Abstract
Replacing the Pro6 in the p6Gag-derived 9-mer “P-E-P-T-A-P-P-E-E” with N-substituted glycine (NSG) residues is problematic. However, incorporation of hydrazone amides (“peptoid hydrazones”) can be readily achieved in library fashion. Furthormore, reduction of these hydrazones to N-substituted “peptoid hydrazides” affords a facile route to library diversification. This approach is demonstrated by application to Tsg101-binding compounds designed as potential HIV budding antagonists.
“Peptoids”1 are polymers of N-substituted glycine residues (NSG 1, Scheme 1), which have emerged as an important class of peptide mimetic that can retain bioactivity while exhibiting resistance to proteolytic degradation.2,3 Peptide-peptoid hybrids containing both peptide and NSG residues have also shown significant utility.4 One example is the replacement of key Pro residues with NSG residues in WW and SH3 domain-binding peptides. These signaling domains specifically recognize Pro-rich sequences and NSG residues can provide greater selectivity and affinity than achieved with parent Pro residues.5 This latter work is particularly relevant to recent findings that efficient budding of HIV from the plasma membrane requires binding of the host Tsg101 protein with the short peptide motif, “Pro-Thr/Ser-Ala-Pro” (“PTAP”) located within the p6 region of the viral Gag protein.6 An NMR solution structure of the PTAP-containing p6Gag-derived 9-mer peptide, “P-E-P-T-A-P-P-E-E” bound to Tsg101 indicated that the most important interactions were derived from the “Ala-Pro” residues, which are in deep depressions reminiscent of the “Xxx-Pro” pockets used by WW and SH3 domains.7 The similarities shared by Tsg101 and SH3 domains in their recognition of key Pro residues suggests that replacement of Pro residues with NSG constructs may also be beneficial for the preparation of PTAP-based Tsg101 binding inhibitors.7
Scheme 1.
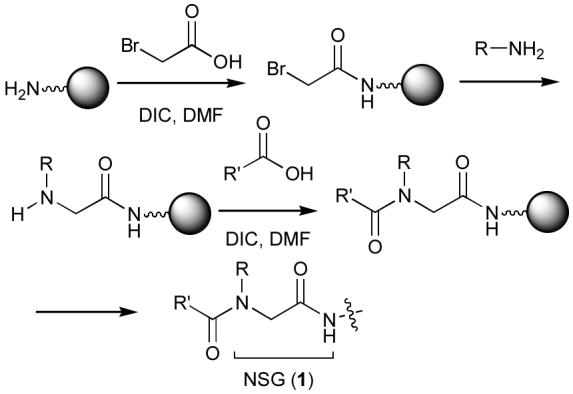
Peptoid synthesis by the “submonomer” approach.
The solid-phase synthesis of NSG-containing libraries is usually achieved by the “submonomer approach,” in which the amino terminus of the growing peptide chain is bromoacetylated, then reacted with amines to yield the corresponding NSG residues (Scheme 1).8 However, this approach can be disadvantageous for long or difficult sequences.9 We found such to be the case with the p6Gag-derived 9-mer sequence, “P-E-P-T-A-P-P-E-E” where the Pro3 residue could be satisfactorily replaced by an NSG residue using the submonomer approach, but replacement of the critical Pro6 residue (changing P-E-P-T-A-P-P-E-E to P-E-P-T-A-[NSG]-P-E-E) proved to be problematic. Although the reasons for this difficulty appeared to be sequence-related and steric in nature, modifications that would be expected to ameliorate steric crowding, such as replacement of the Pro3 residue by Ala (ie, P-E-P-T-A-[NSG]-A-E-E) or resorting to a C-terminal pseudo-proline strategy10 (for P-E-P-T-A-[NSG]-P-E-E-S), failed to overcome the problems. In order to work around the impasse presented above, we envisioned substituting the traditional peptoid NSG unit (1) with hydrazone amides to yield “peptoid hydrazones” of type 4 (Scheme 2).
Scheme 2.
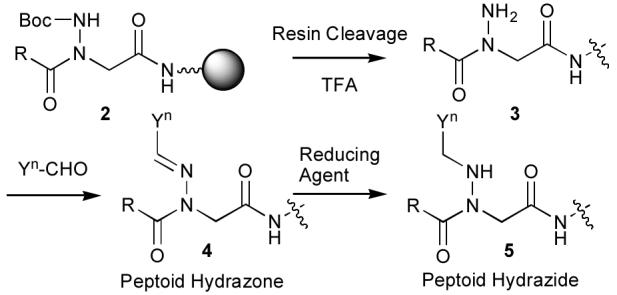
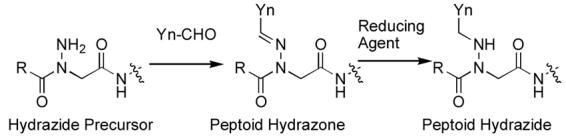
Synthesis of peptoid hydrazones (2) and peptoid hydrazides (4) from a common hydrazide precursor (3).
Peptoid hydrazones would be advantageous over traditional N-alkyl NSG residues for library synthesis, since variation of each N-alkyl NSG residue requires the separate elaboration of the entire peptide sequence amino-terminal of the NSG residue. On the other hand, libraries of peptoid hydrazones could be constructed by reacting a series of aldehydes (Yn-CHO) with a single completed peptide bearing an unsubstituted hyrazide (3). Furthermore, subjecting the resulting peptoid hydrazones 4 to reducing conditions could lead to library diversification by formation of the corresponding N-substitued peptoid hydrazides (5). Although several variations on traditional peptide amide bonds have been reported, including azapeptoids,11 urea peptoids,12 amino-oxypeptoids,13 beta peptoids,14 hydrazine (retro)-peptoids,15 and azapeptides,11 hydrazone amides have found very limited use.15 The current work represents one of the first systematic examinations against an important biological target of mixed peptide-peptoid constructs utilizing hydrazone (2) and hydrazide (4) based NSG residues.
The objective of the current study was to prepare a library of Tsg101-directed peptides of the sequence “FTPP-E-P-T-A-X-P-E-E-amide,” where “X” represents “N(R-C=N)CH2C(O)” and “FTP” stands for amino-terminal “fluorescein thiourea pentanoyl” functionality. The latter would be required for analysis of Tsg101 binding affinities using fluorescence anisotropy.16
The first attempt at solid-phase synthesis used a modified sub-monomer approach that involved reacting tert-butyl carbazate with the N-bromoacetylated peptide chain. However, as we found previously with the unsuccessful sub-monomer synthesis of the corresponding N-alkyl NSG-containing peptides (see above), coupling failed with the next amino acid, N-Fmoc Ala. Therefore, the preformed dipeptide, N-Fmoc Ala-N(NHBoc)CH2CO2H (9), was prepared by solution chemistries (Scheme 3) and incorporated into the growing peptide chain. The remainder of the peptide sequence was constructed using standard Fmoc solid-phase protocols to provide the unsubstituted hydrazide 10 following cleavage from the resin and HPLC purification. This served as a common precursor for hyrazone library construction (Figure 1). Although a similar dipeptide protocol could be used to circumvent problems associated with N-alkyl NSG incorporation (see below), the approach would be extremely tedious by requiring unique dipeptide construction, incorporation and subsequent chain elongation for each N-alkyl NSG-containing final product.
Scheme 3.
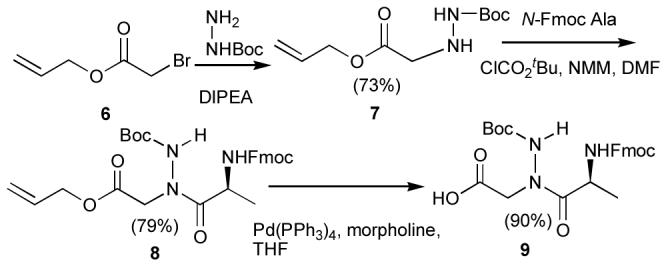
Synthesis of protected dipeptide 9.
Figure 1.
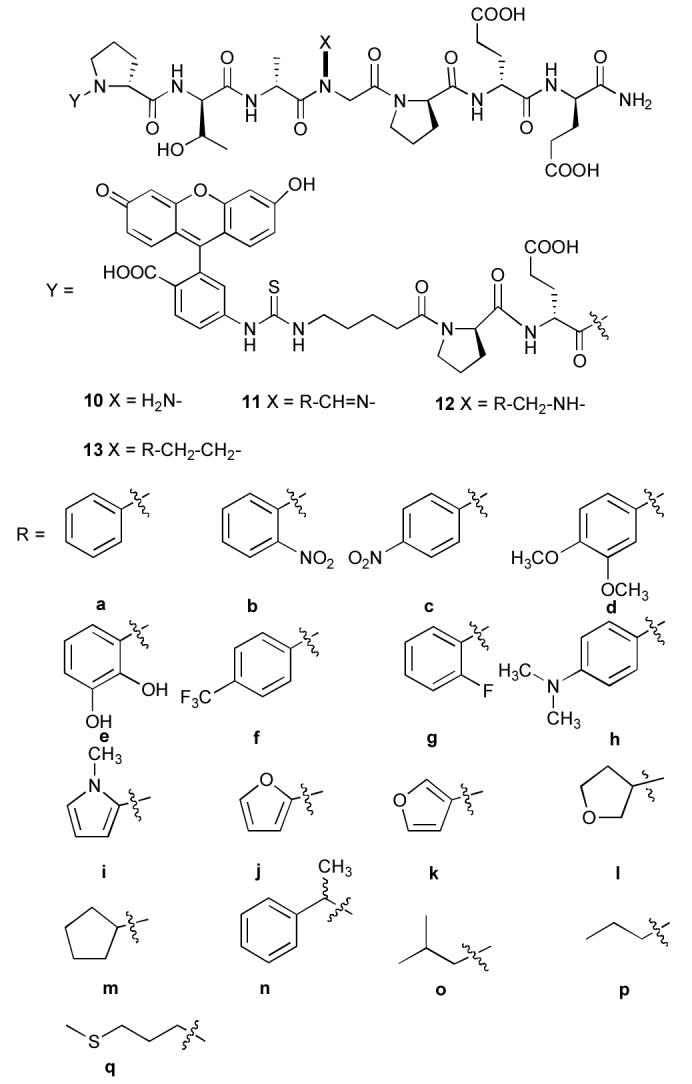
intermediate hydrazide 10 and final products 12 and 13.
With the unsubstituted hydrazide 10 in hand, condensation with a variety of aromatic and aliphatic aldehydes was achieved by gently shaking 10 with excess aldehyde (10 equivalents) in 50% aqueous acetonitrile (overnight). Direct purification by reverse phase HPLC
(0.03% TFA acetonitrile/H2O) gave the peptide hydrazones 11a – 11q (Figure 1). Adequate yields were obtained for all aldehydes examined and good HPLC separation of product hydrazones 11 from other reaction components was achieved in facile fashion.The peptoid hydrazones were stable when stored at −20° C and stability was adequate for biochemical analysis of Tsg101 binding affinities using a fluorescence anisotropy assay.17,18 Treatment of the peptoid hydrazones 11 in 50% aqueous acetonitrile (0.1% TFA) with a 20-fold excess of NaBH3CN at room temperature overnight yielded the hydrazides 12a – 12q (Figure 1), which were evaluated for their Tsg101-binding affinities.17
The Tsg101 binding affinity of the parent PTAP-containing 9-mer is relatively weak (Kd = 50 μM, Table 1). Binding constants for the highest affinity peptoid-hydrazones (11q and 11p) and peptoid-hydrazides (12m, 12k, 12q and 12p) are also shown in Table 1. The highest affinity peptoid-hydrazone was found to be the n-butyl-containing 11p, which exhibited a roughly 5-fold enhancement in affinity relative to the parent PTAP 9-mer (Kd = 10 μM, Table 1). The highest affinity peptoid-hydrazide was 12q.
Table 1.
Tsg101 binding affinities of select compounds.
| compound | Kd (μM)a | compound | Kd (μM)a |
|---|---|---|---|
| 11q | 17.53 ± 1.81 | 12q | 25.27 ± 8.06 |
| 11p | 9.83 ± 1.25 | 13d | >>500 |
| 12k | 46.30 ± 7.41 | 13p | 200 |
| 12m | 85.55 ± 13.08 | PTAPb | 53.5 ± 6.48 |
| 12p | 60.3 ± 16.12 |
Means ± SD determined from at least three independent experiments using at least two batches of protein.
Wild-type peptide.
Peptide-peptoid hybrids based on NSG hydrazones and hydrazides are highly advantageous over traditional N-alkyl NSG constructs in that libraries can be rapidly constructed following solid-phase peptide synthesis. This allows a large number of structural variations to be easily examined. Because peptoid-hydrazones and peptoid-hydrazides present structural features that are not contained in their N-alkyl NSG counterparts, their biological activities may be different. Accordingly, it was of interest to prepare the N-alkyl NSG-containing 13p as a homologue of the most potent peptoid-hydrazone (11p) in order to compare their Tsg101-binding affinities. Peptoid 13p has an n-pentyl-containing N-alkyl NSG residue, which is the same chain length as found with the n-butyl-hydrazone in 11p. The analogue containing an N-3,4-dimethoxyphenethyl NSG residue (13d) was also prepared as a “negative control” based on the poor affinities of 11d and 12d.
Since the synthesis of 13d and 13p by sub-monomer methodology was not possible (see above), the more labor-intensive dipeptide approach was used as already described above. Here, the N-Fmoc-protected peptoid dimers 16a and 16b were first prepared by solution chemistries (Scheme 4), then these N-alkyl NSG-containing dipeptide units were inserted at the appropriate point during solid-phase peptide synthesis to yield the desired 13d and 13p. It was found that 13d bound negligibly (Kd >>500 μM), consistent with the poor affinities of the corresponding peptoid hydrazone 11d and the peptoid hydrazide 12d. On the other hand, 13p was found to bind with higher affinity (Kd 200 μM), than 13d, consistent with the good affinities of 11p and 12p, although, the absolute affinity of 13p was less than its hydrazone and hydrazide congeners (Table 1).
Scheme 4.
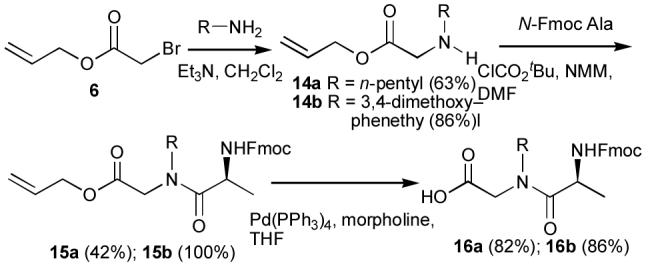
Synthesis of peptoid intermediates 16a and 16b.
In conclusion, reported herein is the facile construction of libraries of peptide-peptoid hybrids based on NSG hydrazones and hydrazides. This approach has several advantages over traditional N-alkyl NSG-containing peptide-peptoid hybrids. First, the structure of the NSG residue is varied in a final step. This is in contrast to N-alkyl NSG-containing peptide-peptoid hybrids where variation of each N-alkyl NSG residue requires the separate elaboration of the entire peptide sequence amino-terminal of the NSG residue. Second, it affords an alternative approach for long or difficult sequences were insertion of N-alkyl NSG residues may be problematic. Finally, it provides products that present structural features not found in N-alkyl NSG-containing peptide-peptoid hybrids. As in the current Tsg101 study, this may result in higher affinities than the corresponding N-alkyl NSG-containing homologues. This approach may be useful in a variety of biological applications.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 with SAIC-Frederick, Inc. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supporting Information Available: Procedures and binding data for Tsg101 FA binding assays and synthetic experimentals for dipeptides 8a, 16a and 16b and hydrazones 11a – 11r and hydrazides 12a – 12r are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, Spellmeyer DC, Tan R, Frankel AD, Santi DV, Cohen FE, Bartlett PA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patch JA, Barron AE. Curr. Opin. Chem. Biol. 2002;6:872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 3.Patch JA, Kirshenbaum K, Seurynck SL, Zuckermann RN, Barron AE. Pseudo-Peptides in Drug Discovery. 2004:1–31. [Google Scholar]
- 4.(a) Holder JR, Bauzo RM, Xiang Z, Scott J, Haskell-Luevano C. Bioorg. Med. Chem. Lett. 2003;13:4505–4509. doi: 10.1016/j.bmcl.2003.08.078. [DOI] [PubMed] [Google Scholar]; (b) Kruijtzer JAW, Nijenhuis WAJ, Wanders N, Gispen WH, Liskamp RMJ, Adan RAH. J. Med. Chem. 2005;48:4224–4230. doi: 10.1021/jm0490033. [DOI] [PubMed] [Google Scholar]
- 5.(a) Nguyen JT, Porter M, Amoui M, Miller WT, Zuckermann RN, Lim WA. Chem. Biol. 2000;7:463–473. doi: 10.1016/s1074-5521(00)00130-7. [DOI] [PubMed] [Google Scholar]; (b) Nguyen JT, A Turck CW, Cohen FE, Zuckermann RN, Lim WA. Science. 2002;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 6.Demirov DG, Freed EO. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Pornillos O, Alam SL, Davis DR, Sundquist WI. Nat. Struct. Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 8.(a) Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Am. Chem. Soc. 1992;114:10646–10647. [Google Scholar]; (b) Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 9.Fara MA, Diaz-Mochon JJ, Bradley M. Tetrahedron Lett. 2006;47:1011–1014. [Google Scholar]
- 10.Haack T, Mutter M. Tetrhedron Lett. 1992;33:1589–1592. [Google Scholar]
- 11.Gibson C, Goodman SL, Hahn D, Hoelzemann G, Kessler H. J. Org. Chem. 1999;64:7388–7394. [Google Scholar]
- 12.Kruijtzer JAW, Lefeber DJ, Liskamp RMJ. Tetrahedron Lett. 1997;38:5335–5338. [Google Scholar]
- 13.(a) Murphy JE, Uno T, Hamer JD, Cohen FE, Dwarki V, Zuckermann RN. Proc. Natl. Acad. Sci. USA. 1998;95:1517–1522. doi: 10.1073/pnas.95.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shin I, Park K. Org. Lett. 2002;4:869–872. doi: 10.1021/ol010251s. [DOI] [PubMed] [Google Scholar]
- 14.Hamper BC, Kolodziej SA, Scates AM, Smith RG, Cortez E. J. Org. Chem. 1998;63:708–718. doi: 10.1021/jo971675w. [DOI] [PubMed] [Google Scholar]
- 15.Aubin S, Martin B, Delcros J-G, Arlot-Bonnemains Y, Baudy-Floc'h M. J. Med. Chem. 2005;48:330–334. doi: 10.1021/jm049455f. [DOI] [PubMed] [Google Scholar]
- 16.(a) Huff S, Matsuka YV, McGavin MJ, Ingham KC. J. Biol. Chem. 1994;269:15563–15570. [PubMed] [Google Scholar]; (b) Li D-H, Zhu Q-Z, Ye D, Fang Y, Xu J-G. Anal. Chem. Acta. 1999;389:85–88. [Google Scholar]; (c) Johnson DA. Biophys. Chem. 2005;116:213–218. doi: 10.1016/j.bpc.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 17.A summary of the experimental procedure and a plot of FA binding data are provided in the Supporting Information.
- 18.Peptoid hydrazones 11 underwent slow decomposition when exposed to air at room temperature (approximately 50% decomposition after 4 days as indicated by HPLC analysis).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


