Abstract
Mammalian mitochondrial ribosomes contain two prokaryotic-like rRNAs, 12S and 16S, both encoded by mitochondrial DNA. As opposed to cytosolic ribosomes, however, these ribosomes are not thought to contain 5S rRNA. For this reason, it has been unclear whether 5S rRNA, which can be detected in mitochondrial preparations, is an authentic organellar species imported from the cytosol or is merely a copurifying cytosol-derived contaminant. We now show that 5S rRNA is tightly associated with highly purified mitochondrial fractions of human and rat cells and that 5S rRNA transcripts derived from a synthetic gene transfected transiently into human cells are both expressed in vivo and present in highly purified mitochondria and mitoplasts. We conclude that 5S rRNA is imported into mammalian mitochondria, but its function there still remains to be clarified.
INTRODUCTION
Mitochondria are organelles present in virtually all eukaryotic cells, responsible for most of the energy production required for normal cellular homeostasis. Human mitochondria possess their own DNA (mtDNA), which encodes the two RNA species present in mitochondrial ribosomes (12S and 16S rRNAs), a full set of transfer RNAs (tRNAs) (22 genes) required for protein synthesis (O’Brien et al., 1990), and 13 polypeptides, all constituents of respiratory chain complexes (Anderson et al., 1981).
Because mitochondria possess a fully functional genetic apparatus capable of replication, transcription, and translation, they are often considered to be intracellular organelles endowed with a partial autonomy. This autonomy, however, is more apparent than real; in addition to the components encoded by mtDNA, all of the remaining enzymes required for proper functioning of the mitochondrion’s genetic machinery (such as DNA and RNA polymerases, ribosomal proteins, aminoacyl tRNA synthetases, etc.) are encoded by nuclear DNA (nDNA), synthesized in the cytosol, and imported into the organelle (Schatz and Dobberstein, 1996; Neupert, 1997). The same is true for all enzymes involved in the myriad metabolic pathways that take place in the mitochondrial environment.
Interestingly, at least two mitochondrial enzymes, RNase MRP (a site-specific endoribonuclease involved in primer RNA metabolism in mammalian mitochondria [Chang and Clayton, 1987; Topper and Clayton, 1990; Li et al., 1994]) and RNase P (an endoribonuclease involved in tRNA processing [Doerson et al., 1985]), are ribonucleoproteins that contain an RNA moiety that is encoded by nDNA and is imported into the organelle. However, unlike the mechanisms for protein import into mitochondria, the mechanisms of RNA import into mitochondria are poorly understood.
The importation of RNA into mitochondria was first postulated over 30 years ago, as a corollary to mitochondrial protein synthesis and the lack of a full set of tRNA genes in the mitochondrial genome of Tetrahymena (Suyama and Eyer, 1967). This postulate was proven recently (Rusconi and Cech, 1996), and the import of tRNAs into mitochondria has now been observed in a variety of biological systems, including plants (Phaseolus vulgaris, Solanum tuberosum, Triticum vulgaris, Zea mays, Marchantia polymorpha, and Chlamydomonas reinhardtii), fungi (Saccharomyces cerevisiae), and protozoa (Tetrahymena pyriformis, Paramecium aurelia, Plasmodium falciparum, Trypanosoma brucei, and Leishmania tarentolae) (Schneider, 1994; Kazakova et al., 1996; Tarassov and Martin, 1996, and references therein). Mammalian mitochondria do not appear to import tRNAs, but in addition to the RNA moieties of RNase P and RNase MRP, one other RNA species was recently observed to be associated with mammalian mitochondria: 5S rRNA, which was isolated from preparations of bovine mitochondria (Yoshionari et al., 1994). Moreover, an RNA species with a size consistent with that of 5S rRNA has also been found associated with purified human (King and Attardi, 1993) and mouse (Wong and Clayton, 1986) mitochondria, but neither the exact identity of this species nor its presence as an authentic mitochondrial RNA was established.
The presence of some nDNA-encoded RNA species within mitochondria has been controversial (Kiss and Filipowicz, 1992; King and Attardi, 1993) for a number of reasons. First, preparing highly purified subcellular fractions presents obvious technical difficulties. Second, the amount of nDNA-encoded RNA species detected in mitochondrial preparations is generally very low and might therefore be attributable to nothing more than a low level of contamination in the preparation. Finally, the electrochemical gradient across the inner mitochondrial membrane can be as high as 240 mV (negative inside), which creates a severe electrophoretic hurdle for the passive transfer of ribonucleic acids from the cytosol into the organellar matrix. Although it is now generally accepted that both the RNase P and RNase MRP RNAs are imported into mitochondria, the situation regarding 5S rRNA is still unclear.
We present here evidence that 5S rRNA is a true organellar species in mitochondrial fractions purified from mammalian cells.
MATERIALS AND METHODS
Plasmids
To construct plasmid pT7.5S.Dra, we amplified human DNA encoding the 121-nucleotide (nt) 5S rRNA from 143B.TK− cells (Bacchetti and Graham, 1977) with primers 5S-F (5′-GTCTACGGCCATACCACCCTG-3′) and 5S-R (5′-AAAGCCTACAGCACCCGGTAT-3′), by the use of Pwo DNA polymerase (Boehringer Mannheim, Indianapolis, IN), and cloned this DNA into SmaI-digested pUC19 (plasmid p5S). By the use of primers T7.5S-F (5′-taatacgactcactataGTCTACGGCCATACCACCC-3′ [T7 promoter in lowercase and 5S RNA sequence in uppercase]) and 5S.Dra-R (5′-tttAAAGCCTACAGCACCCGG-3′ [DraI half-site in lowercase with the DraI site underlined]) and with p5S as template, a 141-bp fragment was amplified and cloned into EcoRV-digested pZErO-2.1 (Invitrogen, San Diego, CA). This plasmid (pT7.5S.Dra) contains 17 bp immediately upstream of the 5S rRNA gene, which directs T7 RNA polymerase to begin transcription at position +1 of the gene. The reverse primer added three As to the 3′ end of the gene; thus, when digested with DraI, the plasmid is linearized at the exact terminus of the 5S rRNA gene.
To construct plasmid p5SSE, we used primers Eco5S-F (5′-gaattcgaattcGGATCCAAAACGCTGCCT-3′ [EcoRI sites in lowercase]) and 5S-R to amplify the 5′ region of the insert of pHU5S1, which harbors a 640-bp BamHI-SacI human DNA fragment containing the 121-bp 5S rRNA gene and its flanking regions (Nielsen et al., 1993), and primers Sma5S-F (5′-GACCGCCTGGGAATcCCGGGT-3′ [the introduced A–C change, in lowercase, creates a SmaI site, underlined]) and Eco5S-R (5′-gaattcgaattcGAGCTCCAGACCATCCCG-3′) to amplify the 3′ region. The two fragments were mixed and amplified with primers Eco5S-F and Eco5S-R. Because of regions of high GC content, all amplifications were performed in the presence of DMSO. The resulting fragment was purified, digested with EcoRI, and ligated into EcoRI-digested pUC19. The insert of a clone differing from that of pHU5S1 only at the SmaI site, as determined by sequencing, was subcloned into EcoRI-digested pSV2neo, yielding p5SSE.
Purification of Mitochondrial Fractions
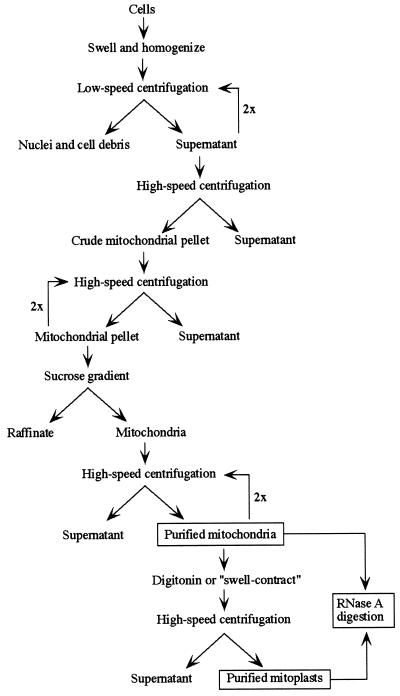
The purification scheme is outlined in Figure 1. Human cell lines 143B.TK− and 293T (DuBridge et al., 1987) were grown by standard procedures. Mitochondria were isolated from human cells, or from adult female Wistar rat liver, by the method of Tapper et al. (1983). Cells were resuspended in 10 mM NaCl, 1.5 mM CaCl2, and 10 mM Tris·HCl, pH 7.5, allowed to swell for ∼4–5 min, and briefly homogenized; sucrose was adjusted to 250 mM by addition of 2 M sucrose and T10E20 (10 mM Tris·HCl, 20 mM EDTA, pH 7.6); nuclei and cell debris were removed by two 3 min sequential centrifugations at low speed (∼1300 × g); mitochondria were collected by high-speed centrifugation (∼15,000 × g for 10 min) and washed three times with 250 mM sucrose and T10E20; the mitochondrial fraction, in 250 mM sucrose and T10E20, was layered on a discontinuous sucrose gradient consisting of 1.0 and 1.7 M sucrose in T10E20 buffer; and after centrifugation at 70,000 × g for 40 min at 4°C, the mitochondria were retrieved from the interface, diluted in 250 mM sucrose and T10E20, washed twice, and collected by high-speed spin. Protein was determined with the Bio-Rad Protein Assay Kit II (Richmond, CA).
Figure 1.
Scheme for the stringent purification of mitochondrial fractions. See text for details.
Mitoplasts were prepared by the use of two procedures. In the “swell-contract” method (Murthy and Pande, 1987), gradient-purified mitochondria were resuspended in 20 mM potassium phosphate, pH 7.2, containing BSA and allowed to swell for 20 min at 0°C, after which ATP and MgCl2 were added to 1 mM each and the incubation was prolonged for an additional 5 min. In the digitonin method (Greenawalt, 1974), the purified mitochondria were treated with ∼0.1 mg of digitonin per milligram of mitochondrial protein for 15 min at 0°C. Mitoplasts prepared by either method were recovered by high-speed spin.
Purified mitochondria and mitoplasts were treated with RNase A essentially as described (Adhya et al., 1997). Mitochondria or mitoplasts were resuspended in 250 mM sucrose and T10E1 (10 mM Tris·HCl, 1 mM EDTA, pH 7.6) and incubated for 30 min at 25°C with RNase A present at a concentration of 0.1 mg/ml; excess enzyme was removed by washing twice in 1 ml of 250 mM sucrose and T10E20, and recovery was by centrifugation. As controls, RNase A–treated mitochondria or mitoplasts were lysed by addition of SDS to 0.5% and incubated for an additional 15 min before the washing step.
Northern Blot Analysis
Total mitochondrial nucleic acids were prepared from highly purified rat liver mitochondria (see Figure 1) and electrophoresed through a 1.4% agarose-methylmercuric hydroxide gel (Attardi and Montoya, 1983). Nucleic acids were transferred onto nylon ZetaProbe membranes (Bio-Rad) and hybridized at 65°C with a PCR-generated probe amplified from plasmid pHU5S1, by the use of primers 5S-F and 5S-R in the presence of [α-32P]dATP.
Reverse Transcription-PCR
Human RNA was isolated by the guanidinium isothiocyanate method with minor modifications. Total RNA from 143B.TK− cells was treated with DNase I and subjected to reverse transcription (RT)-PCR (Titan RT-PCR System [Boehringer-Mannheim]) to amplify 5S rRNA (primers 5S-F and 5S-R), cytochrome c oxidase (COX) I mRNA (primers at positions 6559–6577 and 6769–6749 [Anderson et al., 1981]), and COX VIIc mRNA (forward primer [5′-gcagagcttccagcggctatgttgg-3′] and reverse primer [5′-gacaaacatatctagtatggcatat-3′]). Total RNA from rat liver and from rat liver mitochondria was isolated as described (Attardi and Montoya, 1983), treated with DNase I, and subjected to RT-PCR (SuperScript II Preamplification System [Life Technologies, Gaithersburg, MD]) to amplify 5S rRNA (primers 5S-F and 5S-R), COX I mRNA (primers at positions 6744–6763 and 6960–6941), and 5.8S rRNA (primers 5.8S-F [5′-cgactcttagcggtggatc-3′] and 5.8S-R [5′-agcgacgctcagacaggc-3′]). Total RNA from 293T cells that had been transfected transiently with p5SSE or pSV2neo was subjected to RT-PCR with either primers 5S-F and 5Sm-R (5′-AAAGCCTACAGCACCCG-3′) or primers 5S-C (5′-GGCCTGGTTAGTACTTGG-3′) and 5Sm-R, followed by SmaI digestion, labeling, and electrophoresis through a nondenaturing polyacrylamide gel. Organellar RNA was treated with DNase I as described (Dilworth and McCarrey, 1992) before RT-PCR.
RNA Expression Assays
Total RNA was isolated from 293T cells that had been transfected transiently (Life Technologies; Lipofectamine method) with p5SSE or pSV2neo, treated with DNase I to ensure complete removal of contaminating DNA, subjected to RT-PCR, digested with SmaI, labeled (with [α-32P]dATP in the presence of Klenow enzyme [see Figure 4B] or with [γ-32P]ATP in the presence of T4 polynucleotide kinase [see Figure 4C]), and electrophoresed through nondenaturing polyacrylamide gels. Detection of labeled fragments was performed with the Molecular Imager (model GS-363; Bio-Rad) with the aid of the Molecular Analyst 1.5 software package. All other reagents were from Sigma (St. Louis, MO).
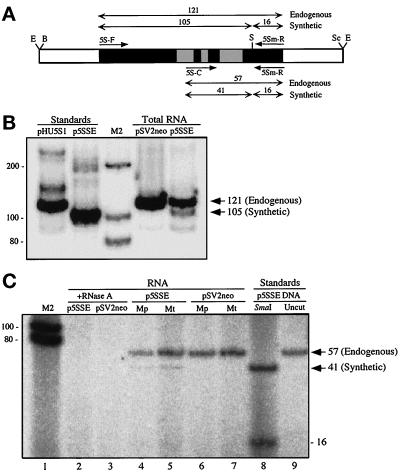
Figure 4.
Importation of synthetic 5S rRNA into human mitochondria. (A) Map of the insert of p5SSE. The two sets of primer pairs (short single-headed arrows) used to analyze the endogenous and synthetic variants and their corresponding SmaI-digested PCR products (double-headed arrows with the expected sizes above the arrows, in bp) are indicated. The 5S rRNA gene is in black with the known Pol III control elements (Willis, 1993) in the shaded boxes. The relevant restriction sites are indicated as follows: B, BamHI; E, EcoRI; S, SmaI; and Sc, SacI. (B) RT-PCR and RFLP analysis of total RNA from 293T cells that had been transfected transiently with p5SSE or pSV2neo, by the use of primers 5S-F and 5Sm-R, and electrophoresed through a 12% nondenaturing polyacrylamide gel. Note that the sizes of the SmaI digestions of RT-PCR products correspond exactly to the PCR products from pHU5S1 (wild-type) and p5SSE (A–C transversion) standards. Sizes of bands, in bp, are shown on the right. Sizes of M2 markers, in bp, are shown on the left. The 16-bp fragment is not resolved in this gel system. (C) RT-PCR and RFLP analysis of RNA isolated from highly purified mitochondria (Mt) and mitoplasts (Mp; obtained by the swell-contract method) from 293T cells transfected with p5SSE or pSV2neo, by the use of primers 5S-C and 5Sm-R, and electrophoresed through a 20% nondenaturing polyacrylamide gel. Note that the diagnostic 41-bp fragment is present in the p5SSE samples (lanes 4 and 5; the 16-bp fragments were diffuse and barely visible but were discernible in extremely dark exposures), with a size identical to that obtained in a PCR and RFLP from the p5SSE standard (lane 8), but is absent in the pSV2neo control samples (lanes 6 and 7). All RT-PCR species were abolished when mitochondria were treated with RNase A in the presence of SDS (lanes 2 and 3).
RESULTS
Northern Blot Analysis
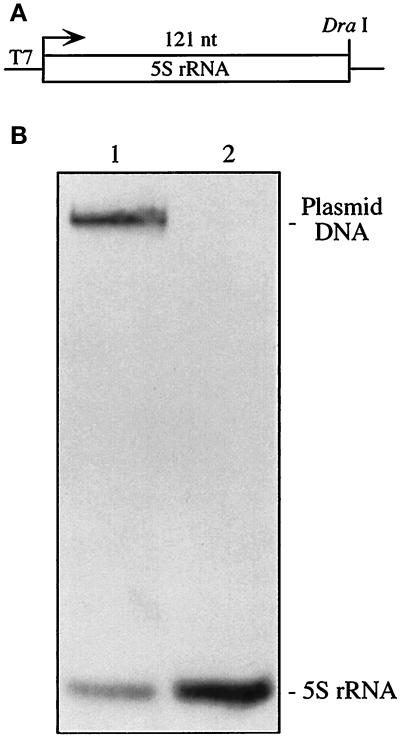
Full-length 5S rRNA was synthesized by in vitro transcription of DraI-linearized plasmid pT7.5S.Dra (Figure 2A) and was used as a standard in the Northern blot analysis of rat mitochondrial nucleic acids separated in high-resolution methylmercuric hydroxide gels (Attardi and Montoya, 1983). A full-length 5S rRNA DNA probe hybridized specifically to a single mitochondrial species, with a size indistinguishable from that of the 5S rRNA standard (Figure 2B).
Figure 2.
(A) Map of the insert of plasmid pT7.5S.Dra. (B) Northern analysis. Total mitochondrial nucleic acids from highly purified rat liver mitochondria (lane 2) were electrophoresed in parallel with T7 runoff transcripts from DraI-digested pT7.5S.Dra (lane 1) and were hybridized with a probe specific for 5S rRNA.
Detection of 5S rRNA in Purified Mitochondrial Fractions
There are at least three potential sources of 5S rRNA contamination in crude mitochondrial preparations: microsomes, cytosolic ribosomes associated with the outer mitochondrial membrane (OMM), and free cytosolic 5S rRNA molecules trapped between the inner mitochondrial membrane and the OMM during the isolation procedure (Attardi et al., 1969; Tapper et al., 1983). Our purification procedure, which is outlined in Figure 1, was designed to address all three problems. Extensive washing in the presence of a low concentration of EDTA (1 mM) has been shown to promote the removal of microsomes and adhering cytoplasmic ribosomes from mitochondrial fractions (Attardi et al., 1969). Our purification was performed in the presence of higher concentrations of EDTA (20 mM), which had the added potential benefit of facilitating the disaggregation of 5S rRNA from its ribosomal location (Hayes and Guérin, 1987). The problem of contamination of 5S rRNA in the intermembrane space (IS) was dealt with by preparing purified mitoplasts. Although none of the available methods for the preparation of mitoplasts is capable of removing the OMM in its entirety (Lazarus et al., 1987; Kang et al., 1992), this procedure nevertheless frees any RNAs trapped in the IS. Purified organellar preparations were treated with RNase A (Adhya et al., 1997) to digest any RNA species potentially adhering to the OMM (in mitochondria) or liberated from the IS (in mitoplasts).
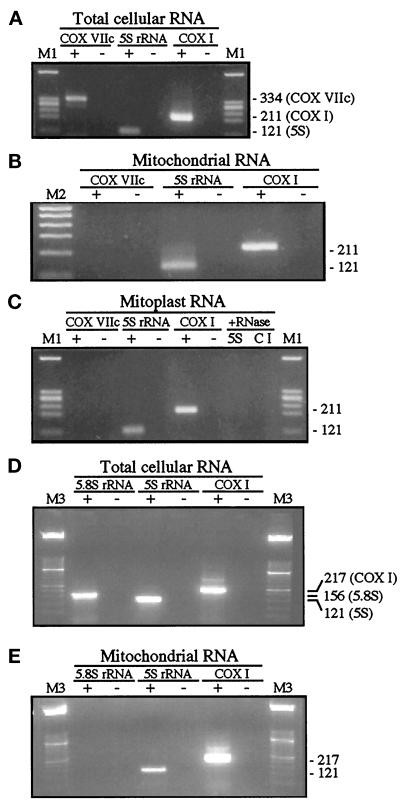
The progress of the purification of human mitochondrial fractions was monitored by following the presence of three RNA species: 5S rRNA, which is encoded by nDNA; COX I mRNA, an mtDNA-encoded transcript specifying subunit I of complex IV of the mitochondrial respiratory chain (cytochrome c oxidase or COX); and COX VIIc mRNA, a nDNA-encoded transcript specifying subunit VIIc of COX that is imported into mitochondria (nuclear-encoded subunits of COX are useful controls for cytosolic contamination because their messages are translated in the vicinity of mitochondria [our unpublished observations]). By the use of primers specific for these three transcripts, RT-PCR of total cellular RNA isolated before the subfractionation of human osteosarcoma-derived 143B cells produced the three expected products (Figure 3A), confirming the validity of the assay. On the other hand, in RNA from both mitochondria and mitoplasts (purified as outlined in Figure 1 and subsequently treated with RNase A), the RT-PCR signal for COX VIIc was absent, whereas the bands for both COX I and 5S rRNA were still present (Figure 3, B and C, respectively).
Figure 3.
RT-PCR analyses. (A) Analysis of 5S rRNA, COX I mRNA, and COX VIIc mRNA transcripts in total cellular RNA isolated from human 143B.TK− cells. RNA was subjected to RT-PCR in the presence (+) or absence (-) of reverse transcriptase activity. M1, markers of HaeIII-digested ΦX174 DNA. Expected sizes of RT-PCR products, in bp, are shown on the right. (B) Analysis described in A except that RNA from highly purified RNAse A–treated mitochondria was used as the template after DNase I treatment. M2, 100-bp ladder (Genosys Biotechnologies, The Woodlands, TX). (C) Analysis described in B except that RNA from highly purified RNase A–treated mitoplasts (obtained by the digitonin method) was used as the template. Mitoplasts were also lysed with 0.5% SDS in the presence of RNase A and subjected to RT-PCR to amplify 5S rRNA (5S) and COX I mRNA (CI). (D) Analysis of 5S rRNA, COX I mRNA, and 5.8S rRNA transcripts in total RNA isolated from rat liver. Methods and notation are described in A. M3, marker XIII (Boehringer Mannheim). (E) Analysis described in D except that RNA from highly purified RNAse A–treated rat liver mitochondria was used as the template.
To confirm these results, we also performed similar RT-PCR analyses on highly purified rat liver mitochondria, using 5.8S rRNA instead of COX VIIc mRNA as the marker for potential cytosolic RNA contamination. Although COX VIIc mRNA is a good marker, we believed that another RNA constituent of ribosomes would also be appropriate. We rejected the use of larger rRNAs (18S and 28S) and focused our efforts on 5.8S rRNA because 1) it has a size (156 nt) similar to that of 5S rRNA (121 nt), 2) it is present in amounts equimolar to 5S rRNA in cytosolic ribosomes, and 3) like 5S, it is highly structured. We obtained the same results, namely, that all three RNA species (5S rRNA, COX I mRNA, and 5.8S rRNA) were present in total cellular RNA (Figure 3D), but only 5S rRNA and COX I mRNA (and not 5.8S rRNA) were present in highly purified mitochondria (Figure 3E). The identity of all rRNA species after RT-PCR was confirmed by DNA sequencing (our unpublished data). We obtained identical results when the RNA samples were treated with RNase-free DNase I (our unpublished data).
Transient Expression of 5S rRNA
We expressed transiently a synthetic 5S rRNA gene that was constructed to allow us to distinguish it from the endogenous pool of human 5S rRNA. Using plasmid pHU5S1, containing the 121-bp wild-type 5S rRNA gene (Nielsen et al., 1993), we introduced an A–C transversion at position 103 in the 5S rRNA gene and subcloned it into pSV2neo (plasmid p5SSE). This mutation is located outside the known pol III transcriptional control elements (Willis, 1993) and also creates a SmaI site useful in restriction fragment length polymorphism (RFLP) analysis of PCR and RT-PCR products (Figure 4A).
Total RNA isolated from cell lysates of human kidney-derived 293T cells transfected transiently either with pSV2neo (control) or p5SSE (test) was amplified by RT-PCR, and the products were then digested with SmaI. Cells transfected with vector only (i.e., no insert) yielded a single 121-bp band originating from the endogenous 5S rRNA transcripts, whereas those transfected with the “synthetic” gene yielded two bands of the expected sizes, one of 121 bp (derived from the endogenous 5S rRNA) and the other of 105 bp (derived from the transfected gene) (Figure 4B).
Detection of Transiently Expressed 5S rRNA in Mitochondrial Fractions
RT-PCR analyses were then performed on highly purified mitochondria and mitoplasts (see Figure 1) from other transiently transfected 293T cells. Using different primers to accentuate the relative difference in the RFLP products between the wild-type and synthetic variants, we again detected only one band (57 bp, as expected) in control transfections but two bands of the predicted sizes (57 and 41 bp) in the genuine transfections (Figure 4C).
DISCUSSION
All cytoplasmic ribosomes studied to date, whether of prokaryotic or eukaryotic origin, possess 5S rRNA (Bogdanov et al., 1995). Moreover, this RNA is a component of the mitoribosomes of flowering plants, algae, and at least one protist (Lang et al., 1996). Fungal and animal mitoribosomes, however, are not thought to contain this RNA (Curgy, 1985; Bogdanov et al., 1995), even though preparations of nucleic acids from their purified mitochondria generally yield an easily distinguishable species with a size compatible with that of 5S rRNA (Tapper et al., 1983). The concept that these mitochondrial ribosomes are devoid of 5S rRNA has also been reinforced by the fact that animal and fungal mtDNAs do not encode 5S rRNA, whereas they do encode 12S and 16S rRNAs.
The presence of 5S rRNA in mitochondria has therefore been controversial and has been considered by some to be a contaminating species in mitochondrial fractions. For example, the 5S rRNA that was observed in preparations of highly purified human mitochondrial tRNAs (Wong and Clayton, 1986; King and Attardi, 1993) had been deemed to be a copurifying cytosolic contaminant, in spite of the fact that those preparations were devoid of cytosolic tRNAs (King and Attardi, 1993).
To verify the identity of this molecule, we synthesized full-length 5S rRNA by in vitro transcription of a linearized plasmid and used it as a standard in Northern blot analysis of mitochondrial nucleic acids isolated from rat liver. A full-length 5S rRNA DNA probe hybridized specifically to a single mitochondrial species, with a size indistinguishable from that of the 5S rRNA standard (Figure 2). We concluded that this mitochondrion-associated species is indeed 5S rRNA.
This result, however, did not resolve the question as to whether the 5S rRNA in mitochondrial fractions is indeed an authentic component of mitochondria or is merely a cytosolic contaminant. This is not a trivial point because there is a paucity of evidence supporting RNA import in mammalian systems, in part because, unlike yeast (Entelis et al., 1996) and Leishmania (Adhya et al., 1997), no in vitro mitochondrial RNA import system exists. Nevertheless, there is a growing body of evidence indicating that RNAs are imported into mammalian mitochondria, although the mechanism(s) by which this occurs is unknown. Besides the RNA moieties of mitochondrial RNase P and RNase MRP, transcripts derived from human immunodeficiency virus have been found in human mitochondria (Somasundaran et al., 1994). Moreover, analysis of the complete mitochondrial genome of the wallaroo, an Australian marsupial, failed to detect any gene for a true tRNALys (Janke et al., 1997), which led the authors to hypothesize that, as in yeast (Entelis et al., 1996), this tRNA is imported into the organelle. Finally, there is evidence that bovine mitochondria contain 5S rRNA (Yoshionari et al., 1994).
The detection of 5S rRNA in highly purified mitochondrial fractions devoid of contamination with potentially adhering cytosolic RNAs would go a long way toward resolving the question of 5S rRNA import. We therefore adopted a stringent multistep procedure to do just that (Figure 1), followed by RT-PCR of selected RNA species from isolated mammalian organellar RNAs. We found that, like mtDNA-encoded COX I mRNA, nDNA-encoded 5S rRNA (but, significantly, neither nDNA-encoded COX VIIc mRNA nor nDNA-encoded 5.8S rRNA) was tightly associated with the mitochondrial fraction (Figure 3). These results imply that a fraction of the total 5S rRNA pool is imported into the organelle.
Our transient expression results were consistent with this conclusion. Specifically, analysis of RNA isolated from purified mitochondria and mitoplasts from human cells transfected with an engineered gene, constructed to allow us to distinguish it from the endogenous pool of 5S rRNA, yielded two RT-PCR products, one derived from the endogenous 5S rRNA and the other derived from the transfected gene. Thus, we were not only able to express and detect an engineered 5S rRNA gene in mammalian cells (Figure 4B) but were also able to demonstrate that this transcript is imported into mitochondria (Figure 4C).
The amount of imported synthetic 5S rRNA detected in all of our experiments was relatively low (note the different intensities of the 57- and 41-bp fragments shown in Figure 4C). This is not surprising for three reasons. First, only ∼10–20% of cells normally express a transiently transfected construct; second, the introduced A–C transversion may have affected both the efficiency of importation and the turnover of the RNA; and finally, studies in yeast in vitro have shown that the mitochondrial importation efficiency for an exogenously added tRNA is <0.5% (Entelis et al., 1996). Thus, these experiments were fundamentally qualitative in nature and did not allow us to obtain an accurate estimate of the fraction of the 5S rRNA pool that is imported into mitochondria. Such quantitative analyses will likely require experiments with stably transformed cells.
Taken together, the Northern, RT-PCR, and transient expression results imply, first, that 5S rRNA is an authentic component of mammalian mitochondria (in agreement with more recent observations regarding the presence of 5S rRNA in bovine mitochondria [Yoshionari et al., 1994]) and, second, that an engineered transcript similar to 5S rRNA can also be imported into mitochondria. We note that the ability to import a synthetic transcript into human mitochondria may allow for the development of new approaches to the treatment of human mitochondrial diseases associated with maternally inherited mutations in mtDNA (Schon et al., 1997).
The mechanisms for the importation of RNAs into mitochondria are unknown but are most likely to be specific for the imported species of RNAs. Although there are no data on this point in human mitochondria, both the requirement for specific RNA sequences and for the presence of mitochondrial receptors for the import of selected small RNAs, in particular tRNATyr, into the organelle have been shown in Leishmania (Mahapatra et al., 1994, 1998; Mahapatra and Adhya, 1996; Adhya et al., 1997). Furthermore, only one of three tRNAGln isoforms is imported into Tetrahymena mitochondria (Rusconi and Cech, 1996), and only tRNALys, and no other tRNA, is imported into yeast mitochondria (Entelis et al., 1996), most likely on the basis of sequence-specific determinants (Entelis et al., 1998). Similarly, deletion experiments imply that mouse MRP RNA also contains importation determinants (Li et al., 1994). Thus, it is reasonable to assume that a similar importation specificity applies to mammalian 5S rRNA.
The role that 5S rRNA plays in mitochondria is still unclear. Even though it is believed that fungal and mammalian mitochondrial ribosomes do not contain 5S rRNA, it is not clear whether ribosomes in general can perform translation in its absence (Camier et al., 1995). Clearly, further work is required to elucidate the role of this small rRNA in mammalian mitochondria.
ACKNOWLEDGMENTS
We thank S. Frederiksen (University of Copenhagen, Copenhagen, Denmark) for plasmid pHU5S1, E. A. Shoubridge (Montreal Neurological Institute, Montreal, Quebec, Canada) and S. Goff (Columbia University, New York, NY) for the cell lines 143B.TK− and 293T, respectively, and M. Hirano and C. Briani for critical and encouraging comments. This work was supported by grants from the National Institutes of Health (NS-28828, NS-32527, NS-11766, and HD-32062) and the Muscular Dystrophy Association.
REFERENCES
- Adhya S, Ghosh T, Das A, Bera SK, Mahapatra S. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J Biol Chem. 1997;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Attardi B, Cravioto B, Attardi G. Membrane bound ribosomes in HeLa cells. I. Their proportion to total cell ribosomes and their association with messenger RNA. J Mol Biol. 1969;44:47–70. doi: 10.1016/0022-2836(69)90404-5. [DOI] [PubMed] [Google Scholar]
- Attardi G, Montoya J. Analysis of human mitochondrial RNA. Methods Enzymol. 1983;97:435–469. doi: 10.1016/0076-6879(83)97154-9. [DOI] [PubMed] [Google Scholar]
- Bacchetti S, Graham FL. Transfer of the gene for thymidine kinase to thymidine kinase-deficient human cells by purified herpes simplex viral DNA. Proc Natl Acad Sci USA. 1977;74:1590–1594. doi: 10.1073/pnas.74.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol. 1995;73:869–876. doi: 10.1139/o95-094. [DOI] [PubMed] [Google Scholar]
- Camier S, Dechampesme AM, Sentenac A. The only essential function of TFIIIA in yeast is the transcription of 5S rRNA genes. Proc Natl Acad Sci USA. 1995;92:9338–9342. doi: 10.1073/pnas.92.20.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Curgy J-J. The mitoribosomes. Biol Cell. 1985;54:1–38. doi: 10.1111/j.1768-322x.1985.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Dilworth DD, McCarrey JR. Single-step elimination of contaminating DNA prior to reverse transcriptase PCR. PCR Methods Appl. 1992;1:279–282. doi: 10.1101/gr.1.4.279. [DOI] [PubMed] [Google Scholar]
- Doerson CJ, Guerrier-Takada C, Altman S, Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. J Biol Chem. 1985;260:5942–5949. [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong P-M, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis NS, Kieffer S, Kolesnikova OA, Martin RP, Tarassov IA. Structural requirements of tRNALys for its import into yeast mitochondria. Proc Natl Acad Sci USA. 1998;95:2838–2843. doi: 10.1073/pnas.95.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis NS, Krasheninnikov IA, Martin RP, Tarassov IA. Mitochondrial import of a yeast cytoplasmic tRNA (Lys): possible roles of aminoacylation and modified nucleosides in subcellular partitioning. FEBS Lett. 1996;384:38–42. doi: 10.1016/0014-5793(96)00259-1. [DOI] [PubMed] [Google Scholar]
- Greenawalt JW. The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 1974;31:310–323. doi: 10.1016/0076-6879(74)31033-6. [DOI] [PubMed] [Google Scholar]
- Hayes F, Guérin MF. 5S RNA-protein complexes released by EDTA treatment of 60S ribosomal subunits of Tetrahymena thermophila. Biochimie. 1987;69:975–982. doi: 10.1016/0300-9084(87)90231-8. [DOI] [PubMed] [Google Scholar]
- Janke A, Xu X, Arnason U. The complete mitochondrial genome of the wallaroo (Macropus robustus) and the phylogenetic relationship among Monotremata, Marsupialia, and Eutheria. Proc Natl Acad Sci USA. 1997;94:1276–1281. doi: 10.1073/pnas.94.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Fujiwara T, Takeshige K. Ubiquinone biosynthesis by mitochondria, sonicated mitochondria, and mitoplasts of rat liver. J Biochem. 1992;111:371–375. doi: 10.1093/oxfordjournals.jbchem.a123764. [DOI] [PubMed] [Google Scholar]
- Kazakova EA, Tarasov IA, Entelis NS. Import of RNA into mitochondria (a review) Mol Biol (Mosk) 1996;30:438–443. [PubMed] [Google Scholar]
- King MP, Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J Biol Chem. 1993;268:10228–10237. [PubMed] [Google Scholar]
- Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7–2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- Lang BF, Goff LJ, Gray MW. A 5S rRNA gene is present in the mitochondrial genome of the protist Reclinomonas americana but is absent from red algal mitochondrial DNA. J Mol Biol. 1996;261:407–413. doi: 10.1006/jmbi.1996.0486. [DOI] [PubMed] [Google Scholar]
- Lazarus GM, Henrich JP, Kelly WG, Schmitz SA, Castora FJ. Purification and characterization of a type I DNA topoisomerase from calf thymus mitochondria. Biochemistry. 1987;26:6195–6203. doi: 10.1021/bi00393a036. [DOI] [PubMed] [Google Scholar]
- Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, Williams RS. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J Biol Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh S, Bera SK, Ghosh T, Das A, Adhya S. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 1998;26:2037–2041. doi: 10.1093/nar/26.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy MSR, Pande SV. Malonyl-CoA binding site and the overt carnitine palmitoyltransferase activity reside on opposite sides of the outer mitochondrial membrane. Proc Natl Acad Sci USA. 1987;84:378–382. doi: 10.1073/pnas.84.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nielsen JN, Hallenberg C, Fredericksen S, Sørensen PD, Lomholt B. Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res. 1993;21:3631–3636. doi: 10.1093/nar/21.16.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TW, Denslow ND, Anders JC, Courtney BC. The translation system of mammalian mitochondria. Biochim Biophys Acta. 1990;1050:174–178. doi: 10.1016/0167-4781(90)90162-u. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Cech TR. Mitochondrial import of only one of three nuclear-encoded glutamine tRNAs in Tetrahymena thermophila. EMBO J. 1996;15:3286–3295. [PMC free article] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schneider A. Import of RNA into mitochondria. Trends Cell Biol. 1994;4:282–286. doi: 10.1016/0962-8924(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Schon EA, Bonilla E, DiMauro S. Mitochondrial DNA mutations and pathogenesis. J Bioenerg Biomembr. 1997;29:131–149. doi: 10.1023/a:1022685929755. [DOI] [PubMed] [Google Scholar]
- Somasundaran M, Zapp ML, Beattie LK, Pang L, Byron KS, Bassell GJ, Sullivan JL, Singer RH. Localization of HIV RNA in mitochondria of infected cells: potential role in cytopathogenicity. J Cell Biol. 1994;126:1353–1360. doi: 10.1083/jcb.126.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama Y, Eyer J. Leucyl tRNA and leucyl tRNA synthetase in mitochondria of Tetrahymena pyriformis. Biochem Biophys Res Commun. 1967;28:746–751. doi: 10.1016/0006-291x(67)90379-8. [DOI] [PubMed] [Google Scholar]
- Tapper DP, Van Etten RA, Clayton DA. Isolation of mammalian mitochondrial DNA and RNA and cloning of the mitochondrial genome. Methods Enzymol. 1983;97:426–434. doi: 10.1016/0076-6879(83)97153-7. [DOI] [PubMed] [Google Scholar]
- Tarassov IA, Martin RP. Mechanisms of tRNA import into yeast mitochondria: an overview. Biochimie. 1996;78:502–510. doi: 10.1016/0300-9084(96)84756-0. [DOI] [PubMed] [Google Scholar]
- Topper JN, Clayton DA. Characterization of human MRP/Th RNA and its nuclear gene: full length MRP/Th RNA is an active endoribonuclease when assembled as an RNP. Nucleic Acids Res. 1990;18:793–799. doi: 10.1093/nar/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM. RNA polymerase III. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- Wong TW, Clayton DA. DNA primase of human mitochondria is associated with structural RNA that is essential for enzymatic activity. Cell. 1986;45:817–825. doi: 10.1016/0092-8674(86)90556-8. [DOI] [PubMed] [Google Scholar]
- Yoshionari S, Koike T, Yokogawa T, Nishikawa K, Ueda T, Miura K-I, Watanabe K. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS Lett. 1994;338:137–142. doi: 10.1016/0014-5793(94)80351-x. [DOI] [PubMed] [Google Scholar]