Abstract
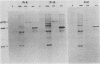
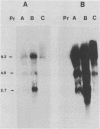
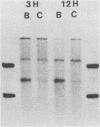
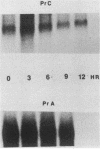
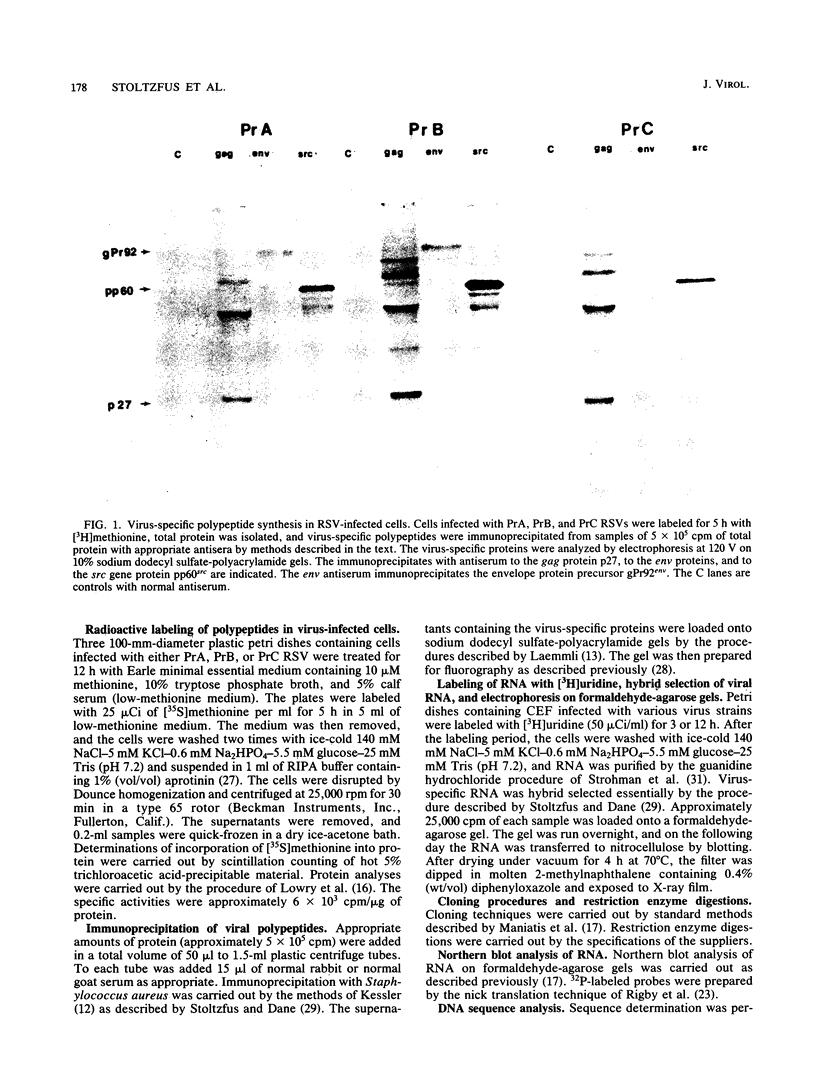
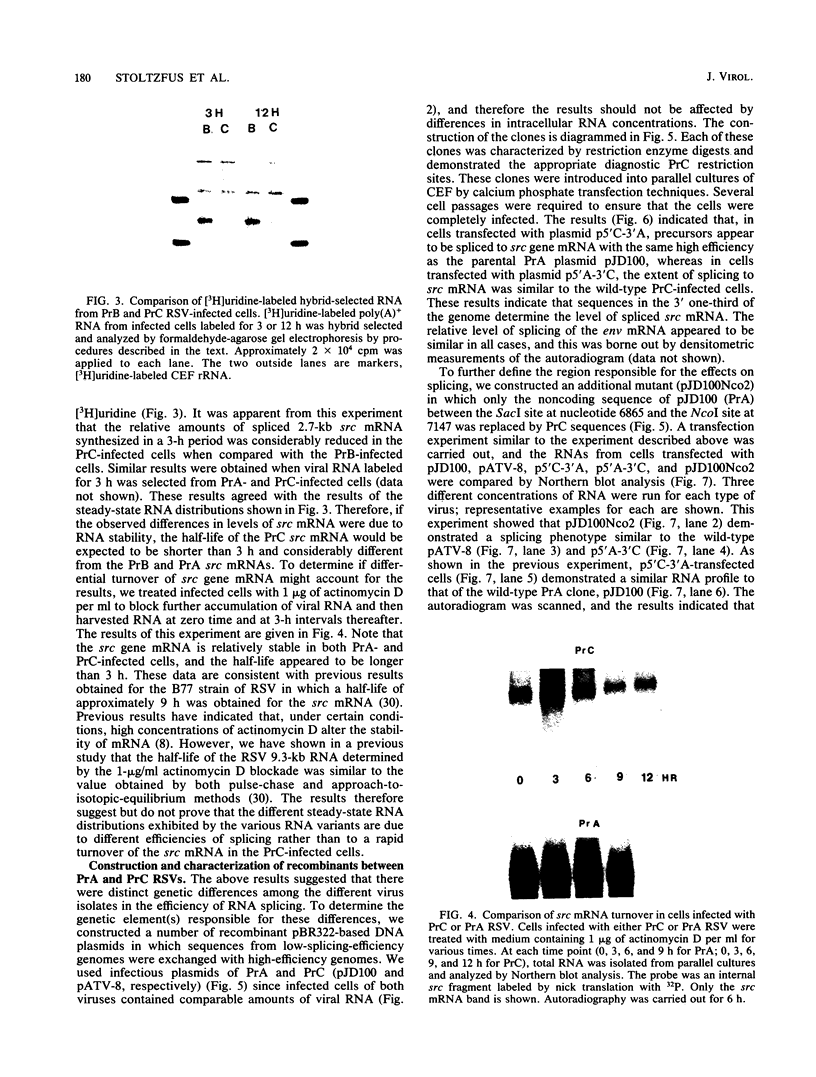
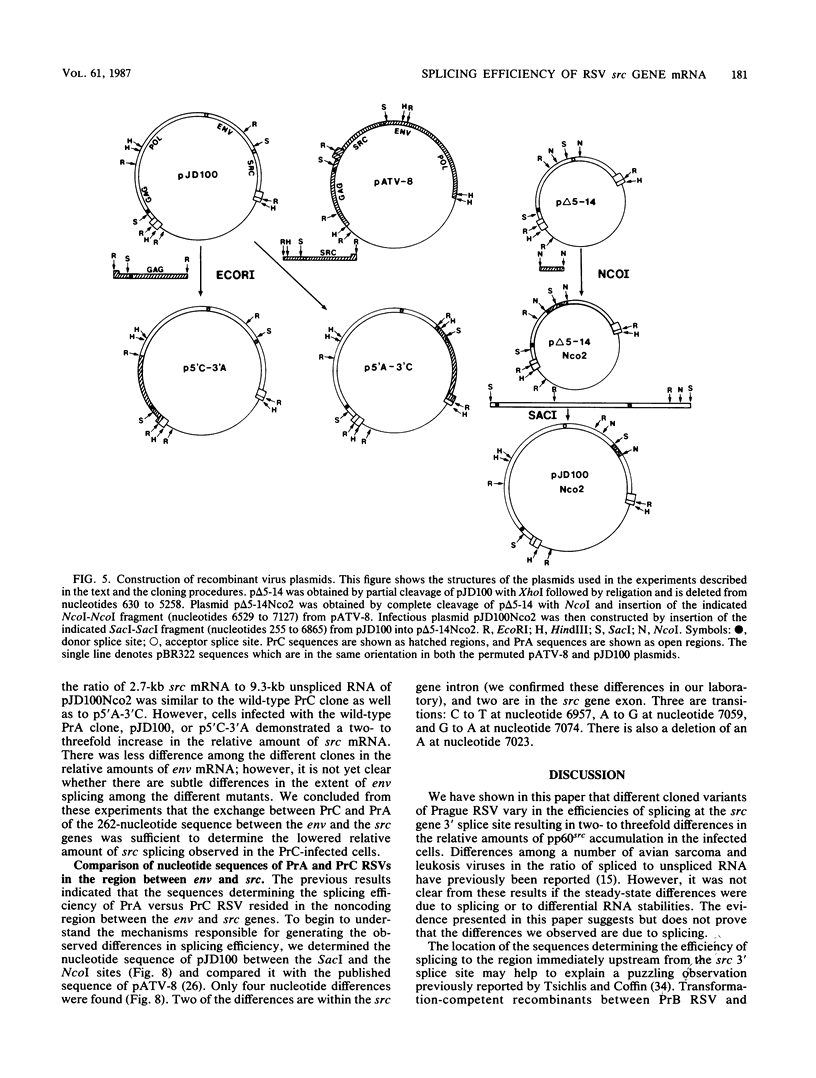
Viral RNA and proteins in chicken embryo fibroblasts infected with different cloned variants of the Prague strain Rous sarcoma virus (RSV) were analyzed. The ratio of immunoprecipitated pp60src to the gag gene product p27 in Prague A (PrA) and Prague B (PrB) RSV-infected cells was two to three times that in Prague C (PrC) RSV-infected cells. A significant increase in the steady-state ratio of spliced 2.7-kilobase src gene mRNA to unspliced 9.3-kilobase genome-size RNA was observed in PrA- and PrB- compared with PrC-infected cells, consistent with the differences in the ratios of the gag to src gene protein products. Similar results were obtained when hybrid-selected RNA, which had been labeled for 3 h with [3H]uridine, was analyzed on formaldehyde-agarose gels, suggesting that the observed differences were due to splicing rather than RNA stability. Recombinant plasmids from infectious molecular clones of PrA and PrC were constructed to localize the regions responsible for the effects on src gene splicing. The substitution in place of the corresponding PrA region of the 262-base-pair region between the env gene and the src gene coding sequences from the PrC clone into the infectious PrA plasmid conferred the low src splicing efficiency of the PrC strain. The nucleotide sequence of this region of the PrA plasmid was determined and compared with the sequence of the PrC strain. Only four nucleotide differences were found; two changes were within the intron sequence, and two were in the exon sequence. The possible role of these differences in determining the extent of viral RNA splicing is discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang L. J., Stoltzfus C. M. Cloning and nucleotide sequences of cDNAs spanning the splice junctions of Rous sarcoma virus mRNAs. J Virol. 1985 Mar;53(3):969–972. doi: 10.1128/jvi.53.3.969-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czernilofsky A. P., Levinson A. D., Varmus H. E., Bishop J. M., Tischer E., Goodman H. Corrections to the nucleotide sequence of the src gene of Rous sarcoma virus. Nature. 1983 Feb 24;301(5902):736–738. doi: 10.1038/301736b0. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Spahr P. F. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J Mol Biol. 1982 Sep 15;160(2):147–161. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- Dimock K., Stoltzfus C. M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977 Feb 8;16(3):471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Ficht T. A., Chang L. J., Stoltzfus C. M. Avian sarcoma virus gag and env gene structural protein precursors contain a common amino-terminal sequence. Proc Natl Acad Sci U S A. 1984 Jan;81(2):362–366. doi: 10.1073/pnas.81.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hackett P. B., Swanstrom R., Varmus H. E., Bishop J. M. The leader sequence of the subgenomic mRNA's of Rous sarcoma virus is approximately 390 nucleotides. J Virol. 1982 Feb;41(2):527–534. doi: 10.1128/jvi.41.2.527-534.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman R. C., Penman S. Multiple decay rates of heterogeneous nuclear RNA in HeLa cells. Biochemistry. 1977 Jul 26;16(15):3460–3465. doi: 10.1021/bi00634a026. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol. 1985 Sep;5(9):2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Omer C. A., Weis J. H., Mitsialis S. A., Faras A. J., Guntaka R. V. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J Virol. 1982 Apr;42(1):346–351. doi: 10.1128/jvi.42.1.346-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leis J. P., McGinnis J., Green R. W. Rous sarcoma virus p19 binds to specific double-stranded regions of viral RNA: effect of p19 on cleavage of viral RNA by RNase III. Virology. 1978 Jan;84(1):87–98. doi: 10.1016/0042-6822(78)90220-9. [DOI] [PubMed] [Google Scholar]
- Leis J. P., Scheible P., Smith R. E. Correlation of RNA binding affinity of avian oncornavirus p19 proteins with the extent of processing of virus genome RNA in cells. J Virol. 1980 Sep;35(3):722–731. doi: 10.1128/jvi.35.3.722-731.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. Insertion of several different DNAs in reticuloendotheliosis virus strain T suppresses transformation by reducing the amount of subgenomic mRNA. J Virol. 1986 Apr;58(1):75–80. doi: 10.1128/jvi.58.1.75-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méric C., Darlix J. L., Spahr P. F. It is Rous sarcoma virus protein P12 and not P19 that binds tightly to Rous sarcoma virus RNA. J Mol Biol. 1984 Mar 15;173(4):531–538. doi: 10.1016/0022-2836(84)90396-6. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Sefton B. M., Beemon K., Hunter T. Comparison of the expression of the src gene of Rous sarcoma virus in vitro and in vivo. J Virol. 1978 Dec;28(3):957–971. doi: 10.1128/jvi.28.3.957-971.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dane R. W. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. J Virol. 1982 Jun;42(3):918–931. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus C. M., Dimock K., Horikami S., Ficht T. A. Stabilities of avian sarcoma virus RNAs: comparison of subgenomic and genomic species with cellular mRNAs. J Gen Virol. 1983 Oct;64(Pt 10):2191–2202. doi: 10.1099/0022-1317-64-10-2191. [DOI] [PubMed] [Google Scholar]
- Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977 Feb;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Parker R. C., Varmus H. E., Bishop J. M. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1983 May;80(9):2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya T., Hanafusa H. Structure and sequence of the cellular gene homologous to the RSV src gene and the mechanism for generating the transforming virus. Cell. 1983 Mar;32(3):881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Coffin J. M. Recombinants between endogenous and exogenous avian tumor viruses: role of the C region and other portions of the genome in the control of replication and transformation. J Virol. 1980 Jan;33(1):238–249. doi: 10.1128/jvi.33.1.238-249.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]