Abstract
Ubiquitously expressed focal adhesion kinase (FAK), a critical component in transducing signals from sites of cell contacts with extracellular matrix, was named after its typical localization in focal adhesions. A nuclear localization of FAK has been also reported and its scaffolding role in nucleus and requirement for p53 ubiquitination were only recently described. Whereas FAK nuclear localization signal (NLS) was found in F2 lobe of FERM domain, nuclear export signal (NES) sequences were has not been yet determined. Here we demonstrate that FAK has two NES sequences, NES1 in F1 lobe of FERM domain and NES2 in kinase domain. Although, both NES1 and NES2 are evolutionary conserved, as well as in FAK-related protein kinase Pyk2, only NES2 demonstrates full biological nuclear export activity.
Keywords: FAK, nuclear export signal, Pyk2
FAK is ubiquitously expressed signaling scaffold protein, indispensable for transduction of signals from supramolecular complexes localized at contact sites of cell with extracellular matrix (Cohen and Guan, 2005; Mitra et al., 2005; Schlaepfer et al., 2004). Although FAK was named according to its localization at focal adhesions (Hanks et al., 1992; Schaller et al., 1992), several groups reported nuclear localization of FAK (Golubovskaya et al., 2005; Kadare et al., 2003; Levkau et al., 1998; Lobo and Zachary, 2000; Stewart et al., 2002; Yi et al., 2003). Only recently, the mechanism(s) that promote or regulate FAK nuclear accumulation and the biological role of nuclear FAK were somewhat revealed (Lim et al., 2008).
Classic nuclear localization sequences, typically containing clusters of basic amino acids, were detected in FAK N-terminal band 4.1, ezrin, radixin, moesin-homology (FERM) F2 lobe. The largest patch consists of residues lysine (K) 190, K191, K216, K218, arginine (R) R221, and K222 whereas a second smaller basic patch is comprised of R204, R205, and K209. In addition, residues R177 and R178 are partially exposed within the FAK FERM F2 lobe structure. Mutations of lysine into alanine (A) in the largest basic patch (K190/191A or K216/218A) blocked, whereas mutations in a second smaller patch (K204/R205A) slightly reduced FERM nuclear accumulation. R177/R178A mutations also prevented FERM nuclear localization (Lim et al., 2008).
Since it is found in both cytoplasm and nucleus, FAK obviously has to have a mechanism that enables nucleocytoplasmic shuttling. Leucine-rich Nuclear Export Signal (NES) sequences often mediate protein export from the nucleus to the cytoplasm (Kutay and Guttinger, 2005; la Cour et al., 2003; Moroianu, 1999; Terry et al., 2007). The first NES were identified in human immunodeficiency virus, type I-coded Rev protein (Fischer et al., 1995) and protein kinase A inhibitor of cAMP-dependent protein kinase (Wen et al., 1995). NES sequences consist of 4–5 hydrophobic residues within a region of about 10 amino acids. These hydrophobic residues are predominantly leucine (L), but may also be isoleucine (I), valine (V), methionine (M) and phenylalanine (P). However, only less than 50% of identified leucine-rich NESs fit into generally accepted loose consensus L-x(2,3)-[LIVFM]-x(2,3)-L-x-[LI] (Bogerd et al., 1996; la Cour et al., 2003). We found that FAK has two putative NES sequences, one within FERM and one within kinase domain.
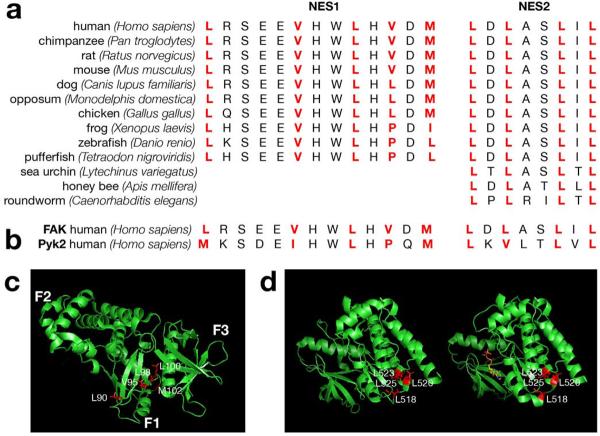
In human FAK, the first cluster, termed NES1, consists of hydrophobic amino acid residues: 90, 95, 98, 100, and 102 (UniProtKB/Swiss-Prot database accession number Q05397). The pattern is conserved through the vertebrate species (Fig. 1a), as well as present in FAK-related protein kinase Pyk2 (Fig. 1b). However, it is not conserved in FERM domains of other proteins and it is likely to be specific for FAK family. The second cluster in human FAK, termed NES2, which consists of hydrophobic residues 518, 520, 523, and 525, can also be found in Pyk2 and it is even more evolutionary conserved than NES 1. NES2, and not NES1, was detected in deduced sequences of invertebrate FAK proteins that are rather divergent from the highly conserved sequences of vertebrates (Fig. 1) (Garcia et al., 2004; Hens and DeSimone, 1995).
Figure 1.
FAK has two potential NES sequences. (a) Whereas NES1, a hydrophobic patch of amino acids in F1 lobe of FERM domain, is conserved in vertebrates, kinase domain's NES2 is preserved in some invertebrates too. (b) Both NES1 and NES2 are present also in FAK-related protein kinase Pyk2. (c) NES1 sequences highlighted in structure of FERM domain. (d) NES2 sequences highlighted in structure of inactive (left) and active (right) FAK kinase domain. Nonhydrolyzable ATP analog AMP-PNP bound to active site is shown in stick representation. Structural images were presented using PyMOL™ program (DeLano Scietific, LLC, Palo Alto, CA) based on reported crystal structure of FERM domain (Ceccarelli et al., 2006) and inactive vs. active FAK (Lietha et al., 2007).
Three FERM subdomains F1, F2, and F3 intimately associate with one another forming a compact structure with the overall shape of a cloverleaf (Ceccarelli et al., 2006). NES1 sequences localized within F1 lobe, which spans residues 33-127, are not as easy accessible as NES2 (Fig 1c, d). Structural changes between autoinhibited and catalytically active FAK do not affect position of NES2 sequences (Lietha et al., 2007). NES2 patch is prominently exposed and easy to access regardless of FAK kinase activity suggesting that NES2 might have more prominent regulatory role in nucleocytoplasmic shuttling of FAK (Fig 1c).
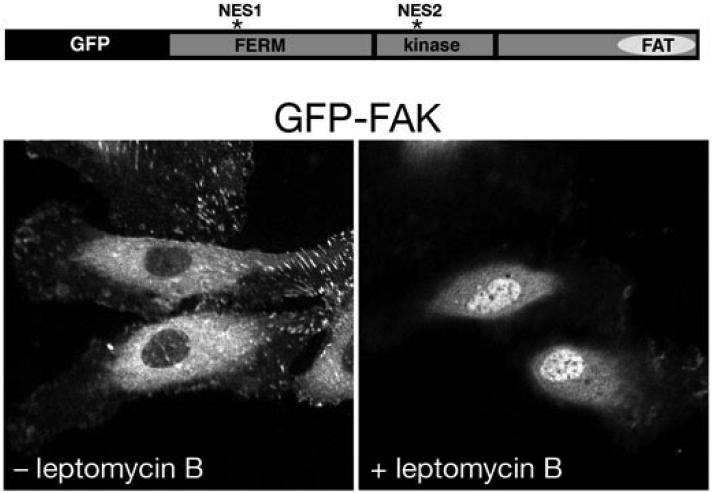
To determine whether FAK nucleocytoplasmic shuttling is in fact NES-dependent, we transduced HUVEC cells with GFP-FAK and then exposed to leptomycin B. Leptomycin B, an unsaturated, branched-chain fatty acid, known as a potent and specific nuclear export inhibitor that alkylates and inhibits chromosomal region maintenance 1 (CRM1)/exportin 1 (XPO1), a transporter required for nuclear export of NES-containing proteins (Fornerod et al., 1997; Fukuda et al., 1997; Ossareh-Nazari et al., 1997). We hypothesized that if FAK contains biologically functional NES sequences, leptomycin B would prevent FAK export from the nucleus. Indeed, HUVECs transduced with GFP-FAK retained GFP-FAK in the nucleus upon exposure to leptomycin B (Fig. 2).
Figure 2.
FAK nucleocytoplasmic shuttling is NES-dependent. Live cell imaging was used to follow GFP-FAK distribution before and 4 h after exposure of GFP-FAK-expressing HUVECs to 10 ng/ml leptomycin B as described (Lim et al., 2008).
To validate whether these two hydrophobic amino acid clusters can function as NES sequences, we created a series of constructs where we inserted combinations of multiple FAK NES sequences aiming to pull out exogenously expressed green fluorescent protein (GFP) tagged with triple NLS from SV40 large T antigen (DPKKKKRKV) (Fig 3a). Primary mouse embryonic fibroblasts (MEF) were analyzed for GFP localization 48 h post transfection. Triple NLS tag (NLS)3 added in frame to C-terminus of GFP [GFP-(NLS)3] was sufficient to restrict GFP localization to the nucleus (Fig. 3b). Adding a single FAK NES1 or NES2 cassette between GFP and triple NLS tag was not enough to pull the fusion GFP-NES1-(NLS)3 or GFP-NES2-(NLS)3 protein out from the nucleus (data not shown). Double cassettes, either two NES1 [GFP-NES1-NES1-(NLS)3], two NES2 [GFP-NES2-NES2-(NLS)3] or combination of one NES1 and one NES2 [GFP-NES1-NES2-(NLS)3] fused between GFP and triple NLS tag provided enough binding sites for exporting molecules and were able to translocate GFP partially or completely out to the cytoplasm (Fig. 3b). In each of three separate experiments, we monitored by fluorescence microscopy 100 transfected cells for each of constructs and assorted these cells based on GFP localization into three groups: 1) nuclear only, 2) nuclear and cytoplasmic, and 3) cytoplasmic only. GFP was localized only in cytoplasm in about 46% of cells expressing GFP-NES1-NES1-(NLS)3 fusion protein. Substitution of one NES1 with NES2 cassette, increased cytoplasmic localization of GFP-NES1-NES2-(NLS)3 fusion protein to 60-62%, whereas double NES2 had even higher nuclear export index (86% % of GFP-NES2-NES2-(NLS)3 fusion protein expressing cells). On the other hand, whereas both GFP-NES1-NES1-(NLS)3 and GFP-NES1-NES2-(NLS)3 fusion proteins had solely nuclear localization in about 20% cells, GFP-NES2-NES2-(NLS)3 was found in nucleus only in 4.4% of transfected cells (Fig. 3c). Finally, mutation of hydrophobic residues into alanines (A) in both NES1 [GFP-mNES1-mNES1-(NLS)3] and NES2 [GFP-mNES2-mNES2-(NLS)3] obliterated their nuclear export function. Neither GFP-mNES1-mNES1-(NLS)3 nor GFP-mNES2-mNES2-(NLS)3 were able to translocate GFP-(NLS)3 from the nucleus into cytoplasm (Fig. 3). These data suggest that cluster of hydrophobic residues in both putative NES sequences of FAK could interact with exporting proteins and that they can be biologically functional.
Figure 3.
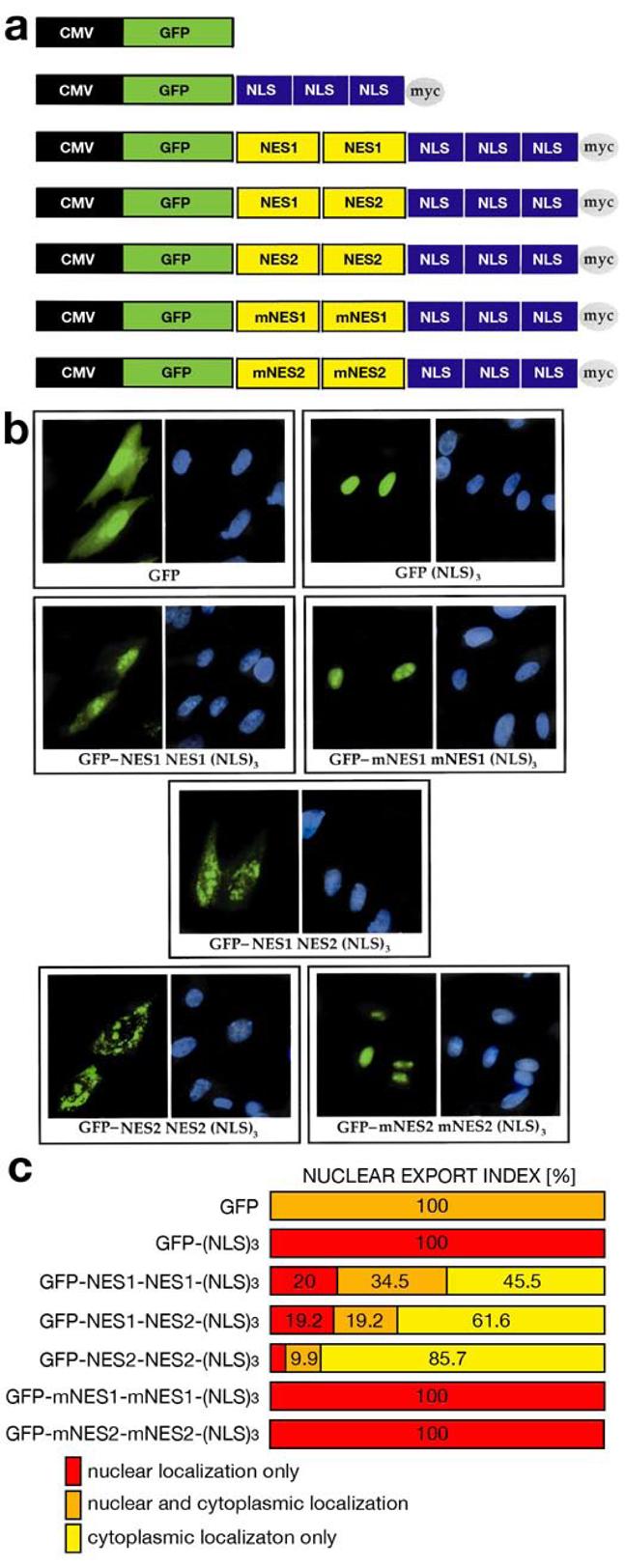
Both NES1 and NES2 sequences in FAK are biologically functional. (a) Diagram of pShooter plasmid (Invitrogen) constructs used to determine functionality of FAK NES sequences. CMV, immediate-early cytomegalovirus promoter; myc, c-myc epitope (EQKLISEEDL); NES1, FAK fragment encoding amino acid sequence RLSHLRSEEVHWLHVDMGV; NES2 FAK fragment encoding amino acid sequence FLQVRKYSLDLASLILYAYQL, mNES1, mutated FAK fragment encoding amino acid sequence RLSHARSEEAHWAHADMGV (mutated amino acids are in bold); mNES2, mutated FAK fragment encoding amino acid sequence FLQVRKYSADAASAIAYAYQL (mutated amino acids are in bold); NLS, nuclear localization signal from SV40 large T antigen (DPKKKKRKV). (b) Nuclear or cytoplasmic localization of GFP or GFP fusion proteins is determined by presence of NLS and functional NES sequences. (c) Quantification of GFP and GFP fusion protein localization. Whereas untagged GFP is uniformly distributed in nucleus and cytoplasm, triple NLS pulls GFP strongly into the nucleus. By adding functional NES1 or NES2, GFP is transferred into the cytoplasm. Mutation of hydrophobic residues in NES1 (mNES1) or NES2 (mNES2) aborts cytoplasmic translocation.
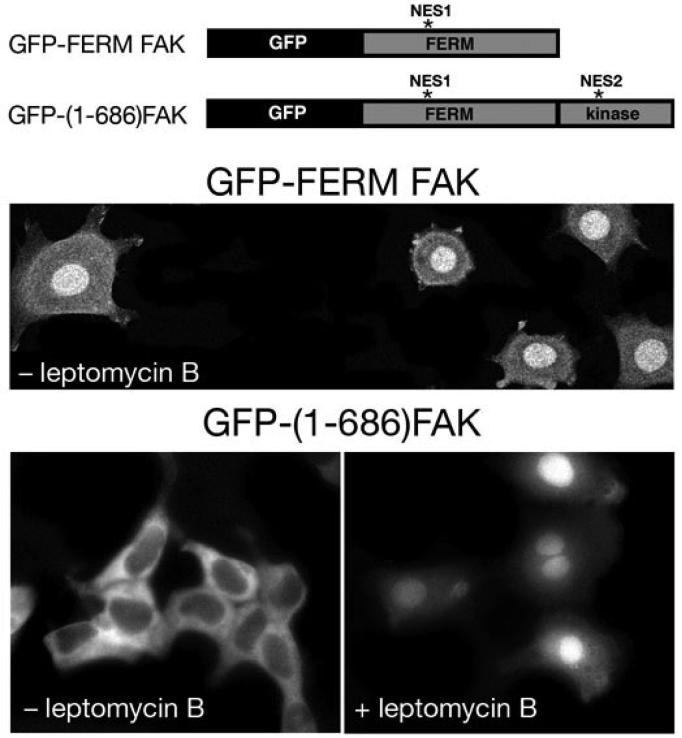
To determine whether biological activity of NES1 and NES2 in intact FAK protein correlates to our data shown in the Figure 3, we transfected HUVECs with GFP fused with N-terminal truncated FAK (GFP-FERM FAK), a construct that contains intact NLS and NES1, but not NES2 sequences (Lim et al., 2008). The GFP-FERM FAK fusion protein was predominantly localized in the nucleus indicating that NES1 is not sufficiently strong to overcome potent NLS sequence (Fig 4). On the other hand, a construct containing intact NLS, NES1, and NES2 (GFP-(1-686)FAK), was localized mainly in perinuclear cytoplasm, whereas in the presence of leptomycin B it was retained in the nucleus. Remaining C-terminal domain of FAK that corresponds to FAK-related non-kinase (FRNK) when fused to GFP was never seen in the nucleus, with or without leptomycin B in the culture medium (data not shown). Taken together our data suggest that both NES sequences are required to keep FAK out of the nucleus (Fig. 4).
Figure 4.
Both NES sequences are required to keep FAK out of the nucleus. GFP-FERM FAK that contains intact NLS and NES1, but not NES2 sequences is predominantly localized in the nucleus, whereas full length FAK containing intact NLS, NES1, and NES2 (GFP-FAK) is localized mainly in perinuclear cytoplasm and in focal contacts. Construction of GFP-FAK and GFP-FERM FAK were described elsewhere (Ilic et al., 1998; Lim et al., 2008). HUVECs expressing GFP-FERM FAK or FAK−/−p53−/− mouse embryonic fibroblasts expressing GFP-(1-686)FAK were visualized by confocal microscopy (Bio-Rad Radiance 2100) as described (Lim et al., 2008).
Weak nuclear export activity of FAK NES1 might be due to the fact that the leucines are more buried in core of domain than exposed, making them practically almost inaccessible to solvent (Fig 1c). Similar phenomenon was reported for proposed NES residues in abl kinase (Hantschel et al., 2005; Taagepera et al., 1998)
Double NES sequence of which one is within a kinase domain is not unique for FAK. Cdc7 serine/threonine kinase, that regulates G1/S transition and initiation of DNA replication also has two NES sequence: NES1 at 458-467 within kinase insert III, and NES2 at 545-554 within the kinase IX domain (Kim et al., 2007). The localization of NES sequence in kinase domain raises an interesting possiblilty to play a double role in regulation of FAK function. The binding of nuclar export proteins to NES may not have a role only in nucleocytoplasmic shuttling, but also in down-regulation of kinase activity by masking FAK kinase domain.
FAK-related protein kinase Pyk2 is also shuttling between nucleus and cytoplasm in NES-dependent manner (Aoto et al., 2002; Arcucci et al., 2006; Faure et al., 2007). Even though the homology between two proteins is only about 40% in FERM and about 60% in the central kinase domain, NLS and both NES clusters are conserved between FAK and Pyk2 (Fig. 1A) (Lim et al., 2008). Interestingly, Pyk2/FAK chimeras, in which FERM and kinase domain of Pyk2 were fused to C-terminal region of FAK with the FAT-containing L1034S mutation, which aborts interaction with paxillin and focal adhesion localization, were also localized in cytoplasm (Klingbeil et al., 2001). These data taken together suggests that FAK and Pyk2 likely use the same mechanism for shuttling in and out of the nucleus.
Recently we have shown that FAK FERM nuclear translocation is pre-requirement for FERM-mediated binding to p53, and FERM-enhanced Mdm2-depenedent p53 ubiquitination (Lim et al., 2008). Golubovskaya et al. localized the site of FAK binding to a 7 amino-acid region (amino acids 65-71) in the N-terminal proline-rich domain of human p53 and shown that mutating this site and targeting the site with peptides affected p53 functioning and viability in the cells (Golubovskaya et al., 2008). Whether Pyk2 interacts also directly with p53 and plays a role in p53 degradation it remains to be determined. Furthermore, interplay between FAK/Pyk2 and p53 family members p63 and p73 should be also a subject of future investigation.
Acknowledgments
This work was supported by grants from NIH to David Schlaepfer (CA102310) and Dusko Ilic (CA087652).
References
- Aoto H, Sasaki H, Ishino M, Sasaki T. Nuclear translocation of cell adhesion kinase beta/proline-rich tyrosine kinase 2. Cell Struct Funct. 2002;27:47–61. doi: 10.1247/csf.27.47. [DOI] [PubMed] [Google Scholar]
- Arcucci A, Montagnani S, Gionti E. Expression and intracellular localization of Pyk2 in normal and v-src transformed chicken epiphyseal chondrocytes. Biochimie. 2006;88:77–84. doi: 10.1016/j.biochi.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–14. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem. 2006;281:252–9. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Guan JL. Mechanisms of focal adhesion kinase regulation. Curr Cancer Drug Targets. 2005;5:629–43. doi: 10.2174/156800905774932798. [DOI] [PubMed] [Google Scholar]
- Faure C, Corvol JC, Toutant M, Valjent E, Hvalby O, Jensen V, et al. Calcineurin is essential for depolarization-induced nuclear translocation and tyrosine phosphorylation of PYK2 in neurons. J Cell Sci. 2007;120:3034–44. doi: 10.1242/jcs.009613. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–83. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–11. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Garcia MG, Toney SJ, Hille MB. Focal adhesion kinase (FAK) expression and phosphorylation in sea urchin embryos. Gene Expr Patterns. 2004;4:223–34. doi: 10.1016/j.modgep.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Golubovskaya V, Finch R, Zheng M, Kurenova EV, Cance WG. The 7 amino-acid site in the proline-rich region of the N-terminal domain of p53 is involved in interaction with FAK and is critical for p53 functioning. Biochem J. 2008 doi: 10.1042/BJ20071657. [DOI] [PubMed] [Google Scholar]
- Golubovskaya VM, Finch R, Cance WG. Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J Biol Chem. 2005;280:25008–21. doi: 10.1074/jbc.M414172200. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Calalb MB, Harper MC, Patel SK. Focal adhesion protein-tyrosine kinase phosphorylated in response to cell attachment to fibronectin. Proc Natl Acad Sci U S A. 1992;89:8487–91. doi: 10.1073/pnas.89.18.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O, Wiesner S, Guttler T, Mackereth CD, Rix LL, Mikes Z, et al. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol Cell. 2005;19:461–73. doi: 10.1016/j.molcel.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Hens MD, DeSimone DW. Molecular analysis and developmental expression of the focal adhesion kinase pp125FAK in Xenopus laevis. Dev Biol. 1995;170:274–88. doi: 10.1006/dbio.1995.1214. [DOI] [PubMed] [Google Scholar]
- Ilic D, Almeida EA, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular matrix survival signals transduced by focal adhesion kinase suppress p53-mediated apoptosis. J Cell Biol. 1998;143:547–60. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, et al. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem. 2003;278:47434–40. doi: 10.1074/jbc.M308562200. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Kim SY, Lee H. Identification and characterization of human cdc7 nuclear retention and export sequences in the context of chromatin binding. J Biol Chem. 2007;282:30029–38. doi: 10.1074/jbc.M703705200. [DOI] [PubMed] [Google Scholar]
- Klingbeil CK, Hauck CR, Hsia DA, Jones KC, Reider SR, Schlaepfer DD. Targeting Pyk2 to beta 1-integrin-containing focal contacts rescues fibronectin-stimulated signaling and haptotactic motility defects of focal adhesion kinase-null cells. J Cell Biol. 2001;152:97–110. doi: 10.1083/jcb.152.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–4. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–6. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkau B, Herren B, Koyama H, Ross R, Raines EW. Caspase-mediated cleavage of focal adhesion kinase pp125FAK and disassembly of focal adhesions in human endothelial cell apoptosis. J Exp Med. 1998;187:579–86. doi: 10.1084/jem.187.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–87. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, et al. Nuclear FAK Promotes Cell Proliferation and Survival through FERM-Enhanced p53 Degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo M, Zachary I. Nuclear localization and apoptotic regulation of an amino-terminal domain focal adhesion kinase fragment in endothelial cells. Biochem Biophys Res Commun. 2000;276:1068–74. doi: 10.1006/bbrc.2000.3547. [DOI] [PubMed] [Google Scholar]
- Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- Moroianu J. Nuclear import and export pathways. J Cell Biochem. 1999;32-33(Suppl):76–83. doi: 10.1002/(sici)1097-4644(1999)75:32+<76::aid-jcb10>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–4. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Schaller MD, Borgman CA, Cobb BS, Vines RR, Reynolds AB, Parsons JT. pp125FAK a structurally distinctive protein-tyrosine kinase associated with focal adhesions. Proc Natl Acad Sci U S A. 1992;89:5192–6. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Stewart A, Ham C, Zachary I. The focal adhesion kinase amino-terminal domain localises to nuclei and intercellular junctions in HEK 293 and MDCK cells independently of tyrosine 397 and the carboxy-terminal domain. Biochem Biophys Res Commun. 2002;299:62–73. doi: 10.1016/s0006-291x(02)02547-0. [DOI] [PubMed] [Google Scholar]
- Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, et al. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci U S A. 1998;95:7457–62. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–6. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–73. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Yi XP, Wang X, Gerdes AM, Li F. Subcellular redistribution of focal adhesion kinase and its related nonkinase in hypertrophic myocardium. Hypertension. 2003;41:1317–23. doi: 10.1161/01.HYP.0000072772.74183.5F. [DOI] [PubMed] [Google Scholar]