Abstract
In their natural environment, cells need to extract useful information from complex temporal signals that vary over a wide range of intensities and time scales. Here, we study how such signals are processed by Escherichia coli during chemotaxis by developing a general theoretical model based on receptor adaptation and receptor–receptor cooperativity. Measured responses to various monotonic, oscillatory, and impulsive stimuli are all explained consistently by the underlying adaptation kinetics within this model. For exponential ramp signals, an analytical solution is discovered that reveals a remarkable connection between the dependence of kinase activity on the exponential ramp rate and the receptor methylation rate function. For exponentiated sine-wave signals, spectral analysis shows that the chemotaxis pathway acts as a lowpass filter for the derivative of the signal with the cutoff frequency determined by an intrinsic adaptation time scale. For large step stimuli, we find that the recovery time is determined by the constant maximum methylation rate, which provides a natural explanation for the observed recovery time additivity. Our model provides a quantitative system-level description of the chemotaxis signaling pathway and can be used to predict E. coli chemotaxis responses to arbitrary temporal signals. This model of the receptor system reveals the molecular origin of Weber's law in bacterial chemotaxis. We further identify additional constraints required to account for the related observation that the output of this pathway is constant under exponential ramp stimuli, a feature that we call “logarithmic tracking.”
Keywords: adaptation kinetics, bacterial chemotaxis, signal processing, Monod-Wyman-Changeux model
Most studies of the kinetics of biological signaling pathways are based on measuring responses to simple controllable stimuli, such as a sudden change of ligand concentration. For example, bacterial chemotaxis is studied by subjecting cells to a step-function change in chemo-effector concentration and measuring the fraction of time that the flagellar motor spins counterclockwise (CCW) or clockwise (CW), for tethered cells (1), or more recently, by measuring the activity of the response regulator (CheY-P) by using FRET (2). Measurements of responses to these step-function stimuli over a range of ambient concentrations have been very useful in revealing the underlying signaling pathway for Escherichia coli chemotaxis (see refs. 3–5 for recent reviews). However, such a simple temporal stimulus is unlikely to be the typical signal encountered by E. coli cells in their natural environment, which changes/fluctuates continuously in time. How does an E. coli cell processes complex time-varying signals to obtain useful information? This is the question we try to address in this article by using a modeling approach tested by comparison with relevant existing experiments. A quantitative model based on microscopic pathway kinetics serves as a natural bridge between the experimentally measured responses to simple signals and predictions of the cell's responses to more realistic complex signals.
Our article aims to establish the connection between the underlying adaptation kinetics and the kinase responses of the cell to various time-varying signals. This connection not only provides a microscopic mechanism for the observed responses it allows us to learn about the (microscopic) internal pathway kinetics (such as receptor methylation/demthylation) from the (macroscopic) response measurements. The approach taken here for studying the bacterial chemotaxis pathway as a biochemical signal processor might provide a useful framework for understanding other biological systems in their natural environments.
General Model for Kinetics of Bacterial Chemotaxis
The primary focus of our study is on the system-level properties of the signaling pathway. Therefore, we use a mean-field approach without explicitly modeling the molecular details, in contrast to the pioneering approach of Bray and colleagues (6, 7), who laid the foundations for computational modeling of this pathway and in particular the detailed function of receptor clusters (8, 9). At this coarse-grained level, the E. coli chemosensory system and its environment can be described by three dynamic variables: the ligand concentration [L](t) (the input), the average kinase activity a(t) (the output), and the average methylation level of the receptors m(t) (the memory), as shown in Fig. 1A. The kinase activity is inhibited by binding of attractant ligand, and the system adapts by receptor covalent modification (methylation/demethylation), which is itself controlled by the receptor activity (feedback). The time scales for ligand binding (τl), kinase response (τk), and receptor covalent modification (τm) are well separated: τm ≫ τk ≫ τl. Here, we focus on studying pathway kinetics at the methylation time scale, the time scale most relevant for bacterial motion (run time ≈1 s) and treat the receptor kinase activity and ligand binding with a quasi-equilibrium approximation. The general model can then be written as:
The kinase activity function G can be expressed by using a simple two-state model:
where the dependence on m and [L] can be accounted for by specifying the functional form of ft(m, [L]), the free-energy difference between the active and inactive state of the receptor cluster. We use the Monod-Wyman-Changeux (MWC) (10) allosteric model, which has recently been shown to describe well the effects of receptor cooperativity on kinase activity (11–15), to prescribe a specific form for ft that is an additive function of two linearly independent terms,
where fm(m) depends on methylation and determines the kinase activity in the absence of ligand, while fL([L]) = ln(1 + [L]/KI) − ln(1 + [L]/KA) is the ligand-dependent free-energy (per binding site) characterized by the dissociation constants KI and KA, for the inactive and active receptors. The ratio of these binding constants, C ≡ KI/KA(<1), determines the maximum effect of ligand on kinase activity, because in the limit [L] → ∞, fL reaches a limiting value, fL([L]) → − ln C. The third parameter N corresponds to the number of ligand-binding units (in the context of bacterial chemotaxis, receptor homodimers) in the functional MWC cluster.
Fig. 1.
The structure and the characteristics of the simple kinetic model for chemotaxis. (A) The structure of the model. (B) The dependence of kinase activity G([L],m) on ligand concentration [L] for increasing methylation levels m (from left to right). (C) Some examples of the possible forms of the function F(a), which determines the dependence of the time rate of change in methylation on the activity a. For perfect adaptation, F(a) must be monotonically decreasing, but its detailed shape can still vary.
The methylation-dependent part of the energy difference is given by fm(m), and has been found recently to depend approximately linearly on m (T.S.S., unpublished work; see also ref. 16 for similar results on amidation modifications). Therefore, we take the simple linear form fm(m) = α(m0 − m) with constants α = 2 and m0 = 1 used to set the scale and the initial value (in zero ligand background) of the methylation level. Other parameters for the Tar receptor [in response to α-methyl-dl-aspartate (MeAsp)] are a0 = 1/3 (see below for definition of a0), N ≈6, KI ≈18 μM, and C ≈0.0062 as determined in ref. 15 from fitting the MWC-type model to the in vivo FRET data (2); these parameters are assumed to be constant, for simplicity. The dependence of kinase activity on ligand (MeAsp) concentration for different receptor methylation levels is illustrated in Fig. 1B.
The net methylation rate F (methylation rate − demethylation rate) depends on the details of the methylation/demethylation kinetics in vivo, which are not well understood quantitatively at present. However, from the system's observed near perfect adaptation (at least to MeAsp) (17–20), the function F should have very weak dependence on [L]. Here, generalizing the linear integral control model by Yi et al. (19), we assume the net methylation rate F depends explicitly only on receptor activity (a) as a nonlinear, monotonically decreasing, and single-valued function of a as illustrated in Fig. 1C. Perfect adaptation follows naturally as the dynamics of activity-dependent methylation, dm/dt = F(a), has a single fixed point at a = a0 with a0 given by F(a0) = 0. This fixed point is globally stable as guaranteed by the conditions F′(a) < 0 and da/dm > 0: regardless of the level of background ligand concentration, the kinase activity always adapts to the same level a0 at steady state.
Despite its simplicity, our model captures the essential features (receptor cooperativity, effects of receptor methylation on kinase activity, perfect adaptation) of the underlying pathway at the appropriate resolution for studying system-level properties of bacterial chemotaxis. The validity of the model and the assumptions made will be justified by existing experiments. Testable predictions will be made from our model as we apply it to study and understand responses of the cell to various time-varying signals.
Results
Response to Exponential Ramps: An Analytical Solution for Activity Shift.
In 1983, Block, Segall, and Berg (21) published measurements of responses (changes in CCW bias) of WT E. coli cells to both up and down exponential ramps with different ramp rates and to exponentiated sine-waves with different frequencies, both made with the nonmetabolizable attractant MeAsp. These are still the only systematic measurements of E. coli responses to time-varying signals. It is now known that MeAsp binds the serine receptor Tsr, as well as Tar, but the affinity of Tsr for MeAsp is very low [KD ≈100 mM (2)], so the latter binding can be ignored in the range of MeAsp concentrations used in these experiments. Here, we model E. coli's response to these time-varying signals and compare theory with experiment (21).
For the exponential ramps, as the ligand concentration changes exponentially with rate r: [L] = [L]0ert, the methylation level of the receptors m(t) increases in response, trying to bring the activity back to the preferred level at a0. However, as [L] keeps increasing, m(t) always lags behind, leading to a steady-state activity shifted from a0. This simple observation suggests a solution (ansatz) to this nonlinear time-varying dynamical system, wherein the kinase activity of the system settles to a constant value ac ≠ a0 and the receptor methylation level increases linearly with rate F(ac) ≠ 0:
This ansatz is valid if the effect of the linearly increasing methylation level can balance that of the exponentially increasing ligand concentration to maintain a constant level of activity. For ligand concentrations in the ramp between the two dissociation constants KI ≪ [L] ≪ KA, fL ≈ ln([L]/KI) ≈ rt + ln([L]0/KI), the expression for the kinase activity can be simplified:
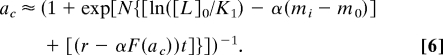 |
Eqs. 5 and 6 can be solved self-consistently to obtain the constant kinase activity level ac and the projected intercept methylation level mi:
 |
where F−1 is the inverse function of F. From our analytical solution, after an initial transient the kinase activity settles to a constant level ac = F−1(r/α) that depends on the ramp rate r, in agreement with experiments (21). This behavior is confirmed by direct simulation of our model for both up and down ramps as shown in Fig. 2 A and B.
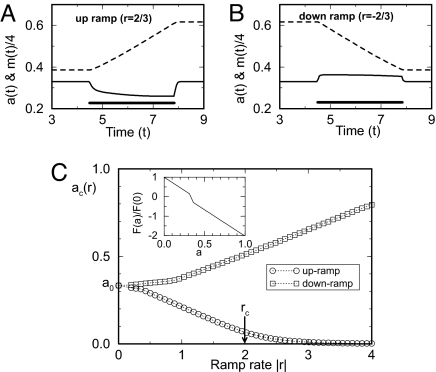
Fig. 2.
The activity shifts for different exponential ramp rates. (A and B) The time dependence of methylation level (dotted lines) and the kinase activity (solid lines) are shown for a case of up ramp with r = 2/3 (A) and a case of down ramp with r = −2/3 (B). The solid bars indicate the duration of the ramp. (C) The steady-state kinase activities ac(r) for different exponential ramp rates for both up ramp (the lower curve) and the down ramp (the upper curve) from direct model simulation are shown. The critical up ramp rate rc = αF(0) = 2, separates the two types of responses to the fast (r > rc) and the slow (r < rc) ramps. (Inset) The piecewise linear form for the methylation rate function F(a) used in the simulations of Figs. 2 and 3: a linear section around a = a0 with slope s1 = −f1/(a0ε1) in the region a0(1 − ε1) ≤ a ≤ a0(1 + ε2) and two linear sections (1 ≥ a > a0(1 + ε2) and a0(1 − ε1) > a ≥ 0) outside of this region with slope s2 = −(1 − f1)/(a0(1 − ε1)). Note F(0) = 1 is used to set the time and frequency units in Figs. 2 and 3. All of the simulations shown in this article are done with the choice ε1 = 0.05, ε2 = 0.1, and f1 = 0.15. This yields a small region near a0 where F(a) has a steeper slope, a feature that is required to reproduce the response thresholds near a0 observed in the exponential ramp experiments (21). This shape of F(a), and our choice for the steady-state activity a0 = 1/3 dictates that F(1) = −2. The asymmetry in the response thresholds results from the choice ε1 ≠ ε2.
The dependence of the steady-state activity ac on the ramp rate r, discovered by the analytical solution Eq. 7, provides a remarkable connection between the (“microscopic”) methylation kinetics and the (“macroscopic”) exponential ramp responses (21). The measured responses (shifted activity) for the up (r > 0) and down (r < 0) ramps can thus be used to determine the full functional form of F(a) in the regimes F > 0 and F < 0, respectively. In Fig. 2C, the kinase responses for different ramp rates with a specific choice of F(a) are shown. This simple piece-wise linear form of F(a) (Fig. 2C Inset) was chosen here to obtain qualitative agreement with the Block et al. (21) experiments. The steep region of F(a) near a0 is required to reproduce the response thresholds for up- and down-ramps observed at small ramp rates (figure 6 in ref. 21). If one were to quantitatively fit these data obtained through measurements of motor switching, the ultrasensitive response of the motor to the intracellular signal would require an even steeper region in F(a) near a0 to reproduce the response thresholds. Full quantitative determination of F(a) awaits more refined ramp experiments measuring the kinase activity directly.
For ramp rates faster than a critical rate rc ≡ αF(0) [or αF(1) for down ramps], even the maximum methylation rate F(0) cannot keep up/balance the fast increase in attractant concentration; as a result, the kinase activity decreases to zero for r > rc as shown in Fig. 2C. For r ≫ rc, the dynamics of the kinase activity become independent of r and is controlled (limited) by the constant (maximum) methylation rate F(0). More discussion on responses to fast ramps will be given later as we study recovery time for large step stimuli, which can be considered as the extreme case of r → ∞.
The predicted different behaviors of kinase responses and methylation rates for slow and fast exponential ramps: a ≈ F−1(r/α), dm/dt ≈ r/α for r < rc; a ≈ 0, dm/dt ≈ F(0) for r ≫ rc should be tested by future ramp experiments measuring kinase activity and receptor methylation level directly.
Response to Exponentiated Sine Waves: Spectral Analysis.
Block et al. (21) also measured responses of E. coli to periodic changes in MeAsp concentration between two values, [L]l ≈50 μM and [L]h ≈500 μM, varying the logarithm of concentration ln[L] sinusoidally over a range of frequencies ν: [L](t) = [L]0 exp[ALsin(2 πνt)], where AL = 1/2ln([L]h/[L]l) is the amplitude of the oscillation in ln[L] and [L]0 = . Our model with the functional form of F(a) determined by the exponential ramp responses can be used to predict the response to these periodic signals and understand the underlying adaptation kinetics in response to signals with different time scales.
When the stimulus variation is relatively small and well within the most sensitive regime of the sensory system: KA ≫ [L]h > [L]l ≫ KI, the change in ligand-dependent free energy ΔfL ≡ fL([L](t)) − fL([L]0) follows exactly the logarithm of ligand concentration, ΔfL(t) = ALsin(2πνt), and the analytical form of the kinase responses as well as the corresponding methylation dynamics can be obtained by linearizing our dynamical model around a = a0:
where Δa ≡ a − a0 and Δm ≡ m − mi with mi ≡ m0 + (αN)−1ln[a0/(1 − a0)] + α−1fL([L]0). Expressing Δm(t) and Δa(t) as: Δm = Re(Am exp(2πiνt)), Δa = Re (Aa exp(2πiνt)), where i = and Re stands for real part, we can solve for Am and Aa:
where ca ≡ −N a0 (1 − a0), cm ≡ α−1 = −(dfm/dm)−1, and the characteristic frequency νm ≡ −(αF′(a0) N a0(1 − a0))/2π are all constants that do not depend on the applied signal frequency ν. |Am| and |Aa| are the amplitudes of the responses, while the phases of Am and Aa determine the time lags between the responses and the stimulus.
Two distinctive types of responses are found from Eq. 10, separated by νm, or equivalently an intrinsic time scale τm ≡ (2πνm)−1. For low-frequency signals with ν ≪ νm, we have:
The kinase activity is proportional to the derivative of the signal (iνAL in frequency space) as the methylation level follows (adapts to) the signal without much delay. For high-frequency signals with ν ≫ νm, we have:
The methylation kinetics cannot keep up with the fast signal, so the methylation level follows the average of the signal (AL/iν in frequency space) and decreases with the frequency of the signal; as a result, the kinase activity is determined by the instantaneous signal: the system loses its ability to calculate the derivative.
The results from simulation of the full nonlinear model in both the high- and the low-frequency regimes, as shown in Fig. 3 A and B, agree with the linear analysis. The amplitude dependence on the frequency (the Bode plot) as shown in Fig. 3C, agrees qualitatively with the experimental results (figure 8 in ref. 21). However, in the experiment, the steep response of the motor to the intracellular signal (22) would be expected to result in saturation of the CCW bias at frequencies below the true value of νm, which, in turn, could vary significantly from cell to cell (23). We can also obtain the phase shift of the kinase response, which is harder to measure experimentally. We find that for a low-frequency signal, the phase of the kinase response lags that of the signal by roughly π/2, a clear indication that the system is calculating the derivative of the signal. For higher frequencies, the phase lag becomes π, indicating that adaptation fails and the kinase activity passively follows the stimulus. This linear spectral analysis agrees well with the noise-filtering properties observed in simulations of the full nonlinear model (see supporting information (SI) Text and Fig. S1).
Fig. 3.
Responses to exponentiated sine-waves. (A and B) The kinase activity (thick solid lines) and the receptor methylation level (thick dotted lines) are shown in response to a low-frequency signal (thin solid lines) with ν = 0.2 (A) and a high-frequency signal (thin solid line) with ν = 2.0 (B). (C and D) The amplitudes and the phase shifts (with respect to the signal) of the kinase response for different signal frequencies are shown.
Response to Impulsive Stimuli: The Effect of Dephosphorylation Kinetics.
At short time scales (≤1 s), the dynamics of pathway activity is controlled by the phosphatase enzyme CheZ. The dynamics of the CheY-P concentration [Y] can be described by a simple equation:
where ka is an effective phospho-transfer rate from active CheA to CheY, and τZ is the dephosphorylation time. From this and Eqs. 8 and 9, the CheY-P response AY in frequency space can be obtained:
where νZ ≡ (2πτZ)−1 ≫ νm is the dephosphorylation frequency. At low frequency ν ≪ νZ, AY follows the behavior of Aa given in Eq. 10. However, at high frequency ν ≫ νZ, AY ≈ ca ka/(2πiν)AL decays as 1/ν. Thus the dephosphorylation frequency provides an upper cutoff to the pathway response, as shown in Fig. 4A where the amplitude of the combined frequency space response function R(ν) ≡ iν/(νm + iν) × νZ/(νZ + iν) is plotted. The CheY-P response in time space can be obtained from Eq. 14:
 |
with the linear response function (the Green's function in physics) R(t) given by:
where R0 = (−ca)kaτZτm/(τm−τZ) > 0, and R(t < 0) = 0. The response function has a negative regime followed by a positive regime (overshoot) with the total integrated area being zero. The response function given by Eq. 16 can be readily used to explain the response of E. coli to impulsive stimuli (24, 25). Fig. 4B shows the (normalized) CheY-P concentration change (thick line) in response to a short square wave pulse (thin line). The overshoot response is evident in Fig. 4B; the cross-over time τ1 defined by R(τ1) = 0 can be determined from Eq. 16: τ1 = τmτZ/(τm−τZ)ln(τm/τZ). In Fig. 4, we choose τm = 3 s as estimated from refs. 24 and 25, and τZ = 0.5 s as measured in ref. 26, thus the size of the negative lobe is τ1 ≈ 1.1 s, in agreement with experiments (24, 25).
Fig. 4.
Response to impulsive stimuli at the level of CheY-P. (A) Frequency-domain representation of the impulse response of CheY-P to ligand stimuli, R(ν) = iν/(νm+iν) × νZ/(νZ+iν). The asympotic slopes for the low-frequency (ν ≪ νm) and high-frequency (ν ≫ νZ) regimes are indicated by dotted lines. The high-frequency cutoff results from the phosphorylation dynamics, and its location is determined by the dephosphorylation frequency, νz. (B) Simulated changes in the CheY-P level Δ[Y] caused by a short square-wave pulse of attractant started at t = 0 with duration tp = 0.2 s. The plot of Δ[Y] (normalized by its peak value Δ[Y]max) clearly demonstrates an overshoot after the recovery from the initial, downward response. The time constants used here are: methylation time τm = 3 s, dephosphorylation time τZ = (2πνz)−1 = 0.5 s.
Response to Large Step Stimuli: Additivity in Recovery Time.
A step function stimulus can be considered as an infinitely fast ramp (r → ± ∞). The kinase activity is suppressed to zero for large increases in attractant concentration, and the time to recover to its prestimulus level depends on the change in attractant concentration. It was discovered by Spudich and Koshland (27) and Berg and Tedesco (28) that the recovery time is additive; that is, the recovery time for a step stimulus from [L]1 to [L]3 is the sum of the recovery times for step stimuli from [L]1 to [L]2 and from [L]2 to [L]3.
From our model, the kinase activity is suppressed to zero (a = 0) for large sudden increase in ligand concentration, i.e., ([L]2/[L]1)N ≫ 1 and ([L]3/[L]2)N ≫ 1. As a result, the receptor methylation level increases with the (constant) maximum methylation rate F(0): dm/dt = F(0). Therefore, for a large stimulus jump, e.g., from [L]1 to [L]2, the recovery time τR can be written as:
where ms([L]) is the steady-state methylation level upon complete adaptation to stimulus concentration [L]. From Eq. 17, additivity of recovery time follows naturally: τR([L]1,[L]2) + τR([L]3,[L]2) = τR([L]3,[L]1). A demonstration of the recovery time additivity in our model is shown in Fig. S2 a and b.
As the perfect adaptation condition fm + fL = const. is satisfied, and fm(m) is a linear function of m, we have (ms([L]2) − ms([L]1)) = (fL([L]2) − fL([L]1))/α. We can use this relation to compute the recovery time in our model by substituting into Eq. 17 and evaluating fL with the parameters KI = 18.2 μM, C = 0.0062 (from ref. 15). The result of such a calculation is plotted in Fig. S2c, demonstrating excellent quantitative agreement with the Berg and Tedesco (28) data without any fitting parameters. From the measured maximum recovery time τmax ≈350 s (28) and our model result, Eq. 17), we have (αF(0))−1 ≈70 s, which leads to an estimate of the maximum methylation rate F(0) (with α = 2): F(0) ≈ 7 × 10−3 s−1.
Summary and Discussion
In this article, we have studied chemotactic responses to various time-varying signals by using a minimal model that captures the essential features of the underlying microscopic pathway. Despite its simplicity, the model produces results that are in quantitative agreement with the existing experimental data for both exponential ramp and exponentiated sine-wave stimuli. An intriguing observation made long ago, the additivity of recovery time for large stimulus changes, is also explained naturally by the model. The agreements with this diverse set of experiments validate the model and its assumptions. More importantly, the relevant biological features captured by the model enable us to look beyond the behaviors for their molecular mechanisms and lead to predictions that can be tested by future experiments. A global description of the chemotaxis pathway as a biochemical signal processor emerges from our study and provides a solid base for further studying/understanding complex chemotaxis behaviors in realistic environments. The pathway acts as a lowpass filter for the derivative of the signal and a highpass filter for the signal itself, with the two regimes separated by an intrinsic adaptation time scale. The pathway senses (processes) the ligand concentration in logarithmic scale, analogous to Weber's law known from other sensory systems. We discuss these findings and their possible implications in the following.
Logarithmic Tracking and Weber's Law: Their Molecular Origins.
From the observation that the shift in the kinase activity during exponential ramps reaches a constant, i.e.,
it is clear that the bacterial chemotaxis system tracks and responds to the gradient of the logarithm of the ligand concentration (dln[L]/dt = [L]−1d[L]/dt) instead of the gradient of the ligand concentration (d[L]/dt) itself. This logarithmic tracking phenomenon reminds us of Weber's law as applied to bacterial chemotaxis (29), which states that the response of a cell to a sudden, small change in stimulus Δ[L] after it has adapted to a stimulus [L]0 should be proportional to the relative change of the ligand concentration, i.e.,
where k is constant. From our linearized model, the kinase response can be obtained Δa = − a0(1 − a0) (∂ft/∂ln[L])Δ[L]/[L]0. Therefore, Weber's law (Eq. 19) emerges given two conditions: (i) the system exhibits perfect adaptation to constant stimuli, so that a0 is a constant independent of [L]0, and (ii) there exists a significant range of [L]0 over which the free energy ft is proportional to ln[L], i.e., ∂ft/∂ln[L] is a constant. These two conditions are satisfied in our model and the Weber-Fechner constant can be determined analytically: k = − Na0(1 − a0), which also demonstrates the amplifying role of the receptor cluster size N.
The requirements for logarithmic tracking (Eq. 18) are more stringent: the basic requirement, from Eq. 3, is that the total free energy remains constant in time during the exponential ramp, i.e., dft/dt = 0. Combined with conditions i and ii, this results in the additional requirement that (iii) the methylation-dependent part of the free energy is a linear function, i.e., dfm/dm is constant. The necessity for this condition is readily seen in the general solution of our model for the constant activity in Eq. 18: factoring dft/dt = 0, we get
which yields
The first two factors in the argument of the inverse function are constant given condition ii and the exponential ramp stimulus, and F(a) is single-valued to satisfy condition i, so ac is constant iff dfm/dm is constant. When this condition is satisfied, ac in Eq. 21 further reduces to the simple form given in Eq. 7. For more complicated (nonlinear) forms of fm(m) in our model, Weber's law (Eq. 19) still holds, but logarithmic tracking (Eq. 18) breaks down.
The existing explanation for Weber's law and logarithmic tracking for E. coli chemotaxis was based on the assumption that the response depends directly on the time rate of change of receptor ligand occupancy (24, 27, 29, 30). Our model suggests an alternative view: the fundamental origin for Weber‘s law and logarithmic tracking in E. coli chemotaxis is the perfect adaptation kinetics (condition i) and the specific characteristics of the free-energy functions. The logarithmic form of the ligand-dependent free energy, fL([L]) ≈ ln[L] (condition ii) ensures that Weber's law holds and further requiring the linear form of the methylation-dependent free energy, fm(m) ≈ −αm (condition iii) leads to logarithmic tracking behavior. Thus, the balance of the two free energies (achieved by adaptation) implies a logarithmic mapping between ligand concentration and methylation: m ∼ α−1 ln[L], the methylation level of the receptors, as the memory of the system, records and tracks the ligand concentration on a logarithmic scale. This view is supported by recent in vivo experiments (2, 31) showing that increasing the receptor methylation level shifts the kinase dose–response curve to higher ligand concentrations in a semilog plot. Functionally, this logarithmic transformation of the external signal condenses the range of the input significantly, as required if the cell is to function in a wide range of environmental conditions with only a finite number of memory (methylation) levels. This desirable ability of logarithmic tracking may provide a clue as to why E. coli has evolved to have the observed linear methylation energy.
Chemotaxis Pathway as a Signal Processor: A Lowpass Filter for the Derivative of the Signal.
At the systems level, all biological signaling pathways can be considered as signal processors designed to obtain useful information from a noisy background, e.g., refs. 32 and 33. Based on their measured dependence of the amplitude of the motor response on the frequencies of the exponentiated sine-wave stimuli (the Bode plot), Block et al. (21) described the bacterial chemotaxis system as a highpass filter of the signal itself. Indeed, for high-frequency signals, adaptation is too slow to process the signal and the kinase response simply follows the signal. However, from the linear analysis and the simulation of the full nonlinear model (Figs. 3 and 4), it is clear now that for the low-frequency signal the system calculates the derivative of the signal, as shown from both the amplitude and the phase of the response. Because the desired function of the chemotaxis pathway is gradient sensing, it is important to emphasize that the bacterial chemotaxis signal transduction pathway is a lowpass filter for the time derivative of the signal. We believe that both the direct sensing in the high-frequency regime and the gradient sensing in the low-frequency regime as described by Eq. 10 are necessary for bacterial chemotaxis in different environments. It would be interesting to study how the adaptation time scale τm, which separates the two regimes, affects the cell's average drift velocity in different spatial ligand profiles.
Besides the upper bound for the direct sensing regime caused by the dephosphorylation kinetics considered in this article, there could also be a lower bound for the gradient sensing regime, resulting from the finite precision of adaptation. Intuitively, any inaccuracy in the memory of the cell caused by imperfect adaptation introduces errors in the cell's computation of the derivative. This eventually destroys the cell's gradient-sensing ability for low-frequency signals, where the gradient is small. Quantitatively, the effect of imperfect adaptation can be modeled (phenomenologically) by including a term −βln[L] (for [L] ≫ K) in the general methylation rate function F(a, m, [L]) with a small constant β, which leads to a weak dependence of the adapted activity on the ligand concentration aadp ≈ a0 + β/F′(a0)ln [L]. A simple spectral analysis of this modified kinetic equation results in a small frequency cutoff νc ≈ αβ/2π in the kinase response function. Measurement of this low cutoff can be used to test our model and quantitatively determine the precision of adaptation.
The Slope of F(a) at a = a0: The Relaxation Dynamics Near Steady State.
The net methylation rate function F(a) in our model describes the receptor methylation/demethylation reactions facilitated by the enzymes CheR/CheB, respectively. From our study, F(a) can be determined from the exponential ramp experiments. In ref. 21, the activity shifts were found to be extremely small for a range of small ramp rates, implying that the methylation rate function F(a) is steep near the preferred activity a0. On the microscopic side, assuming that CheR and CheB only act on inactive or active receptors respectively, F(a) can be written as: F(a) = kcatR[R](1 − a)/(1 − a + KMR) − kcatB[B]a/(a + KMB), where [R] and [B] are the CheR and CheB concentrations, kcatR(B) and KMR(B) are the catalytic rates and the Michaelis Menten constants for CheR and CheB, respectively (normalized by to the total receptor concentration to match the nondimensional units in which a is measured). This form of F(a) yields a flat region near a0 when the values of KMR and KMB are small, as pointed out recently in ref. 23. We note, however, that this view is at odds with the results of the ramp responses measured in ref. 21, where the response thresholds observed at small ramp rates imply a steep region in F(a), according to our analysis.
What could increase the steepness of F(a) near a0? One possible mechanism is the phosphorylation of CheB in competition with CheY. CheB only becomes active (in terms of its demethylation activity) when phosphorylated, and CheB competes with CheY for the same histidine kinase CheA (34). However, CheB is at a considerable disadvantage because it is outnumbered by CheY by 30-fold (35) and it binds less strongly with CheA than CheY (34). Because CheY gets phosphorylated more readily than CheB, the amount of CheB-P is small at small receptor kinase activity a. As the receptor kinase activity increases and CheY-P becomes saturated, the extra CheA-P will phosphorylate CheB. Because of the small CheB concentration (compared with both CheY and CheA) and its slow autodephosphorylation rate (as compared with CheY-P dephosphorylation by CheZ), CheB-P level could increase abruptly and become saturated in a small window of a near a0, which can increase the steepness of F(a) near a0.
But the experiments of ref. 21 were based on motor responses of relatively few cells. Given that cell-to-cell variability is expected to be significant (36), it would be useful to extend this work by measuring the in vivo kinase activity (the CheY-P level) directly by FRET, using the methods of ref. 2.
Supplementary Material
Acknowledgments.
We thank the referees and Ady Vaknin for valuable suggestions and Victor Sourjik and Junhua Yuan for critical reading of the manuscript. Y.T. thanks Dr. Jeremy Rice for interesting discussions on control theory. The work was partially funded by National Institutes of Health Grants GM081747 (to Y.T.) and AI016478 (to H.C.B.) and National Institutes of Health Postdoctoral Fellowship AI063747 (to T.S.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807569105/DCSupplemental.
References
- 1.Larsen SH, Reader RW, Kort EN, Tso WW, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature. 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 2.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bren A, Eisenbach M. How signals are heard during bacterial chemotaxis: Protein–protein interactions in sensory signal propagation. J Bacteriol. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Morton-Firth CJ, Simizu TS, Bray D. A free-energy-based stochastic simulation of the tar receptor complex. J Mol Biol. 1999;286:1059–1074. doi: 10.1006/jmbi.1999.2535. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu TS, Aksenov SV, Bray D. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J Mol Biol. 2003;329:291–309. doi: 10.1016/s0022-2836(03)00437-6. [DOI] [PubMed] [Google Scholar]
- 8.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the E. coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 9.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 10.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 11.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 12.Rao CV, Frenklach M, Arkin AP. An allosteric model for transmembrane signaling in bacterial chemotaxis. J Mol Biol. 2004;343:291–303. doi: 10.1016/j.jmb.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 13.Mello BA, Tu Y. An allosteric model for heterogeneous receptor complexes: Understanding bacterial chemotaxis response to multiple stimuli. Proc Natl Acad Sci USA. 2005;102:17354–17359. doi: 10.1073/pnas.0506961102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in E. coli: Two regimes in two-state receptors. Proc Natl Acad Sci USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mello BA, Tu Y. Effects of adaptation in maintaining high sensitivity over a wide range of backgrounds for E. coli chemotaxis. Biophys J. 2007;92:2329–2337. doi: 10.1529/biophysj.106.097808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg HC, Brown DA. Chemotaxis in E. coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 18.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 19.Yi T-M, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mello BA, Tu Y. Perfect and near perfect adaptation in a model of bacterial chemotaxis. Biophys J. 2003;84:2943–2956. doi: 10.1016/S0006-3495(03)70021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Block SM, Segall JE, Berg HC. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 23.Emonet T, Cluzel P. Relation between cellular response and behavioral variability in bacterial chemotaxis. Proc Natl Acad Sci USA. 2008;105:3304–3309. doi: 10.1073/pnas.0705463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Block SM, Segall JE, Berg HC. Impulse responses in bacterial chemotaxis. Cell. 1982;83:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 25.Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sourjik V, Berg HC. Binding of the E. coli response regulator CheY to its target is measured in vivo by fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spudich JL, Koshland DE. Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acac Sci USA. 1975;72:710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg HC, Tedesco PM. Transient response to chemotactic stimuli in E. coli. Proc Natl Acad Sci USA. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesibov R, Ordal G, Adler J. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. J Gen Physiol. 1973;62:203–223. doi: 10.1085/jgp.62.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown DA, Berg HC. Temporal stimulation of chemotaxis in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:1388–1392. doi: 10.1073/pnas.71.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu TS, Delalez N, Pichler K, Berg H. Monitoring bacterial chemotaxis by bio-luminescence resonance energy transfer: Absence of feedback from the flagellar motor. Proc Natl Acad Sci USA. 2006;103:2093–2097. doi: 10.1073/pnas.0510958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews BW, Yi T-M, Iglesias PA. Optimal noise filtering in the chemotactic response of Escherichia coli. PLoS Comput Biol. 2006;2:e154. doi: 10.1371/journal.pcbi.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi T-M, Andrews BW, Iglesias PA. Control analysis of bacterial chemotaxis signaling. Methods Enzymol. 2007;422:123–140. doi: 10.1016/S0076-6879(06)22006-8. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Swanson RV, Simon MI, Weis RM. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry. 1995;34:14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korobkova E, Emonet T, Vilar JMG, Shimizu TS, Cluzel P. From molecular noise to behavioral variability in a single bacterium. Nature. 2004;428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.