Abstract
The initiation of eukaryotic DNA replication is preceded by the assembly of prereplication complexes (pre-RCs) at chromosomal origins of DNA replication. Pre-RC assembly requires the essential DNA replication proteins ORC, Cdc6, and Cdt1 to load the MCM DNA helicase onto chromatin. Saccharomyces cerevisiae Noc3 (ScNoc3), an evolutionarily conserved protein originally implicated in 60S ribosomal subunit trafficking, has been proposed to be an essential regulator of DNA replication that plays a direct role during pre-RC formation in budding yeast. We have cloned Schizosaccharomyces pombe noc3+ (Spnoc3+), the S. pombe homolog of the budding yeast ScNOC3 gene, and functionally characterized the requirement for the SpNoc3 protein during ribosome biogenesis, cell cycle progression, and DNA replication in fission yeast. We showed that fission yeast SpNoc3 is a functional homolog of budding yeast ScNoc3 that is essential for cell viability and ribosome biogenesis. We also showed that SpNoc3 is required for the normal completion of cell division in fission yeast. However, in contrast to the proposal that ScNoc3 plays an essential role during DNA replication in budding yeast, we demonstrated that fission yeast cells do enter and complete S phase in the absence of SpNoc3, suggesting that SpNoc3 is not essential for DNA replication in fission yeast.
The timely and faithful duplication of large and discontinuous eukaryotic genomes is achieved by the coordinated initiation of DNA replication from hundreds to thousands of chromosomal origins of DNA replication during S phase. The initiation of DNA replication is a stepwise process involving (i) the assembly of prereplication complexes (pre-RCs) at replication origins during G1 phase, (ii) pre-RC activation by conserved S-phase-promoting kinases, and (iii) the establishment of bidirectional DNA replication forks (the replisome) (reviewed in references 6, 24, 34, and 47). Genetic and biochemical approaches have revealed many, if not all, of the factors required for pre-RC assembly (ORC, Cdc6, Cdt1, MCM, and Mcm10), pre-RC activation (CDK and DDK), and replisome assembly (Cdc45, Sld2, Sld3, GINS, RPA, and DNA polymerases). However, the precise functions of many of these proteins await elucidation.
Although the size and sequence composition of chromosomal origins vary widely among eukaryotes (reviewed in reference 12), ORC is universally required to recruit Cdc6 and Cdt1 to chromosomal replication origins prior to S phase (10, 11, 28). In turn, Cdc6 and Cdt1 cooperate with ORC to mediate the ATP-dependent loading of the heterohexameric MCM complex (Mcm2-7) onto chromatin to establish a functional pre-RC (16, 19, 22, 23, 28, 35, 44, 48, 49). The periodic expression and chromatin association of both Cdc6 and Cdt1 serve to restrict MCM loading and pre-RC formation to the G1 phase and ensure that DNA replication occurs only once per cell cycle (2, 20, 21, 25, 35, 36, 39).
Results from experiments in budding yeast suggest that Saccharomyces cerevisiae Noc3 (ScNoc3), a protein essential for large (60S) ribosomal subunit biogenesis (31), plays a direct role in pre-RC formation and is essential for the initiation of DNA replication (52). Budding yeast ScNoc3 was demonstrated to interact with ScMCM and ScORC proteins, while the inactivation of ScNoc3 impaired the chromatin binding of ScCdc6 and ScMCM proteins and resulted in delayed S phase entry. These observations were particularly interesting in the context of a concurrent report that demonstrated a link between another protein required for ribosome biogenesis in budding yeast, ScYph1, and components of the pre-RC (17), suggesting that proteins involved in ribosome production may also function to coordinate the duplication of the genome with cell growth.
ScNoc3 is a member of the family of four nucleolar complex-associated proteins (ScNoc1 to ScNoc4) required for the maturation of small and large ribosomal subunits in budding yeast (31, 32). Heterodimeric subcomplexes of ScNoc1-ScNoc2 and ScNoc2-ScNoc3 associate with different preribosomal particles and localize to different subnuclear compartments. ScNoc1 and ScNoc2 associate primarily with 90S and 66S preribosomes in the nucleolus, while ScNoc2 and ScNoc3 are complexed with 66S preribosomal particles primarily in the nucleoplasm. Consistent with these observations, ScNoc1 and ScNoc2 are required for the transport of pre-60S ribosomal subunits from the nucleolus to the nucleoplasm, whereas the ScNoc2-ScNoc3 complex functions in the transport of pre-60S subunits from the nucleoplasm to the cytoplasm. These results suggest that dynamic interactions between different ScNoc1-ScNoc3 proteins mediate distinct functions during ribosome maturation and transport: the release of pre-60S ribosomal particles from the nucleolus (ScNoc1-ScNoc2) and the acquisition of competence for nucleocytoplasmic transport (ScNoc2-ScNoc3).
We have been interested in understanding the molecular mechanisms that regulate pre-RC assembly and activation using the fission yeast Schizosaccharomyces pombe as a model eukaryotic system. We therefore cloned the fission yeast S. pombe noc3+ (Spnoc3+) gene and examined the requirement for the SpNoc3 protein during ribosome biogenesis, cell cycle progression, and DNA replication. We demonstrated that fission yeast Spnoc3+ is an essential gene and SpNoc3 protein activity is required for the accumulation of large ribosomal subunits, consistent with the requirement for ScNoc3 during ribosome biogenesis in budding yeast. We also observed that SpNoc3 inactivation in fission yeast results in the accumulation of septated and binucleated cells, suggesting that SpNoc3 is also required for normal cell division. However, contrary to the proposed essential role for ScNoc3 during budding yeast DNA replication, we demonstrated that fission yeast cells not expressing SpNoc3 are capable of entering and completing S phase, suggesting that SpNoc3 does not have a direct and essential role in regulating fission yeast DNA replication.
MATERIALS AND METHODS
Cloning of Spnoc3+.
A search of the S. pombe genome database (S. pombe Genome Project at the Sanger Institute [www.sanger.ac.uk/Projects/S.pombe]) identified a single protein-coding sequence (GenBank accession number SPBC887.03C) sharing significant amino acid homology with ScNoc3 (E value of 6 × 10−73). PCR primers were designed to amplify the entire 2,241-bp SPBC887.03C cDNA (herein referred to as Spnoc3+) from an S. pombe cDNA library (a gift of G. Hannon, Cold Spring Harbor Laboratory, Plainview, NY).
S. pombe plasmids and strains.
Strain CHY86, a heterozygous Spnoc3+/Spnoc3Δ:ura4+ diploid strain, was generated by using standard gene replacement methods (33). For this purpose, a DNA fragment containing Spura4+ flanked by 1-kb genomic fragments from both the 5′ and 3′ sides of the Spnoc3+ open reading frame was introduced into diploid strain YJ18. Heterozygous Spnoc3+/Spnoc3Δ:ura4+ diploids were identified among ura+ transformants by colony PCR.
SpNoc3 shutoff strains expressing SpNoc3-hemagglutinin (HA) from a plasmid under the regulation of the attenuated thiamine-repressible Spnmt1+ promoter (5) in a Spnoc3Δ:ura4+ haploid were constructed as follows. Plasmid pRCE29 was derived from pSLF372 (18) by replacing the Spura4+ gene with the ScLEU2 gene. The Spnoc3+ cDNA was cloned into pRCE29 in frame with the triple HA epitope tag to generate plasmid pCRH303. Strain CHY108 was generated by transforming the heterozygous Spnoc3+/Spnoc3Δ:ura4+ diploid (CHY86) with pCRH303 and selecting ura+ and leu+ haploids. Strain CHY108 was crossed by standard genetic methods with YMF111 and TK177 to generate strains CHY160 and CHY161, respectively.
The SpNoc3 IIPGY-to-AAAAA alteration (Spnoc3ts) was introduced into the Spnoc3+ locus to generate strain CHY222 as follows. A DNA fragment containing the Spnoc3+ coding sequence flanked by 1-kb genomic fragments from both the 5′ and 3′ sides of the Spnoc3+ open reading frame was PCR amplified from genomic DNA and cloned as a NotI fragment into the NotI sites of plasmid pARG3MYC to generate plasmid pCRH349. The Spnoc3+ sequence in pCRH349, encoding amino acid residues 271 to 275, was changed to alanine residues by PCR mutagenesis to generate plasmid pCRH350. The NotI DNA fragment was isolated from plasmid pCRH350 and introduced into strain CHY161, and clones that were 5-fluoroorotic acid resistant were selected. Several independent clones of the resultant strain, CHY222, that lacked the pCRH303 rescue plasmid and were viable at 25°C but not viable at 36°C were identified.
S. pombe strains used in this study can be found in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| YJ18 | h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 |
| TK17 | h− cdc10-129 ura4-D18 ade6-M210 |
| GBY118 | h+ ura-D18 leu1-32 cdc25-22 |
| YMF111 | h+ ura-D18 leu1-32 ade6-M210 cdc10-129 |
| TK177 | h+ ura4-D18 leu1-32 his3-D1 arg3-D4 |
| CHY82 | h− cdc10-129 ura4-D18 ade6-M210 noc3+::3HA[ura4+] |
| CHY86 | h+/h− noc3+/Δnoc3::ura4+ leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 |
| CHY108 | h− Δnoc3::ura4+ leu1-32 ura4-D18 ade6-M210 pCRH303 |
| CHY154 | h+ ura4-D18 leu1-32 arg3-D4 |
| CHY159 | h− Δnoc3::ura4+ leu1-32 ura4-D18 cdc25-22 pCRH303 |
| CHY160 | h− Δnoc3::ura4+ leu1-32 ura4-D18 cdc10-129 pCRH303 |
| CHY161 | h+ Δnoc3::ura4+ leu1-32 ura4-D18 arg3-D4 pCRH303 |
| CHY202 | h+ Δnoc3::ura4+ leu1-32 ura4-D18 arg3-D4 pCRH303 pARG3HA |
| CHY203 | h+ Δnoc3::ura4+ leu1-32 ura4-D18 arg3-D4 pCRH303 pCRH320 |
| CHY212 | h+ Δnoc3::ura4+ leu1-32 ura4-D18 arg3-D4 pCRH33 pCRH336 |
| CHY222 | h+ leu1-32 ura4-D18 arg3-D4 noc3ts |
S. pombe media and manipulations.
Standard methodologies (33) were used for maintenance, plasmid transformation, genetic crosses, tetrad analysis, and selective spore germination of S. pombe strains. Edinburgh minimal medium (EMM) (Bio 101) was supplemented with 250 μg/ml of each adenine, leucine, uracil, lysine, and histidine and 1 mg/ml arginine. EMM6S-leu, EMM6S-ura, and EMM6S-arg are EMM6S medium without the addition of leucine, uracil, and arginine, respectively. EMM-N is EMM lacking nitrogen (Bio 101). Where indicated, 5 μg/ml thiamine was added to plates or liquid medium. Cells were grown at 30°C unless otherwise indicated. Yeast extracts were prepared for Western blot analysis as described previously (9).
HU arrest of strains CHY154 and CHY222.
For the hydroxyurea (HU) arrest and release of strains CHY154 and CHY222, cells were grown in EMM6S at 36°C to early log phase, followed by the addition of 25 mM HU for 4 h at 36°C. Cells were analyzed by flow cytometry for DNA content or by phase-contrast microscopy or fluorescence microscopy of DAPI (4′,6′-diamidino-2-phenylindole)-stained cells.
Ribosome profiles of strains CHY154 and CHY222.
The relative concentration of 40S and 60S ribosomal subunits was determined using low-Mg2+ conditions as previously described (3). Briefly, 10 A260 units of cell extracts were layered onto 5% to 45% sucrose gradients prepared in polysome lysis buffer (20 mM Tris-HCl [pH 7.5], 40 mM EDTA, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/ml cycloheximide, 0.2 mg/ml heparin) and centrifuged for 4 h at 39,000 rpm and 4°C in a Beckman SW41 rotor. The gradients were then fractionated by upward displacement with 55% (wt/vol) sucrose using a gradient fractionator (Brandel Inc.) connected to a UA-6 UV monitor (Teledyne Isco) for continuous measurement of the absorbance at 254 nm.
RESULTS
Spnoc3+ is an essential gene.
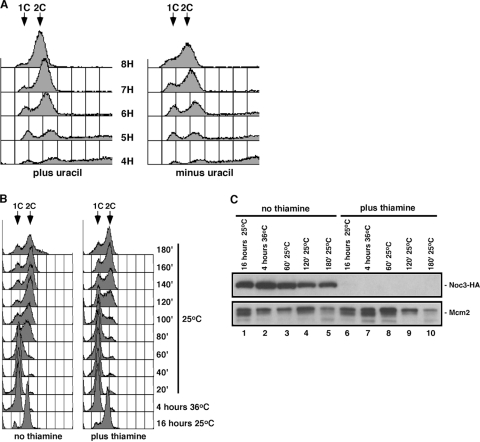
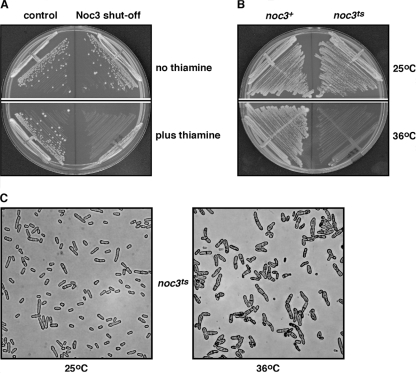
Analysis of the Schizosaccharomyces pombe genome database identified a single protein-coding sequence (GenBank accession number SPBC887.03C) (herein referred to as Spnoc3+) predicted to encode a 747-amino-acid polypeptide (SpNoc3) that shares significant amino acid sequence homology with ScNoc3 (E value of 1 ×10−73) and human HsNoc3 (E value of 6 ×10−48). We generated a Spnoc3+/Spnoc3Δ:ura4+ heterozygous diploid (strain CHY86) to determine whether Spnoc3+ is an essential gene. Tetrad analysis of the Spnoc3+/Spnoc3Δ:ura4+ heterozygous diploid demonstrated that Spnoc3Δ:ura4+ cells are inviable (data not shown). We also generated a fission yeast haploid SpNoc3 “shutoff” strain (CHY108) in which the Spnoc3Δ:ura4+ disruption was complemented by a plasmid expressing SpNoc3-HA under the control of the thiamine-repressible nmt1+ promoter. Thiamine represses the expression of SpNoc3-HA, with no protein detected 16 h after thiamine addition (see Fig. 4C, lane 6). Wild-type cells (control) and cells expressing SpNoc3-HA in the SpNoc3 shutoff strain were capable of forming colonies on plates lacking thiamine (Fig. 1A). In contrast, SpNoc3 shutoff cells grown in the presence of thiamine (Fig. 1A) did not form single colonies. These results confirm that Spnoc3+ is an essential gene and that the expression of SpNoc3 is required for viability in fission yeast.
FIG. 4.
SpNoc3 is not essential for DNA replication. (A) Flow cytometric analysis of the DNA content of a selective germination of spores derived from the Spnoc3+/Spnoc3Δ:ura4+ heterozygous diploid strain CHY86, in which only Spnoc3Δ:ura4+ spores are capable of germinating in the medium lacking uracil (note that the more rapid accumulation of cells with a 2C DNA content in medium containing uracil [plus uracil] than in spores grown in the absence of uracil [minus uracil] is the result of both Spnoc3+ and Spnoc3Δ:ura4+ spores germinating in medium containing uracil). (B) The SpNoc3 shutoff strain (CHY160) was grown for 16 h at 25°C in the absence (no thiamine) or presence (plus thiamine) of thiamine, shifted to 36°C for 4 h to arrest cells in G1, and released from the G1 block at 25°C for 3 h. Samples were taken every 20 min for flow cytometric analysis of DNA content. (C) Western blot analysis of expression of SpNoc3-3HA and SpMcm2 (in cells from B) after 16 h at 25°C (lanes 1 and 6), at 4 h at 36°C (lanes 2 and 7), and at 60 min (lanes 3 and 8), 120 min (lanes 4 and 9), and 180 min (lanes 5 and 10) after return to 25°C in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of thiamine.
FIG. 1.
Spnoc3+is an essential gene. (A) Wild-type cells (control) and cells expressing SpNoc3-HA in the Spnoc3Δ:ura4+ disruption haploid strain (SpNoc3 shutoff) were examined for growth on plates in the presence or absence of thiamine. (B) Wild-type Spnoc3+ (CHY154) and Spnoc3ts mutant (CHY222) strains were examined for growth on plates at 25°C and 36°C. (C) Microscopic analysis of the Spnoc3ts mutant strain grown on plates for 72 h at 25°C and 36°C.
To identify essential domains required for SpNoc3 function in fission yeast cells, we introduced small (3 to 6 amino acids) alanine substitution mutations in conserved SpNoc3 protein-coding regions and examined the ability of the resultant Spnoc3 mutant alleles, ectopically expressed from a plasmid under the control of the Spnoc3+ promoter, to restore viability to the SpNoc3 shutoff strain (CHY108) grown in the presence of thiamine. We identified one Spnoc3 mutant allele (alanine substitutions at amino acids 271 to 275 [IIPGY]) that restored cell viability at 25°C but not at 36°C (data not shown), indicating that this mutant allele of Spnoc3 (Spnoc3ts) conferred a temperature-sensitive phenotype to fission yeast.
The Spnoc3ts mutant allele was integrated at the chromosomal Spnoc3+ locus to generate the Spnoc3ts mutant strain (CHY222). The Spnoc3ts mutant was able to grow on plates at 25°C but was not viable at 36°C (Fig. 1B), consistent with the temperature-sensitive phenotype conferred when the Spnoc3ts mutant allele was ectopically expressed from a plasmid. The ectopic expression of the wild-type SpNoc3 protein from a plasmid was able to rescue the viability of the Spnoc3ts mutant strain at 36°C (data not shown), confirming that the temperature-sensitive phenotype of the Spnoc3ts mutant strain was due to the integrated Spnoc3ts mutant allele. After incubation at the restrictive temperature for 72 h, the Spnoc3ts mutant strain arrested with a variety of abnormal morphologies including multiseptated, elongated, and branched cells (Fig. 1C). Similar results were observed using the SpNoc3 shutoff strain (data not shown). However, inactivation of SpNoc3 protein function in the Spnoc3ts mutant was not lethal, as demonstrated by the ability of the mutant, after incubation at 36°C for 72 h, to form colonies of cells with normal morphologies when returned to the permissive temperature (data not shown). Together, these results demonstrate that the fission yeast Spnoc3+ gene is required for cell viability and that the loss of SpNoc3 protein function results in a pleiotropic phenotype with gross, but reversible, growth defects and morphologies.
SpNoc3 is required for 60S ribosomal subunit maturation.
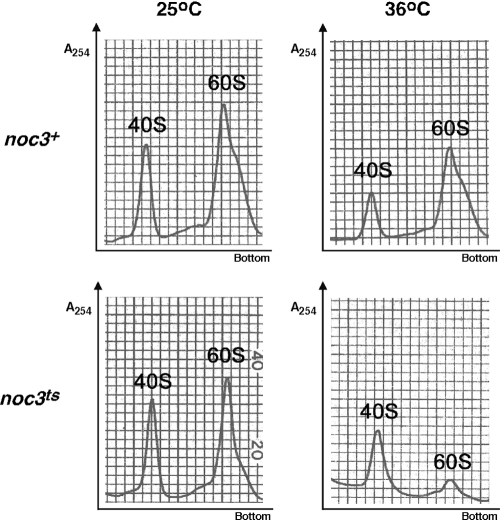
To determine whether SpNoc3 is required for ribosome biosynthesis in fission yeast, ribosome profiling experiments were carried out using the Spnoc3ts mutant strain (CHY222) and an isogenic Spnoc3+ wild-type control strain (CHY154) grown at the permissive and restrictive temperatures. Cellular extracts were prepared under a low-Mg2+ ion concentration that causes all cellular ribosomes to dissociate into separate subunits. As can be seen in Fig. 2, the Spnoc3ts strain grown at 25°C as well as the control strain grown at 36°C displayed similar 40S:60S ribosomal subunit ratios. In contrast, the Spnoc3ts mutant strain grown at the restrictive temperature resulted in a significant decrease in 60S ribosomal subunit levels, whereas the level of 40S ribosomal subunits did not change. These results are consistent with findings reported previously for budding yeast demonstrating an increase in the 40S:60S ratio in extracts prepared from an ScNoc3 mutant strain (31). These results demonstrate that budding yeast ScNoc3 and fission yeast SpNoc3 are functionally homologous proteins essential for the complete maturation of large ribosomal subunits.
FIG. 2.
SpNoc3 is required for 60S ribosomal subunit maturation. Ribosome profiling experiments were carried out to examine the production of 40S and 60S ribosomal particles in the wild-type Spnoc3+ strain and the Spnoc3ts mutant strain grown at the permissive (25°C) and restrictive (36°C) temperatures.
SpNoc3 is required for normal cell division.
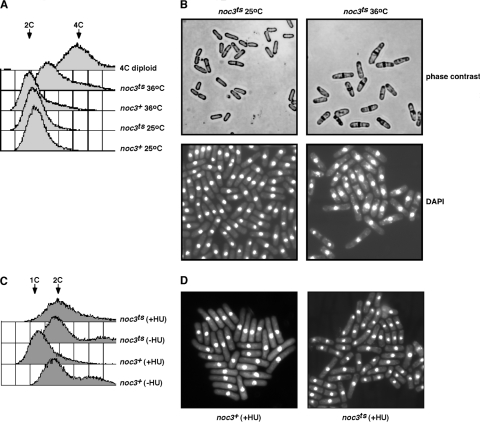
By flow cytometric analysis of DNA content, we observed that the Spnoc3ts mutant cells grown at the nonpermissive temperature for 20 h in liquid medium completed S phase before arresting with a DNA content greater than 2C (but less than 4C) (Fig. 3A). The rightward shift in DNA content of the Spnoc3ts mutant is typical for arrested S. pombe cells and is likely due to mitochondrial DNA synthesis that occurs in the absence of nuclear DNA replication (43). Microscopic analysis revealed the appearance of binucleated Spnoc3ts mutant cells containing thickened septa by 10 h at the restrictive temperature, and by 20 h, approximately 30% of the Spnoc3ts mutant cells were septated (Fig. 3B) and binucleated (DAPI), compared to 5% of the wild-type Spnoc3+ cells. These results suggest that fission yeast cells do enter and complete S phase in the absence of functional SpNoc3 but are defective in completing normal cell division.
FIG. 3.
SpNoc3 is required for normal cell division. (A) The DNA contents of the wild-type Spnoc3+ strain and the Spnoc3ts mutant strain grown for 20 h at the permissive (25°C) and restrictive (36°C) temperatures were determined by flow cytometric analysis. (B) Microscopic examination of Spnoc3ts mutant cells grown in liquid medium at 36°C for 20 h. (C) Flow cytometric analysis of the DNA content of wild-type Spnoc3+ (CHY154) and Spnoc3ts mutant (CHY222) cells grown in liquid media at 36°C for 4 h and then incubated in the absence (−HU) or presence (+HU) of HU for an additional 4 h at 36°C. (D) Microscopic examination of wild-type Spnoc3+ and Spnoc3ts mutant cells grown in liquid medium at 36°C for 4 h and then incubated in the presence of HU for an additional 4 h at 36°C.
We employed a second approach to examine whether the inactivation of SpNoc3 protein function results in cell division defects in fission yeast. Normally, fission yeast cells grown in the presence of HU, an inhibitor of DNA replication, will undergo mitosis and cell division and arrest in early S phase with a 1C DNA content. We reasoned that if SpNoc3 was not required for completing cell division, Spnoc3ts mutant cells would complete cell division at the restrictive temperature and arrest with a 1C DNA content in the presence of HU. Alternatively, if SpNoc3 inactivation does prevent normal cell division, then we expected that Spnoc3ts mutant cells with a 2C DNA content would be evident. To distinguish between these two possibilities, wild-type Spnoc3+ and Spnoc3ts mutant cells were grown for 4 h at the nonpermissive temperature of 36°C in the absence of HU (Fig. 3C), followed by the addition of HU to the medium and growth for an additional 4 h at 36°C. As expected, the wild-type Spnoc3+ cells did arrest in early S phase with a 1C DNA content (Fig. 3C) and with single nuclei (Fig. 3D) in the presence of HU. In contrast, the Spnoc3ts mutant cells treated with HU accumulated as septated and binucleated cells (Fig. 3D) with a 2C DNA content (Fig. 3C), supporting our observations that SpNoc3 activity is required for normal cell division in fission yeast.
SpNoc3 is not essential for DNA replication.
Results from a previous study led to the suggestion that budding yeast ScNoc3 plays a direct role in facilitating pre-RC formation and is essential for DNA replication. To examine the requirement for SpNoc3 during DNA replication in fission yeast, a selective spore germination experiment of the Spnoc3+/Spnoc3Δ:ura4+ heterozygous diploid (strain CHY86) was performed in medium lacking uracil. Under these conditions, only Spnoc3Δ:ura4+ spores (uracil prototrophs) were able to germinate, while Spnoc3+ spores (uracil auxotrophs) remain as ungerminated spores. Results obtained from flow cytometric analysis of DNA content indicated that the majority of the germinating Spnoc3Δ:ura4+ spores do undergo a complete round of chromosomal duplication (minus uracil) (Fig. 4A), similar to the Spnoc3+ spores germinated in medium containing uracil, suggesting that SpNoc3 is not essential for DNA replication in fission yeast. However, we cannot rule out the possibility that Spnoc3Δ:ura4+ spores do complete DNA replication due to maternally contributed wild-type SpNoc3 protein.
We therefore employed a second approach to examine if SpNoc3 is essential for fission yeast DNA replication by assaying the ability of haploid cells not expressing SpNoc3 to enter S phase after release from a block in G1 phase. We introduced the temperature-sensitive cdc10-129 allele (which blocks cells in G1 phase at 36°C) into the SpNoc3 shutoff strain (CHY108) to generate the cdc10-129/SpNoc3 shutoff strain (CHY160). The expression of the SpNoc3-HA protein in this strain was undetectable by Western blot analysis after 16 h of growth in medium containing thiamine (Fig. 4C, lane 6). The cdc10-129/SpNoc3 shutoff strain was grown for 16 h at the permissive temperature of 25°C in the presence (SpNoc3 not expressed) or absence (SpNoc3 expressed) of thiamine before shifting to the nonpermissive temperature of 36°C for 4 h to block cells in G1 phase. Cells were returned to the permissive temperature in the presence or absence of thiamine, and samples were taken every 20 min for 3 h for flow cytometric analysis of cellular DNA content to monitor cell cycle progression after release from the G1 block.
In either the presence or the absence of thiamine, CHY160 cells blocked normally in G1 with a 1C DNA content after 4 h at the restrictive temperature of 36°C (Fig. 4B). Upon release from the G1 block, CHY160 cells expressing SpNoc3-HA (Fig. 4C, lanes 1 to 5) entered S phase between 60 and 80 min and completed DNA synthesis by approximately 120 min, as expected (no thiamine) (Fig. 4B). Similarly, cells lacking SpNoc3-HA (Fig. 4C, lanes 6 to 10) also initiated DNA synthesis and completed S phase albeit with a delay of approximately 40 to 60 min (plus thiamine) (Fig. 4B) relative to cells expressing SpNoc3-HA. Similar results (delayed replication initiation and completion of S phase) were observed after release from a block in G2 phase induced by the cdc25-22 mutation (data not shown). These results suggest that SpNoc3 is not essential for the initiation of DNA replication or progression through S phase in fission yeast. The 40- to 60-min S-phase entry delay observed in CHY160 cells not expressing SpNoc3-HA is likely the result of defects in ribosome production and decreased levels of expression of essential DNA replication proteins and/or a general cellular response to ribosome and protein synthesis defects (see Discussion).
DISCUSSION
We demonstrated that fission yeast SpNoc3 and budding yeast ScNoc3 proteins are functional homologs required for cell viability (Fig. 1A and B) and the maturation of the 60S ribosomal subunit (Fig. 2). We also found that fission yeast not expressing SpNoc3 accumulates as binucleated cells with thickened septa (Fig. 3), while a prolonged inactivation of SpNoc3 results in multiseptated, elongated, and branched cells (Fig. 1C), indicating that SpNoc3 is also required for completing normal cell division. Cell separation in fission yeast requires a number of proteins: the SpEgn1 and SpAgn1 glucanases for hydrolysis of the primary septum and old cell wall, respectively; SpSep1 and SpAce2, which regulate the expression of SpEgn1 and SpAgn1; the exocyst complex that targets SpEng1 and SpAgn1 to division sites; SpRho3, which modulates exocyst function; and SpMid2 and the SpSpn1 to SpSpn4 septins, which position the exocyst at the division site (1, 14, 15, 29, 30, 50, 51). The phenotypes observed in fission yeast mutants of these genes are very similar to those observed when fission yeast SpNoc3 is inactivated and suggest that the expression of SpNoc3 and/or ribosome production is required for completing cell separation. Similar phenotypes (binucleated cells with thickened septa and multiseptated, elongated, and branched cells) have also been observed in fission yeast in the absence of functional SpSkb15, a protein required for 60S ribosomal subunit biogenesis (26, 42), suggesting that defects in completing cell separation may be a general consequence of ribosome biogenesis defects.
Although defects in ribosome production likely affect global protein synthesis, the pronounced cell separation defect observed in fission yeast when SpNoc3 is not expressed may be attributed to three factors: (i) the G2 phase occupies approximately 70% of the fission yeast cell cycle, (ii) ribosomal protein genes are expressed in late G2 prior to the expression of genes required for cell separation (37, 40), and (iii) the completion of cell division requires new protein synthesis after mitosis. In support of this, the inhibition of protein synthesis by cycloheximide or the inactivation of the RNA polymerase II subunit SpRpb4 in fission yeast specifically affects the expression of SpSep1 and SpAce2 and results in defects in completing cell separation but not mitosis or cell septation (4, 38, 45). Additionally, the SpWee1 protein kinase is required to prevent cell separation in fission yeast in response to protein synthesis defects (46), suggesting that preventing cell division may be an active response to defects in ribosome biogenesis. Although the cell separation defect observed in the absence of SpNoc3 function in fission yeast may be indirect, resulting from defects in ribosome biogenesis, we cannot rule out a direct involvement of SpNoc3 during cell division. In fact, genetic interactions between ScNOC3 and ScCDC12, a component of the septin ring required for cytokinesis in S. cerevisiae, have been observed in budding yeast (13).
ScNOC3 was independently identified in a genetic screen as a multicopy suppressor of the budding yeast mcm5/cdc46-1 DNA replication mutant (52). The loss of ScNoc3 protein function in budding yeast resulted in defects in the association of ScCdc6 and ScMCM proteins with chromosomal DNA and delayed entry into S phase after release from a G1 block. From these studies, it was proposed that budding yeast ScNoc3 is essential for the initiation of DNA synthesis by directly facilitating prereplication complex assembly. However, results obtained using purified proteins derived from both budding yeast and Xenopus laevis suggested that ORC, Cdc6, Cdt1, MCM, and nucleoplasmin (in Xenopus) are sufficient for pre-RC formation in vitro (19, 22) without any apparent requirement for Noc3. We therefore characterized the requirements of SpNoc3 in regulating DNA replication in fission yeast.
Our results suggest that SpNoc3 does not have a direct and essential role during the initiation of DNA replication in fission yeast. We demonstrated that fission yeast cells not expressing SpNoc3 do initiate and complete DNA replication after release from a G1 block (Fig. 4B). Although S phase entry is delayed, it must be emphasized that fission yeast does enter and complete S phase in the absence of functional SpNoc3. Second, germinating fission yeast Spnoc3Δ spores initiate DNA synthesis and complete chromosomal DNA replication (Fig. 4A). Third, thermosensitive Spnoc3ts fission yeast cells complete DNA replication before arresting with a 2C DNA content (Fig. 3A). Taken together, these three independent lines of evidence suggest that SpNoc3 does not play a direct and essential role during DNA replication in fission yeast.
Zhang and colleagues previously presented results from a single experiment that directly addressed the issue of whether budding yeast ScNoc3 is essential for chromosomal DNA replication (52). They demonstrated that ScNOC3-td (thermosensitive degron) cells delay S-phase entry after release from a G1-phase block in the absence of functional ScNoc3. However, it is evident from their data that the ScNOC3-td cells do enter and complete S phase after release from the G1 block. In contrast, thermosensitive degron budding yeast mutants of the essential replication proteins ScOrc6 (ScORC6-td) and all six MCM proteins (ScMCM2-td to ScMCM7-td) are not able to enter S phase after release from a block in G1 phase (8, 27). Therefore, budding yeast ScNoc3 is not essential for DNA replication, as claimed by those authors. We suggest that the delayed S-phase entry observed in ScNOC3-td budding yeast as well as the minor effects on the association of pre-RC components with chromatin are likely indirect consequences of defects in ribosome production. This hypothesis is supported by observations that defects in ribosome biogenesis in budding yeast delay DNA replication and S-phase entry in an ScSwe1- and ScWhi5-dependent manner (7, 41). Such an active-response mechanism may also function in fission yeast to delay S-phase entry in response to defects in ribosome biogenesis. In the present study, we have provided the results from three independent experiments that functionally addressed the role of SpNoc3 during fission yeast chromosomal DNA replication. Our results demonstrated in each case that fission yeast cells do enter and complete DNA replication in the absence of functional SpNoc3. Although Noc3 may be indirectly required for DNA replication in both yeasts, it is evident from both our results and those from budding yeast that Noc3 does not have a direct and essential role in DNA replication in either organism, as previously claimed.
Acknowledgments
We thank Greg Hannon for the S. pombe cDNA library used to clone Spnoc3+, Ray Chuang for the generation of plasmid pRCE29, and Yongjie Xu for the generation of strain YJ18.
This work was supported by a grant from the National Institute of General Medical Sciences to T.J.K. and by a CIHR grant, MOP84237, to F.B. F.B. is a New Investigator of the CIHR.
Footnotes
Published ahead of print on 7 July 2008.
REFERENCES
- 1.Alonso-Nunez, M. L., H. An, A. B. Martin-Cuadrado, S. Mehta, C. Petit, M. Sipiczki, F. del Rey, K. L. Gould, and C. R. de Aldana. 2005. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 162003-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, E. E., and J. C. Walter. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19114-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachand, F., and P. A. Silver. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 232641-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahler, J. 2005. A transcriptional pathway for cell separation in fission yeast. Cell Cycle 439-41. [DOI] [PubMed] [Google Scholar]
- 5.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123131-136. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71333-374. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, K. A., F. Bleichert, J. M. Bean, F. R. Cross, and S. J. Baserga. 2007. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol. Biol. Cell 18953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S., M. A. de Vries, and S. P. Bell. 2007. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 212897-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang, R., and T. J. Kelly. 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA 962656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 27716920-16927. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, T. R., P. B. Carpenter, and W. G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 8753-63. [DOI] [PubMed] [Google Scholar]
- 12.Cvetic, C., and J. C. Walter. 2005. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 16343-353. [DOI] [PubMed] [Google Scholar]
- 13.Davierwala, A. P., J. Haynes, Z. Li, R. L. Brost, M. D. Robinson, L. Yu, S. Mnaimneh, H. Ding, H. Zhu, Y. Chen, X. Cheng, G. W. Brown, C. Boone, B. J. Andrews, and T. R. Hughes. 2005. The synthetic genetic interaction spectrum of essential genes. Nat. Genet. 371147-1152. [DOI] [PubMed] [Google Scholar]
- 14.Dekker, N., A. de Haan, and F. Hochstenbach. 2006. Transcription regulation of the alpha-glucanase gene agn1 by cell separation transcription factor Ace2p in fission yeast. FEBS Lett. 5803099-3106. [DOI] [PubMed] [Google Scholar]
- 15.Dekker, N., D. Speijer, C. H. Grun, M. van den Berg, A. de Haan, and F. Hochstenbach. 2004. Role of the alpha-glucanase Agn1p in fission-yeast cell separation. Mol. Biol. Cell 153903-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donovan, S., J. Harwood, L. S. Drury, and J. F. Diffley. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 945611-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, Y. C., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109835-848. [DOI] [PubMed] [Google Scholar]
- 18.Forsburg, S. L., and D. A. Sherman. 1997. General purpose tagging vectors for fission yeast. Gene 191191-195. [DOI] [PubMed] [Google Scholar]
- 19.Gillespie, P. J., A. Li, and J. J. Blow. 2001. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 215-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann, J. F., and D. Beach. 1994. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 13425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jallepalli, P. V., G. W. Brown, M. Muzi-Falconi, D. Tien, and T. J. Kelly. 1997. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 112767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki, Y., H. D. Kim, A. Kojima, T. Seki, and A. Sugino. 2006. Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro. Genes Cells 11745-756. [DOI] [PubMed] [Google Scholar]
- 23.Kearsey, S. E., S. Montgomery, K. Labib, and K. Lindner. 2000. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 191681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69829-880. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, T. J., G. S. Martin, S. L. Forsburg, R. J. Stephen, A. Russo, and P. Nurse. 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74371-382. [DOI] [PubMed] [Google Scholar]
- 26.Kim, H. W., P. Yang, Y. Qyang, H. Lai, H. Du, J. S. Henkel, K. Kumar, S. Bao, M. Liu, and S. Marcus. 2001. Genetic and molecular characterization of Skb15, a highly conserved inhibitor of the fission yeast PAK, Shk1. Mol. Cell 71095-1101. [DOI] [PubMed] [Google Scholar]
- 27.Labib, K., J. A. Tercero, and J. F. Diffley. 2000. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 2881643-1647. [DOI] [PubMed] [Google Scholar]
- 28.Maiorano, D., J. Moreau, and M. Mechali. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404622-625. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Cuadrado, A. B., E. Duenas, M. Sipiczki, C. R. Vazquez de Aldana, and F. del Rey. 2003. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 1161689-1698. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Cuadrado, A. B., J. L. Morrell, M. Konomi, H. An, C. Petit, M. Osumi, M. Balasubramanian, K. L. Gould, F. Del Rey, and C. R. de Aldana. 2005. Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol. Biol. Cell 164867-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105499-509. [DOI] [PubMed] [Google Scholar]
- 32.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn, S. Schutz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2784072-4081. [DOI] [PubMed] [Google Scholar]
- 33.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194795-823. [DOI] [PubMed] [Google Scholar]
- 34.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells 7523-534. [DOI] [PubMed] [Google Scholar]
- 35.Nishitani, H., Z. Lygerou, T. Nishimoto, and P. Nurse. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404625-628. [DOI] [PubMed] [Google Scholar]
- 36.Nishitani, H., and P. Nurse. 1997. The cdc18 protein initiates DNA replication in fission yeast. Prog. Cell Cycle Res. 3135-142. [DOI] [PubMed] [Google Scholar]
- 37.Oliva, A., A. Rosebrock, F. Ferrezuelo, S. Pyne, H. Chen, S. Skiena, B. Futcher, and J. Leatherwood. 2005. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 3e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polanshek, M. M. 1977. Effects of heat shock and cycloheximide on growth and division of the fission yeast, Schizosaccharomyces pombe. With an appendix. Estimation of division delay for S. pombe from cell plate index curves. J. Cell Sci. 231-23. [DOI] [PubMed] [Google Scholar]
- 39.Rialland, M., F. Sola, and C. Santocanale. 2002. Essential role of human CDT1 in DNA replication and chromatin licensing. J. Cell Sci. 1151435-1440. [DOI] [PubMed] [Google Scholar]
- 40.Rustici, G., J. Mata, K. Kivinen, P. Lio, C. J. Penkett, G. Burns, J. Hayles, A. Brazma, P. Nurse, and J. Bahler. 2004. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36809-817. [DOI] [PubMed] [Google Scholar]
- 41.Saracino, F., J. Bassler, D. Muzzini, E. Hurt, and M. L. A. Carbone. 2004. The yeast kinase Swe1 is required for proper entry into cell cycle after arrest due to ribosome biogenesis and protein synthesis defects. Cell Cycle 3648-654. [PubMed] [Google Scholar]
- 42.Saveanu, C., J. C. Rousselle, P. Lenormand, A. Namane, A. Jacquier, and M. Fromont-Racine. 2007. The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors. Mol. Cell. Biol. 272897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97509-516. [DOI] [PubMed] [Google Scholar]
- 44.Seki, T., and J. F. Diffley. 2000. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 9714115-14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma, N., S. Marguerat, S. Mehta, S. Watt, and J. Bahler. 2006. The fission yeast Rpb4 subunit of RNA polymerase II plays a specialized role in cell separation. Mol. Genet. Genomics 276545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suda, M., S. Yamada, T. Toda, T. Miyakawa, and D. Hirata. 2000. Regulation of Wee1 kinase in response to protein synthesis inhibition. FEBS Lett. 486305-309. [DOI] [PubMed] [Google Scholar]
- 47.Teer, J. K., and A. Dutta. 2006. Regulation of S phase. Results and problems in cell differentiation. 4231-63. [DOI] [PubMed] [Google Scholar]
- 48.Tsuyama, T., S. Tada, S. Watanabe, M. Seki, and T. Enomoto. 2005. Licensing for DNA replication requires a strict sequential assembly of Cdc6 and Cdt1 onto chromatin in Xenopus egg extracts. Nucleic Acids Res. 33765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waga, S., and A. Zembutsu. 2006. Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts. J. Biol. Chem. 28110926-10934. [DOI] [PubMed] [Google Scholar]
- 50.Wang, H., X. Tang, and M. K. Balasubramanian. 2003. Rho3p regulates cell separation by modulating exocyst function in Schizosaccharomyces pombe. Genetics 1641323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H., X. Tang, J. Liu, S. Trautmann, D. Balasundaram, D. McCollum, and M. K. Balasubramanian. 2002. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell 13515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., Z. Yu, X. Fu, and C. Liang. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109849-860. [DOI] [PubMed] [Google Scholar]