Abstract
The Cdc6 protein of budding yeast and its homologues in other species play an essential role in the initiation of DNA replication. A cDNA encoding a human homologue of Cdc6 (HsCdc6) has been cloned and expressed as a fusion protein in a soluble and functionally active form. The purified protein bound specifically to ATP and slowly hydrolyzed it, whereas HsCdc6 mutants containing amino acid substitutions in the Walker A or B motifs were defective. The mutant proteins retained the ability to bind HsOrc1 and HsCdc6 but displayed aberrant conformations in the presence of nucleotides. Microinjection of either mutant protein into human cells in G1 inhibited DNA replication, suggesting that ATP binding and hydrolysis by HsCdc6 are essential for DNA replication.
INTRODUCTION
Chromosomal DNA replication in eukaryotic cells is tightly controlled and coordinated with other events in the cell cycle to ensure that the genome is duplicated only once before cell division. It is believed that this control is exerted primarily at the initiation of DNA synthesis. Studies on Escherichia coli, bacteriophage, viral, and yeast DNA replication (reviewed in Stillman, 1996; Dutta and Bell, 1997; Baker and Bell, 1998; Waga and Stillman, 1998) have led to a model that eukaryotic DNA synthesis is initiated by sequence-specific recognition of an origin of DNA replication, or replicator element, by an initiator protein or protein complex, which nucleates the assembly of other prereplication proteins and subsequently recruits replication initiation proteins such as DNA polymerase α-primase.
The assembly of eukaryotic initiator proteins on replicator elements is best understood in the yeast Saccharomyces cerevisiae (reviewed in Dutta and Bell, 1997; Newlon, 1997; Baker and Bell, 1998). DNase I footprinting on yeast origins revealed that these sequence elements are occupied by a six-subunit protein complex, called the origin recognition complex (ORC), throughout the cell cycle. However, additional factors associate with the ORC on the origin during G1 to form a prereplicative complex (preRC) that persists until DNA replication is initiated in S phase (Diffley and Cocker, 1992; Diffley et al., 1994). These factors include Cdc6, members of the mini-chromosome maintenance (MCM) family of proteins, Cdc45, and Cdc7/Dbf4, all of which are conserved in higher eukaryotes including humans (reviewed in Dutta and Bell, 1997). Although it is clear that the formation of the preRC is an essential step preceding origin firing, we are only beginning to understand how these components are assembled and what their functions are.
A large number of studies has established a critical role for the Cdc6 protein in the initiation of DNA replication in yeast and in Xenopus extracts (reviewed in Dutta and Bell, 1997). S. cerevisiae Cdc6 and its homologue in Schizosaccharomyces pombe Cdc18 interact with the ORC (Li and Herskowitz, 1993; Liang et al., 1995; Leatherwood et al., 1996; Lopez-Girona et al., 1998) and with the Cdc28/cdc2 kinase (Elsasser et al., 1996; Leatherwood et al., 1996; Brown et al., 1997; Jallepalli et al., 1997; Lopez-Girona et al., 1998). Some of these interactions have also been confirmed with the frog and mammalian homologues of Cdc6 (Coleman et al., 1996; Saha et al., 1998; Petersen et al., 1999). Cdc6/Cdc18 is essential for entry into S phase (Kelly et al., 1993; Piatti et al., 1995; Muzi Falconi et al., 1996; Detweiler and Li, 1997), affects the frequency of the initiation of DNA synthesis (Liang et al., 1995), and is required for the formation of prereplicative complexes at yeast origins (Cocker et al., 1996; Santocanale and Diffley, 1996; Donovan et al., 1997; Tanaka et al., 1997). Immunodepletion of the Cdc6 protein (Xcdc6) from Xenopus extracts inhibits the initiation of DNA replication (Coleman et al., 1996), apparently by interfering with MCM loading onto the chromatin (Coleman et al., 1996; Hua and Newport, 1998). Together, these studies suggest that one function of Cdc6/Cdc18 is to load the MCMs near the site where the ORC is bound, a critical step in licensing the origins for activation. During the activation process, phosphorylation of Cdc6/Cdc18 by the Cdc28/Cdc2 kinase targets the protein for ubiquitin-mediated degradation (Elsasser et al., 1996; Drury et al., 1997; Jallepalli et al., 1997, 1998; Kominami and Toda, 1997; Hua and Newport, 1998), which in turn prevents rereplication by inhibiting further recruitment of the MCMs onto the origin. Recently, a protein related to yeast Cdc6/Cdc18 was identified in humans (Williams et al., 1997; Hateboer et al., 1998; Saha et al., 1998). Although this protein, like its yeast and Xenopus counterparts, performs at least one essential function before DNA synthesis initiates (Saha et al., 1998; Yan et al., 1998; Petersen et al., 1999), it remains unclear how it performs this function(s).
All Cdc6-related proteins identified so far contain a putative purine nucleoside triphosphate–binding site consisting of two sequence elements called the Walker A and B motifs (Walker et al., 1982; Koonin, 1993). On the basis of studies of known purine nucleotide–binding proteins and ATP- and GTPases, the Walker A motif is thought to contact the triphosphate moiety of the nucleotide, whereas the Walker B motif coordinates Mg2+ via a water molecule (Saraste et al., 1990; Story and Steitz, 1992). Although purine nucleotide binding and hydrolysis by Saccharomyces cerevisiae Cdc6p has been reported (Zwerschke et al., 1994), it remains controversial (Elsasser et al., 1996; Weinreich et al., 1999). Nevertheless, it seems likely that ATP plays a critical role in regulating the function of yeast Cdc6, because some missense mutations in the Walker A and B motifs of yeast Cdc6 cause cell inviability (Elsasser et al., 1996; Perkins and Diffley, 1998; DeRyckere et al., 1999; Wang et al., 1999; Weinreich et al., 1999). On the other hand, the mutational analysis of the Walker A and B motifs has led to conflicting conclusions on the functional role of ATP binding and hydrolysis in Cdc6 function in yeast.
In this study, we have investigated the ability of purified human Cdc6 (HsCdc6), expressed in the baculovirus system as a GST-tagged recombinant protein, to bind and hydrolyze ATP. We show that HsCdc6 bound specifically to ATP and displayed ATPase activity. A mutation in the Walker A motif of HsCdc6 almost abolished nucleotide binding and hydrolysis by the fusion protein, whereas a mutation in the Walker B motif strongly reduced the ATPase activity without detectable loss of nucleotide-binding activity. The mutant proteins retained the ability to form protein–protein complexes with HsCdc6 and HsOrc1 at levels equivalent to that of the wild-type protein. However, their ability to undergo a nucleotide- induced conformational change, detectable with the wild-type protein by altered protease sensitivity, was impaired. Microinjection of the Walker A and B mutant proteins into human cells during G1 disrupted DNA replication, whereas injection of the mutant proteins into cells in G1/S did not. Kinetic experiments demonstrate that cells lost their susceptibility to the Walker A mutant earlier in G1 compared with the Walker B mutant and that they arrested earlier in the cell cycle. Injection of wild-type Cdc6 protein bound to a poorly hydrolyzable ATP analogue resulted in a phenocopy of the Walker B mutant. The results suggest that ATP binding and hydrolysis by HsCdc6 are essential for the initiation of human chromosomal DNA replication and progression through S phase.
MATERIALS AND METHODS
Cloning of HsCdc6
A search of the expressed sequence tag database (dbEST, National Center for Biotechnology Information) revealed the partial cDNA sequence of a human protein closely related to SpCdc18 and ScCdc6 (AA045217). With the information from this sequence, a 3′-rapid amplification of cDNA ends was performed using human cDNA, obtained by reverse transcription of total RNA from 293 cells, as a template. The resulting 1479-bp fragment was sequenced, revealing 516 bp of coding sequence as well as 963 bp of 3′-untranslated region. Using the cDNA sequence of Xcdc6 (Coleman et al., 1996), another search in the dbEST identified a partial cDNA sequence highly homologous to the N-terminal region of Xcdc6 (H59203). Using primers based on both EST sequences, PCR was performed on human 293 cDNA, resulting in a 1026-bp cDNA fragment corresponding to the central region of the HsCdc6 gene. The 5′-end of the gene was obtained by PCR amplification using oligonucleotides based on the sequences of dbEST clone H59203 and the 5′-untranslated region of human p62cdc6 that by then became available in GenBank (U77949). All three fragments were joined via internal restriction sites and cloned into pBluescript KS II+ (Stratagene, La Jolla, CA). The resulting cDNA was sequenced for verification. The resulting vector (pBS-Cdc6-fl) encodes a 2763-bp cDNA of HsCdc6, starting at the internal BamHI restriction site. The cDNA cloned encodes a predicted 64-kDa protein that is identical to the human p62cdc6 (Williams et al., 1997) and hCdc18 (Saha et al., 1998) identified previously.
Construction of Baculovirus Transfer Vectors
To generate a baculovirus transfer vector encoding full-length HsCdc6 protein with GST fused to its N-terminal end, we created a BamHI restriction site immediately upstream of the start codon of HsCdc6 by PCR. The 5′-primer used was 5′-CCGGATCCATGCCTCAAACCCGATCCC-3′, which is complementary to the start of the HsCdc6 open reading frame (BamHI restriction site is underlined), whereas the 3′-primer was 5′-CAGTGGTTTGAGAATAGTCTGCAGAC-3′, which is located 1242 bp downstream of the initiation codon. The template for the PCR was pBS-Cdc6-fl. The PCR amplification product was digested with BamHI/DrdI and used to replace the BamHI/DrdI fragment in pBS-Cdc6-fl (pBS-Cdc6-3). To remove most of the 3′-untranslated region, we digested pBS-Cdc6-3 with BamHI/SspI and subcloned the resulting fragment into pVL1393 (Invitrogen, San Diego, CA) that had been digested with BamHI/SmaI (pVL-Cdc6). The BamHI/NotI fragment of pVL-Cdc6 was then subcloned into BamHI/NotI–digested pBS-GST (provided by C. Rehfuess), resulting in pBS-GST-Cdc6, a plasmid in which the cDNA of GST-2T was located immediately upstream of the initiation codon of HsCdc6. The full-length GST-HsCdc6 cDNA was transferred as a HincII/NotI fragment into pVL1392 (Invitrogen), digested with SmaI/NotI (pVL1393-GST-Cdc6). The mutation of Lys208 to Ala208 was generated by mismatch PCR using the forward primer 5′-TTTGAATTCTCCTCGTGTAAAAGCCCTG-3′, the backward primer 5′-GGCTTAAGCAGGCAGTTGCTCCAGTTCC-3′, and pBS-Cdc6-fl as a template. The PCR amplification product was digested with ApaI/XbaI, and the resulting fragment was used to replace the ApaI/XbaI fragment in pBS-GST-Cdc6. The full-length GST-HsCdc6 (K208A) cDNA was transferred as a HincII/NotI fragment into pVL1393 (Invitrogen), which had been digested with SmaI/NotI (pVL1393-GST-Cdc6 [K208A]). The mutation of Glu285 to Gln285 was generated by mismatch PCR using the forward primer 5′-AAGGGCCCCATGATTGTGTTGGTATTGGACCAGATGGATC-3′ , the backward primer 5′-CAGTGGTTTGAGAATAGTCTGCAGAC-3′, and pBS-Cdc6-fl as a template. The PCR amplification product was digested with AflII, and the resulting fragment was used to replace the AflII fragment in pBS-GST-Cdc6. The full-length GST-HsCdc6 (E285Q) cDNA was transferred as a HincII/NotI fragment into pVL1393 (Invitrogen), which had been digested with SmaI/NotI (pVL1393-GST-Cdc6 [E285Q]). The cDNA of GST was subcloned from pBS-GST into pVL1392 (Invitrogen) as a EcoRI/BamHI fragment (pVL1392-GST). Recombinant baculoviruses were generated according to the manufacturer’s instructions (Invitrogen).
Expression and Purification of GST-HsCdc6 Fusion Proteins
Hi-5 insect cells (3.6 × 108 cells; Invitrogen) were infected with each recombinant baculovirus at 10 pfu/cell for 48 h. Cells were lysed in 10 ml of buffer A (100 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 0.5% Nonidet P-40, 1 mM DTT, 10 mM NaF, 1 mM EGTA, 2 mM EDTA, 1 mM PMSF, and 1 μg/ml each aprotinin and leupeptin) using a Dounce homogenizer. After the cell debris was removed by centrifugation, the supernatant was gently mixed with 0.5 ml of glutathione-agarose suspension (Sigma, St. Louis, MO) equilibrated in buffer A for 1 h at 4°C. The resin was recovered by centrifugation and washed twice with PBS, washed once with PBS containing 1 M NaCl, washed once with PBS containing 1 M NaCl and 0.1% (vol/vol) Nonidet P-40, and then reequilibrated in PBS. For some experiments, the resin was washed twice with PBS, washed once with PBS containing 1.5 M NaCl for 10 min, washed once with PBS containing 1.5 M NaCl and 0.1% (vol/vol) Nonidet P-40 for 10 min, and then reequilibrated in PBS. This wash step removed a contaminating ATPase activity that was associated with the GST fusion proteins but not with GST. GST fusion proteins were eluted in batch with 0.3 ml of buffer B (50 mM Tris-HCl [pH 8] and 100 mM reduced glutathione; Sigma). The eluted fractions were dialyzed overnight against 2 l of buffer C (25 mM HEPES-KOH [pH 7.5], 50 mM NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol) and stored at −80°C until use.
ATP-binding Assay
ATP binding was performed essentially as described (Klemm et al., 1997) with the following modifications. Reactions contained 1 pmol of protein in 50 μl of ATP-binding buffer (50 mM HEPES-KOH [pH 7.5], 1 mM EDTA, 1 mM EGTA, 5 mM Mg-acetate, 150 mM KCl, 10% glycerol, 0.02% [vol/vol] Nonidet P-40, and 0.15 mg/ml BSA). Radiolabeled [α-32P]NTPs and [α-32P]dNTPs were used at a specific activity of 50,000 cpm/pmol except for [γ-35S]ATPγS, which was used at 20,000 cpm/pmol. Except in the ATP titration (see Figure 1B), all nucleotides were used at a concentration of 2.5 μM. All fusion proteins used for this experiment were prepared at a concentration of 75 ng/μl or greater. This reduced the amount of a contaminating ATP-binding activity that was detectable in more dilute GST fusion protein preparations to a level that was not detectable using this assay.
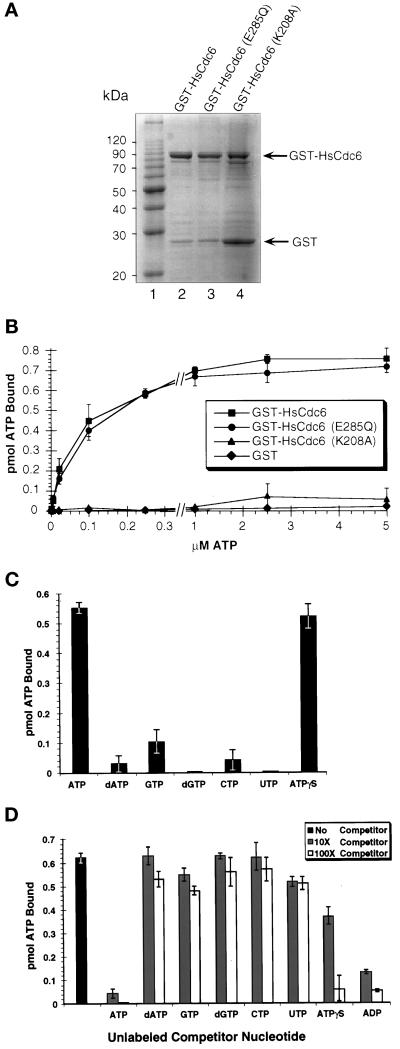
Figure 1.
Activity of wild-type and mutant HsCd6 ATP binding. (A) Purified wild-type HsCdc6 fused to GST and GST-HsCdc6 mutants with single amino acid substitutions in the Walker A or B motifs was separated by 10% SDS-PAGE and stained with Coomassie brilliant blue. (B) One pmol of GST-HsCdc6 (squares), GST-HsCdc6 (K208A) (triangles), GST-HsCdc6 (E285Q) (circles), or GST (diamonds) was incubated with increasing amounts of [α-32P]ATP for 10 min at room temperature. After gel filtration, the amount of ATP bound to the protein in the void volume was determined by scintillation counting. Recovery of protein in the void volume was consistently ∼70%. The values are averages from three separate experiments, and error bars indicate the SD. (C) One pmol of GST-HsCdc6 was incubated with the indicated radiolabeled NTP or dNTP for 10 min at room temperature. Unbound nucleotides were removed by gel filtration, and the amount of NTP or dNTP bound to GST-HsCdc6 in the void volume was determined by scintillation counting (mean of 2 separate experiments). Error bars indicate the SD. (D) GST-HsCdc6 (1 pmol) was incubated with 2.5 μM [α-32P]ATP in the absence (black bar) or presence of the indicated unlabeled competitor nucleotides in a 10-fold (hatched bars) or a 100-fold (white bars) excess. Unbound nucleotides were removed by gel filtration, and the amount of [α-32P]ATP bound to GST-HsCdc6 was determined by scintillation counting (mean of 2 separate experiments). Error bars indicate the SD.
ATPase Assay
Hydrolysis of ATP was measured in ATPase buffer (20 mM Tris-HCl [pH 7.5], 0.1 mg/ml bovine serum albumin, 0.5 mM DTT, 10 mM MgCl2, and 50 μM [γ-32P]ATP [1 Ci/mmol]) with 0.25 pmol of GST fusion proteins as described previously (Podust et al., 1998). The reaction products were separated by TLC on polyethyleneimine-cellulose and developed in 1 M LiCl and 0.5 M formic acid, and the amounts of [γ-32P]ATP hydrolyzed to [32P]orthophosphate were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). ATP hydrolysis was performed in the linear range of protein concentration dependence.
Partial Tryptic Digest of GST-HsCdc6
Tryptic digestion of GST-HsCdc6 and mutant forms of the fusion protein was performed for 10 min at 37°C in a total volume of 20 μl. The reaction contained 0.5 μg of GST-HsCdc6 bound to glutathione-agarose (Sigma) and either no nucleotide, ATP, ATPγS, ADP, or UTP (2 mM) in buffer D (20 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 2 mM DTT, and 8 mM MgCl2). To start the reaction, trypsin was added to a final concentration of 2.5 ng/μl. The reaction was stopped with SDS sample buffer and boiled for 5 min. The proteolytic products were resolved by 15% SDS-PAGE and visualized by silver staining.
In Vitro Protein Interaction Assay
Untagged HsCdc6 and Orc1 were in vitro translated using TNT reticulocyte (Promega, Madison, WI) according to the manufacturer’s instructions. GST fusion proteins immobilized on glutathione-agarose beads were washed twice with 500 μl of 30 mM HEPES-KOH (pH 7.8), 10 mM KCl, and 7 mM MgCl2 in the absence and presence of ATP or ADP (2 mM) and incubated for 1 h at 4°C with 5 μl of either in vitro–translated HsCdc6 or in vitro–translated Orc1 in 250 μl of the same buffer containing 2% nonfat dry milk powder and 1 mM DTT. Where indicated, the reaction also contained ATP or ADP at a concentration of 2 mM. All reactions were performed in the presence of 0.1 U/μl benzon nuclease (EM Science, Gibbstown, NJ) to eliminate the possibility of protein–nucleic acid interactions. The beads were recovered by centrifugation and washed three times with 1 ml of 30 mM HEPES-KOH (pH 7.8), 25 mM KCl, 7 mM MgCl2, 0.25% inositol, 0.25 mM EDTA, and 0.1% Nonidet P-40. The bound proteins were eluted by boiling in one volume of 4× SDS sample buffer, separated by SDS-PAGE, and detected by PhosphorImaging.
Cell Culture and Cell Synchronization
HeLa-S3 cells were grown in monolayer in DMEM (Life Technologies, Gaithersburg, MD) supplemented with antibiotics and 10% fetal bovine serum (FBS; Life Technologies) in a humidified incubator at 37°C and 10% CO2. Exponentially growing HeLa-S3 cells were arrested in G1/S with 2.5 mM thymidine (Sigma), 5 μg/ml aphidicolin (Sigma), or 10 μM hydroxyurea (Sigma) for 24 h. To release the cells into S phase, we aspirated the medium and washed the cells three times with DMEM plus 10% FBS. Exponentially growing HeLa-S3 cells were arrested in G2/M for 16 h with 50 ng/ml nocodazole (Sigma). Cells were released into G1 by gently shaking off mitotic cells from the flask and washing them three times with DMEM plus 10% FBS. Cells were either plated on glass coverslips for microinjection or into tissue culture flasks for propagation or flow cytometry analysis. With one exception (see Figure 9), the medium was supplemented with 10 μM bromodeoxyuridine (BrdU; Sigma) to monitor DNA replication. The cell cycle distribution of the cell populations was determined by flow cytometry after propidium iodide staining of the DNA.
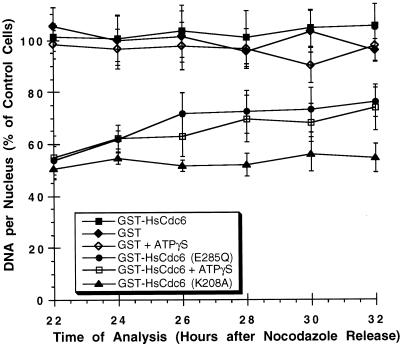
Figure 9.
Walker A mutants of HsCdc6 arrest cells before S phase, and Walker B mutants prevent completion of S phase. HeLa-S3 cells that had been arrested in G2/M with nocodazole for 16 h were released into nocodazole-free medium for 6–8 h. The following proteins at the indicated concentrations were microinjected into the nuclei of the cells: GST (500 ng/μl; filled diamonds), GST-HsCdc6 (61 ng/μl; filled squares), GST-HsCdc6 (K208A) (20 ng/μl; filled triangles), GST-HsCdc6 (E285Q) (30 ng/μl; filled circles), GST-HsCdc6 (109 ng/μl) preincubated with a twofold molar excess of ATPγS (open squares), and GST (500 ng/μl) preincubated with the same concentration of ATPγS used for GST-HsCdc6 (open diamonds). At 12 h after the release, the medium was again supplemented with nocodazole to prevent progression through mitosis. At the indicated times, the cells were stained with anti-GST polyclonal antibody and FITC-conjugated goat anti-rabbit secondary antibody and Hoechst 33258 fluorochrome. Nuclear DNA content was measured by fluorescence microscopy. Nuclear DNA content of injected cells is expressed as a percentage of the nuclear DNA content of uninjected cells in the same field of vision, which was set to 100%. For each time point, the average value obtained from at least 10 cells is shown. The SD of the mean is indicated by error bars.
Microinjection
With one exception (see experiment in Figure 8), HeLa-S3 cells that had been released from a nocodazole block were plated on glass coverslips for 6–8 h, which allowed the majority of the cells to attach to the coverslip. Cells to be synchronized and injected in G1/S were grown on the glass coverslips. Except for GST, all fusion proteins were used at a concentration of 25–100 μg/ml and supplemented with GST at a concentration of 0.5 mg/ml for easier detection of injected cells. Both stringently washed and conventionally washed GST fusion proteins were used in injection experiments and yielded similar phenotypes (our unpublished results). GST alone was used at a concentration of 0.5 mg/ml. Where indicated, GST-HsCdc6 was incubated with a twofold molar excess of ATPγS or ATP over protein in the presence of 5 mM MgCl2 for 10 min at room temperature. GST was incubated similarly, except that the concentration of ATPγS in the sample was equal to that in the GST-HsCdc6 sample. Except for the nucleotide–protein mixtures, all samples were centrifuged before microinjection for 30 min at 14,000 × g. Needles used for microinjection were pulled from glass capillaries (Clark Electromedical Instruments, Reading, UK) on an automatic pipette puller (Zeitz Instruments, Augsburg, Germany). To deliver the samples, a microinjector (model 5246; Eppendorf Scientific, Madison, WI) and a manipulator (model 5171; Eppendorf Scientific) mounted on an inverted microscope (model IM35; Carl Zeiss, Oberkochen, Germany) were used.
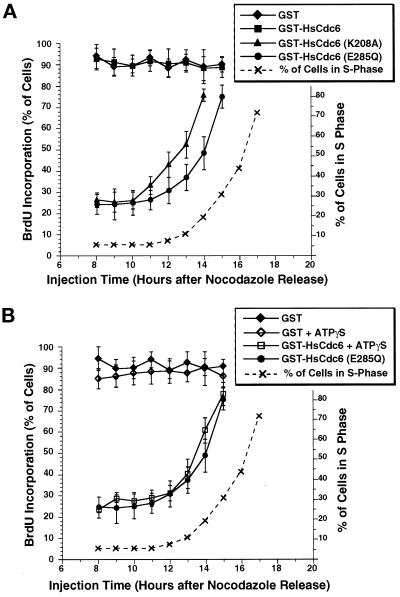
Figure 8.
Requirements for ATP binding and ATP hydrolysis of HsCdc6 in G1 are temporally separated. HeLa-S3 cells were arrested for 16 h in G2/M with nocodazole and released into nocodazole-free medium. (A) At the indicated times, GST-HsCdc6 (61 ng/μl; filled squares), GST-HsCdc6 (K208A) (20 ng/μl; filled triangles), GST-HsCdc6 (E285Q) (30 ng/μl; filled circles), and GST (500 ng/μl; filled diamonds) were microinjected into the nuclei of the cells. Immediately after microinjection, the medium was supplemented with BrdU, and growth was continued until 22 h after the nocodazole release. Cells were then stained with anti-GST and anti-BrdU antibodies and evaluated by indirect immunofluorescence microscopy. For each time point, the average value obtained from at least 70 injected cells is shown; error bars indicate the SD. The dashed line indicates the percentage of uninjected cells in S phase at the indicated times after release from a nocodazole block, as determined by flow cytometry of HeLa-S3 cells blocked and released in the same manner. At 12 h after the release, the medium was supplemented with nocodazole to prevent the cells from passing through mitosis into G1. (B) The same experiment shown in A was performed, except that GST-HsCdc6 (E285Q) (30 ng/μl; filled circles), GST-HsCdc6 (109 ng/μl) preincubated with a twofold molar excess of ATPγS (open squares), GST (500 ng/μl; filled diamonds), and GST (500 ng/μl) preincubated with the same concentration of ATPγS used for GST-HsCdc6 (open diamonds) were microinjected, and DNA replication was analyzed in the same manner described in A.
Immunofluorescence
Cells were washed three times with PBS, fixed with 3% formaldehyde in PBS for 20 min, permeabilized for 20 min using 0.2% Triton X-100, and incubated with 10% FCS in PBS for 1 h. GST was visualized by staining with a rabbit polyclonal anti-GST antibody (provided by R. Weber) at a dilution of 1:100 in PBS plus 10% FBS for 2 h at room temperature, followed by FITC-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:50 in PBS plus 10% FBS for 1 h at room temperature. BrdU incorporated into the DNA was visualized by staining with a mouse monoclonal anti-BrdU antibody (Amersham, Arlington Heights, IL) at a dilution of 1:100 in PBS plus 10% FBS containing 125 U/ml benzon nuclease for 2 h at room temperature, followed by a Cy3-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) at a dilution of 1:100 in PBS plus 10% FBS for 1 h at room temperature. After being washed three times, the cells were incubated for 15 min with Hoechst 33258 fluorochrome (Hoechst, Frankfurt, Germany) at a concentration of 2 μM in PBS. The coverslips were mounted in 90% glycerol containing 0.1 mg/ml paraphenylenediamine in PBS (Johnson and Nogueira Araujo, 1981) and analyzed using a fluorescence microscope (model Axiovert 35; Carl Zeiss).
Quantification of Nuclear DNA
To quantify the nuclear DNA content, we incubated cells that had been stained with rabbit polyclonal anti-GST antibody and the FITC-conjugated goat anti-rabbit secondary antibody (see above) for 1 h in 10 mM Tris-HCl (pH 7.5) containing 1 M NaCl and 2 μM Hoechst 33258 fluorochrome at room temperature (Araki et al., 1987). Images of fluorescent cells were captured at 63× magnification using a digital camera (charge-coupled device camera, model C 4880; Hamamatsu Phototonics, Bridgewater, NJ). The amount of fluorescence emitted at 460 nm in the nucleus of the cell was measured using the Image/MetaMorph Imaging System (Universal Imaging Corporation, West Chester, PA). Fluorescence per nucleus was evaluated for injected and uninjected cells in each field of vision. The nuclear DNA content of injected cells was expressed as a percentage of the nuclear DNA content of uninjected cells in the same field of vision, which was set to 100%.
RESULTS
HsCdc6 Binds Specifically to ATP and Hydrolyzes ATP
To determine whether HsCdc6 binds to and hydrolyzes ATP, we first sought to express HsCdc6 in a tagged form to aid in purification and detection of the protein. Because GST-Cdc18 had been shown to be fully functional in vivo in fission yeast (Brown et al., 1997), we reasoned that a GST tag would be unlikely to impair the function of the protein. Thus, recombinant baculoviruses encoding GST-HsCdc6, GST, and two mutant forms of GST-HsCdc6 were generated. One mutant contained alanine in place of the conserved lysine of the Walker A motif (Lys208→Ala208), which is involved in binding to the β- and γ-phosphates of the ATP molecule (Saraste et al., 1990; Story and Steitz, 1992; Koonin, 1993). The other mutant carried glutamine instead of the conserved glutamic acid of the Walker B motif (Glu285→Gln285), which is involved in coordinating the Mg2+ ion and the water molecule required for the nucleophilic attack on the β-γ bond of the purine nucleotide (Story and Steitz, 1992; Koonin, 1993). Mutating the Walker A motif at the conserved lysine was expected to inactivate ATP binding and hence hydrolysis, whereas the mutation in the Walker B motif would likely impair only the hydrolysis event.
All three proteins, GST-HsCdc6, GST-HsCdc6 (E285Q), and GST-HsCdc6 (K208A), were expressed in insect cells and purified in soluble form (Figure 1A). The major band in each preparation (Figure 1A, lanes 2–4) represents GST-HsCdc6, migrating at the expected molecular weight of 90 kDa, whereas the faint, faster migrating bands are predominantly degradation products, as determined by Western blot analysis using polyclonal antibodies against GST (our unpublished results). In all three protein preparations, GST was observed as a stable degradation product, which was more prominent in the GST-HsCdc6 (K208A) sample than in the other two samples (Figure 1A, compare lanes 2 and 3 with lane 4).
Using a nucleotide-binding assay developed by Klemm et al. (1997) in which nucleotide–protein complexes are separated from free nucleotides by gel filtration, we analyzed the ability of wild-type and mutant forms of GST-HsCdc6 to bind to ATP. Because an α-32P-radiolabeled nucleotide was used for these experiments, binding either to nucleoside triphosphate or, if the nucleotide were hydrolyzed, to stably bound hydrolysis products would be detected. As shown in Figure 1B, GST-HsCdc6 (squares) and GST-HsCdc6 (E285Q) (circles) bound ATP with similar activity at all nucleotide concentrations tested. The ATP-binding activity of GST-HsCdc6 (K208A) (Figure 1B, triangles) was reduced to a low level that did not differ significantly from that measured for GST (diamonds) in three separate experiments. The absence of nucleotide binding to GST demonstrated that the binding observed with the fusion proteins did not arise from the GST protein of the fusion protein. Addition of single- or double-stranded DNA to the reaction mixture did not affect the ATP binding of GST-HsCdc6 or the mutant proteins (our unpublished results). At ATP concentrations that saturated binding to GST-HsCdc6 (2.5 μM or greater), GST-HsCdc6 and GST-HsCdc6 (E285Q) coeluted in the void volume with 0.7 pmol of ATP. After we corrected for retention of ∼30% of the 1 pmol of input GST fusion protein in the gel filtration column during the centrifugation step, binding to ATP appeared to be stoichiometric.
To investigate the specificity of nucleotide binding to HsCdc6, we tested GST-HsCdc6 binding to a variety of nucleotides by the use of the gel filtration assay. As shown in Figure 1C, ATP complexed well with GST-HsCdc6. Strong binding of GST-HsCdc6 to a nonhydrolyzable analogue of ATP (ATPγ35S) was also observed. A small amount of GTP binding was detectable, whereas little or no direct binding to dATP, dGTP, CTP, and UTP was observed.
These results indicate that the interaction between HsCdc6 and ATP or ADP was quite specific. To confirm these data, we performed competition experiments using α-32P-radiolabeled ATP and a 10- or 100-fold molar excess of unlabeled NTPs, dNTPs, or ADP in the gel filtration assay. Only ATP and ADP proved to be efficient competitors at a 10-fold molar excess, suggesting that both nucleotides bound specifically to HsCdc6 (Figure 1D). Although ATPγS competed efficiently for binding to labeled ATP when used at a 100-fold molar excess, a 10-fold molar excess of ATPγS decreased ATP binding to GST-HsCdc6 by only 40%. These results suggest that although ATPγS was able to form a complex with HsCdc6, its affinity for the protein was somewhat lower than that of ATP. In contrast, addition of dGTP, CTP, or UTP did not result in any significant reduction of binding to radiolabeled ATP. Competition with dATP or GTP at a concentration 100-fold that of the radiolabeled ATP reduced binding by ∼10–20%.
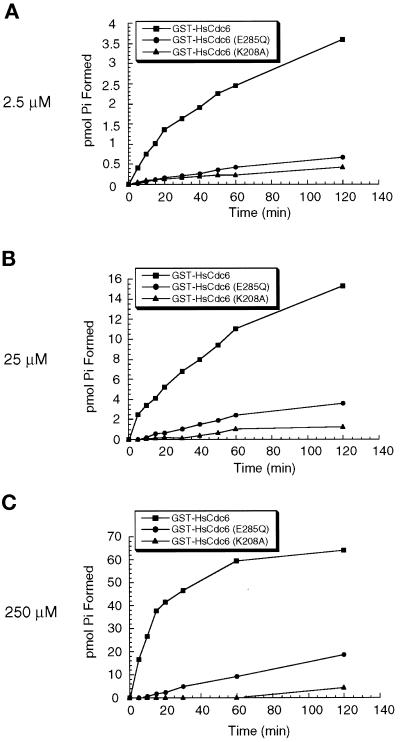
We next tested the ATPase activity of purified wild-type and mutant forms of GST-HsCdc6 using γ-32P-radiolabeled ATP as the substrate at concentrations from 2.5 to 250 μM. GST-HsCdc6 displayed weak but clearly detectable ATPase activity (Figure 2, squares). No hydrolysis of 35S-radiolabeled ATPγS was detectable (our unpublished results). GST alone, purified in an identical manner, displayed no detectable ATPase activity (our unpublished results). Kinetic analysis of ATP hydrolysis revealed that 1 pmol of GST-HsCdc6 hydrolyzed ∼3 pmol of ATP per minute (Figure 2C), with a KM of 14 μM (our unpublished results). In contrast, ATP hydrolysis by GST-HsCdc6 (E285Q) (Figure 2C, circles) was clearly reduced (0.17 pmol/min). ATP hydrolysis by GST-HsCdc6 (K208A) (Figure 2C, triangles) was barely detectable (∼0.03 pmol/min), demonstrating that mutations in either the Walker A or B motif interfered with ATP hydrolysis. The ATPase activity of the GST-HsCdc6 proteins was not affected by single- or double-stranded DNA (our unpublished results).
Figure 2.
ATP hydrolysis. (A) GST-HsCdc6 (0.25 pmol; squares), GST-HsCdc6 (K208A) (0.25 pmol; triangles), or GST-HsCdc6 (E285Q) (0.25 pmol; circles) was incubated with 2.5 μM [γ-32P]ATP for the indicated times at 37°C. Hydrolysis products were separated by TLC, and the amount of phosphate formed was quantified by PhosphorImaging. (B and C) ATP hydrolysis was analyzed as described in A except that 25 μM [γ-32P]ATP (B) or 250 μM [γ-32P]ATP (C) was present in the reaction.
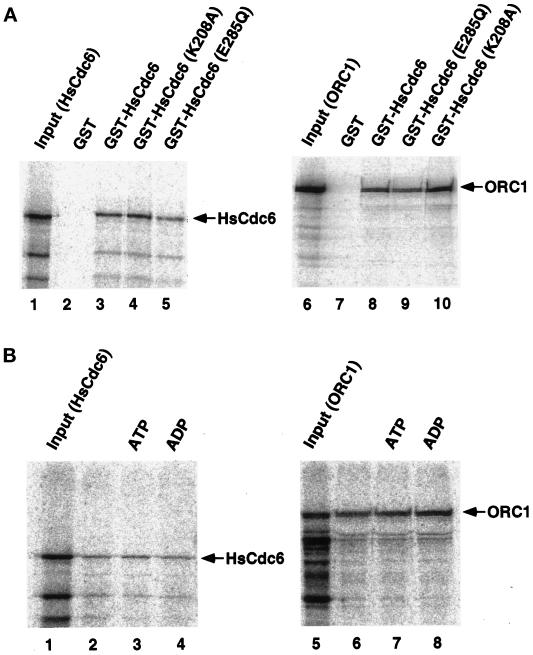
Nucleotide Binding to HsCdc6 Does Not Affect Complex Formation with HsOrc1 or HsCdc6 but Does Alter Its Conformation
The nucleotide-binding and hydrolysis properties of the Walker A and B mutants of HsCdc6 observed in Figures 1 and 2 were consistent with our expectations. However, these results do not eliminate the possibility that the single amino acid substitution in each mutant protein did not interfere specifically with nucleotide binding or hydrolysis but rather caused a more global structural change, which would affect other biochemical properties of the protein as well. To assess this possibility, we tested the ability of wild-type and mutant forms of HsCdc6 to form complexes with HsCdc6 and HsOrc1 proteins (Saha et al., 1998). In vitro–translated 35S-radiolabeled HsCdc6 or HsOrc1 was incubated with glutathione beads containing equal amounts of GST, GST-HsCdc6, GST-HsCdc6 (E285Q), or GST-HsCdc6 (K208A); the beads were washed; and the bound proteins were analyzed by denaturing gel electrophoresis, Coomassie blue staining (our unpublished results), and PhosphorImaging (Figure 3A). Wild-type GST-HsCdc6 bound to radiolabeled HsCdc6 (Figure 3A, lane 3) and HsOrc1 (lane 8), as reported by Saha et al. (1998), whereas binding of HsCdc6 and HsOrc1 to GST was not detectable (lanes 2 and 7). Both the Walker A and B mutant proteins bound to labeled HsCdc6 and HsOrc1 in amounts similar to those observed with the wild-type HsCdc6 protein (Figure 3A, compare lanes 4 and 5 with lane 3 and lanes 9 and 10 with lane 8). This result indicates that the amino acid substitutions at residues 208 and 285 did not affect the HsCdc6- or HsOrc1-binding activity of HsCdc6.
Figure 3.
Wild-type and mutant forms of HsCdc6 bind to Orc1 and to HsCdc6. (A) Glutathione-agarose beads containing equal amounts of GST (lanes 2 and 7), GST-HsCdc6 (lanes 3 and 8), or GST-HsCdc6 with the indicated amino acid substitutions (lanes 4, 5, 9, and 10) were incubated with radiolabeled HsCdc6 protein (lanes 2–5) or Orc1 protein (lanes 7–10) produced by in vitro transcription–translation in rabbit reticulocyte lysates. After washing, the beads were boiled in sample buffer, and the eluted proteins were resolved by SDS-PAGE (10% polyacrylamide gel). Bound proteins were visualized by PhosphorImaging. Lanes 1 and 6 contained one-fifth of the labeled input protein. (B) Glutathione-agarose beads containing GST-HsCdc6 were incubated with labeled HsCdc6 protein (lanes 2–4) or Orc1 protein (lanes 6–8) produced by in vitro transcription–translation in rabbit reticulocyte lysates. Reactions were performed in the absence of nucleotide (lanes 2 and 6) or in the presence of Mg2+-ATP (2 mM; lanes 3 and 7) or Mg2+-ADP (2 mM; lanes 4 and 8). Bound proteins were visualized by SDS-PAGE (10% gel) and PhosphorImaging. Lanes 1 and 5 contained one-fifth of the labeled input protein.
Because the mutations in the Walker A and B motifs did not affect the ability of HsCdc6 to bind to HsCdc6 or HsOrc1, one might expect that nucleotide binding to HsCdc6 might also fail to affect these protein–protein interactions. This prediction was tested by preincubating glutathione beads containing wild-type GST-HsCdc6 with buffer, magnesium ATP, or magnesium ADP and then incubating the beads with radiolabeled HsCdc6 or HsOrc1 (Figure 3B). The amount of HsCdc6 and HsOrc1 bound to HsCDC6, determined as described in Figure 3A, was unaffected by the presence of nucleotides in the reaction (Figure 3B, compare lanes 3 and 4 with lane 2 and lanes 7 and 8 with lane 6). Protein–protein interactions were also unaffected by preincubation of GST-HsCdc6 with EDTA, magnesium alone, or magnesium ATPγS (our unpublished results). Taken together, the results in Figures 1–3 argue that the mutations in the Walker A (K208A) and Walker B (E285Q) motifs of HsCdc6 are likely to cause specific defects in the nucleotide binding (Walker A) and hydrolysis (Walker A and B) properties of HsCdc6 and probably do not grossly distort the global conformation of the protein.
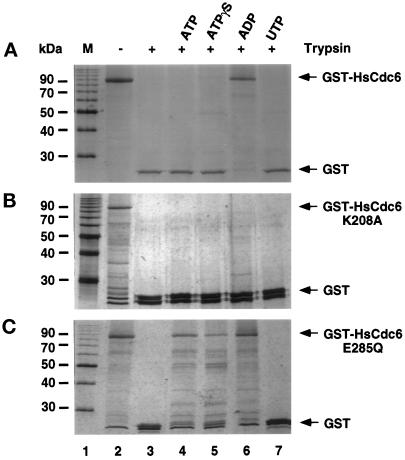
Although nucleotide binding of HsCdc6 does not appear to affect these protein–protein interactions, it is possible that it does alter the conformation of HsCdc6. To address this question, we preincubated GST-HsCdc6 with ATP, ATPγS, ADP, or, as a negative control, UTP and then subjected these incubations to proteolytic digestion. Figure 4A demonstrates that the HsCdc6 portion of the fusion protein was highly sensitive to tryptic digestion (compare lanes 2 and 3). Preincubation of the protein with ADP rendered it primarily resistant to trypsin (Figure 4A, lane 6), whereas ATP, ATPγS, and UTP had no protective effect (lanes 4, 5, and 7). When this experiment was repeated using HsCdc6 (K208A) instead of the wild-type protein, little or no protection against tryptic digestion was observed with any of these nucleotides (Figure 4B, compare lane 3 with lanes 4–7). When HsCdc6 (E285Q) was tested for sensitivity to trypsin, we observed that it became partially resistant to digestion after preincubation with ATP, ATPγS, or ADP but not UTP (Figure 4C, lanes 4–7). The protection afforded by preincubation of wild-type HsCdc6 with ADP suggests that ADP binding induces a conformational change in the HsCdc6 portion of the fusion protein, whereas ATP and ATPγS binding to HsCdc6 failed to induce the change. The inability of nucleotides to protect the Walker A mutant against digestion is consistent with its poor nucleotide-binding activity. The ability of all three adenine nucleotides to protect the Walker B mutant partially against digestion suggests that it responded aberrantly to nucleotide binding.
Figure 4.
ADP binding induces a conformational change in HsCdc6. (A) GST-HsCdc6 (0.5 μg) bound to glutathione-agarose was partially digested with trypsin in the absence of nucleotide (lane 3) or in the presence of 2 mM ATP (lane 4), ATPγS (lane 5), ADP (lane 6), or UTP (lane 7). The reaction was analyzed by SDS-PAGE (15% polyacrylamide gel) and silver staining. No trypsin was added to the reaction shown in lane 2. (B and C) The experiment in A was repeated, except that GST-HsCdc6 (K208A) (B) or GST-HsCdc6 (E285Q) (C) was used instead of wild-type protein. M, 10-kDa marker protein ladder.
HsCdc6 Defective in ATP Binding or Hydrolysis Blocks Chromosomal DNA Replication In Vivo
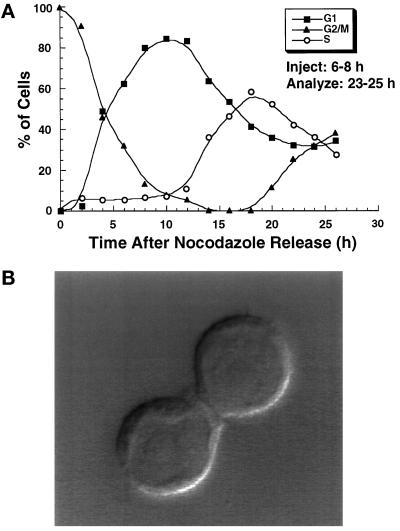
To determine whether ATP binding and hydrolysis by HsCdc6 are required for DNA replication in human cells, we reasoned that the mutant forms of the protein, because they retain the ability of the wild-type protein to form protein–protein complexes (Figure 3), might interfere with the functions of the endogenous wild-type HsCdc6. Thus we microinjected purified wild-type and the two mutant forms of HsCdc6 into HeLa-S3 cells and monitored their ability to undergo DNA replication. Cells to be injected were synchronized either in early G1, presumably before Cdc6 function was required, or, as a control, in very early S phase. To obtain cells synchronized in early G1, we released HeLa-S3 cells that had been arrested with the microtubule inhibitor nocodazole into nocodazole-free medium. After different times of incubation, the DNA content of these cells was examined by flow cytometry (Figure 5A). Cells entered G1 ∼1 h after the release, with the highest percentage in G1 after 10–12 h (Figure 5A, filled squares). As this peak declined, the cells progressed into S phase at 13–14 h after the release (Figure 5A, open circles). S phase was completed ∼20 h after the nocodazole release, when cells in G2/M began to appear (Figure 5A, filled triangles). A small fraction of cells seemed to recover more slowly from the nocodazole block, as seen by the 5–10% of the cells that were still in G2/M at 10–12 h after release. To avoid these slowly responding cells, we chose to inject cells that were completing cytokinesis 6–8 h after the nocodazole release (Figure 5B). Because only one of the two daughter cells was injected, this also allowed us to compare directly the subsequent DNA replication in these two cells. To obtain cells that were synchronized in G1/S of the cell cycle, a thymidine block was performed and released just before injection. After the cells were microinjected, the medium was supplemented with BrdU, and the cells were allowed to grow for an additional 17 h (injected in G1) or 12 h (injected in G1/S). By that time, uninjected cells had reached G2/M (Figure 5A). DNA replication was then detected by immunofluorescent staining against BrdU, and microinjected cells were identified by immunofluorescent staining against GST.
Figure 5.
Analysis of nocodazole-synchronized HeLa-S3 cells by flow cytometry. (A) Asynchronous HeLa-S3 cells were arrested at G2/M for 16 h by the use of the microtubule inhibitor nocodazole. The cells were washed, released into nocodazole-free medium, and analyzed by flow cytometry at different times after release. The percentage of cells in G1 (filled squares), G2/M (filled triangles), and S phase (open circles) was plotted against the time after release from the nocodazole block. (B) Asynchronous HeLa-S3 cells were arrested at G2/M for 16 h, released into nocodazole-free medium for 6 h, and visualized by DIC at a 100× magnification. This pair of cells is an example of those chosen for microinjection in G1, in which one of the two daughter cells was injected while the other remained uninjected for comparison of BrdU incorporation into the DNA or quantification of nuclear DNA content (see Figures 6–9).
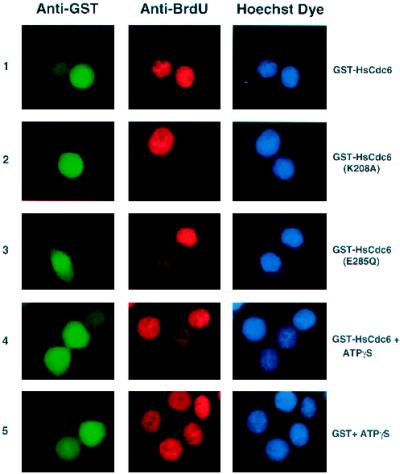
The phenotypes caused by the Walker A and B mutations in HsCdc6 are shown in Figure 6. Cells that had been microinjected in G1 with the indicated GST fusion protein (Figure 6, left column) were analyzed for BrdU incorporation into the DNA (middle column) and stained with Hoechst dye (right column) to detect bulk nuclear DNA. Although GST-HsCdc6 had no visible effect on chromosomal DNA replication (Figure 6, row 1), microinjection of GST-HsCdc6 (K208A) completely blocked DNA synthesis in most cells (row 2). In cells injected with GST-HsCdc6 (E285Q), BrdU incorporation was detectable (Figure 6, row 3), but the level of incorporation was significantly lower than that in neighboring uninjected cells. Because GST-HsCdc6 was able to bind to ATPγS (Figure 1, C and D) but could not hydrolyze this nucleotide (our unpublished results), we reasoned that microinjection of wild-type GST-HsCdc6 complexed to ATPγS into HeLa-S3 cells might cause the same defects in DNA replication as microinjection of GST-HsCdc6 (E285Q), i.e., a phenocopy of the Walker B mutant. As predicted, DNA replication was impaired in cells that had been injected in G1 with GST-HsCdc6 preincubated with ATPγS (Figure 6, row 4). In contrast, GST preincubated with ATPγS as a control did not interfere with DNA replication (Figure 6, row 5). None of the GST-HsCdc6 proteins or GST affected DNA replication in cells that were microinjected at G1/S (our unpublished results, but see Figure 7). These results demonstrate that HsCdc6 deficient in its ability to bind to or hydrolyze ATP specifically inhibited DNA replication, apparently by interfering with the activity of the endogenous HsCdc6.
Figure 6.
Mutations within the Walker A and B motifs of HsCdc6 cause different phenotypes. HeLa-S3 cells were arrested for 16 h in G2/M using nocodazole. At 6–8 h after release into nocodazole-free medium, the following proteins were injected into the nucleus of the cells: GST-HsCdc6 (row 1), GST-HsCdc6 (K208A) (row 2), GST-HsCdc6 (E285Q) (row 3), GST-HsCdc6 preincubated with a twofold molar excessof ATPγS (row 4), and GST preincubated with the same concentration of ATPγS used for GST-HsCdc6 (row 5). After injection, cell growth was continued for 17 h in medium containing BrdU. The cells were stained with anti-GST polyclonal antibody and FITC-conjugated goat anti-rabbit secondary antibody (left column), anti-BrdU monoclonal antibody and Cy3-conjugated goat anti-mouse secondary antibody (middle column), and Hoechst 33258 fluorochrome (right column). Micrographs were taken at a 100× magnification with a digital camera mounted on a fluorescence microscope.
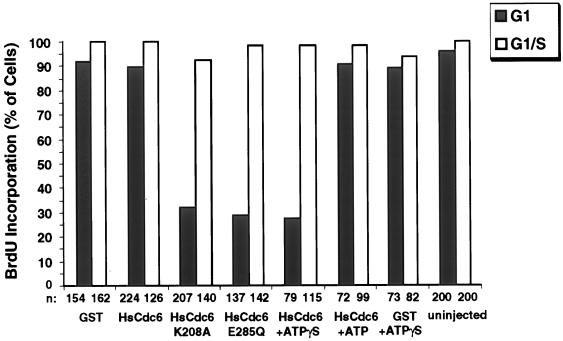
Figure 7.
HsCdc6 mutants deficient in ATP binding and hydrolysis impair DNA replication in human cells. The following proteins at the indicated concentrations were injected into the nuclei of HeLa-S3 cells in G1 or G1/S: GST (500 ng/μl), GST-HsCdc6 (61 ng/μl), GST-HsCdc6 (K208A) (28 ng/μl), GST-HsCdc6 (E285Q) (30 ng/μl), GST-HsCdc6 (61 ng/μl) preincubated with a twofold molar excess of ATPγS, GST-HsCdc6 (61 ng/μl) preincubated with a twofold molar excess of ATP, and GST (500 ng/μl) preincubated with the same concentration of ATPγS used for GST-HsCdc6. The proteins were injected either at 6–8 h after release from nocodazole (shaded bars) or in G1/S, immediately after release from a thymidine block (white bars). After injection of G1 cells, growth was continued for 17 h in medium containing BrdU. Cells injected in G1/S were grown for an additional 12 h in medium containing BrdU. Cells were fixed, stained with antibodies against GST and against BrdU, and analyzed by indirect immunofluorescence microscopy; n represents the number of cells that were successfully injected and analyzed for BrdU incorporation. BrdU-positive cells were defined as those that stained as brightly as uninjected cells, whereas BrdU-negative cells were those not stained or stained weakly (see Figure 6, row 3, for an example of a weakly stained cell).
DNA replication in microinjected and uninjected cells was quantitatively evaluated (Figure 7). Over 90% of the uninjected cells analyzed 23 h after release from the nocodazole block had incorporated BrdU into the DNA, and almost 100% of the uninjected cells that had been released from a thymidine block for 12 h had replicated DNA. GST and GST-HsCdc6 had no effect on DNA replication when injected in G1 or G1/S. In contrast, microinjection of GST-HsCdc6 (K208A) blocked DNA replication in 70% of the cells when injected in G1 (Figure 7, shaded bars) but not when injected in G1/S after DNA replication had initiated (white bars). However, ∼30% of the cells that had been injected with GST-HsCdc6 (K208A) in G1/S incorporated BrdU as efficiently as cells injected with wild-type fusion protein or GST. Significantly, these cells exhibited less intense immunofluorescent nuclear staining with anti-GST antibodies than did cells that failed to incorporate BrdU, suggesting that the amount of fusion protein in the nucleus of these cells may have been lower. Cells that had been injected with GST-HsCdc6 (E285Q) in G1 were also defective in initiating chromosomal DNA replication (Figure 7, shaded bars), as evidenced by weak staining with anti-BrdU in 70% of the injected cells (Figure 7). In contrast, injection of GST-HsCdc6 (E285Q) into cells in G1/S had no effect on DNA replication (Figure 7, white bars). DNA replication was inhibited in cells that had been injected in G1 with GST-HsCdc6 preincubated with ATPγS to mimic the defect caused by GST-HsCdc6 (E285Q), whereas GST-HsCdc6 preincubated with ATP or ADP had no effect. DNA replication remained unaffected when GST-HsCdc6 preincubated with ATPγS was injected into cells in G1/S. Microinjection of GST preincubated with ATPγS also had no detectable effect on DNA replication (Figure 7). These data indicate that mutant HsCdc6 deficient in binding or hydrolysis of ATP or wild-type HsCDC6 bound to a poorly hydrolyzable ATP analogue inhibits DNA replication in human cells when present during G1 but not when introduced after S phase has begun.
The Susceptibility of Cells in G1 to the Walker A and B Mutants Is Extinguished before S Phase
HsCdc6 mutants defective in binding or hydrolyzing ATP interfered with DNA replication when injected in early G1 but not in G1/S (Figure 7). Similarly, wild-type HsCdc6 protein bound to ATPγS interfered with DNA replication when injected in early G1 but not in G1/S (Figure 7). To investigate further when during G1 these mutants interfere with the activities of HsCdc6, we released HeLa-S3 cells from a nocodazole block and microinjected the cells with wild-type and mutant GST-HsCdc6 at different times after the release. BrdU was added to the medium after injection, and incorporation was evaluated by immunofluorescence at 22 h after release from the block. To determine when S phase began, we analyzed cells that had been treated similarly, but not injected, by flow cytometry. As shown in Figure 8A, HeLa-S3 cells entered S phase ∼13 h after their release from a nocodazole block (dashed line). Injection with GST or GST-HsCdc6 between 8 and 15 h after the nocodazole release did not detectably inhibit DNA replication, because >90% of the cells had incorporated BrdU by the time of analysis (Figure 8A, filled diamonds and filled squares). Microinjection of GST-HsCdc6 (K208A) at 8–10 h after the release prevented BrdU incorporation in 75% of the cells (Figure 8A, filled triangles). However, when the cells were injected at 11 h after the release or later, the number of cells that incorporated BrdU increased steadily with the time of injection. GST-HsCdc6 (E285Q) interfered with DNA replication when injected into HeLa-S3 cells at 8–10 h after the nocodazole release (Figure 8A, filled circles), and the number of cells showing BrdU incorporation also increased at later injection times. However, this increase was delayed by 1–1.5 h compared with that in cells injected with GST-HsCdc6 (K208A), and the initial rate of increase was slower. Quantitative evaluation of the data suggests that this temporal difference between the two mutants was significant (Figure 8A). When GST-HsCdc6 complexed with ATPγS was microinjected into HeLa-S3 cell nuclei, its ability to inhibit DNA replication was lost at approximately the same time in G1 as that of the Walker B mutant (Figure 8B, compare open squares and filled circles), whereas GST alone preincubated with ATPγS did not interfere with DNA replication (compare open diamonds and filled diamonds). Microinjection of either wild-type or mutant protein into cells that had been arrested in G1/S with hydroxyurea or aphidicolin did not affect DNA replication after the block was released, because incorporation of BrdU into the DNA of uninjected and injected cells was indistinguishable (our unpublished results). These findings provide additional evidence that ATP binding and hydrolysis by HsCdc6 are needed before DNA replication initiates.
A Mutation in the Walker A Motif Causes a G1 Arrest, whereas a Mutation in the Walker B Motif of HsCdc6 Causes an S Phase Arrest
In the experiments presented in Figures 6–8, BrdU incorporation into the DNA was measured 22–25 h after the nocodazole release, at which time the cells had reached the G2/M phase of the cell cycle (Figure 5A). Although GST-HsCdc6 (K208A) and GST-HsCdc6 (E285Q) appeared to inhibit DNA replication in these experiments, it is also possible that they merely delayed DNA replication, causing it to be undetectable at the time points that were analyzed.
To assess this possibility, HeLa-S3 cells were released from a nocodazole block for 6–8 h and microinjected in the nuclei with wild-type or mutant forms of GST-HsCdc6. We also injected GST as a control to ensure that the effects observed did not arise from the GST portion of the fusion protein. At 12 h after the release, the medium was again supplemented with nocodazole to prevent the cells from passing through mitosis into the following G1 phase. Beginning at 22 h after release, when uninjected cells were expected to be in G2/M (Figure 5A), the DNA content of microinjected cells stained with Hoechst 33258 was quantified by fluorescence microscopy (Araki et al., 1987). Figure 9 shows that the nuclear DNA content of cells that had been injected with GST (filled diamonds) or GST-HsCdc6 (filled squares) was the same as that in uninjected cells, indicating that DNA replication was not affected at any time point tested. In contrast, cells microinjected with GST-HsCdc6 containing the mutated Walker A motif (Figure 9, filled triangles) contained only one-half the amount of nuclear DNA present in uninjected cells at all times of analysis. Therefore, these data strongly suggest that GST-HsCdc6 (K208A) does not cause a delay in DNA replication but instead blocks progression into S phase.
The nuclear DNA content of cells that had been microinjected with GST-HsCdc6 (E285Q) (Figure 9, filled circles) increased slightly between 22 and 26 h after the nocodazole release, consistent with the weak BrdU incorporation observed in most cells injected with this mutant in Figures 6–8. Thus the injected cells entered S phase. However, these injected cells appeared to arrest ∼26 h after the nocodazole release, with a DNA content that was ∼1.5 times that of unreplicated nuclear DNA. These results show that cells injected with the Walker B mutant of GST-HsCdc6 replicated DNA significantly more slowly than did cells injected with the wild-type fusion protein or GST and were unable to complete DNA synthesis at the times tested. Cells that had been microinjected with GST-HsCdc6 preincubated with ATPγS (Figure 9, open squares) replicated nuclear DNA with kinetics similar to that of cells that had been microinjected with GST-HsCdc6 (E285Q), suggesting that the defect observed was caused by the inability of HsCdc6 to hydrolyze bound ATP. When GST preincubated with ATPγS was injected into the cells (Figure 9, open diamonds), the nuclear DNA content remained similar to that of uninjected cells, demonstrating that the amount of nucleotide injected did not interfere with normal cell cycle progression.
DISCUSSION
HsCdc6 Binds ATP and Hydrolyzes It
A number of studies have now demonstrated a requirement for intact Walker A and B motifs of Cdc6 for DNA replication in yeasts (Zwerschke et al., 1994; Elsasser et al., 1996; Perkins and Diffley, 1998; DeRyckere et al., 1999; Wang et al., 1999; Weinreich et al., 1999) and human cells (this article). These results strongly suggest a critical role for binding and hydrolysis of a nucleoside triphosphate in Cdc6 function in vivo. However, the dearth of biochemical characterization of the various Walker A and B mutants in yeast has led to conflicting conclusions as to what the roles of ATP binding and hydrolysis might be.
In this article, we have shown that the human Cdc6 indeed binds specifically to ATP (Figure 1, B–D). A Lys to Ala mutation within the highly conserved Walker A motif of GST-HsCdc6 strongly reduced ATP-binding activity, and GST alone did not bind to ATP (Figure 1), demonstrating that complex formation with ATP is an intrinsic function of HsCdc6. Binding to ATP appeared to be stoichiometric, suggesting that one molecule of HsCdc6 associates with one molecule of ATP under the in vitro conditions tested. The interaction with ATP or ATPγS was highly specific, because none of the other nucleotides tested bound efficiently to HsCdc6 (Figure 1C). ADP competed with ATP, implying that HsCdc6 can exist in an ATP- and in an ADP-bound form (Figure 1D). Unlike S. cerevisiae Cdc6 expressed in E. coli (Zwerschke et al., 1994), HsCdc6 bound poorly to GTP (Figure 1, C and D). Perhaps yeast Cdc6 differs from HsCdc6 in this property, but it is also possible that the bacterially expressed protein behaved aberrantly or that GTP may have bound to a contaminating protein in the preparation.
Like its yeast homologue (Zwerschke et al., 1994), the human Cdc6 protein displayed weak but detectable ATPase activity (Figure 2). This activity appeared to be intrinsic to HsCdc6 and not to the GST portion of the fusion protein, because mutations within the Walker A or B motifs reduced the ability of HsCdc6 to hydrolyze ATP (Figure 2) and GST alone did not show any detectable ATPase activity (our unpublished results). The rate of ATP hydrolysis by HsCdc6 was relatively slow. Although it is possible that the GST-tag affected the ATPase activity of HsCdc6, we believe that this effect is minor, because microinjection of GST-HsCdc6 into human cells in G1 did not affect subsequent DNA replication whereas injection of the mutant forms of GST-HsCdc6 did (Figures 6–9). Moreover, S. pombe Cdc18 can be replaced with GST-Cdc18, demonstrating that GST-Cdc18 is functionally active in vivo (Brown et al., 1997).
Neither ATP binding nor hydrolysis by HsCdc6 appears to be required for protein–protein interactions of HsCdc6 with HsOrc1 or HsCdc6, because both the Walker A and B mutants displayed activities similar to those of the wild-type protein in pull-down assays (Figure 3). Consistent with this interpretation, adenine nucleotides, with or without magnesium, failed to affect the ability of the wild-type protein to form protein–protein complexes. The ability of the wild-type and mutant forms of HsCdc6 to form oligomers in vitro is consistent with the dominant-negative phenotypes displayed by these mutants in vivo (Figures 6–9). There is some evidence that the ability of yeast Cdc6 to bind to Orc1 in vitro may be correlated with its ability to associate with yeast chromatin before initiation of replication in vivo (Wang et al., 1999), suggesting that nucleotide binding and hydrolysis by HsCdc6 may not be required for its association with Orc multimers in prereplication complexes in human cells. However, this question remains to be addressed experimentally.
Although nucleotides did not appear to affect the ability of HsCdc6 to form protein–protein complexes, they did induce alterations in its conformation (Figure 4). Binding of nucleotide to HsCdc6 appeared to be important in inducing an altered conformation, because UTP, a nucleotide unable to bind to HsCdc6, did not protect it against proteolysis. Tryptic digestion did not reveal any conformational change induced by ATP or ATPγS in the wild-type HsCdc6 even though both nucleotides bound quite well to the protein, whereas a marked change was induced in the presence of ADP. We cannot eliminate the possibility that binding of ATP or ATPγS also induced a change in HsCdc6 conformation that was not detected by this method. However, the ATP-bound conformation of HsCdc6 must differ from the ADP-bound form, and it seems likely that ADP release after ATP hydrolysis will be required for HsCdc6 to revert to its original conformation. The apparent inability of the Walker B mutant protein to respond differently to ATP and ADP binding suggests that the conformation of the Walker B protein bound to ATP may, in part, resemble that of the wild-type protein after ATP hydrolysis has occurred and before ADP has been released. It seems likely that these different protein conformations are important in HsCdc6 function, but how they are related to loading of MCM proteins in prereplication complexes or subsequent events is still unclear.
ATP Binding and Hydrolysis by HsCdc6 Are Essential for DNA Replication in Human Cells
The biochemical evidence presented above demonstrates that the Walker A and B mutants of HsCdc6 display specific defects in ATP binding and hydrolysis, or solely in hydrolysis, but not in protein–protein interactions (Figures 1–3). Both mutant proteins failed to exhibit a nucleotide-induced conformational change observed with the wild-type protein in vitro, but their phenotypes differed (Figure 4). Both mutant proteins interfered with the DNA replication activity of the endogenous wild-type HsCdc6 in human cells when introduced in G1 but not when introduced after S phase entry (Figures 6 and 7). The in vivo phenotypes of the two mutants differed, in that the Walker A mutant blocked S phase entry, whereas the Walker B mutant slowed or halted progression through S phase (Figure 9). Moreover, the replication-interfering activity of the Walker A mutant was lost earlier in G1 than was that of the Walker B mutant. Although the temporal difference was only 60–90 min, it was significant as demonstrated by its reproducibility in multiple experiments (Figure 8A). The qualitative differences between the Walker A and B mutant phenotypes in vitro and in vivo suggest strongly that both ATP binding and hydrolysis by HsCdc6 are required for DNA replication. Further support for the interpretation that ATP binding is not sufficient for the replication function of HsCdc6 is provided by the observation that introduction of wild-type HsCdc6 bound to ATPγS into human cells in G1 resulted in a phenocopy of the Walker B mutant (Figures 8B and 9).
In view of the functional conservation among initiation proteins in yeast and Xenopus (Dutta and Bell, 1997), it seems likely that ATP binding and hydrolysis by HsCdc6 are essential to assemble active prereplicative complexes on human origins of replication. It has been suggested that ATP metabolism by the Cdc6 proteins may be required for the loading of the MCMs onto chromatin, similar to the E. coli DnaB-loading factor DnaC (Baker and Bell, 1998). If HsCdc6 is involved in the assembly of the preRC in human cells, the mutant forms of HsCdc6 might associate with and block the ability of endogenous wild-type protein to load the MCMs stably onto chromatin.
Two types of models that would be consistent with the phenotypes observed in vivo with these Walker A and B mutants of HsCdc6 could be proposed for the role of nucleotide binding and hydrolysis in DNA replication. If the ATPase activity of HsCdc6 were primarily responsible for its ability to load MCMs onto chromatin, the near absence of ATPase activity in the Walker A mutant might account for its earlier loss of replication-interfering activity in G1 and for its failure to enter S phase. In this model, cells harboring the Walker B mutant would be predicted to load MCMs at a small number of origins during a limited period in G1, whereas cells harboring the Walker A mutant would fail to load MCMs. After this time window for MCM loading in G1 had passed, the number of prereplication complexes that could be activated in S phase would be directly related to the amount of HsCdc6 ATPase activity that was present during that time window. The slow rate of ATP hydrolysis by HsCdc6 in vitro raises the question of whether the ATPase activity may be stimulated in vivo by other proteins that are involved in the formation of the prereplicative complex. The possibility that ATP hydrolysis of Cdc6 may be activated upon phosphorylation by cyclin-dependent kinases has been suggested (Perkins and Diffley, 1998). However, preliminary data indicate that phosphorylation by recombinant cycE/cdk2 did not affect the ATPase activity of the human Cdc6 protein under our conditions in vitro. Another possibility is that the rate and timing of ATP hydrolysis by HsCdc6 in vivo could be regulated by other events, such as the binding of HsCdc6 to other components of the prereplicative complex such as Orc, MCMs, or Cdc45.
In a second type of model, ATP binding and hydrolysis by HsCdc6 might represent two temporally separable steps in G1. In this scenario, HsCdc6 would function as a switch regulated by nucleotide binding and hydrolysis. If association of HsCdc6 with prereplication complexes in vivo, like its association with HsOrc1 in vitro, does not require ATP binding or hydrolysis, we suggest that both HsCdc6 (K208A) and HsCdc6 (E285Q) may retain the ability to associate with chromatin-bound Orc. Two subsequent steps would then depend on ATP binding and hydrolysis by HsCdc6. For example, if complex formation between HsCdc6 and the MCMs were stabilized by ATP binding to HsCdc6, analogous to the DnaB/DnaC/ATP complex (Wahle et al., 1989), HsCdc6 (K208A) would not be able to interact stably with the MCMs. If only the subsequent delivery of the MCMs onto chromatin required ATP hydrolysis by HsCdc6, the complex of MCMs with HsCdc6 (E285Q) bound to ATP or with wild-type HsCdc6 bound to ATPγS might be stable, but loading of the MCMs onto chromatin would be inhibited. Alternatively, the conformation of the Walker B mutant bound to ATP might mimic that of the ADP-bound wild-type HsCdc6, preventing its association with the MCMs or feigning a completed loading reaction.
In summary, the data presented here provide further evidence that the human Cdc6 protein is essential for DNA replication in human cells (Saha et al., 1998; Yan et al., 1998; Petersen et al., 1999). They also provide the first evidence that ATP binding and hydrolysis are intrinsic activities of human Cdc6 and that both activities are specifically required during G1 for DNA replication in human cells. However, much experimental work remains to elucidate how ATP binding and ATP hydrolysis regulate the molecular interactions of HsCdc6 with its partners in initiation.
ACKNOWLEDGMENTS
We thank Lynda O’Rear for technical assistance, R. Weber for polyclonal antibodies against GST, C. Rehfuess for the GST parent plasmid, B. Stillman for the ORC1 cDNA, and C. Guyer, C. Rehfuess, V. Podust, T. Graham, R. Stein, and the reviewers of our manuscript for constructive criticism. The financial support of the National Institutes of Health (GM-52948), Vanderbilt University, and a Shared Equipment grant from the National Science Foundation (BIR-9419667) is gratefully acknowledged.
REFERENCES
- Araki T, Yamamoto A, Yamada M. Accurate determination of DNA content in single cell nuclei stained with Hoechst 33258 fluorochrome at high salt concentration. Histochemistry. 1987;87:331–338. doi: 10.1007/BF00492587. [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- Brown GW, Jallepalli PV, Huneycutt BJ, Kelly TJ. Interaction of the S phase regulator cdc18 with cyclin-dependent kinase in fission yeast. Proc Natl Acad Sci USA. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the prereplicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- DeRyckere D, Smith CL, Martin GS. The role of nucleotide binding and hydrolysis in the function of the fission yeast cdc18+ gene product. Genetics. 1999;151:1445–1457. doi: 10.1093/genetics/151.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler CS, Li JJ. Cdc6p establishes and maintains a state of replication competence during G1 phase. J Cell Sci. 1997;110:753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto prereplicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Lou F, Wang B, Campbell JL, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Wobst A, Petersen BO, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Tien D, Kelly TJ. sud1(+) targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc Natl Acad Sci USA. 1998;95:8159–8164. doi: 10.1073/pnas.95.14.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Nogueira Araujo GM. A simple method of reducing the fading of immunofluorescence during microscopy. J Immunol Methods. 1981;43:349–350. doi: 10.1016/0022-1759(81)90183-6. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust VN, Tiwari N, Ott R, Fanning E. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J Biol Chem. 1998;273:12935–12942. doi: 10.1074/jbc.273.21.12935. [DOI] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Stillman B. Comparison of DNA replication in cells from prokaryotes and eukaryotes. In: DePamphilis ML, editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor, NY: CSH Press; 1996. pp. 435–460. [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an MCM protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. I. Formation and properties. J Biol Chem. 1989;264:2463–2468. [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Feng L, Hu Y, Huang SH, Reynolds CP, Wu L, Jong AY. The essential role of Saccharomyces cerevisiae CDC6 nucleotide-binding site in cell growth, DNA synthesis, and Orc1 association. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Rottjakob HW, Kuntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]