Abstract
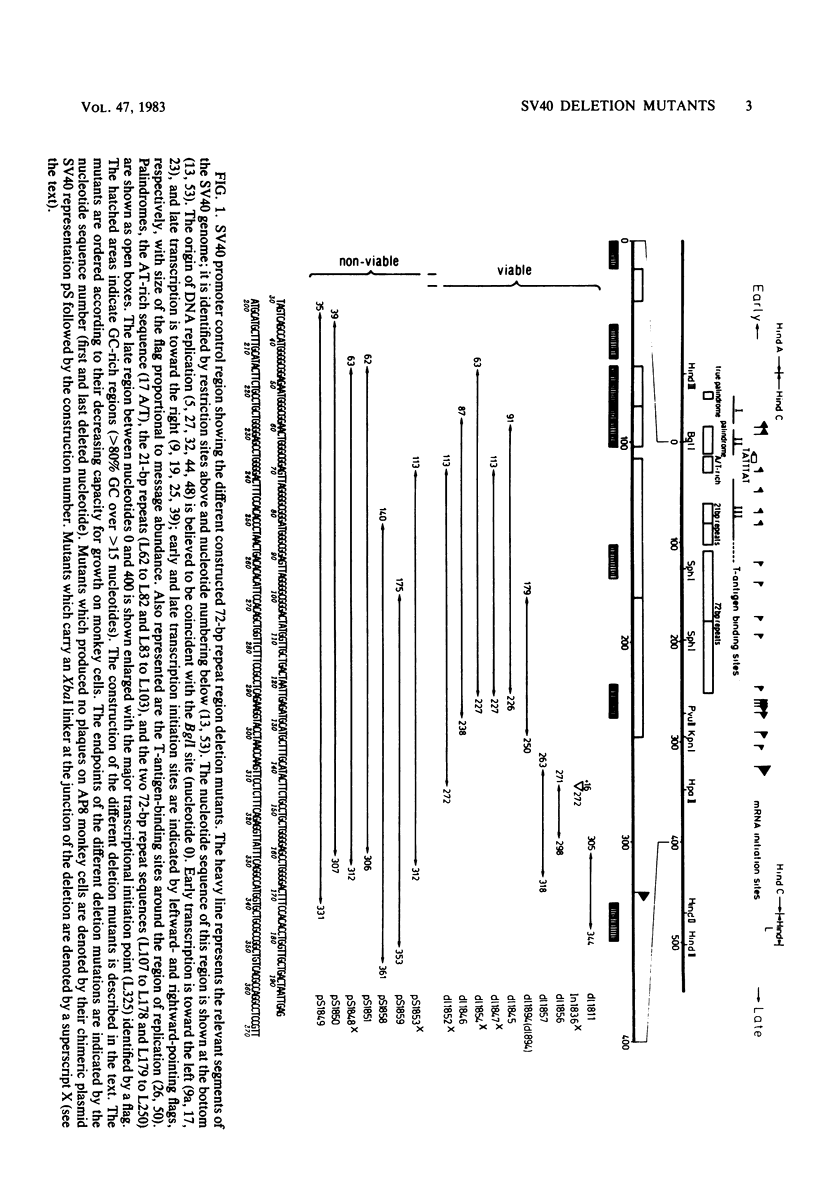
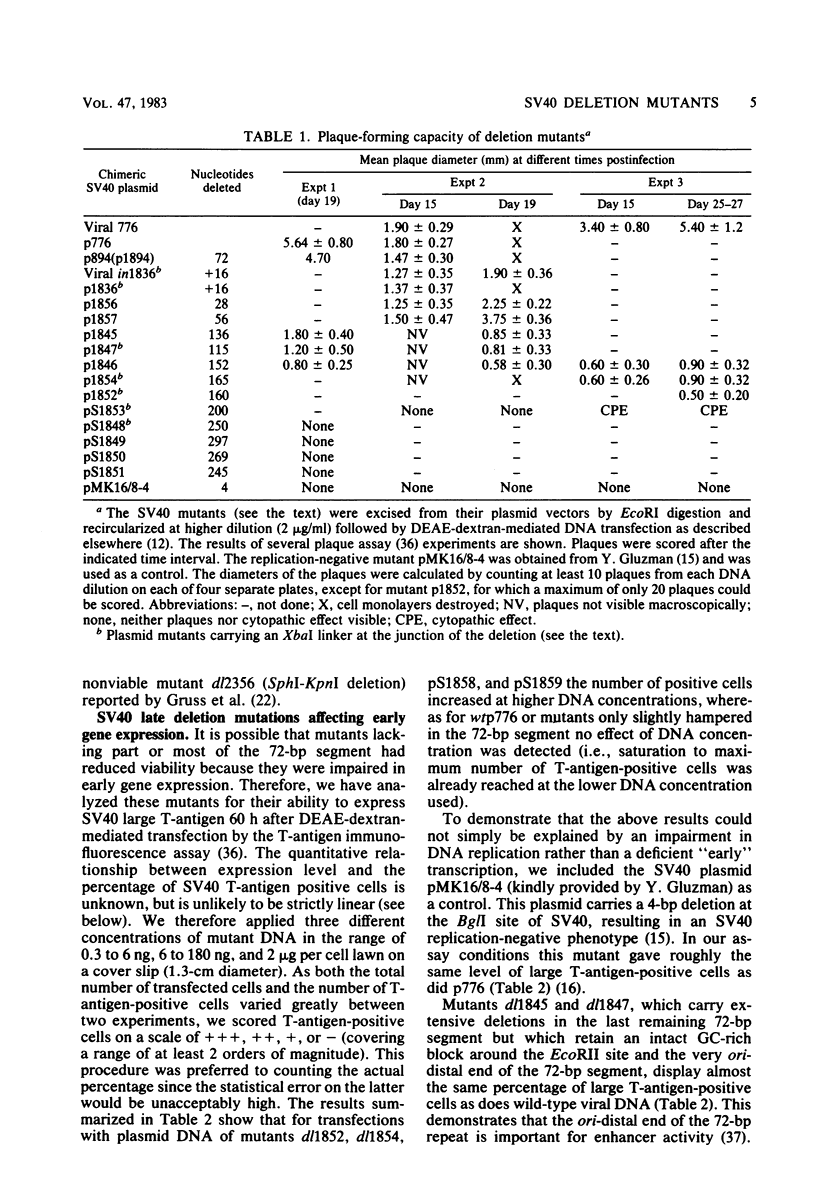
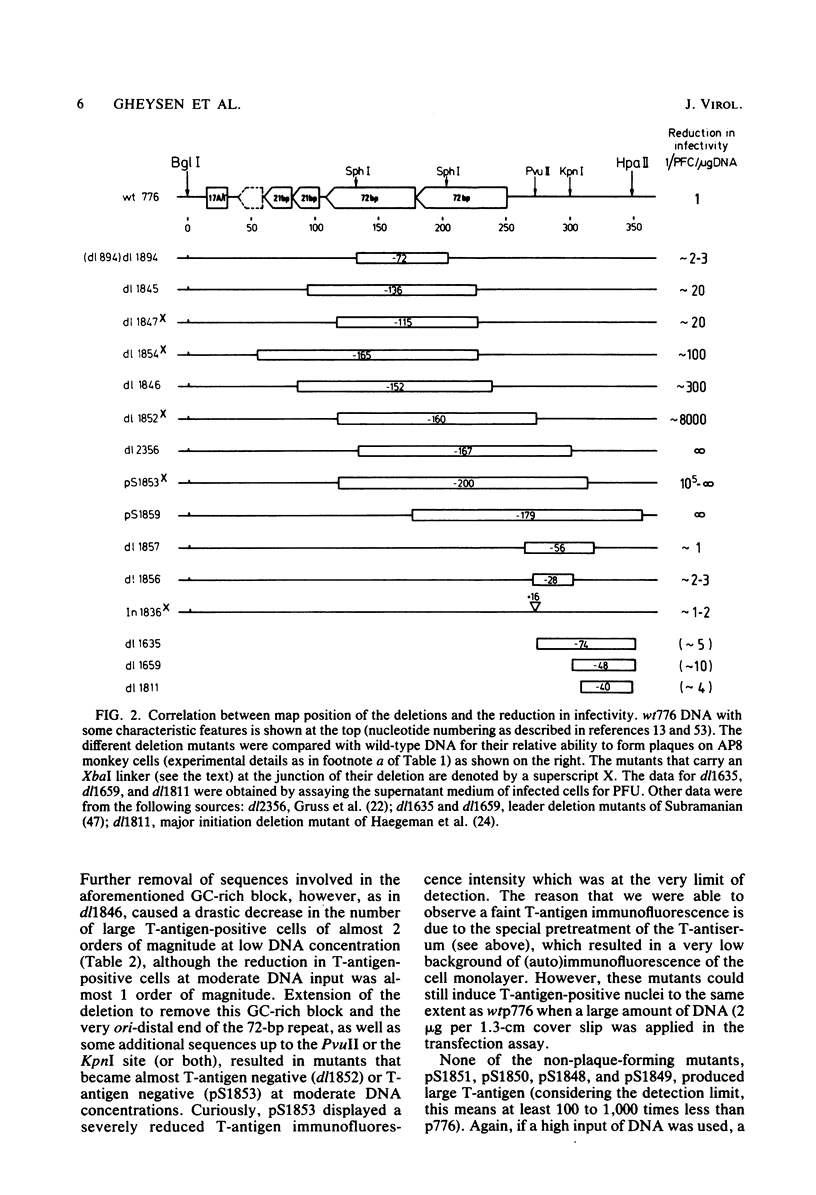
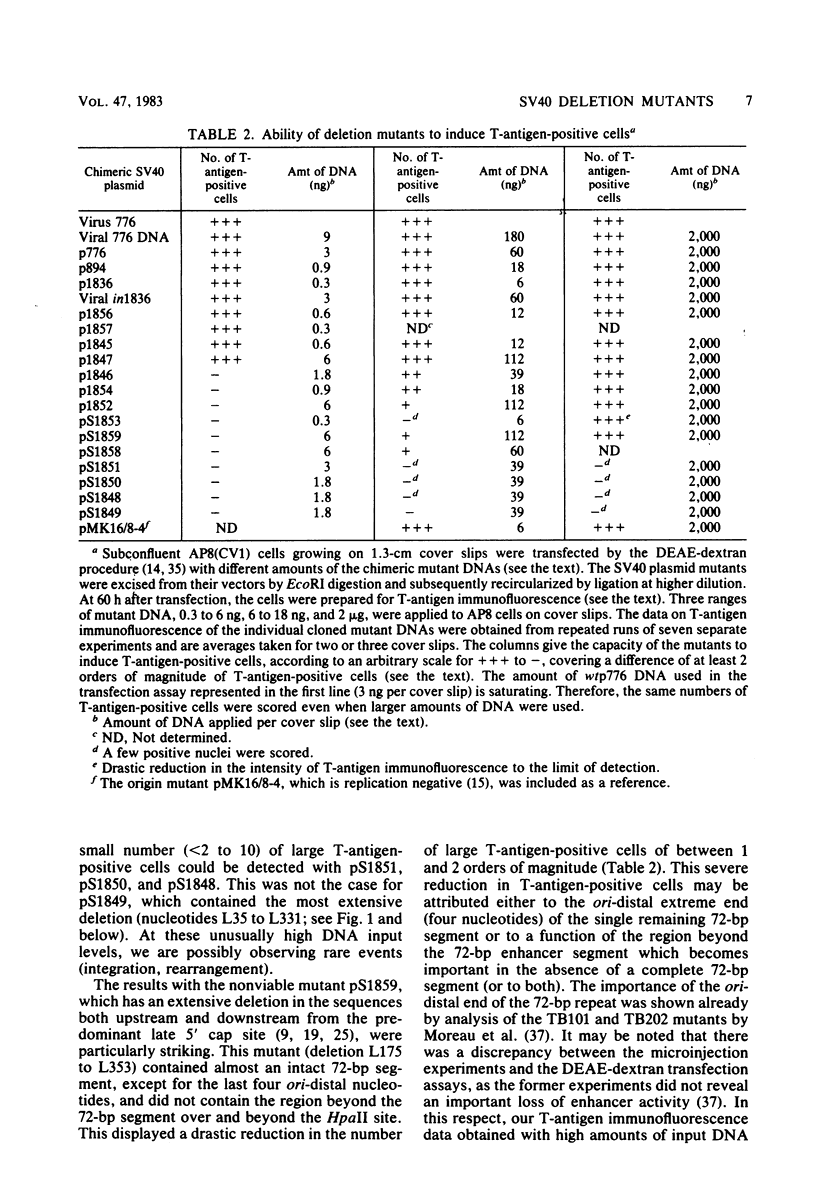
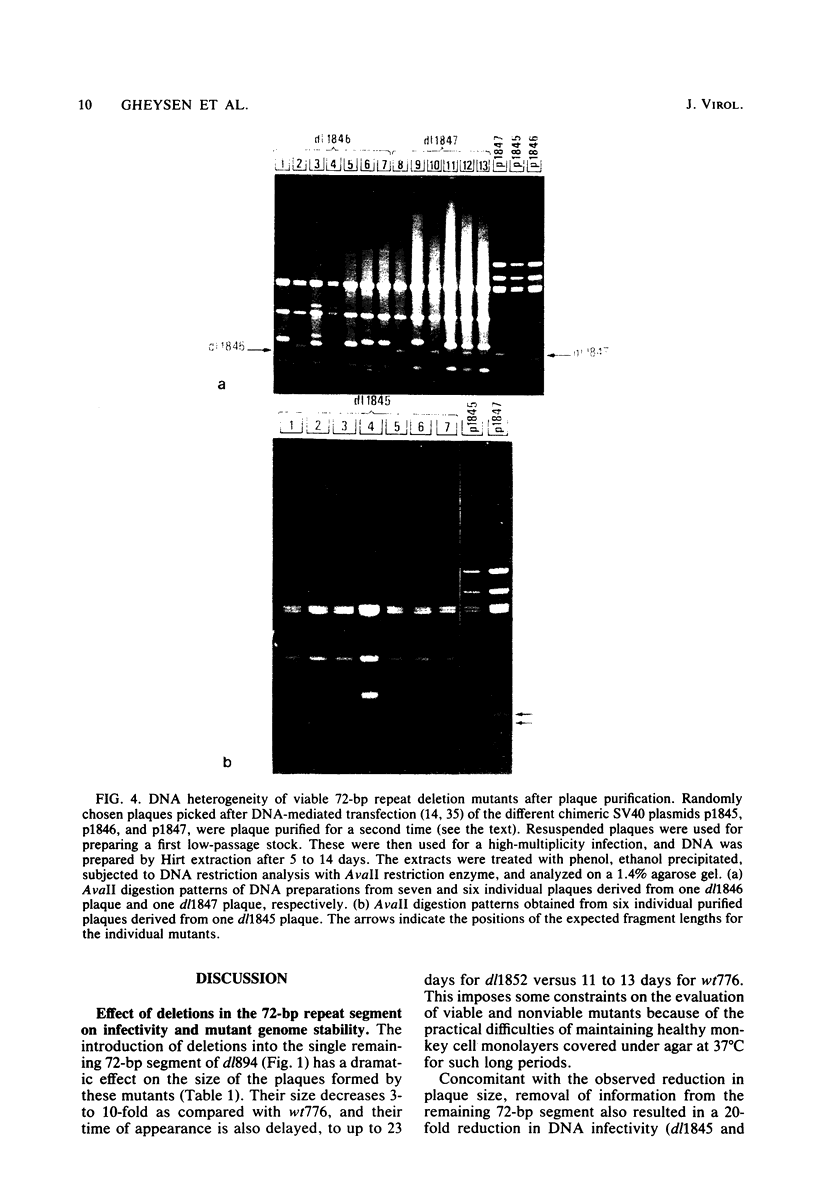
Simian virus 40 mutants were constructed with deletions at the late side of the origin of DNA replication by partial Bal 31 digestion at the SphI site or at the PvuII site. Some of these mutants lost virtually all of both 72-base-pair repeat segments ("enhancer" sequences) and exhibited a decrease in viability from 20-to 300-fold; one particular mutant, dl1852, even showed a reduction of almost 10(4)-fold. The very poorly growing deletion mutants were unstable and gave rise to DNA rearrangements upon further growth. An essential region for viability, at least in the absence of a 72-base-pair repeat, was revealed at the distal side of the 72-base-pair elements (L250 through L272). The effect of the deletions on T-antigen expression was measured, and the decreased viability of the mutants correlated with the impairment of T-antigen expression in all cases. The study of these mutants also revealed that the 72-base-pair repeats are not required for late transcription.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Barkan A., Mertz J. E. DNA sequence analysis of simian virus 40 mutants with deletions mapping in the leader region of the late viral mRNA's: mutants with deletions similar in size and position exhibit varied phenotypes. J Virol. 1981 Feb;37(2):730–737. doi: 10.1128/jvi.37.2.730-737.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. Deletions covering the putative promoter region of early mRNAs of simian virus 40 do not abolish T-antigen expression. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3865–3869. doi: 10.1073/pnas.77.7.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler-White A. J., Humphrey G. W., Pigiet V. Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell. 1980 Nov;22(1 Pt 1):37–46. doi: 10.1016/0092-8674(80)90152-x. [DOI] [PubMed] [Google Scholar]
- Contreras R., Fiers W. Initiation of transcription by RNA polymerase II in permeable, SV40-infected or noninfected, CVI cells; evidence for multiple promoters of SV40 late transcription. Nucleic Acids Res. 1981 Jan 24;9(2):215–236. doi: 10.1093/nar/9.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Gheysen D., Knowland J., van de Voorde A., Fiers W. Evidence for the direct involvement of DNA replication origin in synthesis of late SV40 RNA. Nature. 1982 Dec 9;300(5892):500–505. doi: 10.1038/300500a0. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Mantei N., Weissmann C. DNA sequences preceding the rabbit beta-globin gene are required for formation in mouse L cells of beta-globin RNA with the correct 5' terminus. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1411–1415. doi: 10.1073/pnas.78.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Gheysen D., Fiers W. Expression and excretion of human fibroblast beta 1 interferon in monkey cells after transfection with a recombinant SV40 plasmid vector. J Mol Appl Genet. 1982;1(5):385–394. [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P. Simian virus 40 early mRNA's contain multiple 5' termini upstream and downstream from a Hogness-Goldberg sequence; a shift in 5' termini during the lytic cycle is mediated by large T antigen. J Virol. 1981 Oct;40(1):224–240. doi: 10.1128/jvi.40.1.224-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Frisque R. J., Sambrook J. Origin-defective mutants of SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):293–300. doi: 10.1101/sqb.1980.044.01.033. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Shewmaker C. K., Jat P., Flavell R. A. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981 Jul;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman G., Fiers W. Characterization of the 5'-terminal cap structures of early simian virus 40 mRNA. J Virol. 1980 Sep;35(3):955–961. doi: 10.1128/jvi.35.3.955-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman G., Iserentant D., Gheysen D., Fiers W. Characterization of the major altered leader sequence of late mRNA induced by SV40 deletion mutant d1-1811. Nucleic Acids Res. 1979 Dec 11;7(7):1799–1814. doi: 10.1093/nar/7.7.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman G., van Heuverswyn H., Gheysen D., Fiers W. Heterogeneity of the 5' terminus of late mRNA induced by a viable simian virus 40 deletion mutant. J Virol. 1979 Aug;31(2):484–493. doi: 10.1128/jvi.31.2.484-493.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Tenen D. G., Livingston D. M., Sharp P. A. T antigen repression of SV40 early transcription from two promoters. Cell. 1981 Dec;27(3 Pt 2):603–613. doi: 10.1016/0092-8674(81)90402-5. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., McCready S. J., Cook P. R. RNA is synthesized at the nuclear cage. Nature. 1981 Aug 6;292(5823):552–555. doi: 10.1038/292552a0. [DOI] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Martin R. G., Setlow V. P. The initiation of SV40 DNA synthesis is not unique to the replication origin. Cell. 1980 Jun;20(2):381–391. doi: 10.1016/0092-8674(80)90624-8. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Rio D. C., Robbins A. K., Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981 Aug;25(2):373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- Piatak M., Subramanian K. N., Roy P., Weissman S. M. Late messenger RNA production by viable simian virus 40 mutants with deletions in the leader region. J Mol Biol. 1981 Dec 15;153(3):589–618. doi: 10.1016/0022-2836(81)90409-5. [DOI] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Regulatory mutants of simian virus 40: constructed mutants with base substitutions at the origin of DNA replication. J Mol Biol. 1979 Jul 15;131(4):801–817. doi: 10.1016/0022-2836(79)90202-x. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Subramanian K. N. Segments of simian virus 40 DNA spanning most of the leader sequence of the major late viral messenger RNA are dispensable. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2556–2560. doi: 10.1073/pnas.76.6.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N., Shenk T. Definition of the boundaries of the origin of DNA replication in simian virus 40. Nucleic Acids Res. 1978 Oct;5(10):3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier J., Derynck R., Fiers W. Evidence for a unique human fibroblast interferon (IFN-beta 1) chromosomal gene, devoid of intervening sequences. Nucleic Acids Res. 1981 Feb 11;9(3):461–471. doi: 10.1093/nar/9.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]