Abstract
The color patterns on the wings of butterflies have been an important model system in evolutionary developmental biology. A recent computational model tested genetic regulatory hierarchies hypothesized to underlie the formation of butterfly eyespot foci (Evans and Marcus, 2006). The computational model demonstrated that one proposed hierarchy was incapable of reproducing the known patterns of gene expression associated with eyespot focus determination in wild-type butterflies, but that two slightly modified alternative hierarchies were capable of reproducing all of the known gene expressions patterns. Here we extend the computational models previously implemented in Delphi 2.0 to two mutants derived from the squinting bush brown butterfly (Bicyclus anynana). These two mutants, comet and Cyclops, have aberrantly shaped eyespot foci that are produced by different mechanisms. The comet mutation appears to produce a modified interaction between the wing margin and the eyespot focus that results in a series of comet-shaped eyespot foci. The Cyclops mutation causes the failure of wing vein formation between two adjacent wing-cells and the fusion of two adjacent eyespot foci to form a single large elongated focus in their place. The computational approach to modeling pattern formation in these mutants allows us to make predictions about patterns of gene expression, which are largely unstudied in butterfly mutants. It also suggests a critical experiment that will allow us to distinguish between two hypothesized genetic regulatory hierarchies that may underlie all butterfly eyespot foci.
Keywords: butterfly eyespot, genetic regulatory hierarchy, computational models, Bicyclus anynana, wing patterns
1. Introduction
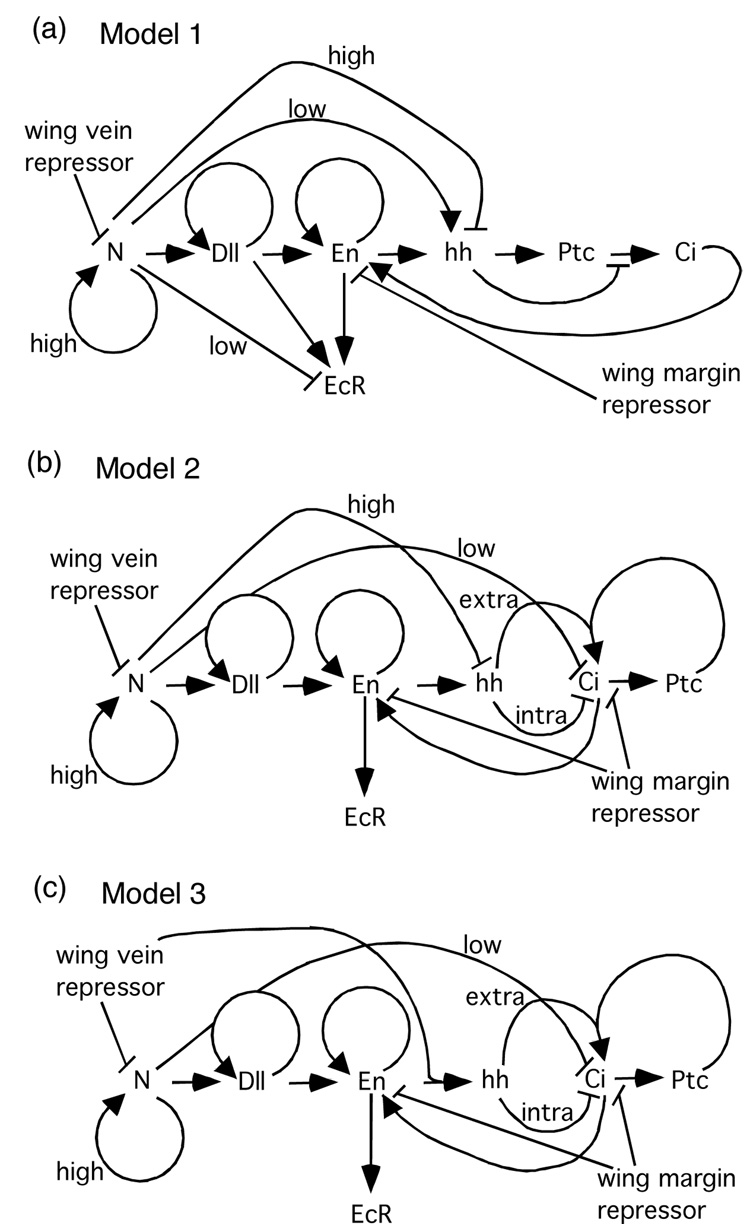
Butterfly wing color patterns are an attractive model system for exploring the relationship between developmental genetics and evolution. Color patterns are suitable for study because they are highly variable, consist of clearly defined subunits, exist in two dimensions, and are structurally simple (Beldade and Brakefield, 2002; Nijhout, 1991), and at least some patterns are clearly associated with fitness benefits associated with natural or sexual selection (Hill and Vaca, 2004; Kingsolver, 1995; Robertson and Monteiro, 2005; Stevens, 2005). Recently, we implemented a model for color pattern development that combined gene expression data from developing wings with computational algorithms that until that point had only been used to model generalized mechanisms of pattern development in butterflies (Evans and Marcus, 2006). This computational approach revealed that a previously proposed hypothesis for the genetic regulatory network underlying eyespot development (Marcus, 2005) was flawed, but revealed two alternative networks that were capable of producing all of the gene expression patterns known from wild-type pre-pupal butterfly eyespots (Fig. 1; Evans and Marcus, 2006).
Fig. 1.
(a) Diagram of Model 1 genetic regulatory network for eyespot focus determination falsified by Evans and Marcus (2006) because it failed to reproduce several known gene expression pattersn. Models of alternative regulatory network models that can reproduce these patterns are shown in (b) and (c). Genetic symbols: N—Notch, Dll—Distal-less, En—Engrailed, hh—hedgehog, Ptc—Patched, Ci—Cubitus interruptus, EcR—Ecdysone Receptor.
Butterfly color pattern mutations and aberrations have played an important role in the development of evolutionary theory (Marcus, et al., ms.). Such mutations have been key examples in discussions of diverse phenomena including industrial melanism (Kettlewell, 1973; Majerus, 1998), the evolution of supergenes (Ford, 1971), the role of veination in color pattern formation (Koch and Nijhout, 2002; Nijhout, 1991), and the identification of wing compartment boundaries (Blanchard and Descimon, 1988; Sibatani, 1980). Their prominence is such that many Lepidopteran aberrations have been named individually.
In spite of this prominence, the incorporation of the study of butterfly mutants into the study of color pattern development is still in its infancy. Several different approaches have been taken thus far. First is the characterization of gene expression patterns in existing spontaneous mutations (Brakefield, et al., 1996; Weatherbee, et al., 1999). Such studies have lead to important insights into the organization of the genetic regulatory hierarchy that underlies eyespot development (Nijhout, 1996). Second is the deliberate generation of new mutations in the laboratory to identify genes that may play a role in color pattern development (Marcus, et al., 2004; Monteiro, et al., 2003; Ramos and Monteiro, 2007). Finally, some studies have taken a modeling approach using computational techniques that implement generalized mechanisms of pattern development to reproduce mutant phenotypes (Dilao and Sainhas, 2004; Koch and Nijhout, 2002; Sekimura, et al., 2000). However, since it is currently very difficult to characterize spontaneous butterfly mutants at the molecular level, obtaining a mechanistic understanding of the role that these genes play in color pattern development has been elusive.
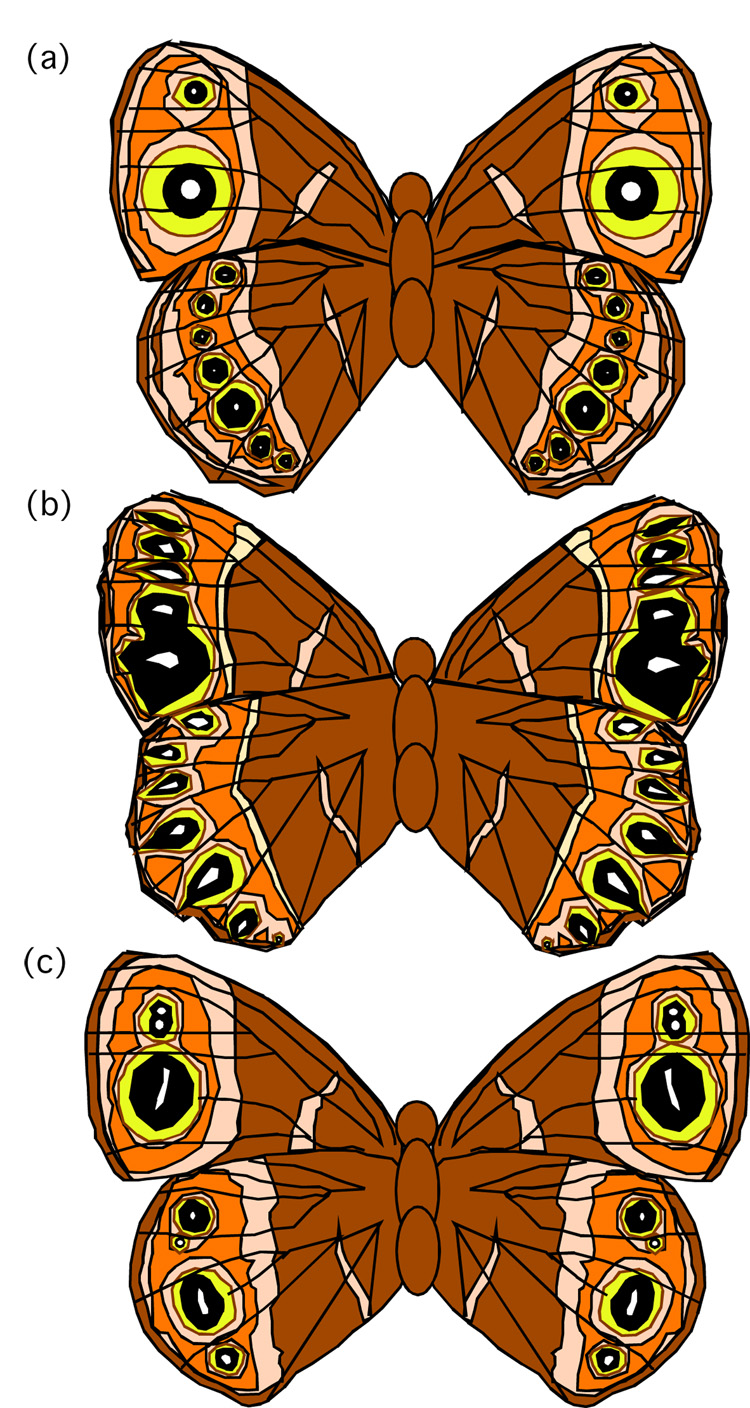
It therefore seemed appropriate for us to extend our computational modeling of gene expression involved in eyespot focus specification to include color pattern mutants, as has been done in other systems (Sharp and Reinitz, 1998). While there are many butterfly color pattern mutants that have been described, many of them were unsuitable for our models because they either affected color patterns other than eyespots (e.g. rosa (Rountree and Nijhout, 1995), Hindsight (Nijhout and Rountree, 1995; Weatherbee, et al., 1999); in these cases our models would be uninformative), they primarily affected numbers of wing-cells that produce eyespots (e.g. Spotty, Missing, 3+4 (Monteiro, et al., 2003); processes that occur prior to the developmental stage when our computational model begins), or they affected the size of the eyespot or its color composition without changing its shape or basic structure (e.g. Goldeneye, Bigeye (Beldade and Brakefield, 2002)); such phenotypes that are generated after eyespot foci are specified and after the developmental stage where our model ends). We selected two squinting bush brown butterfly (Bicyclus anynana) mutants that produce altered eyespot focus shapes as being most appropriate for our analysis: comet and Cyclops (Fig. 2).
Fig. 2.
Ventral wing surfaces of (a) a wild-type squinting bush brown butterfly (Bicyclus anynana), (b) a comet mutant, (c) a Cyclops mutant (Brakefield, 2001; Brakefield, et al., 1996).
The comet mutation appears to produce a modified interaction between factors secreted by the wing margin and the genetic regulatory networks responsible for producing parfocal pattern elements (border chevrons and bands) and eyespot foci (Brakefield, 2001). In homozygotes, the eyespot foci are transformed from circular structures into a series of comet or tear drop-shaped eyespot foci. The dominant Cyclops mutation causes the failure of wing vein formation between two adjacent wing-cells and the subsequent fusion of two adjacent eyespot foci to form a single large elongated focus in heterozygotes (Brakefields, et al., 1996). Homozygous Cyclops mutants are lethal (Brakefield, 2001).
We expanded the computational model first presented in Evans and Marcus (2006) to incorporate the changes associated with these mutant phenotypes. This computational approach to modeling pattern formation in these mutants allows us to make predictions about patterns of gene expression, which with few exceptions (Brakefield, et al., 1996; Weatherbee, et al., 1999) are largely unstudied in butterfly mutants. It also suggests a critical experiment that will allow us to distinguish between two hypothesized genetic regulatory hierarchies that may underlie all butterfly eyespot foci.
2. Materials and method
The experimental basis and general structure of our computer simulation of the developing butterfly wing are described in detail elsewhere (Evans and Marcus, 2006). Briefly, we implemented a set of gene expression threshold equations in a PASCAL application written within the Delphi 2.0 (1996) programming environment in order to place the alternative models for the genetic regulatory hierarchy within a spatial modeling environment. The program in Delphi was written such that a large grid of thirty-five by thirty-five cytological cells, representing a wing-cell (a field of cytological cells bordered by the wing margin and by a series of wing veins) in which an eyespot focus will develop, would display gradients of the different products proposed by the genetic network. The position and relative amount of the products could then be spatially compared to the relative amounts and positions of the products found experimentally within developing butterfly wing imaginal discs.
Three factors, in the genetic model, were selected to diffuse within the wing-cell on the basis of theorized and known diffusion patterns. It has been hypothesized that an as of yet unknown repressor is secreted from the incipient wing veins which participates in patterning the wing-cell (Koch and Nijhout, 2002; Nijhout, 1991; Nijhout, 1994). Similar diffusible repressors are known to participate in wing vein patterning in Drosophila (Biehs, et al., 1998; Marcus, 2001). We hypothesize that the role of this wing vein repressor functions in a direct down-regulation of Notch, such that N is contained within a stalk-like domain of expression in the midline of the wing-cell, the terminus of which later becomes the eyespot focus (Reed and Serfas, 2004). We also hypothesized the existence of a second repressor that diffused from the wing margin, as has been described previously but of which is currently unidentified (Nijhout, 1990). Ligands that diffuse from the wing margin are also known in Drosophila (e.g. wingless; Phillips and Whittle, 1993; Blair, 1994). This wing margin repressor was utilized to define a region of Engrailed expression within the co-expression of Notch and Distal-less of the eyespot similar to that observed in developing wing imaginal discs. The hedgehog gene product is also known to diffuse (Peifer and Bejsovec, 1992), and we tracked intracellular and diffusing extracellular hedgehog, and calculated diffusion gradients for all three factors as described previously (Evans and Marcus, 2006).
Keys et al. (1999) suggested that the hedgehog regulatory circuit as expressed in the developing butterfly eyespot differs in two important ways from the same circuit in Drosophila. First, they showed via whole-mount immunohistochemistry that the Engrailed and/or invected gene products and the Cubitus interruptus gene product are co-expressed simultaneously in the cells that make up the eyespot focus (Keys, et al., 1999), suggesting that Engrailed does not suppress Cubitus interruptus in these cells as it would in Drosophila embryonic segment development (Ingham and Martinez Arias, 1992). This has been incorporated into the Evans and Marcus (2006) models. Keys et al. (1999) also used in-situ hybridization to study the expression of hedgehog mRNA, which appears to be expressed in a bracket-like pattern on either side of the center of the eyespot focus, but the degree of overlap between Engrailed/invected protein expression and hedgehog mRNA expression was not examined directly in the same samples.
The difference between Engrailed/invected protein expression and hedgehog mRNA expression in butterfly eyespots suggested to Keys et al. (1999) a second major change to the circuit, that Engrailed is not the inducer of hedgehog signaling as in Drosophila, but is rather the target of that signaling. They offer no functional data in support of this suggestion, which seems rather unlikely for three reasons. First, the Engrailed-inducing-hedgehog interaction is conserved throughout Drosophila development (Biehs, et al., 1998; Mohler and Vani, 1992) and is also highly conserved phylogenetically; the same mechanism is used in vertebrates (Ingham, 1994; Loomis, et al., 1996). Second, Keys et al. (1999) do not suggest an alternative inducer for hedgehog signaling. Without an identified alternative induction mechanism, it is very difficult to include hedgehog signaling in computational models for this system. Third, such a radical change in the genetic architecture of this highly conserved circuit is unnecessary to explain the experimental data. Rather, all that is needed is for the induction of hedgehog mRNA by Engrailed to be suppressed by high levels of another gene product in the center of the eyespot focus (such as Notch in Model 2, Fig. 1b) or for the induction of hedgehog mRNA by Engrailed to be promoted by a gene product found at the periphery of the eyespot focus (such as the wing vein repressor in Model 3, Fig. 1c). Evans and Marcus (2006) have implemented simulations of both of these alternatives.
By comparing the output of their simulations with known patterns of gene expression, Evans and Marcus (2006) were able to show that one proposed network of genetic interactions (Marcus, 2005) is almost certainly incorrect, but that two alternative networks (Models 2 and 3) were each capable of reproducing almost all known gene expression patterns (including known temporal changes) from the pre-pupal stages of butterfly eyespot development. The PASCAL source code and the executable compiled applications (for Microsoft Windows operating systems) for all of the simulations of color pattern development discussed in this paper may be accessed without restriction at http://bioweb.wku.edu/faculty/Marcus/simulations.html.
To model the effects of the comet mutation, we modified our original wild-type simulation (Evans and Marcus, 2006). The comet phenotype appears to be due to an alteration of the interactions between the wing margin and the developing color patterns (Brakefield, 2001). Since comet affects not only eyespots, but parfocal elements as well, the molecular defect is likely to be in the signals coming from the wing margin, rather than in the responses of the color patterns. In addition, the fact that comet is a recessive mutation suggests that it is a loss of function mutation, probably a hypomorph. Consequently, we focused our attention on the wing margin repressor in our model. To mimic a reduction in signaling, we reduced the amount of wing margin repressor from 1000 arbitrary units in our wild-type simulations to 10 arbitrary units in our comet simulation.
Before we could model the effects of the Cyclops mutant, which affects two adjacent wing-cells, we first had to expand our wild-type model to track two such cells simultaneously. Due to the memory limitations of the Delphi 2.0 compiler, we could not expand the graphical field of our simulation to display two full-sized wing-cells. We therefore had to shrink the field allocated to each wing-cell by half so that each was 17 by 35 pixels, with each pixel representing a cytological cell. We separated the two fields with a wing vein, one pixel wide. In order to accommodate this change in scale, we had to reduce the threshold values for the interactions between gene products in our simulations. These modifications allowed us to place two functioning wing-cells side-by-side that displayed virtually all of the salient gene expression patterns seen in our original wild-type simulations (Evans and Marcus, 2006).
To model the effects of the Cyclops mutant, we modified our wild-type two wing-cell simulation. Since a key feature of the Cyclops phenotype is the failure of the wing vein to form and create a compartment boundary between the two adjacent wing-cells (Brakefield, 2001), our modifications focused on this feature. Two types of changes were made. First, the vein separating the two adjacent wing-cells was altered so that it was no longer a source of wing vein repressor. Second, diffusing molecules (vein repressor, margin repressor, and extracellular hedgehog) that had been prevented from crossing between the wing-cells by the vein in our wild-type model were allowed to diffuse freely across this boundary.
3. Results and discussion
The deficiencies of Model 1 (Fig. 1a) have been thoroughly described elsewhere (Evans and Marcus, 2006) so this model will not be discussed further here. As mentioned previously, Models 2 and 3 (Figs. 1b and 1c) are capable of reproducing the known wild-type expression patterns for all of the genes included in our single wing-cell simulations (Evans and Marcus, 2006).
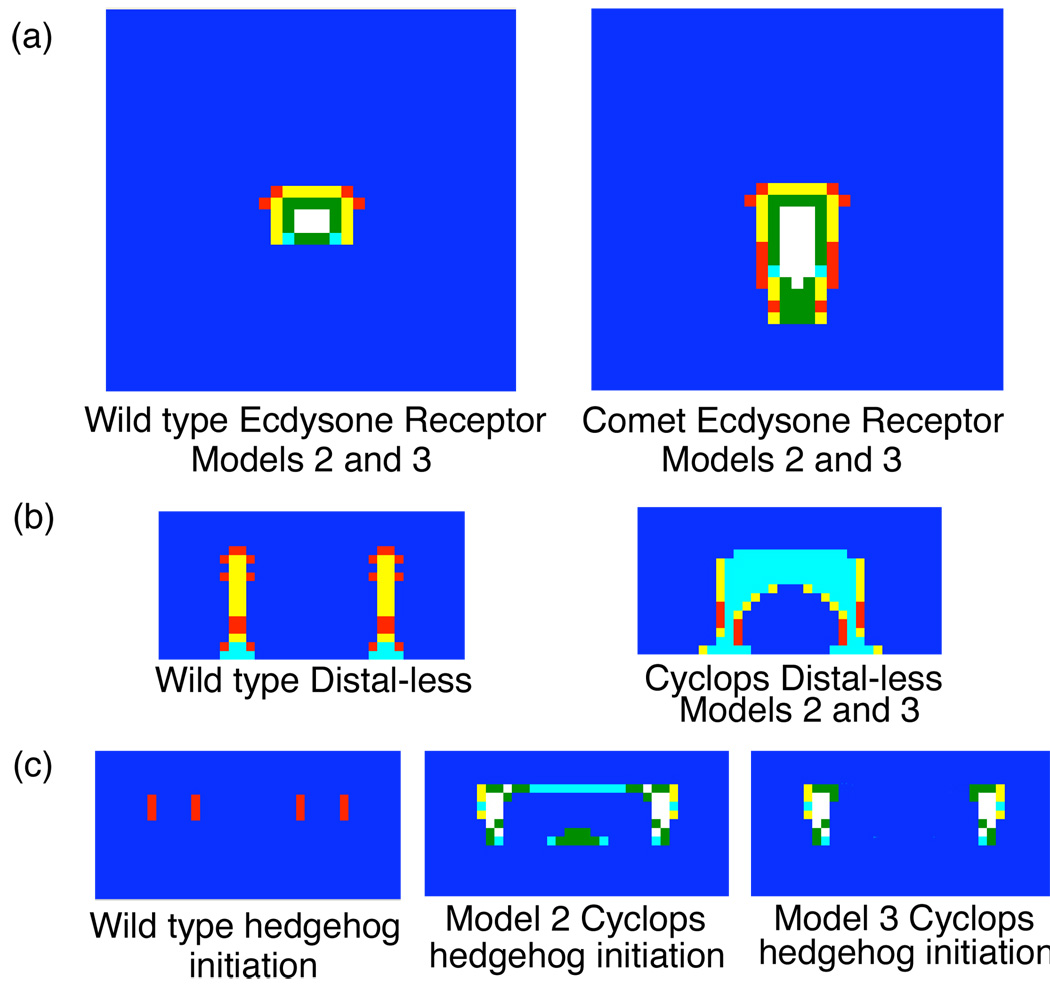
In general, the simulations of expression patterns in comet mutants were generally similar to those produced in simulations wild-type animals. However, gene products that are late in the eyespot focus genetic regulatory hierarchy (e.g. Engrailed, Cubitus interruptus, Ecdysone Receptor) show elongated domains of expression in comet mutants that approach the wing margin more closely than what is observed in wild type (Fig. 3a). This corresponds to the comet-shaped eyespot foci that are characteristic of comet mutants. This supports our hypothesis that the wing margin repressor plays a key role in the formation of the comet phenotype.
Fig. 3.
Simulated gene expression patterns of (a) Ecdysone Receptor in one wing-cell models. This gene product appears to be the last gene product in the regulatory network before the focal signal is released and the shape of the expression pattern corresponds to the shape of the eyespot focus. Comet mutants have enlarged eyespot foci that are tear-drop shaped instead of the circles as is typical of wild-type animals. The narrow end of the tear-drop is oriented towards the wing margin, the bottom of the field in these simulations. The expression of Distal-less (b) in two wing-cell models matches known expression patterns in both wild type and Cyclops mutant genotypes (Brakefield, et al., 1996). To date, no additional expression patterns have been published for Cyclops mutants. Models 2 and 3 produce similar results for all gene products in the regulatory network in Cyclops except for hedgehog (c), which shows radically different patterns of expression. This suggests that examining hedgehog expression in Cyclops mutants is a critical experiment for differentiating between Models 2 and 3.
Our wild type two wing-cell simulations largely replicate the results of the wild type one wing-cell simulations published previously (Evans and Marcus, 2006). After removing the wing vein barrier between the two wing-cells to simulate the Cyclops mutant, expression patterns change considerably. In both Models 2 and 3, the “stalks” of Distal-less expression in the two adjacent wing-cells of the wild-type wing fuse together in the Cyclops mutant. This creates a single large elongate eyespot focus that runs parallel to the wing margin instead of two small circular wing foci (Fig. 3b). This pattern of Distal-less expression matches the known expression pattern of Distal-less in Cyclops mutants (Brakefield, et al., 1996), further supporting our inference that the wing vein repressor plays an important role in producing the Cyclops phenotype. As genomic resources in Bicyclus improve (Beldade, et al., 2006), these mechanistic details may be extremely useful in identifying candidate genes for sequencing from mutant genotypes.
Other expression patterns in our simulations of Cyclops mutants differ from what is seen in wild-type simulations. To date, Distal-less is the only published expression pattern available from Cyclops (Brakefield, et al., 1996), so comparisons between our simulations and what is occurring in vivo will have to wait until the appropriate experiments have been conducted. For other gene products in Cyclops, Models 2 and 3 generally produce very similar expression patterns in our simulations. However, Models 2 and 3 give different predictions for the expression of hedgehog gene product in Cyclops mutants (Fig. 3c). In Model 2, the simulations predict a horseshoe shaped domain of hedgehog expression around the incipient eyespot focus with the open end of the horseshoe facing the wing margin. A second domain of expression is found the middle of the open end of the horseshoe, in between the eyespot focus and the wing margin. In Model 3, the simulations predict two domains of hedgehog expression, immediately anterior and posterior to the eyespot focus. These highly dissimilar predictions of hedgehog expression suggest that the expression of this gene product in Cyclops mutants may be an important tool for differentiating between Models 2 and 3, and allow us to eliminate one of the hypothetical genetic regulatory networks proposed by Evans and Marcus (2006). This enhanced ability to identify key experiments is one of the major advantages of taking computational genetic approaches to the study of genetic regulatory networks.
Unfortunately, our laboratory does not have the containment facilities and USDA-APHIS permits required to import and rear the squinting bush brown butterfly (Bicyclus anynana) in the United States due to the fact that it is a potential crop pest. It is our hope that this paper will encourage other researchers who work with Bicyclus to specifically study hedgehog expression in these butterflies because such experiments will likely yield important information about the nature of the genetic regulatory pathway that generates all butterfly eyespot foci. This will be extraordinarily helpful in further exploiting the butterfly eyespot system, permitting the integrated study of phenotypic traits at many levels of organization from genetics, development, and cell biology, to ecology and evolutionary biology.
Acknowledgements
Thanks to Fred Nijhout for many conversations about eyespot focus determination, the role of modeling in evolutionary developmental biology, and for assistance with Delphi 2.0. Thanks to Aaron Edwards, Rachel Barber, Michelle Dodson, Morgan Foster, Amber Harper, Sarah House, Tia Hughes, Mollie Johnson, Zan Lee, Joseph Marquardt, Amanda Maupin, Brooke Polen, Tara Powell, and Tim Shehan for discussions and comments on the paper. Thanks to Vittoria Ariazi and Joanne, Harry, and Sally Seiff for their encouragement. Support for this research is from the National Science Foundation and the Commonwealth of Kentucky through EPSCoR awards (EPS-0132295 and 0447479), a grant from the US Environmental Protection Agency (X796463906-0), and from a National Institutes of Health and National Center for Research Resources Grant (P20 RR16481).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Delphi Desktop Version 2.0. Borland: 1996. Delphi Desktop Version 2.0. [Google Scholar]
- Beldade P, Brakefield PM. The genetics and evo-devo of butterfly wing patterns. Nat. Rev. Genet. 2002;3:442–452. doi: 10.1038/nrg818. [DOI] [PubMed] [Google Scholar]
- Beldade P, Rudd S, Gruber JD, Long AD. A wing expressed sequence tag resource for Bicyclus anynana butterflies, an Evo-Devo model. BMC Genomics. 2006;7:130. doi: 10.1186/1471-2164-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehs B, Sturtevant MA, Bier E. Boundaries in the Drosophila wing imaginal disc organize vein-specific genetic programs. Development. 1998;125:4245–4257. doi: 10.1242/dev.125.21.4245. [DOI] [PubMed] [Google Scholar]
- Blair SS. A role for the segment polarity gene shaggy-zeste white 3 in the specification of regional identity in the developing wing of Drosophila. Dev. Biol. 1994;162:229–244. doi: 10.1006/dbio.1994.1081. [DOI] [PubMed] [Google Scholar]
- Blanchard R, Descimon H. Hybridization between two species of swallowtails, meiosis mechanism, and the genesis of gynandromorphs. J. Lepid. Soc. 1988;42:94–102. [Google Scholar]
- Brakefield PM. Structure of a character and the evolution of butterfly eyespot patterns. J. Exper. Zool. 2001;291:93–104. doi: 10.1002/jez.1062. [DOI] [PubMed] [Google Scholar]
- Brakefield PM, Gates J, Keys D, Kesbeke F, Wijngaarden PJ, Monteiro A, French V, Carroll SB. Development, plasticity and evolution of butterfly eyespot patterns. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- Dilao R, Sainhas J. Modelling butterfly wing eyespot patterns. Proc. R. Soc. B. 2004;271:1565–1569. doi: 10.1098/rspb.2004.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TM, Marcus JM. A simulation study of the genetic regulatory hierarchy for butterfly eyespot focus determination. Evol. Dev. 2006;8:273–283. doi: 10.1111/j.1525-142X.2006.00098.x. [DOI] [PubMed] [Google Scholar]
- Ford EB. Ecological genetics. Vol. 3. London: Chapman and Hall; 1971. p. 410. Ecological genetics. [Google Scholar]
- Hill RI, Vaca JF. Differential wing strength in Pierella butterflies (Nymphalidae, Satyrinae) supports the deflection hypothesis. Biotropica. 2004;36:362–370. [Google Scholar]
- Ingham PW. Hedgehog points the way. Curr. Biol. 1994;4:345–350. doi: 10.1016/s0960-9822(00)00076-2. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Martinez Arias A. Boundaries and Fields in Early Embryos. CELL. 1992;68:221–235. doi: 10.1016/0092-8674(92)90467-q. [DOI] [PubMed] [Google Scholar]
- Kettlewell B. The Evolution of Melanism: The Study of a Recurring Necessity. Oxford: Oxford University Press; 1973. p. 423. The Evolution of Melanism: The Study of a Recurring Necessity. [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RJ, Gates J, Scott MP, Carroll SB. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–534. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- Kingsolver JG. Viability selection on seasonally polyphenic traits: Wing melanin pattern in western white butterflies. Evolution. 1995;49 doi: 10.1111/j.1558-5646.1995.tb02328.x. [DOI] [PubMed] [Google Scholar]
- Koch PB, Nijhout HF. The role of wing veins in colour pattern development in the butterfly Papilio xuthus (Lepidoptera : Papilionidae) Eur. J. Entomol. 2002;99:67–72. [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst J, Hanks W, Joyner AL. The mouse Engrailed-1 gene and ventral limb paterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Majerus MEN. Melanism: Evolution in Action. Oxford: Oxford University Press; 1998. p. 338. Melanism: Evolution in Action. [Google Scholar]
- Marcus JM. The development and evolution of crossveins in insect wings. J. Anat. 2001;199:211–216. doi: 10.1046/j.1469-7580.2001.19910211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM. Jumping genes and AFLP maps: Transforming Lepidopteran color pattern genetics. Evol. Dev. 2005;7:108–114. doi: 10.1111/j.1525-142X.2005.05012.x. [DOI] [PubMed] [Google Scholar]
- Marcus JM, Ramos DM, Monteiro A. Transformation of the butterfly Bicyclus anynana. Proc. R. Soc. B. Biol. Lett. 2004;27 Suppl.:263–265. doi: 10.1098/rsbl.2004.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM, Harper AL, Hughes TM, Johnson MR, Maupin AB, Polen AB, Powell TB, Shehan TH, Ritland DB, Covell CV. ms. Phylogenetics and hybridization in the North American butterfly genus Limenitis (Nymphalidae) and the origins of the aberrant Limenitis form rubidus (Strecker) In prep. [Google Scholar]
- Mohler JD, Vani K. Molecular organization and embryonic expression of the hedgehog gene involved in cell-cell communication in segmental patterning in Drosophila. Development. 1992;115:957–971. doi: 10.1242/dev.115.4.957. [DOI] [PubMed] [Google Scholar]
- Monteiro A, Prijs J, Hakkaart T, Bax M, Brakefield PM. Mutants highlight the modular control of butterfly eyespot patterns. Evol. Dev. 2003;5:180–187. doi: 10.1046/j.1525-142x.2003.03029.x. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. A comprehensive model for color pattern formation in butterflies. Proc. R. Soc. B. 1990;239:81–113. [Google Scholar]
- Nijhout HF. The development and evolution of butterfly wing patterns. Washington: Smithsonian Institution Press; 1991. p. 297. The development and evolution of butterfly wing patterns. [Google Scholar]
- Nijhout HF. Symmetry systems and compartments in Lepidopteran wings: the evolution of a patterning mechanism. Development. 1994 suppl.:225–233. [Google Scholar]
- Nijhout HF. Focus on butterfly eyespot development. Nature. 1996;384:209–210. doi: 10.1038/384209a0. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Rountree DB. Pattern induction across a homeotic boundary in the wings of Precis coenia (Lepidoptera: Nymphalidae) Int. J. Insect Morphol. Embryol. 1995;24:243–251. [Google Scholar]
- Peifer M, Bejsovec A. Knowing your neighbors: Cell interactions determine intrasegmental patterning in Drosophila. Trends Genet. 1992;8:243–249. [Google Scholar]
- Phillips RG, Whittle JRS. wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development. 1993;118:427–438. doi: 10.1242/dev.118.2.427. [DOI] [PubMed] [Google Scholar]
- Ramos DM, Monteiro A. Transgenic approaches to study wing color pattern development in Lepidoptera. Mol. BioSyst. 2007;3:530–535. doi: 10.1039/b701965n. [DOI] [PubMed] [Google Scholar]
- Reed RD, Serfas MS. Butterfly wing pattern evolution Is associated with changes in a Notch/Distal-less temporal pattern formation process. Curr. Biol. 2004;14:1159–1166. doi: 10.1016/j.cub.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Robertson KA, Monteiro A. Female Bicyclus anynana butterflies choose males on the basis of their dorsal UV-reflective eyespot pupils. Proc. R. Soc. B. 2005;272:1541–1546. doi: 10.1098/rspb.2005.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree DB, Nijhout HF. Genetic control of a seasonal morph in Precis coenia (Lepidoptera: Nymphalidae) J. Insect Physiol. 1995;41:1141–1145. [Google Scholar]
- Sekimura T, Madzvamuse A, Wathen AJ, Maini PK. A model for colour pattern formation in the butterfly wing of Papilio dardanus. Proc. R. Soc. London B. 2000;267:851–859. doi: 10.1098/rspb.2000.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DH, Reinitz J. Prediction of mutant expression patterns using gene circuits. Biosystems. 1998;47:79–90. doi: 10.1016/s0303-2647(98)00014-8. [DOI] [PubMed] [Google Scholar]
- Sibatani A. Wing homeosis in Lepidoptera. A survey. Dev. Biol. 1980;79:1–18. doi: 10.1016/0012-1606(80)90069-x. [DOI] [PubMed] [Google Scholar]
- Stevens M. The role of eyespots as anti-predator mechanisms, principally demonstrated in the Lepidoptera. Biological Reviews. 2005;80:573–588. doi: 10.1017/S1464793105006810. [DOI] [PubMed] [Google Scholar]
- Weatherbee SD, Nijhout HF, Grunert LW, Halder G, Galant R, Selegue J, Carroll S. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr. Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]