Abstract
The kinesin-related protein Kar3 is a minus end-directed molecular motor that plays a multifunctional role in microtubule-directed nuclear movement. Previously, it was shown that Candida albicans Kar3p is critical for nuclear fusion during mating as kar3 mutants were defective in karyogamy. In this study, we confirm that Kar3p is required for nuclear congression in mating but that neither Kar3p nor the dynein motor protein Dyn1p is required for nuclear migration in the mating projection prior to cell fusion. In addition, we show that C. albicans Kar3p plays an important role in the cell and colony morphology of mitotically dividing cells, as evidenced by diminished filamentation of kar3 cells on Spider medium and an increased tendency of mutant cells to form pseudohyphal cells in liquid culture. Loss of Kar3p also led to defects in nuclear division, causing cells to grow slowly and exhibit reduced viability compared to wild-type cells. Slow growth was due, at least in part, to delayed cell cycle progression, as cells lacking Kar3p accumulated in anaphase of the cell cycle. Consistent with a role in mitotic division, Kar3 protein was shown to localize to the spindle pole bodies. Finally, kar3 cells exhibited unstable or aberrant mitotic spindles, a finding that accounts for the delay in cell cycle progression and decreased viability of these cells. We suggest that the altered morphology of kar3 cells is a direct consequence of the delay in anaphase, and this leads to increased polarized growth and pseudohypha formation.
The kinesin superfamily of proteins are ATP-driven molecular motors required for transporting cargo along the microtubule highways that traverse cells. Kinesins represent a large family of proteins involved in the intracellular trafficking of organelles, protein complexes, and mRNAs, as well as in mediating chromosome movement during cell division (25, 41, 61). Most homologs have microtubule plus end-directed motility, although a few kinesins have minus end-directed motility. Sequence analysis and microscopic studies of the minus end-directed kinesins, such as Saccharomyces cerevisiae Kar3p and Drosophila ncd, have indicated that their motility is due to the reversed orientation of the head and tail domains of the protein; while most kinesins contain the motor domain at the N terminus, the motor regions of Kar3p and ncd are at the C terminus (16, 38).
In S. cerevisiae, Kar3p is essential for nuclear fusion (karyogamy) during mating, as well as regulation of mitotic spindle assembly (39, 54). Mutants lacking Kar3p exhibit a range of mitotic phenotypes, including reduced growth rate, decreased cell viability, mitotic spindle instability, and mitotic arrest (39, 52, 54). In addition, it has been demonstrated that S. cerevisiae Kar3p acts as an antagonist to the kinesin-like motors Cin8p and Kip1p, and the dynein motor Dyn1p, during mitosis. Cin8p and Kip1p are nonessential motors that localize to the mitotic spindle and function during spindle assembly (26, 52, 54). Dyn1p is required for accurate positioning of the mitotic spindle at the bud neck prior to anaphase; loss of Dyn1p activity results in stalled nuclear migration, and nuclear division therefore occurs within the mother cell (55). Kar3p is required for an inwardly directed spindle force that regulates elongation and integrity of the anaphase mitotic spindle; overexpression of Kar3p leads to cells exhibiting shorter mitotic spindles (19, 26, 53). Thus, the balanced action of multiple microtubule motor proteins is required for normal spindle morphogenesis during mitotic division.
The N terminus of Kar3p is a nonmotor domain that interacts with other factors to execute cellular functions. For example, S. cerevisiae Kar3p forms a Kar3p-Cik1p heterodimer, which drives nuclear congression during karyogamy (35, 39, 43). Kar3p-Cik1p also functions in mitosis as loss of either protein leads to shortened mitotic spindles and the accumulation of large-budded cells, indicating cell cycle delay (39, 46). Kar3p forms alternative heterodimers with the related protein Vik1p; this complex is specific for mitotically dividing cells, where Vik1p acts to recruit Kar3p to spindle pole bodies during nuclear division (36).
While there is a single Kar3-like protein in S. cerevisiae, Schizosaccharomyces pombe has two kinesins of the Kar3p family that localize to different cellular locations and have distinct functions during mitotic division. One Kar3p homolog, Pkl1p, localizes to the nucleus during interphase and associates with spindle microtubules during mitosis (49, 59). A second Kar3p homolog, Klp2p, localizes to the cytoplasm during interphase but is translocated into the nucleus during mitotic division (59). In the absence of Klp2, mitotic spindles are lengthened while Pkl1p mutants exhibit shortened spindles. In addition, karyogamy failed in triple mutants lacking Klp2, Pkl1p, and the Dyn1p homolog Dhc1p (59). Thus, in S. pombe, Kar3p-like kinesins play important but distinct roles in promoting spindle assembly, cell cycle progression, chromosome segregation, and karyogamy.
The related ascomycete Candida albicans is the most commonly isolated human fungal pathogen, causing debilitating mucosal infections, as well as life-threatening systemic infections. The ability of C. albicans to cause disease is associated with its capacity to grow in multiple morphological forms. C. albicans yeast and pseudohyphal forms resemble those of S. cerevisiae, while the hyphal form is more analogous to true hyphal growth in filamentous fungi (11, 13, 32, 44, 57, 62). There is a growing interest in the proteins that regulate mitotic events in C. albicans, as these represent targets for novel antifungal drugs (14, 63). Recent studies have demonstrated that there is a single Kar3-like protein in C. albicans (8, 31). Despite high conservation with the motor domain of S. cerevisiae Kar3p (64% identity), C. albicans Kar3p shows less homology with the central region of S. cerevisiae Kar3p (25% identity) and contains a unique amino-terminal region not present in the S. cerevisiae protein (31). This suggests that the function of Kar3p may have diverged since C. albicans and S. cerevisiae last shared a common ancestor around 100 to 900 million years ago (20, 24). Similar to S. cerevisiae Kar3p, however, the C. albicans homolog is essential for efficient nuclear fusion during mating (8).
Since Kar3 proteins play pleiotropic roles in nuclear division in both S. cerevisiae and S. pombe, we have investigated the role of C. albicans Kar3p in mitotic division. We demonstrate that C. albicans cells lacking Kar3p exhibit abnormal cellular morphologies, including an increased tendency to form pseudohyphal cells, delayed anaphase progression, and abnormal mitotic spindle morphology. C. albicans Kar3p is also shown to localize to spindle pole bodies, the fungal equivalent of centrosomes in higher organisms. These observations indicate that Kar3p, similar to the case in S. cerevisiae, is required for normal mitotic spindle formation and efficient nuclear division in C. albicans.
MATERIALS AND METHODS
Media and reagents.
Standard laboratory media were prepared as previously described (22, 50). Spider medium contained 1.35% agar, 1% nutrient broth, 0.4% potassium phosphate, and 2% mannitol (pH 7.2) (33). Nourseothricin-resistant transformants were selected on yeast extract-peptone-dextrose (YPD) medium containing 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) as previously described (50). Alpha pheromone (GFRLTNFGYFEPG) was synthesized by Genemed Synthesis. Calcofluor white stain was obtained from Fluka Biochemika.
Strains.
All of the C. albicans strains used in this study are listed in Table 1, and the oligonucleotides used in this study are listed in Table 2. Starting strain SNY152 (leu2/leu2 his1/his1 arg4/arg4) (45) was used for the construction of KAR3 deletion strains. First, a and α derivatives of SNY152 were generated by growth on sorbose medium as previously described (9, 29) to create RBY1132 (a/a strain) and RBY1133 (α/α strain). Similarly, SNY95 (his1/his1 arg4/arg4) (45) was grown on sorbose to generate a and α derivatives RZY53 and RZY56, respectively. The HIS1 gene was added back to RZY53 and RZY56 as previously described (7) to generate strains RBY1175 and RBY1176, respectively. KAR3 deletion strains were gene disruption mutants generated as described previously (45). In brief, 5′ and 3′ homologous flanks were generated by PCR of genomic DNA with oligonucleotides 1/2 and 3/4, respectively. The PCR product designed to target the KAR3 open reading frame contained a selectable marker sequence (C. dubliniensis HIS1 [CdHIS1] or C. maltosa LEU2 [CmLEU2]) that was amplified from plasmid pSN52 or pSN40 with oligonucleotides 5 and 6 (45). Fusion PCR products were generated with oligonucleotides 1 and 4 to amplify 5′ and 3′ flanks together with the marker PCR product. The resulting linear construct contained a CdHIS1 or CmLEU2 sequence flanked by homologous KAR3 sequences. The first KAR3 allele was replaced by homologous recombination with the construct containing the CmLEU2 marker, creating KAR3 heterozygous strains RSY8 and RSY9. Likewise, the second allele was replaced with the construct containing a CdHIS1 marker, thereby generating homozygous KAR3 knockout (Δkar3/Δkar3) strains RSY11 and RSY12. Correct integration of these constructs was verified by PCR across 5′ and 3′ disruption junctions, and loss of the gene was confirmed with primers internal to the open reading frame.
TABLE 1.
Names, genotypes, mating types, and sources of the strains used in this study
| Strain(s) | Genotype | Mating type | Source |
|---|---|---|---|
| RBY1009 | URA3/ura3::imm343 HTB/HTB-YFP::URA3 | a/a | 8 |
| RBY1132 | leu2/leu2 his1/his1 arg4/arg4 | a/a | This study |
| RBY1133 | leu2/leu2 his1/his1 arg4/arg4 | α/α | This study |
| RSY8 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG KAR3/kar3::LEU2 | α/α | This study |
| RSY9 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG KAR3/kar3::LEU2 | a/a | This study |
| RSY11 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::HIS1 | α/α | This study |
| RSY12 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::HIS1 | a/a | This study |
| RSY15, RSY16 | leu2/leu2 his1/his1 arg4/arg4 HTB/HTB-GFP::SAT1 | a/a | This study |
| RSY19 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::LEU2 HTB/HTB-GFP::SAT1 | a/a | This study |
| RSY26 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::LEU2/KAR3::SAT1 | α/α | This study |
| RSY27 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::LEU2/KAR3::SAT1 | α/α | This study |
| RSY31, RSY32 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG TUB2/TUB2-GFP::SAT1 | a/a | This study |
| RSY35, RSY36 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::HIS1 TUB2/TUB2-GFP::SAT1 | a/a | This study |
| RSY39, RSY40 | leu2::hisG/leu2::hisG arg4::hisG/arg4::hisG KAR3/KAR3-GFP::SAT1 | a/a | This study |
| RSY44 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/KAR3-GFP::SAT1 | a/a | This study |
| RSY120, RSY121 | leu2::hisG/leu2::hisG arg4::hisG/arg4::hisG KAR3/KAR3-GFP::SAT1 TUB2/TUB2-RFP::ARG4 | a/a | This study |
| RSY136, RSY138 | ura3::λimm34/ura3::λimm34 his1::hisG/his1::hisG arg4::hisG/arg4::hisG dyn1::URA3/dyn1::HIS1 HHF1/HHF1-GFP::ARG4 MTLa/mtlα::SAT1 | a/a | This study |
| RSY150, RSY155 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG kar3::LEU2/kar3::HIS1 TUB2/TUB2-GFP::SAT1 | a/a | This study |
| RSY156, RSY157 | leu2::hisG/leu2::hisG his1::hisG/his1::hisG arg4::hisG/arg4::hisG kar3::LEU2/kar3::HIS1 TUB2/TUB2-GFP::SAT1 HTB/HTB-RFP::ARG4 | a/a | This study |
TABLE 2.
Oligonucleotides used in this study
| Name | Oligonucleotide sequencea |
|---|---|
| 1 | GCAAGGCTAGTCACGTGCAA |
| 2 | CACGGCGGCGCCTAGCAGCGGGATGGTTGTGACACATTTAG |
| 3 | GTCAGCGGCCGCATCCCTGC |
| 4 | GTCATGTATGTAAACTATTAACGTAG |
| 5 | CCGCTGCTAGGCGCGCCGTGACCAGTGTGATGGATATCTGC |
| 6 | GCAGGGATGCGGCCGCTGACAGCTCGGATCCACTAGTAACG |
| 7 | GCGGCCATGGGCCCACACCACGTTGTGCAACTTT |
| 8 | CAGGCGCGCTCGAGCCAAAACATATATCTGAGCC |
| 9 | AGGTGAATTGGCTAAACATGCTGTTTCTGAAGGTACTAGAGCTGTTACCAAATACTCTTCTGCTTCTAATGGTGGTGGTAGTAAGGGAGAAGAACTTTTC |
| 10 | TATTAATATACAATACAAAATAAAACAACAATAATTTGGAGAAATAAACCATTCATGACAAACCTCTCTCCTGCAGGACCACCTTTGATT |
| 11 | AGTGAATACCAACAATACCAAGAAGCTAGTATTGATGAAGAAGAATTAGAATATGCCGATGAAATCCCATTAGAAGATGCCGCCATGGAGGTGGTGGTAGTAAGGGAGAAGAACTTTTCACTGGAGTT |
| 12 | GTATATATAGAAAAGAGTAAGGAAAATTATAGTTTAAGATAGACAACAAATCAAACTATACAACAGATATTGTGCCGATAAATAATAAAACTGCAGGACCACCTTTGATTGTAAATAGTA |
| 13 | GTGGTCAATGGAGCTGATAC |
| 14 | ACATGTGGTCGCCCAACTCC |
| 15 | CCTTCAATTGCATCGTAAGTACC |
| 16 | GAAGATGACTCAGGTCATGC |
| 17 | GTAAACATATCACCACTTACCAAGGATTTAAATGAAACCATAAACTCTTTGAGGTTTGCAACCAAGGTTAACAATACTAGAATCAATAAAGGTGGTGGTAGTAAGGGAGAAGAACTTTTCACTGGAGTT |
| 18 | CAGTTAAATCATCCAGCAATTTGGCAACAAGCTGTTGAATCAGGAAAACTAATTGATGCTTCGGGATATAAACCAAATGAGAATTTTCTTCTGCAGGACCACCTTTGATTGTAAATAGTA |
| 19 | AGAAGCTAGTATTGATGAAGAAGAATTAGAATATGCCGATGAAATCCCATTAGAAGATGCCGCCATGGAAGGTGGTGGTGATAACACTGAAGATGTTATT |
| 20 | GGAAAATTATAGTTTAAGATAGACAACAAATCAAACTATACAACAGATATTGTGCCGATAAATAATAAAATTGTAAAACGACGGCCAGTG |
| 21 | GGAGCCAAGCTTTATTAAAATGGATAACACTGAAGATG |
| 22 | GGCCGCGGATCCGATATTTCAACGCCTTCCAGC |
| 23 | GGCGCCCTCGAGAAGAACATTTCTGTACCGCA |
| 24 | GGCCGCTCTAGAGATTTGTAAGTTTGATCAGGAT |
| 25 | TATTAATATACAATACAAAATAAAACAACAATAATTTGGAGAAATAAACCATTCATGACAAACCTCTCTCTTGTAAAACGACGGCCAGTG |
| 26 | AGGTGAATTGGCTAAACATGCTGTTTCTGAAGGTACTAGAGCTGTTACCAAATACTCTTCTGCTTCTAATGGTGGTGGTGATAACACTGAAGATGTTATT |
| 27 | GGCGCCGGGCCCTAGACTCTTTGGTCATGCCTT |
| 28 | GCCGGCCTCGAGACCTGAGTTGGCTGCTTTGC |
| 29 | GCCGGCCCGCGGGGCTTGTTCGCTTTCAAACGA |
| 30 | GGCGCCGAGCTCGGTTAGTATGTCTTCATCATTATC |
| 31 | AGGTGAATTGGCTAAACATGCTGTTTCTGAAGGTACTAGAGCTGTTACCAAATACTCTTCTGCTTCTAATGGTGGTGGTGATAACACTGAAGATGTTATT |
| 32 | TATTAATATACAATACAAAATAAAACAACAATAATTTTGGAGAAATAAACCATTCATGACAAAACCTCTCTCTTGTAAAACGACGGCCAGTG |
Underlining indicates the restriction enzyme site.
A wild-type copy of KAR3 was reintroduced into Δkar3/Δkar3 strains for complementation. The wild-type KAR3 sequence and approximately 1 kb of upstream sequence were PCR amplified from genomic DNA with oligonucleotides 7 and 8. At ApaI and XhoI restriction enzyme sites, the KAR3 PCR product was ligated into pSFS2A, which contains a dominant nourseothricin resistance marker (50). This vector was then linearized by HpaI digestion and subsequently integrated into the 5′ KAR3 flanking homologous sequence of Δkar3/Δkar3 strains, creating RSY26 and RSY27. Correct integration of the wild-type allele was confirmed by PCR across the boundary of inserted DNA.
Constructs containing the genes for histone protein B (HTB; orf19.6925) and tubulin 2 (TUB2; orf19.6034) fused to the green fluorescent protein (GFP) gene were introduced into both wild-type and Δkar3/Δkar3 strains by using PCR products with long homologous flanks (21). Oligonucleotides 9/10 and 11/12, containing ∼100 bp of HTB and TUB2 homologous sequences, respectively, were used to amplify the SAT1 and GFP genes from pNIM1 (48). The resulting linear constructs were integrated 3′ of the HTB and TUB2 genes of the wild-type RBY1132 and Δkar3/Δkar3 mutant RSY12 strains. Correct integration was verified by PCR across the junction of inserted DNA. Integration resulted in HTB-GFP-labeled strains RSY15, RSY16, and RSY19. TUB2-GFP integration was similarly carried out to generate strains RSY31, RSY32, RSY35, and RSY36. Strains expressing fluorescently labeled Kar3p were constructed with oligonucleotides 17/18 to amplify SAT1 and GFP sequences from pNIM1. PCR constructs were integrated into the wild-type RBY1132 and KAR3/kar3 heterozygote RSY9 strains by homologous recombination, creating strains RSY39, RSY40, and RSY44.
Plasmid pRS1, containing the red fluorescent protein (RFP) and C. dubliniensis ARG4 genes, was constructed as follows. The RFP sequence was amplified from pMG2169 (a gift from Judith Berman, University of Minnesota) with oligonucleotides 21/22, containing HindIII and BamHI restriction sequences, respectively, and ligated into the pCR-BluntII-TOPO vector (Invitrogen, Carlsbad, CA). A selectable C. dubliniensis ARG4 sequence was amplified from pSN69 (45) with oligonucleotides 23 and 24, containing XhoI and XbaI restriction sequences. The PCR product was ligated into XhoI/XbaI restriction sites downstream of the RFP sequence, thereby creating pRS1. Subsequently, oligonucleotides 19/20, containing sequence homology to TUB2, were used to amplify the RFP-ARG4 sequence from pRS1. This construct was integrated in frame with the 3′ end of TUB2 in strains RSY39 and RSY40, creating strains RSY120 and RSY121. Correct integration of the KAR3-GFP and TUB2-RFP fusion constructs was verified by PCR.
An a/α Δdyn1/Δdyn1 strain (GC17) was received as a gift from J. Wendland (Friedrich-Schiller University, Germany). The MTLα allele was deleted by transformation with plasmid pRB102. pRB102 was created by amplifying 300 bp of homologous sequences upstream and downstream of MTLα with oligonucleotides 27/28 and 29/30, respectively. The 5′ DNA product was ligated into pSFS2A with ApaI and XhoI restriction sites, and subsequently the 3′ DNA product was ligated between the SacII and SacI restriction sites. The vector was linearized by cutting with ApaI and SacI and transformed into GC17, creating strains RSY136 and RSY138. Correct integration of pRB102 was verified by PCR.
HTB-RFP, TUB2-GFP strains were constructed from strains RSY31, RSY32, and RSY35. In brief, oligonucleotides 31 and 32, containing 100 bp of sequence homology to HTB, were used to amplify pRS1. The resulting linear construct was integrated into the 3′ region of the wild-type strain, creating strains RSY150, RSY155, RSY156, and RSY157. Correct integration of the HTB-RFP construct was verified by PCR.
Generation time, cell viability, and colony morphology assays.
The generation times of these cultures were determined by measuring optical density every hour after induction of exponential growth. Strain viability and morphology were assessed by counting cells on a hemocytometer and plating 75 cells from each culture onto YPD medium, YPD medium plus 10% serum, and Spider medium. These cultures were grown for 3 to 11 days at 30°C (37°C for YPD medium plus 10% serum) and assessed for CFU and morphology. Student's t test was used to determine significant differences between the numbers of colonies formed by different strains.
Micromanipulation of mating zygotes for karyogamy assessment.
Liquid cultures of opaque-phase a and α cells were grown overnight in SCD (synthetic complete plus dextrose) medium at 22°C. Cells (3 × 107) of both strains were washed, mixed, and deposited on the surface of Spider medium plates. Cells were allowed to grow for 2 to 3 days at room temperature. Mating zygotes were identified with a Nikon Eclipse E400 microscope (Nikon, Melville, NY) and selected with a glass needle attached to a micromanipulator (Microvideo Instruments Inc., Avon, MA). After 2 days at 30°C, zygotes were streaked for single colonies and tetraploid cells were identified by PCR analysis of the MTL locus with a-specific oligonucleotides 13/14 and α-specific oligonucleotides 15/16.
Mating projection induction.
Opaque-phase a/a C. albicans strains were grown overnight at 22°C in SCD medium. Approximately 9 × 107 cells were removed from each culture, washed, and resuspended in 3 ml of liquid Spider medium. Alpha pheromone (10 mg/ml dissolved in 10% dimethyl sulfoxide) or 10% dimethyl sulfoxide was added to experimental or control cultures, respectively. Cells were incubated at 22°C for 18 h, washed, sonicated to disperse cell clumps, and resuspended in SCD medium. Cells were mounted on poly-l-lysine-coated slides for microscopic analysis (Polysciences Inc., Warrington, PA). The statistical significance of cell measurement differences was determined with Student's t test.
Time-lapse microscopy.
Approximately 700 cells were plated onto thin YPD medium plates. A small slab of culture was removed, inverted, and placed in a Lab-Tek II two-chamber slide (Nalgene Nunc International, Naperville, IL). Time-lapse images were obtained with a Zeiss LSM 510 Meta microscope system (Carl Zeiss Microimaging, Oberkochen, Germany) with a 561-nm laser at ×20 magnification. LSM 510 software was used to program the stage position and capture images at 15.1-min intervals over the course of 6 to 14 h. A PeCon GmbH Tempcontrol 37-2 digital temperature regulator (PeCon, Erbach, Germany) was used to maintain the stage temperature at 30°C over the course of the experiment. Doubling times of cells were calculated by averaging the time from bud emergence to formation of the daughter cell, and the standard error of the mean (SEM) was determined.
Cell sorting and flow cytometry.
C. albicans strains were inoculated into liquid YPD medium and grown in logarithmic phase at 30°C to an optical density of 1 to 2.5. As a control for staining of nonviable cells, an aliquot of the overnight culture was heat killed at 65°C for 1 h. The control sample and experimental cultures were washed in phosphate-buffered saline (PBS; Gibco, Grand Island, NY). Cells were resuspended in a 0.5% eosin Y-0.15 M NaCl solution for 20 min in the dark and washed three times in PBS. Finally, cells were resuspended in PBS to an optical density of 0.5. Cells from the heat-killed control sample were used to positively identify eosin Y-stained dead populations with a FACSAria fluorescence-activated cell sorter (Becton Dickinson, Franklin Lakes, NJ). The observed population of fluorescently labeled cells in the control sample was used to determine gating to allow sorting of live cells in experimental cultures. Experimental cell suspensions were sorted, and approximately 0.8 × 106 to 2 × 106 unstained (live) cells were used for subsequent flow cytometric analysis. Live cells were fixed, stained with SYTOX Green, and prepared for cytometric analysis as previously described (27). A total of 50,000 cells from each experimental condition were analyzed and used to generate histograms by flow cytometry.
Hyphal induction and calcofluor staining.
C. albicans strains were grown overnight in YPD medium at 30°C, and 0.2 ml of each strain was inoculated into 10 ml of prewarmed (37°C) liquid YPD medium supplemented with 10% serum. Cultures were then incubated at 37°C for 6 h with agitation, and the cells washed with water, gently sonicated, and resuspended in distilled H2O. Equal volumes of hyphal cell suspension, 20% calcofluor white solution, and 10% NaOH were placed onto a slide, and cells were analyzed by fluorescence microscopy. The statistical significance of observed morphogenesis was assessed by determining the P value of a Bonferroni error-corrected comparison of two proportions.
Immunofluorescence.
Logarithmic-phase budding yeast cultures were harvested and fixed in 4.5% formaldehyde for 1 h. Cells were washed in PBS, followed by potassium phosphate sorbitol buffer (1.2 M sorbitol, 0.1 M potassium phosphate). C. albicans cell walls were digested with Zymolyase 100T at 37°C and microscopically monitored for spheroplast formation. Cells were then applied to eight-well poly-l-lysine-coated microscope slides and flattened by incubations in methanol and acetone. Cells were blocked in 1% bovine serum albumin-0.1% Tween diluted in PBS for 60 to 90 min. Primary monoclonal antibody YOL1/34 (Accurate Chemical and Scientific Corporation, Westbury, NY) was used at a 1:200 dilution to detect yeast tubulin. The primary antibody was incubated with cells for 1 h at room temperature. Cells were then washed in PBS and incubated for 1 h in the dark with a fluorescein isothiocyanate-conjugated donkey anti-rat secondary antibody (1:500 dilution; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Finally, cells were washed with PBS, treated with Fluoromount-G (Southern Biotech, Birmingham, AL), and imaged.
Fluorescence and DIC microscopy.
Differential interference contrast (DIC) and fluorescent images were obtained with a Zeiss Axioplan 2 microscope (Carl Zeiss Microimaging, Oberkochen, Germany) equipped with a Hamamatsu-ORCA camera (Hamamatsu Photonics, Hamamatsu City, Japan). Captured DIC and fluorescent images were merged and deconvoluted with Openlab software (Improvision Inc., Lexington, MA). Pictures of C. albicans colonies were obtained with a Zeiss stemi 2000C stereo microscope (Carl Zeiss Microimaging, Oberkochen, Germany) equipped with an Infinity 1 camera (Zarbeco, LLC, Creative Imaging Solutions, Randolph, NJ). Captured images of colonies were processed by Micron Imaging software (Westover Scientific, Mill Creek, WA).
RESULTS
C. albicans KAR3 is necessary for karyogamy.
In previous studies, we demonstrated that karyogamy in C. albicans was dependent upon the KAR3 gene (8). In particular, mating between Δkar3/Δkar3 a and α strains failed to generate stable tetraploid a/α cells. However, these strains were constructed with selectable markers (URA3 and ADE2) that are known to influence growth, filamentation, and virulence in C. albicans (12, 28, 45). For the present study, in which we investigated the role of KAR3 in both mating and mitotic divisions, we constructed an independent series of kar3 mutant strains with genetic markers (HIS1, LEU2, and ARG4) that do not effect growth and virulence of C. albicans (45).
To confirm that the new series of Δkar3/Δkar3 strains were also defective in karyogamy, mating assays were performed with opaque a and α strains. C. albicans a and α cells can switch between two alternative heritable states, known as the white and opaque phases, but only in the opaque phase do they undergo efficient mating (30, 42, 56). Individual zygotes were micromanipulated from wild-type and Δkar3/Δkar3 crosses, and the resulting colonies were analyzed by PCR to determine the configuration of genes at the MTL (see Materials and Methods). Only those cells that have undergone nuclear fusion generate tetraploid cells that contain both MTLa and MTLα alleles (8). The results of this experiment were consistent with the published studies; while 19 (63%) of 30 tetraploid zygotes were obtained from crosses between wild-type a and α strains, no tetraploids were obtained from crosses between Δkar3/Δkar3 strains (21 zygotes analyzed). Karyogamy was restored to mating between Δkar3/Δkar3 a and α strains by the reintroduction of one copy of the wild-type KAR3 gene with 5 (33%) out of 15 zygotes undergoing nuclear fusion (Table 3).
TABLE 3.
Analysis of karyogamy in C. albicansa
| MTLa × MTL α | % Tetraploid | % Diploid |
|---|---|---|
| Wild type a × wild type α | 63 | 37 |
| Δkar3/Δkar3a × Δkar3/Δkar3 α | 0 | 100 |
| Δkar3/Δkar3+KAR3a × Δkar3/Δkar3 α | 33 | 67 |
Shown are the percentages of tetraploid cells formed following mating crosses between the opaque wild-type (RBY1132 and RBY1132), Δkar3/Δkar3 (RSY12 and RSY11), and Δkar3/Δkar3 strains with the Δkar3/Δkar3+KAR3 (RSY12 and RSY26) strains. Mating zygotes were picked by micromanipulation and analyzed by PCR of the MTL locus as described in Materials and Methods.
Role of Kar3p and Dyn1p in nuclear positioning during mating.
During mating in C. albicans, long polarized mating projections are formed by opaque a and α cells as they sense mating pheromones secreted by the opposite cell type (9, 34, 42, 47). After cell fusion of opaque mating partners, karyogamy takes place within the resulting zygote cell (8). In addition to Kar3p, Dyn1p has been shown to play a role in nuclear migration in C. albicans, as it is required for movement and positioning of the nucleus at the bud neck during metaphase (37). To determine the role of Kar3p and Dyn1p during mating projection germination, we used nucleus-labeled Δkar3/Δkar3 or Δdyn1/Δdyn1 strains to visualize nuclear dynamics. The mating response was induced in a-type opaque cells by treatment with alpha pheromone for 18 h in Spider medium at 25°C, and nuclear positioning was determined by fluorescence microscopy, as shown in Fig. 1.
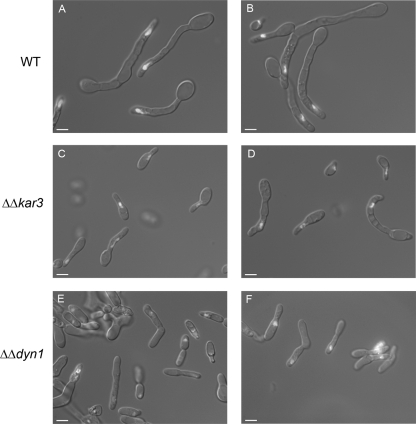
FIG. 1.
Nuclear positioning within C. albicans mating projections. Mating projections were induced in opaque a cells with alpha pheromone. The HTB-GFP-tagged wild-type (WT; RBY1009) or Δkar3/Δkar3 (RSY19) strain or a Δdyn1/Δdyn1 (RSY136) strain expressing HHF1-GFP was incubated in Spider medium containing alpha pheromone for 18 h at 25°C. Representative live cells were photographed, and fluorescent nuclear images were superimposed over DIC images. Cells from each strain produced mating projections containing nuclei that migrated close to the tip of the mating projection (A and B, C and D, and E and F, respectively). Bars, 5.0 μm.
The dimensions of cells responding to mating pheromone were measured, and wild-type, Δkar3/Δkar3, and Δdyn1/Δdyn1 cells all produced mating projections that were often several times the length of the original opaque cell. Compared to wild-type strains, however, the Δkar3/Δkar3 and Δdyn1/Δdyn1 strains produced mating projections that were more variable in length and morphology. The average length of mating projections produced by wild-type strains was 28.2 μm, whereas that of mating projections produced by Δkar3/Δkar3 strains was 14.0 μm (Fig. 1, compare A and B with C and D; Table 4). Mating projections produced by Δkar3/Δkar3 strains were also narrower, having an average diameter of 2.2 μm, in comparison to a diameter of 2.8 μm in wild-type strains. Mating projections produced by Δdyn1/Δdyn1 strains were even shorter than those of Δkar3/Δkar3 strains, having an average length of 11.2 μm, although they were similar in width to wild-type mating projections. Nuclear positioning within the mating projections appeared similar between the wild-type and Δkar3/Δkar3 strains (Table 4). However, the nuclei in Δdyn1/Δdyn1 strains migrated closer to the projection tip, with an average distance of only 3.6 μm (Fig. 1, compare A and B with E and F; Table 4).
TABLE 4.
Nuclear migration during C. albicans matinga
| Strain | Range (mean ± SEM) of:
|
||
|---|---|---|---|
| Mating projection length (μm) | Mating projection width (μm) | Distance of nucleus from end of mating projection (μm) | |
| Wild type | 7.5-61.7 (28.2 ± 1.2) | 1.3-6.5 (2.8 ± 0.1) | 0.7-14.4 (4.3 ± 0.2) |
| Δkar3/Δkar3 | 3.1-48.1 (14.0 ± 0.6)b | 1.2-7.2 (2.2 ± 0.1)b | 1.3-36.0 (5 ± 0.3) |
| Δdyn1/Δdyn1 | 1.5-38.2 (11.2 ± 0.4)b | 2.2-10.8 (2.6 ± 0.1) | 0-15.2 (3.6 ± 0.2)c |
Compared are mating projection morphology and nuclear positioning within mating projections of wild-type (RBY1009) and Δkar3/Δkar3 (RSY19) strains expressing HTB-GFP and a Δdyn1/Δdyn1 (RSY136) strain expressing HHF1-GFP. Opaque cells of each strain were treated with alpha factor in Spider medium for 18 h at 25°C as described in Materials and Methods. A total of 600 cells were microscopically analyzed.
Statistically significantly different from the wild-type strain (P < 0.005).
Statistically significantly different from the wild-type strain (P < 0.05).
These experiments demonstrate that both Kar3p and Dyn1p, while not essential for mating projection formation, are required for the normal morphogenesis of mating projections. Nuclear movement was not significantly compromised by the absence of Kar3p or Dyn1p, however, as nuclei migrated close to the growing tip of the mating projection in both mutant strains.
Disruption of KAR3 influences cellular morphogenesis.
C. albicans is capable of growing as budding yeast, in which round cells give rise (through budding) to similar daughter cells or can grow as polarized pseudohyphae or even true hyphae (filamentous cells) in response to environmental changes. Each morphological form is recognized as a distinct developmental state with discrete gene expression patterns (11, 57). C. albicans cells primarily form budding yeast when grown in SCD or YPD medium at 30°C but can be induced to form hyphae upon culturing in Spider (nutrient-limiting) medium at 30°C or in YPD medium supplemented with 10% serum at 37°C. We first addressed whether the Kar3 protein plays a role in regulating morphogenesis in C. albicans by culturing Δkar3/Δkar3 cells under these alternative conditions.
Wild-type and Δkar3/Δkar3 cells were grown in SCD medium at 30°C, and cell morphology was assessed by microscopy. Under these conditions, both wild-type and Δkar3/Δkar3 cells grew mainly as yeast cells, although Δkar3/Δkar3 cells were generally larger than their wild-type counterparts (Fig. 2A). A subset of Δkar3/Δkar3 cells exhibited a hyperpolarized phenotype in which they formed large elongated cells resembling pseudohyphal cells (Fig. 2A, right panel) (10, 11, 57). C. albicans cells experiencing a delay in the cell cycle have previously been shown to enter a constitutive polarized growth program in which significantly elongated cells are formed that have characteristics in common with pseudohyphal cells (57). In addition, we observed an increased number of mother cells with large daughter buds in Δkar3/Δkar3 strains compared to the wild-type or reconstituted (Δkar3/Δkar3+KAR3) strains (Fig. 2A and Table 5). The dimensions of wild-type and Δkar3/Δkar3 mother cells and daughter buds were also compared before nuclear division, during nuclear division (anaphase), and after nuclear division (but prior to cytokinesis) with strains expressing Htb-GFP fusion proteins. We observed that Δkar3/Δkar3 mother and daughter cells were consistently larger than wild-type cells examined at all three stages of the cell cycle (Table 5). For example, the average length of the mother cell from wild-type strains was 4.5 μm during anaphase while that of daughter cells was approximately 2.9 μm. In contrast, mother cells from Δkar3/Δkar3 strains were 6.2 μm long and daughter cells were 4.6 μm long (Table 5). Similarly, the volume of Δkar3/Δkar3 yeast cells was also significantly larger than that of wild-type yeast cells, with mutant cells averaging 60 μm3, which was twice that of wild-type cells (30 μm3).
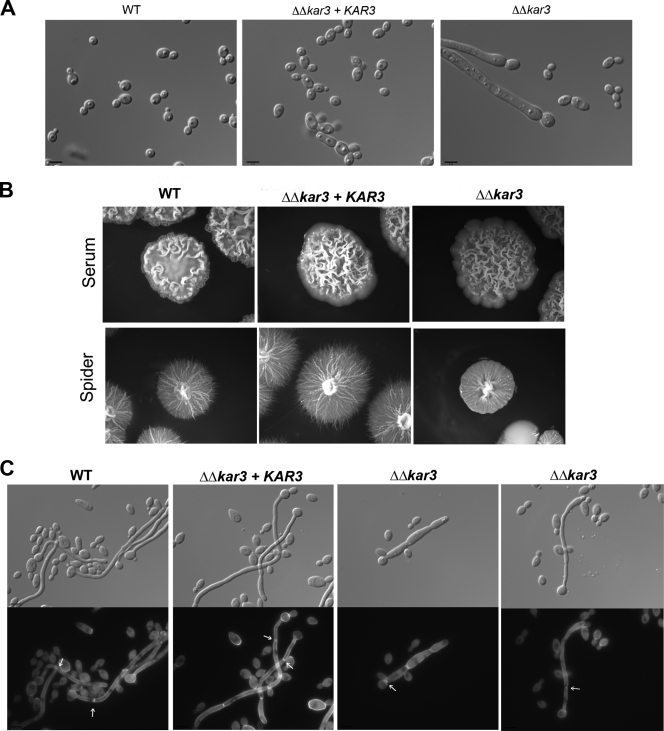
FIG. 2.
C. albicans kar3 mutants display altered cell and colony morphogenesis. (A) Logarithmic growth of wild-type (WT; RBY1132, RBY1133), KAR3/Δkar3 (RSY8, RSY9), Δkar3/Δkar3 (RSY11, RSY12), and Δkar3/Δkar3+KAR3 (RSY26, RSY27) cells. C. albicans cells were cultured in liquid SCD medium at 30°C and imaged. Bars, 5.0 μm. (B) Wild-type (RBY1132), Δkar3/Δkar3 (RSY11), and Δkar3/Δkar3+KAR3 (RSY26) strains plated for single colonies on Spider medium or on YPD medium supplemented with 10% serum. Cells on Spider medium plates were grown at 30°C, and those on plates containing YPD medium plus 10% serum were grown at 37°C. After 10 days of culture, representative colonies were photographed. (C) Overnight liquid cultures were diluted into prewarmed YPD medium supplemented with 10% serum and grown at 37°C for 6 h. Cells were fixed, stained with calcofluor white, and photographed. Pseudohyphal cells are characterized by septum formation at the bud neck, whereas true hyphal cells form septa within germ tubes having parallel walls (11). The first septum is indicated by an arrow.
TABLE 5.
Sizes of mother cells and daughter buds prior to and following nuclear divisiona
| Strain and stage | Size (μm) range (mean ± SEM) of:
|
|
|---|---|---|
| Mother cells | Daughter buds | |
| Wild type | ||
| Before mitosis | 2.0-4.3 (4.2 ± 0.1) | 2.3-6.8 |
| Anaphase | 3.5-5.5 (4.5 ± 0.1) | 1.8-4.3 (2.9 ± 0.1) |
| After mitosis | 3.4-5.7 (4.6 ± 0.1) | 2.2-5.4 (3.6 ± 0.1) |
| Δkar3/Δkar3 | ||
| Before mitosis | 2.9-11.6 (6.3 ± 0.2)b | 1.0-6.5 |
| Anaphase | 2.9-10.1 (6.2 ± 0.2)b | 2.3-7.8 (4.6 ± 0.2)b |
| After mitosis | 4.3-10.0 (6.3 ± 0.1)b | 2.8-7.6 (4.6 ± 0.1)b |
The wild-type (RSY15) and Δkar3/Δkar3 (RSY19) HTB-GFP strains were grown to logarithmic phase in liquid YPD medium at 30°C. A total of 400 cells were measured along the longest axis and classified by the presence of one or two distinct nuclei or as being in the process of nuclear division.
Statistically significantly different from the wild-type strain (P < 0.005).
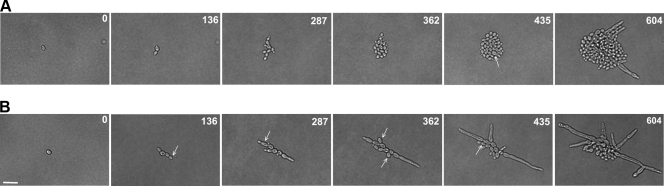
We assessed hypha formation in wild-type and Δkar3/Δkar3 strains by growing cells under hypha-inducing conditions. Both wild-type and mutant strains formed colonies with characteristic surface wrinkling, indicating the formation of filamentous cells (Fig. 2B). Filamentous growth was confirmed by microscopic analysis of cells from the wrinkled colonies (data not shown). Only wild-type and Δkar3/Δkar3+KAR3 colonies, however, exhibited a halo of invasive growth when cultured on Spider medium, indicating defective filamentation by Δkar3/Δkar3 cells on this medium (Fig. 2B). Cells of wild-type and kar3 mutant strains were also induced to form filaments in liquid YPD medium plus 10% serum at 37°C to more closely examine their hyphal morphology. True hyphae are identified by the formation of the first septum within the germ tube and have parallel walls with no obvious constrictions at sites of septation. Pseudohyphal cells more closely resemble yeast cells in that they form septa across the bud neck of the basal cell and nuclei divide across the neck. Pseudohyphae also typically show constrictions at the sites of septation and have nonparallel cell walls that are wider than those of true hyphae (11, 57). To differentiate between these morphological states, septa were stained with calcofluor white after hyphal induction.
Wild-type strains produced cultures that were predominantly composed of true hyphal cells in YPD medium plus 10% serum, with characteristic formation of the first septum within the parallel-walled germ tubes (Fig. 2C). In contrast, cells of Δkar3/Δkar3 strains formed cultures composed primarily of pseudohyphal cells; cells had constrictions at the neck with septum formation across the bud neck and often did not contain parallel cell walls (Fig. 2C). Δkar3/Δkar3+KAR3 strains had an intermediate phenotype containing both hyphal and pseudohyphal populations (Fig. 2C and 3).
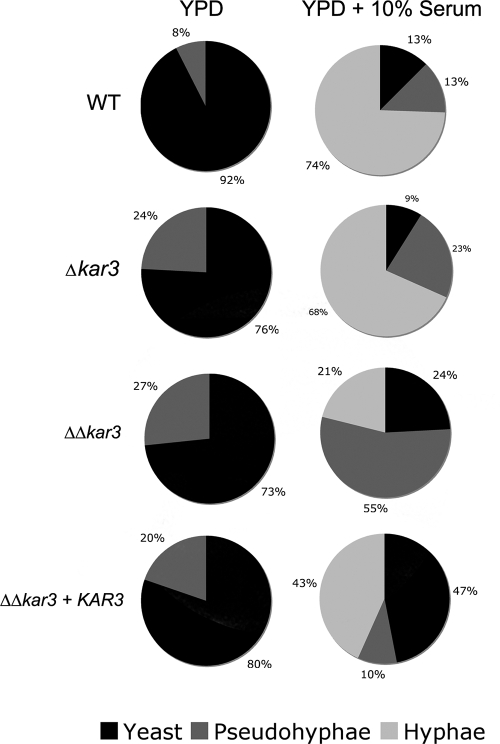
FIG. 3.
Quantification of different cell morphologies under standard (YPD medium) and filamentation-inducing (YPD medium plus 10% serum) growth conditions. Cultures of wild-type (WT; RBY1132), KAR3/Δkar3 (RSY8), Δkar3/Δkar3 (RSY11), and Δkar3/Δkar3+KAR3 (RSY26) cells were incubated for 6 h in YPD medium at 30°C or in YPD medium plus 10% serum at 37°C. A total of 3,001 cells were assessed for morphology and classified as budding yeast, pseudohyphae, and hyphae, and the results are expressed as percentages in pie charts. A statistically significant difference in the percentage of cells forming hyphae was observed between the wild-type strain and either the Δkar3/Δkar3+KAR3 strain or the Δkar3/Δkar3 strain (P < 0.005). Note that cells within germ tubes or within chains of pseudohyphal cells were counted as distinct cells.
Quantification of budding yeast, pseudohyphae, and hyphae was carried out with calcofluor white-stained cells grown in both YPD medium and YPD medium plus 10% serum. As expected, strains grown in YPD medium were mainly budding yeast, although Δkar3/Δkar3 strains showed an increased percentage of pseudohyphal cells (20% versus 8% in wild-type cells; Fig. 3). Under filamentation-inducing conditions, wild-type and KAR3/Δkar3 cultures were primarily composed of true hyphae, which represented 74% and 68% of the respective populations (Fig. 3). However, cells of Δkar3/Δkar3 strains contained only 21% hyphal filaments and instead showed a high fraction of pseudohyphal cells, with 55% of the cells exhibiting a pseudohyphal morphology. Reconstituted Δkar3/Δkar3+KAR3 strains had an intermediate phenotype, although 47% of these cells were not induced to form hyphal cells but remained as budding yeast (Fig. 3). To confirm the correct morphological assignment of yeast, pseudohyphal, and hyphal cells of the different strains, the morphology index (MI) was also calculated for these populations. The MI is based on a mathematical formula that relates cell length, diameter, and septal width to characterize cells as budding yeast, pseudohyphae, or hyphae (40). There was greater than 90% agreement between cell types determined by MI and those characterized by microscopic analysis of septal positioning and cell shape (11, 57).
Together, these experiments reveal that Kar3p is required for normal budding yeast and hyphal morphogenesis in mitotically dividing cells. Notably, Δkar3/Δkar3 cells were larger than wild-type cells and exhibited an increased tendency to form pseudohyphal cells, particularly under filament-inducing conditions, both of which are suggestive of a delay in cell cycle progression (discussed below).
Role of C. albicans Kar3p in mitotically dividing cells.
The experiments described above reveal that Kar3p plays an important role in regulating cell morphogenesis. We next sought to determine if C. albicans Kar3p, similar to ScKar3p, is required for normal mitotic division. Viability and generation time assays were performed with wild-type and Δkar3/Δkar3 strains to determine if the absence of Kar3p negatively influenced cell division.
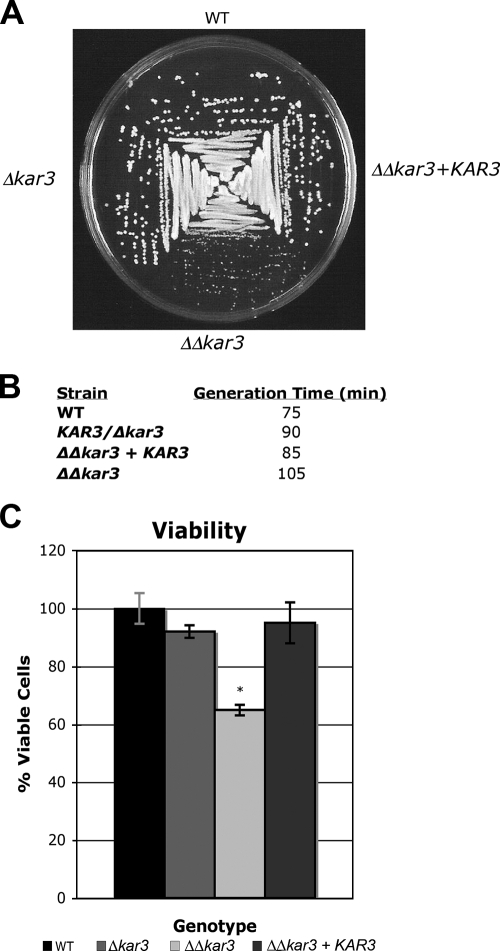
Analysis of wild-type and Δkar3/Δkar3 strains streaked for single colonies on YPD plates revealed that kar3 mutant strains consistently produced smaller colonies of more variable size than those of wild-type, KAR3/Δkar3, and Δkar3/Δkar3+KAR3 (reconstituted) strains (Fig. 4A). Cultures were also grown in logarithmic phase in liquid YPD medium at 30°C, and generation times were deduced. Figure 4B shows that wild-type strains had a doubling time of approximately 75 min, while KAR3/Δkar3 and Δkar3/Δkar3+KAR3 strains had doubling times of 90 and 85 min, respectively. The Δkar3/Δkar3 strain had the longest generation time, requiring 105 min for doubling of the population.
FIG. 4.
C. albicans Kar3p function in mitotically dividing cells. The colony formation, generation times, and viability of the wild-type (WT; RBY1132, RBY1133), KAR3/Δkar3 (RSY8, RSY9), Δkar3/Δkar3 (RSY11, RSY12), and Δkar3/Δkar3+KAR3 (RSY26, RSY27) strains were analyzed. (A) Cells of each strain were streaked onto YPD medium, incubated at 30°C for 2 days, and photographed. (B) Generation times of wild-type and kar3 mutant cells grown in YPD medium at 30°C. Doubling times were averaged from experiments done in triplicate, as described in Materials and Methods. (C) Cells were counted and plated in triplicate (75 cells per plate) on YPD medium and incubated at 30°C for 3 days. CFU were counted and averaged and are expressed as percentages. Error bars represent means ± standard deviations. *, P < 0.05.
A viability assay was also performed to determine if increased cellular death contributed to the lengthened generation times of Δkar3/Δkar3 strains. In this assay, equal numbers of cells were plated onto YPD medium for quantification of CFU. This experiment revealed that there was a significant reduction in the viability of Δkar3/Δkar3 cells in comparison to wild-type cells, with only 65% of the mutant cells giving rise to colonies in these plating assays (Fig. 4C). Again, the reconstituted strain (Δkar3/Δkar3+KAR3) exhibited a phenotype close to that of the wild-type strain, with 95% of the cells able to form viable colonies.
To further evaluate the pattern of cell division in kar3 mutants, time-lapse microscopy was performed on wild-type and Δkar3/Δkar3 cells. Wild-type cells grown on YPD medium at 30°C divided every 72 ± 2 min as budding yeast cells. In contrast, Δkar3/Δkar3 cells showed two distinct modes of growth on YPD medium. First, many Δkar3/Δkar3 cells divided as budding yeast, although the doubling time (84 ± 3 min) was longer than that of wild-type cells (Fig. 5A). In addition, some Δkar3/Δkar3 cells grew initially as budding yeast but switched to growing as pseudohyphal cells. In each case, the switch to pseudohyphal growth followed an extended cell cycle that lasted an average of 154 ± 10 min (Fig. 5B). After the delayed first division, pseudohyphal cells continued to grow as pseudohyphae with a doubling time close to that of Δkar3/Δkar3 yeast cells (82 ± 4 min).
FIG. 5.
Dynamics of cell division in kar3 mutants. Time-lapse microscopy images of Δkar3/Δkar3 (RSY11) cells growing on YPD medium at 30°C were acquired at 15-min intervals. (A) Some populations of cells divided primarily as budding yeast over the time course. (B) Other populations began dividing as budding yeast but, following an extended delay in cell division, underwent a switch to grow as pseudohyphal cells. Arrows indicate budding cells, where they are clearly evident, that switched to growing as pseudohyphal cells. Time is shown in minutes. Bar, 15 μm.
Based on these observations, we conclude that Kar3p plays an important role in mitotically dividing cells of C. albicans. Cells lacking Kar3p grow more slowly than wild-type cells, exhibit decreased viability, and also switch to grow as pseudohyphal cells following a delay in cell cycle progression.
Kar3p is required for normal progression through anaphase.
Mutant kar3 strains grow more slowly than wild-type strains, exhibit decreased viability, and display an increase in cell size and pseudohypha formation. As discussed earlier, prior studies of C. albicans have indicated that conditions that delay or arrest the cell cycle result in polarized growth and, in particular, increased formation of pseudohyphal cells (1, 5, 6, 11, 23). To further define the role of Kar3p in cell cycle progression, we analyzed the distribution of cells in G1 and G2/mitosis and also directly examined cells for defects in nuclear migration during mitotic division.
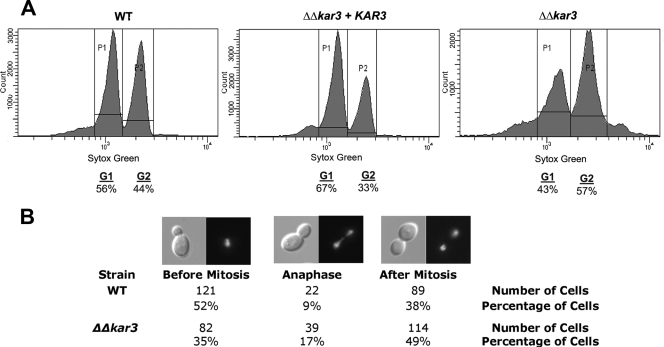
Flow cytometric analysis was performed to compare cell cycle distributions in populations of cells derived from wild-type, Δkar3/Δkar3, and Δkar3/Δkar3+KAR3 strains. As loss of Kar3p causes a significant proportion of Δkar3/Δkar3 cells to be nonviable (Fig. 4C), live cells from log-phase cultures were selected with the dye eosin Y. Eosin Y stains dead C. albicans cells (15) and allowed the sorting of live (unstained) cells, which were then subsequently fixed and stained with the DNA stain SYTOX Green. Cell cycle stages were followed by using flow cytometry (as described in Materials and Methods). For simplicity, cells were identified as being in the G1 or G2/M phase of the cell cycle (Fig. 6A). Under our experimental conditions, an average of 56% of the wild-type cells were in G1 of the cell cycle, with 44% being in G2/M. The distribution of cells between G1 and G2/M was significantly different in Δkar3/Δkar3 strains, however, with only 43% of the cells in G1 and 57% in G2/M. The reconstituted Δkar3/Δkar3+KAR3 strain, similar to the wild-type strain, contained the majority (67%) of the population in G1 of the cell cycle. Similar results were obtained with propidium iodide as an alternative stain to select for viable cells in the population (data not shown). These results demonstrate that populations of Δkar3/Δkar3 cells have an increased tendency to accumulate in the G2/M phases of the cell cycle.
FIG. 6.
Cell cycle analysis of wild-type and kar3 mutant cells. (A) Wild-type (WT; RBY1132), Δkar3/Δkar3 (RSY11), and Δkar3/Δkar3+KAR3 (RSY26) cultures grown in logarithmic phase were sorted to select live cells, stained with the DNA stain SYTOX Green, and prepared for flow cytometry as described in Materials and Methods. Fifty thousand cells of each strain were used to generate histograms. For simplicity, the resulting percentages were normalized and cells were assigned to either the G1 (DNA unreplicated) or the G2/mitosis (DNA replicated) phase of the cell cycle. (B) Mitotic division in budding yeast forms of the wild type and the Δkar3/Δkar3 mutant. Htb-GFP-tagged wild-type (RSY15) and Δkar3/Δkar3 (RSY19) strain cells were grown in logarithmic phase in YPD medium and analyzed by fluorescence microscopy. Only mother cells with attached daughter buds were assessed for nuclear positioning. Cells were categorized as follows: cells containing a single nucleus within the mother bud, cells in anaphase with the DNA stretched between mother and daughter cells, and cells containing two nuclei where nuclear division was completed. The presence of a larger fraction of cells of the Δkar3/Δkar3 strain undergoing nuclear division is indicative of a cell cycle delay in anaphase. For all values, P < 0.05.
Microscopic evaluation of Htb-GFP nucleus-labeled strains was also used to directly visualize patterns of nuclear division between mother and daughter cells in wild-type and kar3 strains. Cells were grown in log phase for 4 h to allow for several rounds of cell division and subsequently evaluated microscopically. Budding yeast cells containing a single nucleus were considered not to have entered mitosis and therefore represented cells in G1, S, or G2 phase. Cells having nuclei spanning between mother and daughter cells were in the process of undergoing anaphase in mitosis. Finally, cells having discrete nuclei in both mother and daughter cells had completed mitosis. In wild-type strains, 52% of the population was in G1, S, or G2 phase; 38% of the cells had completed mitosis; and the remaining 9% were in anaphase (Fig. 6B). In comparison, Δkar3/Δkar3 cells showed a markedly different cell cycle distribution, in which 35% had not begun mitosis, 49% had completed mitosis, and 17% were in anaphase, still undergoing nuclear division (Fig. 6B). These microscopic observations confirm the flow cytometric analysis in showing that a subpopulation of Δkar3/Δkar3 cells accumulate in anaphase due to delayed progression through this phase of the cell cycle. However, nuclear segregation occurred correctly in Δkar3/Δkar3 strains, as nuclear division took place across the mother-daughter bud neck (Fig. 6B). This is in contrast to Δdyn1/Δdyn1 mutants, where nuclear division often takes place within the mother cell of yeast cells, although the daughter nucleus can subsequently segregate to the daughter bud prior to septum formation (18, 37).
C. albicans Kar3p localizes to spindle pole bodies.
Localization studies have shown that S. cerevisiae Kar3p associates primarily with spindle pole bodies, as well as attached and unattached kinetochores in dividing and interphase cells, respectively (39, 58, 60). We therefore sought to determine if Kar3p associates with the mitotic spindle apparatus in C. albicans, based on the evidence that anaphase progression is delayed in the absence of Kar3p.
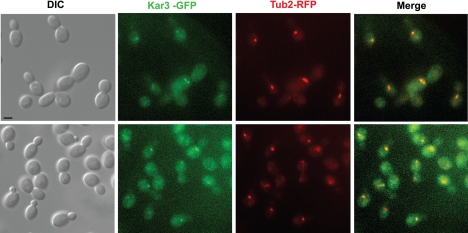
We constructed a Kar3-GFP-labeled strain to determine Kar3p localization, an approach that has successfully been used in studies on S. cerevisiae Kar3p (60). In complementation tests, the Kar3-GFP construct was able to restore normal growth to Δkar3/Δkar3 strains, demonstrating that the Kar3-GFP protein encodes a functional Kar3p activity (data not shown). The Kar3-GFP-labeled strain also coexpressed a tubulin 2-RFP (Tub2-RFP) fusion protein to compare the localization of these proteins in wild-type cells. The Tub2-RFP fusion protein labels the β subunit of tubulin and has previously been used to accurately monitor microtubules, mitotic spindles, and spindle pole bodies in C. albicans (17, 18, 23). Kar3-GFP showed a faint but reproducible pattern of fluorescence, with one or two spots evident in many of the cells (Fig. 7). When the pattern of Kar3-GFP fluorescence was compared to that of Tub2-RFP, there was considerable overlap between GFP and RFP fluorescence, as shown in Fig. 7. These results demonstrate that C. albicans Kar3-GFP colocalizes with Tub2 to the spindle pole bodies in mitotically dividing cells.
FIG. 7.
Kar3p localizes to spindle pole bodies in C. albicans. Wild-type cells expressing RFP-tagged Tub2p and GFP-tagged Kar3p (RSY120 and RSY121) were grown to logarithmic phase in YPD medium at 30°C and imaged by fluorescence microscopy to determine the subcellular localization of Kar3p. DIC images are provided at the left, the Kar3-GFP signal is shown in green, the Tub2-RFP signal is shown in red, and colocalization of Kar3p and Tub2p is represented by a yellow signal in the merged image. Bar, 5.0 μm.
Spindle defects in kar3 mutants.
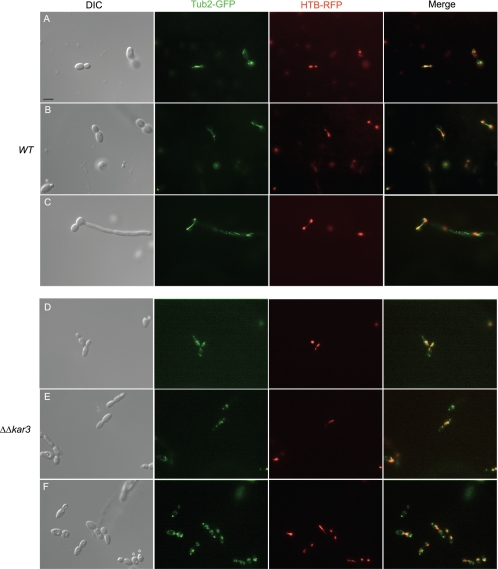
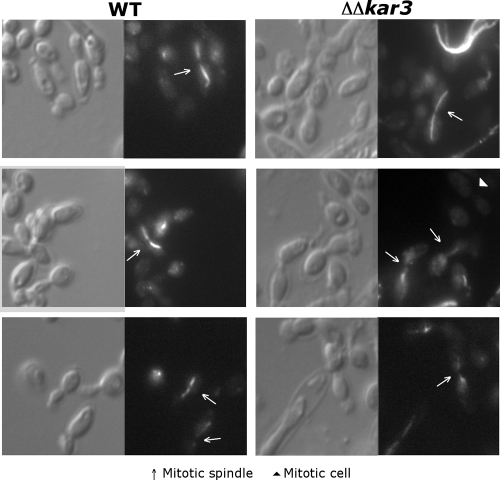
S. cerevisiae Kar3p and the S. pombe homologs Pkl1 and Klp2 are required for normal formation of the mitotic spindle apparatus, and spindle structures were therefore compared in C. albicans wild-type and Δkar3/Δkar3 strains. With Tub2-GFP-labeled strains, we visualized the mitotic spindle in dividing cells and found that while the spindle varied in length in wild-type strains, it consistently spanned the distance between mother and daughter cells, as shown in Fig. 8A to C. In contrast, well-defined mitotic spindles were missing in strains lacking Kar3p, as Tub2-GFP fluorescence invariably failed to show the characteristic pattern of spindle formation between mother and daughter cells (Fig. 8D to F). Instead, Tub2-GFP often showed an irregular pattern of fluorescence in cells undergoing nuclear division, with multiple spots of fluorescence evident, indicating breakdown of the mitotic spindle.
FIG. 8.
Comparison of spindle morphologies of C. albicans wild-type and Δkar3/Δkar3 cells. Htb-RFP- and Tub2-GFP-tagged wild-type (WT; RSY150) and Δkar3/Δkar3 (RSY155) strain cells in logarithmic phase were imaged by live fluorescence microscopy. The cells in rows A, B, D, and F were grown in YPD at 30°C, and the cells in row C were cultured in YPD medium plus 10% serum at 37°C. Wild-type cells contained mitotic spindles in cells where the DNA was stretched between mother and daughter cells (A to C). In contrast, normal mitotic spindles were absent in Δkar3/Δkar3 cells (Tub2-GFP pattern in panels D to F), even in cells that were undergoing anaphase (Htb-RFP pattern). Despite the absence of normal mitotic spindles in kar3 mutants, cells still underwent nuclear division, evidenced by DNA spanning between mother and daughter cells (D to F). Bar, 5.0 μm.
Consistent with prior studies, both wild-type and Δkar3/Δkar3 strains expressing Tub2-GFP fusion constructs grew significantly slower than those containing only untagged tubulin (data not shown), suggesting that the Tub2 fusion protein is not fully functional in these strains (17, 23). It is possible, therefore, that the absence of a stable mitotic spindle in Δkar3/Δkar3 strains is due to an additive effect of both loss of Kar3p and expression of GFP-tagged tubulin. To test this possibility, an indirect immunofluorescence assay was performed to detect mitotic spindles in untagged wild-type and Δkar3/Δkar3 strains. A purified anti-α-tubulin antibody was used which could detect both spindle pole bodies in interphase cells and mitotic spindles in dividing cells (51). Mitotic spindles were clearly visible in medium- and large-budded wild-type cells that were undergoing cell division (Fig. 9). In contrast, cells of Δkar3/Δkar3 strains typically contained no clear mitotic spindle. However, unlike the tubulin-tagged strains, rare mitotic spindles could be detected in a small number of Δkar3/Δkar3 cells by the immunofluorescence assay (Fig. 9).
FIG. 9.
Assessment of mitotic spindle formation in wild-type and Δkar3/Δkar3 cells by immunofluorescence assay. Wild-type (WT; RBY1132) and Δkar3/Δkar3 (RSY11, RSY12) cells were grown in YPD medium in logarithmic phase and prepared for immunofluorescence as described in Materials and Methods. Staining with a rat YOL1/34 anti-tubulin antibody was detected with an fluorescein isothiocyanate-conjugated anti-rat secondary antibody. Mitotic spindles and spindle pole bodies can be seen in both wild-type and Δkar3/Δkar3 cells. However, mitotic spindles were rarely visible in Δkar3/Δkar3 cells and often exhibited diffuse fluorescence or a reduced fluorescence intensity, as exemplified by arrows in the bottom two panels. In addition, many large-budded Δkar3/Δkar3 cells presumed to be undergoing mitosis did not exhibit clearly defined mitotic spindles (triangle).
DISCUSSION
These results present the first detailed picture of the role of C. albicans Kar3p both in mating cells and during mitotic division. During mating, we have confirmed that Kar3p is necessary for karyogamy in strains derived from SC5314, the standard laboratory strain of C. albicans. Following fusion of mating a and α cells, nuclear congression does not occur in the absence of Kar3p (this study and reference 8).
Nuclear positioning within cells responding to mating pheromone was largely unaffected by the loss of Kar3p or the dynein motor protein Dyn1p. Mutants lacking either protein produced shorter mating projections than wild-type strains, but in both cases, nuclei were able to migrate to the tip of the mating projection. This result contrasts sharply with nuclear positioning in hyphal cells of C. albicans, where the absence of Dyn1p was found to often block nuclear migration into the growing hyphal cell (18, 37). The lack of nuclear migration in dyn1 mutants prevented efficient growth of hyphal cells and resulted in a marked defect in filamentation. The results described here demonstrate that, despite similarities in morphology and overlapping gene expression profiles (9, 34), C. albicans mating projections and hyphal cells carry out long-range nuclear migration by distinct mechanisms.
The present study also addresses the role of Kar3p in mitotic division in C. albicans. Cells lacking Kar3p displayed a significant defect in mitotic growth, as evidenced by both reduced viability and an increased doubling time. Mutant cells tended to accumulate in anaphase as large-budded yeast or pseudohyphal cells undergoing nuclear division. This delay is presumably a direct result of defective mitotic spindle morphogenesis, as kar3 cells exhibited aberrant spindles or, more often, lacked visible mitotic spindles altogether. This result is reminiscent of that obtained with S. cerevisiae, where the loss of Kar3p was found to reduce the stability of the mitotic spindle (39, 46, 53, 54).
The lack of normal mitotic spindles was particularly striking in C. albicans kar3 cells colabeled with a Tub2-GFP construct. In these cells, it is likely that tagging the tubulin protein, in combination with the absence of Kar3p, caused a synergistic defect in anaphase spindle integrity. This hypothesis agrees with prior studies that showed that fluorescently labeled tubulin strains exhibited delayed elongation of the mitotic spindle during anaphase, indicating that spindle function had been compromised in these strains (17, 23). Altogether, our observations demonstrate that Kar3p plays an important role in stabilizing mitotic spindles in C. albicans, and in strains expressing a tagged tubulin protein, this role is of even greater importance to spindle integrity.
Consistent with a role in mitotic spindle assembly and chromosome segregation, Kar3 protein localized to spindle pole bodies in mitotically dividing cells. In S. cerevisiae, Kar3p localized primarily to the spindle pole bodies in mitotic cells but could also be found at kinetochores, particularly if the kinetochores had become detached from spindle microtubules (58, 60). In S. pombe, the two homologs of Kar3p showed distinct localization patterns; Klp2p localized mainly at or near the kinetochores, while Pkl1p localized to the nucleus during interphase and to the spindle during mitosis (49, 59). It therefore appears that C. albicans Kar3p most closely resembles S. cerevisiae Kar3p in its subcellular localization. While S. cerevisiae Kar3p acts in concert with Cik1p and Vik1p, orthologs of these genes have yet to be identified in C. albicans, due in part to the large family of kinesin-like genes in the sequenced C. albicans genome (www.candidagenome.org) (2, 18). Indeed, probing of C. albicans extracts with an antibody against the motor domain of S. cerevisiae Kar3p (gift of Susan Gilbert, University of Pittsburgh) revealed the presence of multiple proteins with an antigenicity similarity to that of ScKar3p (data not shown). Clearly, it is of considerable interest to see if C. albicans Kar3p function is also regulated by the formation of heterodimers with Cik1- or Vik1-related proteins.
C. albicans kar3 mutants also exhibited altered cell and colony morphologies, particularly under filamentation-inducing conditions. Δkar3/Δkar3 colonies grown on Spider (nutrient-poor) medium lacked the usual pattern of peripheral filamentation, and when grown in liquid medium containing serum, kar3 cells failed to efficiently form true hyphal cells, instead forming mainly pseudohyphal cells. Pseudohyphae and hyphae are now recognized as distinct morphological states of C. albicans that can both originate from yeast form cells but rarely undergo interconversion with each other (11, 57). These forms are distinguished by the pattern of septation; pseudohyphal cells contain a restriction at the bud neck, where the nucleus undergoes division, while hyphal cells do not have constrictions and undergo nuclear division within the hyphal tube.
It is now apparent that a number of conditions that arrest or delay the cell cycle can induce polarized growth in C. albicans (11). For example, treatment of cells with agents that block cells in S phase or in mitosis results in hyperpolarized cells (4, 6). Similarly, mutant strains that are delayed in cell cycle progression also show increased polarized growth. Mutants delayed in G1, such as Δcdc4/Δcdc4, demonstrate increased hyphal growth, while mutants delayed in S, G2, or M phase, such as Δrad52/Δrad52 or Δclb4/Δclb4, display increased pseudohyphal growth (1, 3, 10, 11). In the case of C. albicans Δkar3/Δkar3 cells, we suggest that a delay in anaphase contributes to the tendency of these cells to form pseudohyphal cells. In support of this, kar3 mutants were shown to accumulate as large-budded cells in the process of undergoing nuclear division. In addition, time-lapse microscopy showed that cells switched from growing as yeast cells to growing as pseudohyphal cells following an extended delay in cell division. These observations resemble those on Δdyn1/Δdyn1 mutants of C. albicans, which also grow as a mixture of yeast and pseudohyphal cells (18). In this case, pseudohyphal growth was a consequence of a delay in anaphase mediated by the Bub2 checkpoint pathway. It thus appears that the loss of distinct molecular motors can lead to polarized growth due to delays in cell cycle progression.
Recent studies have highlighted the potential for novel antifungal drugs that target different aspects of the microtubule apparatus. Xu et al. demonstrated that several new antifungal compounds behaved similarly to known microtubule inhibitors in C. albicans and also induced the characteristic pseudohyphal morphology of cells undergoing G2/M cell cycle arrest (63). In addition, Chua et al. investigated the role of the kinesin Kip1p in C. albicans and showed that loss of Kip1p led to aberrant rounds of spindle pole body duplication, again accompanied by mitotic delay and elongated growth (14). Kip1p was also inhibited by an aminobenzothiazole compound, which caused Kip1p to bind to microtubules in a rigor-like mechanism resulting in cell death. These studies reveal that drugs that target nonessential components of the microtubule apparatus can produce fungicidal effects, presumably due to arrest of the mitotic spindle, and thus are effective antifungal agents (14). It remains to be seen if similar studies of other kinesins, including Kar3p, will provide additional targets for antifungal drug discovery.
Acknowledgments
We particularly thank Ken Finley and Judy Berman for the gift of plasmids and for technical assistance with microscopy of live cells. We are also grateful to Susan Gilbert and Laurent Brossay for gifting antibodies, the Wendland laboratory for sharing strains, and Chui-Sun Yap for help with statistical analysis. Tracy Rosebrock, Kevin Alby, Anthony Choi, and Dana Schaefer provided helpful discussions and comments on the manuscript.
This work was supported in part by grants to R.J.B. from the Rhode Island Foundation and a Richard B. Salomon Faculty Research Award and by support of R.K.S. on NIH training grant 5T32GM007601.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Andaluz, E., T. Ciudad, J. Gomez-Raja, R. Calderone, and G. Larriba. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 591452-1472. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atir-Lande, A., T. Gildor, and D. Kornitzer. 2005. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 162772-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachewich, C., A. Nantel, and M. Whiteway. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57942-959. [DOI] [PubMed] [Google Scholar]
- 5.Bachewich, C., D. Y. Thomas, and M. Whiteway. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 142163-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai, C., N. Ramanan, Y. M. Wang, and Y. Wang. 2002. Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 4531-44. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62100-119. [DOI] [PubMed] [Google Scholar]
- 8.Bennett, R. J., M. G. Miller, P. R. Chua, M. E. Maxon, and A. D. Johnson. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 551046-1059. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, R. J., M. A. Uhl, M. G. Miller, and A. D. Johnson. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 238189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bensen, E. S., A. Clemente-Blanco, K. R. Finley, J. Correa-Bordes, and J. Berman. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 163387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman, J. 2006. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 9595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand, A., D. M. MacCallum, A. J. Brown, N. A. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9327-335. [DOI] [PubMed] [Google Scholar]
- 14.Chua, P. R., D. M. Roof, Y. Lee, R. Sakowicz, D. Clarke, D. Pierce, T. Stephens, M. Hamilton, B. Morgan, D. Morgans, T. Nakai, A. Tomasi, and M. E. Maxon. 2007. Effective killing of the human pathogen Candida albicans by a specific inhibitor of non-essential mitotic kinesin Kip1p. Mol. Microbiol. 65347-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantino, P. J., D. E. Budd, and N. F. Gare. 1995. Enumeration of viable Candida albicans blastospores using tetrabromofluorescein (eosin Y) and flow cytometry. Cytometry 19370-375. [DOI] [PubMed] [Google Scholar]
- 16.Endow, S. A., S. J. Kang, L. L. Satterwhite, M. D. Rose, V. P. Skeen, and E. D. Salmon. 1994. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 132708-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finley, K. R., and J. Berman. 2005. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot. Cell 41697-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finley, K. R., K. J. Bouchonville, A. Quick, and J. Berman. 2008. Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J. Cell Sci. 121466-476. [DOI] [PubMed] [Google Scholar]
- 19.Gardner, M. K., J. Haase, K. Mythreye, J. N. Molk, M. Anderson, A. P. Joglekar, E. T. O'Toole, M. Winey, E. D. Salmon, D. J. Odde, and K. Bloom. 2008. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J. Cell Biol. 18091-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gargas, A., P. T. DePriest, M. Grube, and A. Tehler. 1995. Multiple origins of lichen symbioses in fungi suggested by SSU rDNA phylogeny. Science 2681492-1495. [DOI] [PubMed] [Google Scholar]
- 21.Gola, S., R. Martin, A. Walther, A. Dunkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 201339-1347. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 23.Hazan, I., M. Sepulveda-Becerra, and H. Liu. 2002. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13134-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedges, S. B., J. E. Blair, M. L. Venturi, and J. L. Shoe. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirokawa, N., and R. Takemura. 2004. Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 30150-59. [DOI] [PubMed] [Google Scholar]
- 26.Hoyt, M. A., L. He, L. Totis, and W. S. Saunders. 1993. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 13535-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 2851271-1275. [DOI] [PubMed] [Google Scholar]
- 28.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289307-310. [DOI] [PubMed] [Google Scholar]
- 29.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 955150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, A. 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1106-116. [DOI] [PubMed] [Google Scholar]
- 31.Kim, M. K., Y. M. Lee, W. Kim, and W. Choi. 2005. Complete sequence of a gene encoding KAR3-related kinesin-like protein in Candida albicans. J. Microbiol. 43406-410. [PubMed] [Google Scholar]
- 32.Liu, H. 2002. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292299-311. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 2661723-1726. [DOI] [PubMed] [Google Scholar]
- 34.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddox, P. S. 2005. Microtubules: Kar3 eats up the track. Curr. Biol. 15R622-R624. [DOI] [PubMed] [Google Scholar]
- 36.Manning, B. D., J. G. Barrett, J. A. Wallace, H. Granok, and M. Snyder. 1999. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 1441219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, R., A. Walther, and J. Wendland. 2004. Deletion of the dynein heavy-chain gene DYN1 leads to aberrant nuclear positioning and defective hyphal development in Candida albicans. Eukaryot. Cell 31574-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald, H. B., R. J. Stewart, and L. S. Goldstein. 1990. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 631159-1165. [DOI] [PubMed] [Google Scholar]
- 39.Meluh, P. B., and M. D. Rose. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 601029-1041. [DOI] [PubMed] [Google Scholar]
- 40.Merson-Davies, L. A., and F. C. Odds. 1989. A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 1353143-3152. [DOI] [PubMed] [Google Scholar]
- 41.Miki, H., Y. Okada, and N. Hirokawa. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15467-476. [DOI] [PubMed] [Google Scholar]
- 42.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110293-302. [DOI] [PubMed] [Google Scholar]
- 43.Molk, J. N., E. D. Salmon, and K. Bloom. 2006. Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J. Cell Biol. 17227-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro-García, F., M. Sanchez, C. Nombela, and J. Pla. 2001. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25245-268. [DOI] [PubMed] [Google Scholar]
- 45.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Page, B. D., L. L. Satterwhite, M. D. Rose, and M. Snyder. 1994. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J. Cell Biol. 124507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panwar, S. L., M. Legrand, D. Dignard, M. Whiteway, and P. T. Magee. 2003. MFα1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot. Cell 21350-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, Y. N., and J. Morschhauser. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 41328-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pidoux, A. L., M. LeDizet, and W. Z. Cande. 1996. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol. Biol. Cell 71639-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341119-127. [DOI] [PubMed] [Google Scholar]
- 51.Sanyal, K., and J. Carbon. 2002. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc. Natl. Acad. Sci. USA 9912969-12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders, W., D. Hornack, V. Lengyel, and C. Deng. 1997. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J. Cell Biol. 137417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saunders, W., V. Lengyel, and M. A. Hoyt. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell 81025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunders, W. S., and M. A. Hoyt. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70451-458. [DOI] [PubMed] [Google Scholar]
- 55.Saunders, W. S., D. Koshland, D. Eshel, I. R. Gibbons, and M. A. Hoyt. 1995. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soll, D. R., S. R. Lockhart, and R. Zhao. 2003. Relationship between switching and mating in Candida albicans. Eukaryot. Cell 2390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12317-324. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, K., E. Kitamura, Y. Kitamura, and T. U. Tanaka. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Troxell, C. L., M. A. Sweezy, R. R. West, K. D. Reed, B. D. Carson, A. L. Pidoux, W. Z. Cande, and J. R. McIntosh. 2001. pkl1+ and klp2+: two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol. Biol. Cell 123476-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tytell, J. D., and P. K. Sorger. 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172861-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vale, R. D., and R. J. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13745-777. [DOI] [PubMed] [Google Scholar]
- 62.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7350-357. [DOI] [PubMed] [Google Scholar]
- 63.Xu, D., B. Jiang, T. Ketela, S. Lemieux, K. Veillette, N. Martel, J. Davison, S. Sillaots, S. Trosok, C. Bachewich, H. Bussey, P. Youngman, and T. Roemer. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3e92. [DOI] [PMC free article] [PubMed] [Google Scholar]