski, and D. G. Wolf. 2006. Structural changes in human cytomegalovirus cytoplasmic assembly sites in the absence of UL97 kinase activity. Virology 35469-79. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 756865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 9714506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britt, W. 2007. CMV maturation and egress, p. 311-323. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 7.Britt, W. J., and D. Auger. 1985. Identification of a 65000 Dalton virion envelope protein of human cytomegalovirus. Virus Res. 431-36. [DOI] [PubMed] [Google Scholar]
- 8.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, M., S. A. Rudolph, B. Plachter, B. Barrell, and G. Jahn. 1989. Identification of the major capsid protein gene of human cytomegalovirus. J. Virol. 631345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 26010-16. [DOI] [PubMed] [Google Scholar]
- 11.Compton, T., and A. Feire. 2007. Early events in human cytomegalovirus infection, p. 231-240. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 12.Das, S., A. Vasanji, and P. E. Pellett. 2007. Three dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 8111861-11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, S., N. Costantino, and D. L. Court. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379109-115. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 10014223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39389-400. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 972692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 761591-1601. [DOI] [PubMed] [Google Scholar]
- 18.Hertel, L., S. Chou, and E. S. Mocarski. 2007. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 3e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130118-133. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 781488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krzyzaniak, M., M. Mach, and W. J. Britt. 2007. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J. Virol. 8110316-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landini, M. P., B. Severi, L. Badiali, E. Gonczol, and G. Mirolo. 1987. Structural components of human cytomegalovirus: in situ localization of the major glycoprotein. Intervirology 27154-160. [DOI] [PubMed] [Google Scholar]
- 24.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 792160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 26.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7980-990. [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski, E. S. 2007. Betherpesvirus genes and their functions, p. 202-228. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 28.Mocarski, E. S. 2007. Comparative analysis of herpesvirus-common proteins, p. 44-58. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 29.Mocarski, E. S., Jr., T. Shenk, and R. F. Pass. 2006. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 30.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 31.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 7915494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roby, C., and W. Gibson. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59714-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 792754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly J. Virol. 74975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmolke, S., H. F. Kern, P. Drescher, G. Jahn, and B. Plachter. 1995. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 695959-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 8011658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo, J. Y., and W. J. Britt. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 816536-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo, J. Y., and W. J. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 805611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 9851-64. [DOI] [PubMed] [Google Scholar]
- 40.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 7710594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomason, L. C., D. L. Court, M. Bubunenko, N. Costantino, H. Wilson, S. Datta, and A. Oppenheim. 2003. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 20031.16.11-11.16.16. [DOI] [PubMed] [Google Scholar]
- 42.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60163-178. [PubMed] [Google Scholar]
- 43.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 732181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf, D. G., C. T. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Distinct and separate roles for herpesvirus-conserved UL97 kinase in cytomegalovirus DNA synthesis and encapsidation. Proc. Natl. Acad. Sci. USA 981895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]