Abstract
Mammalian circadian clocks consist of regulatory loops mediated by Clock/Bmal1-binding elements, DBP/E4BP4 binding elements, and RevErbA/ROR binding elements. As a step toward system-level understanding of the dynamic transcriptional regulation of the oscillator, we constructed and used a mammalian promoter/enhancer database (http://promoter.cdb.riken.jp/) with computational models of the Clock/Bmal1-binding elements, DBP/E4BP4 binding elements, and RevErbA/ROR binding elements to predict new targets of the clock and subsequently validated these targets at the level of the cell and organism. We further demonstrated the predictive nature of these models by generating and testing synthetic regulatory elements that do not occur in nature and showed that these elements produced high-amplitude circadian gene regulation. Biochemical experiments to characterize these synthetic elements revealed the importance of the affinity balance between transactivators and transrepressors in generating high-amplitude circadian transcriptional output. These results highlight the power of comparative genomics approaches for system-level identification and knowledge-based design of dynamic regulatory circuits.
Keywords: comparative genomics, promoter and enhancer database, synthetic biology, systems biology, transcription
The rapidly expanding number of sequenced mammalian genomes (1–3), annotated and cloned full-length cDNAs (4–6), transcriptional starts sites (TSSs) (7–9) and transcription factor binding sites (TFBSs) (10–12) has provided new opportunities to unravel the control of dynamic transcriptional programs. Comparative genomics approaches applying these resources have been used to identify target genes of specific biological pathways. These efforts used consensus sequence searches (13, 14), positional weight matrices (15), hidden Markov models (HMMs) (16). and specifically tailored algorithms (17, 18) to define candidate response elements and target genes in raw genomic sequence. Additionally, post hoc analysis employing evolutionary conservation (15, 16, 18) together with positional information of TSSs (15) and/or translational start sites (16) has helped to further define candidate elements and genes and greatly expanded our knowledge of transcriptional output regulation.
The mammalian circadian clock is an ideal system to apply these tools as it consists of integrated transcriptional regulatory loops that direct output through at least three types of transcriptional regulatory elements, the Clock/Bmal1-binding elements (E-box) (CACGTG) (19–21), DBP/E4BP4 binding elements (D-box) (TTATG[T/C]AA) (21–23), and RevErbA/ROR binding elements (RRE) ([A/T]A[A/T]NT[A/G]GGTCA) (15, 21, 24, 25). Several groups, including our own, have shown that approximately 5–10% of mammalian genes display circadian expression in central and peripheral clock tissues (26). However, for the most part, the transcriptional regulation of these thousands of clock-controlled genes has remained uncharacterized. We and others have used comparative genomics approaches to analyze E-box (21, 27, 28), D-box (21), and RRE (15, 21), highlighting the importance of both their core consensus and flanking sequences (15, 21, 27, 28) in circadian gene control. In this study, we further extend comparative genomics approaches toward a system-level understanding of the dynamic transcriptional regulations of the mammalian circadian clock.
Results and Discussion
Prediction of Direct Clock Targets Through Utilization of the Mammalian Promoter/Enhancer Database.
To generate a resource that facilitates identification of clock-controlled genes, we constructed a mammalian promoter/enhancer database (http://promoter.cdb.riken.jp/) by integrating information sources such as conserved non-coding regions, TSSs and TFBSs [supporting information (SI) Fig. S1 and SI Appendix]. Although excellent and similar databases exist such as DBTSS (8), CisView (29) and ECRbase (30), none were tailored to specifically identify clock gene targets and having local control of the database facilitated manipulation of the underlying data (see also SI Appendix). We then developed a comparative genomics strategy employing this database and profile HMMs using the HMMER software package (31). Profile HMMs are powerful tools to extract the statistical properties of input sequences by representing multiple sequences as a transition probability matrix marching from one position to the neighboring position. HMMs were built and calibrated on known functional clock-controlled elements experimentally verified in our previous (15, 21) or current studies (Fig. S2 and Table S1), consisting of 12 E-boxes, 10 D-boxes and 15 RREs (Table S2). Profile HMM searches to identify new clock-controlled elements from conserved non-coding regions between human and mouse identified 1,108 E-boxes, 2,314 D-boxes, and 3,288 RREs candidate elements (see the circadian section of the mammalian promoter/enhancer database: http://promoter.cdb.riken.jp/circadian.html for element lists). To set appropriate reporting thresholds, we used the match scores of known functional clock-controlled elements (Material and Methods). Predicted clock-controlled elements exhibited an un-biased distribution of chromosomal position spread over whole mouse genome (Fig. 1A and http://promoter.cdb.riken.jp/circadian.html).
Fig. 1.
Computational prediction of clock-controlled elements using HMMs. (A) Chromosomal distributions of predicted conserved clock-controlled elements of conserved non-coding regions mapped on the mouse genome. Chromosomal positions of the 100 most significant hits for E-boxes, D-boxes, and RREs are shown (red). (B) Plots of false discovery rates (FDRs) against match scores of HMM searches in three conditions: (1) searches for conserved elements within conserved non-coding regions (red, “Conserved element”), (2) searches for mouse elements within conserved or non-conserved non-coding regions (blue, “Non-coding region”) and (3) searches in the entire genome relaxing both element conservation and search space (orange, “Whole genome”). FDRs in “Conserved elements” search are plotted against the average match score of human and mouse elements. (C) The distributions of distance from transcriptional starts sites (TSSs) for predicted conserved clock-controlled elements of conserved non-coding regions (1,108 E-boxes, 2,314 D-boxes, and 3,288 RREs). The E-box displays a biased distribution of distance from TSSs, while the D-box and RRE show unbiased, near random distributions (“Random E-box,” “Random D-box,” and “Random RRE,” see also SI Appendix). (D) The average expression of transcripts harboring each element exhibit circadian rhythms in the liver. The average expressions of 36 genes with E-boxes, 29 genes with D-boxes, and 34 genes with RREs exhibited significant circadian oscillations (P = 0.01 for E-box, P = 0.0005 for D-box, and P = 0.001 for RRE). Data were normalized so that the average signal intensity over 12-point time courses is 1.0. Estimated peak times of circadian oscillation were also indicated.
However, the match score on its own does not give a good estimate of accuracy and false discovery from HMM searches. To better estimate the prediction for each HMM, we searched each against randomized genome sequence to generate a background distribution of false positive occurrences (see SI Appendix for detail). We found that the value of the false discovery rate (FDR) is inversely proportional to the match score of the HMM, which is a representation of the statistical significance of the candidate element (Fig. 1B). Importantly, we found the accuracy of the HMM-based prediction as measured by the FDR is dependent on the initial search conditions. HMM searches in conserved non-coding regions (the original condition) had the lowest FDR, while higher rates were observed in conserved or non-conserved non-coding regions, or in searches of raw genome sequence (Fig. 1B). These results demonstrate the value of using human/mouse conservation and a confined search space for generation of the most accurate response element predictions.
Several intriguing features resulted from this analysis. Interestingly, like cAMP-responsive elements (CREs) (16), putative E-box displayed a biased distance distribution from TSSs, while putative D-box and RRE had random localization distributions in the genome and were not more likely to be near TSSs (Fig. 1C). This result is consistent with and extends an earlier report that described the preferential localization of CpG containing transcription factor binding sites including the E-box and CRE to proximal promoter regions of housekeeping genes (32). In addition to housekeeping genes, we see circadian E-box sequences present in many genes with specific functions such as enzymes and signaling molecules.
Gratifyingly, we noted a significant enrichment of previously identified clock-controlled genes (15, 33) in our predicted clock-controlled elements (the 100 most significant sequences for each HMM search, E-box, D-box, and RRE, respectively). After removing the 21 clock-controlled genes used for HMM generation and training, we found an additional 19 putative clock-controlled genes (out of the 6,195 genes common in our mammalian promoter/enhancer database and U74 mouse microarray) versus the expected 10.67 that would have arisen from chance, a significant enrichment of clock-controlled genes (P < 0.01, see also SI Appendix).
The presence of statistically significant clock-controlled elements in the promoters of these genes suggests that their message levels may oscillate. To examine this possibility, we selected the 100 most significant putative clock-controlled elements for each model (E-box, D-box, and RRE) and determined their gene expression levels from previously obtained liver data (15). The average expression of 36 E-box containing genes, 29 D-box genes, and 34 RRE genes exhibited significant circadian oscillations (P = 0.01 for E-box, P = 0.0005 for D-box, and P = 0.001 for RRE) with a surprisingly consistent peak time of expression (E-box = 8.8, D-box = 10.8, and RRE = 13.6, Fig. 1D). Taken in sum, these data show that our HMM models and strategy identifies elements and genes that oscillate with a circadian period with peak phases of expression that are consistent with the previously reported literature.
In Vitro Validation of Putative Clock Controlled Genes.
To provide further validation of these predictions, we used an in vitro system of the autonomous circadian clock to empirically test candidate elements in circadian transcriptional output assays. In brief, we used a cell culture system (15, 34) that allowed the monitoring of circadian transcriptional dynamics using a destabilized luciferase reporter (dLuc) driven by known or putative clock-controlled response elements (Fig. 2A). We selected the ten most significant sequences for each HMM search, E-box, D-box, and RRE respectively, and located within 1kb of the TSS (Table S3). We constructed reporter vectors in which three predicted elements were inserted in front of the SV40 basic promoter driving a dLuc reporter (see Material and Methods). We transfected these constructs into cultured NIH 3T3 fibroblasts, stimulated them with forskolin to synchronize circadian rhythms of individual cells, and measured the sum of their transcriptional activity by monitoring bioluminescence over several days; 40% of putative E-boxes, 70% of D-boxes, and 60% of RREs generated strong circadian reporter gene activity (P < 0.01 and high-amplitude) in phase with the Per1 E-box, Per3 D-box, and Arntl RRE, respectively (Fig. 2A and Fig. S3 A–D). The remaining sequences generated weak, low amplitude circadian transcriptional activity or were arrhythmic (Fig. S3 E–G). To supplement our observed 40% prediction success, we constructed 14 reporters containing conserved low-scoring E-boxes and found only one exhibited high-amplitude oscillations (Fig. S3H and Table S3). This result indicates that our observed 40% prediction success is suggestively higher than expected (P = 0.075, Fisher's exact test). Taken in sum, these results demonstrate utility of this approach in finding elements within structural genes that dictate rhythmic transcription.
Fig. 2.
Experimental validation of HMM-based predictions at cellular and organismal levels. (A) Circadian rhythms of bioluminescence from the predicted clock-controlled elements fused to the SV40 basic promoter driving a dLuc reporter in NIH 3T3 fibroblasts. Three known clock-controlled elements from clock genes (E-box of Per1, D-box of Per3, and RRE of Arntl) are used as positive controls. The bioluminescence data were detrended in baseline and amplitude and normalized so that their maximum, minimum, and average were set to 1, −1, and 0, respectively. The colors in descending order from magenta to black to green represent the detrended bioluminescence. Columns represent time points and rows represent the predicted elements on the designated genes. (B) Circadian rhythms of temporal mRNA expression profiles of the predicted clock-controlled genes in mouse seven tissues (‘A’, ‘B’, ‘H’, ‘K’, ‘Li’, ‘Lu’ and ‘M’ for aorta, bone, heart, kidney, liver, lung, and muscle, respectively). An estimated peak time with color of type of predicted clock-controlled element (green, red, and blue for E-box, D-box, and RRE, respectively) is also indicated. The colors in descending order from magenta to black to green represent the normalized data (the average and standard deviation over 12-point time courses are 0.0 and 1.0, respectively). Columns represent time points, and rows represent the predicted clock-controlled genes in the designated tissues.
If these in vitro validated 17 elements (4 E-boxes, 7 D-boxes, and 6 RREs) play a prominent role in gene regulation in vivo, we would predict that the endogenous transcripts for these genes would likely oscillate in a circadian fashion. To test this, we harvested mRNA from seven tissues (aorta, bone, heart, kidney, liver, lung, and muscle) isolated from mice entrained to a 12:12 light:dark cycle and then released to free run in constant darkness. Using quantitative PCR assays, we measured expression profiles from our predicted clock-controlled elements, and evaluated their rhythmicity using a statistical method based on analysis of variance (ANOVA) followed by curve fitting to a cosine wave. These experiments revealed that circadian expression profiles (P < 0.03) for 13 genes (76%): 3 E-box controlled genes, 4 D-box controlled genes and 6 RRE controlled genes, respectively, with a consistent order of peak time (4.1, 15.5, and 18.8 for mean value of the peak time of putative E-box, D-box, and RRE-controlled genes, respectively) (Fig. 2B; See also http://promoter.cdb.riken.jp/circadian.html for detailed data). For those genes that did not confirm circadian rhythmicity, the average level of expression was lower, implying mRNA detection was limiting for these genes. Collectively, these in vitro and in vivo experiments suggest that many predicted E-box, RRE, and D-box containing genes are bona fide first-order clock-controlled genes.
Design and Validation of the Synthetic Regulatory Elements.
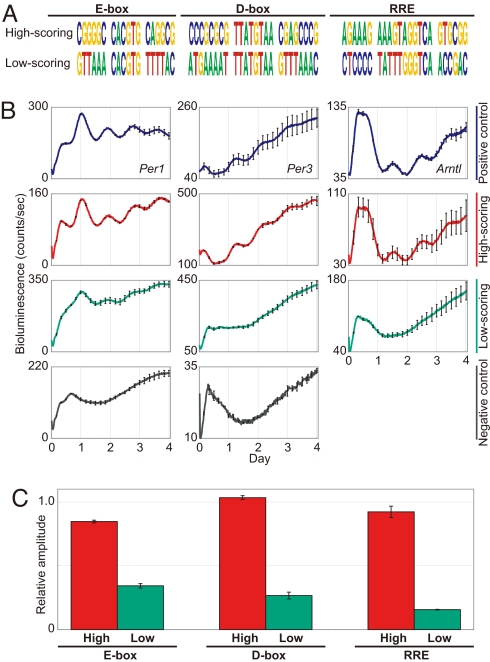
One of the goals of systems biology is the synthesis of knowledge and the generation of testable (and tested) hypotheses. We reasoned that if our HMMs truly represented the functional response elements of these three transcription factor complexes, then synthetic regulatory elements derived from these models should mediate rhythmic transcription as well. To test this idea, we emitted sequences from the E-box, D-box, and RRE models, respectively, and filtered out those that naturally exist in either the human or mouse genomes. Furthermore, to not unduly focus our attention on outliers, we required that all candidates adhere to the consensus rules for each element, CACGTG for the E-box (19), TTATGTAA for the D-box (22), and [A/T]A[A/T]NT[A/G]GGTCA for the RRE (24). For the remaining sequences, we chose each one of the highest and lowest scoring synthetic representatives for three types of elements and named them “high-scoring” and “low-scoring” elements, respectively (Fig. 3A). We tested these elements in a synthetic reporter system as above (Fig. 3B). All three “high-scoring” elements showed high-amplitude circadian transcriptional activity equivalent to known elements from canonical clock genes (E-box of Per1, D-box of Per3, and RRE of Arntl are used as 1.0, respectively) (21) (Fig. 3C). On the other hand, the “low-scoring” elements emitted from the HMMs showed very low- amplitude transcriptional activity, despite the presence of “consensus” E-box, RRE, or D-box core sequences (Fig. 3C). These results show the utility of this comparative genomics approach in synthetic design of dynamic cis-acting elements, as well as highlight the contribution of flanking sequences in generating high-amplitude rhythmicity.
Fig. 3.
Computational design and experimental characterization of high-amplitude circadian transcriptional activity. (A) HMM-based design of the high- and low-scoring elements. Match scores of synthetic elements are 19.84 (high-scoring E-box), −1.36 (low-scoring E-box), 22.17 (high-scoring D-box), −4.33 (low-scoring D-box), 24.45 (high-scoring RRE) and −0.46 (low-scoring RRE), respectively. All elements were filtered to identify those that do not exist in either the human or mouse genomes. (B) Bioluminescence from synthetic elements inserted into the SV40 basic promoter driving a dLuc reporter (SV40-dLuc) in NIH 3T3 fibroblasts. SV40-dLuc containing known clock-controlled elements (E-box of Per1, D-box of Per3, or RRE of Arntl) and SV40-dLuc with no insert are the positive and negative controls, respectively. The negative control of the D-box element was also used for the RRE as experiments for these elements were performed at the same time. (C) The amplitude of bioluminescence activity driven by synthetic elements relative to that of positive controls. The relative amplitude of positive controls is 1.0. Error bars indicate SEM determined from independent experimental duplicates.
Investigating the Contribution of Flanking Sequences.
Using these synthetic elements, we next attempted to explore the contribution of E-box flanking regions to identify critical residues that modulate amplitude and rhythmicity. We clustered their nucleotide sequences, and, interestingly, found two patterns of high-amplitude E-box flanking sequences adjacent to the core CACGTG element (Fig. S4). However, these positions do not absolutely dictate high-amplitude rhythmicity, as some elements that meet these rules exhibit lower-amplitude oscillations, possibly because they exhibit much higher GC content. In either case, these experimental results also imply that amplitude information is encoded in specific residues adjacent to the core consensus element and further strengthen the previous reports by other groups on the importance of flanking sequence of E-box (27, 28, 35, 36). Interestingly, the identified patterns in this study partly overlap with the computational models based on the evolutionarily conserved E-box structure from insects to mammals (27).
High-Amplitude Oscillations Require Appropriate Affinity Balance Between Activators and Repressors.
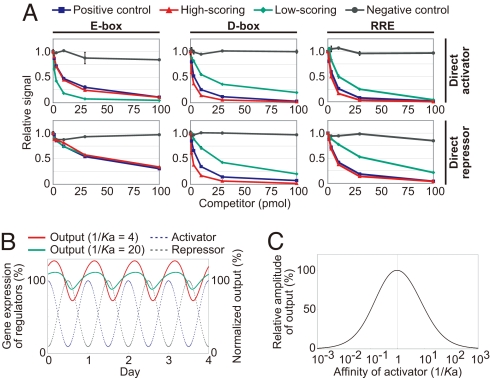
To explore the properties of these elements that result in high amplitude oscillations, we took a simplified molecular modeling and experimental approach. First, we assumed concentrations of activators and repressors were within similar ranges (see also SI Appendix Discussion in more general cases). We further hypothesized that flanking region DNA sequence impacted DNA-binding affinity of clock gene regulators and therefore altered amplitude. We further hypothesized that tightly binding sequences would have higher amplitudes of circadian oscillation. To test this notion, we analyzed the DNA-binding affinity of activators and repressors to these response elements using competitive binding assays (Fig. 4A and Fig. S5A). For the D-box and RRE elements, “high-scoring” elements showed approximately the same DNA-binding affinity for their activators and repressors, while “low-scoring” elements of D-box and RRE showed relatively weak affinity, confirming this hypothesis. Surprisingly, in the case of E-box, “low-scoring” sequences had a higher affinity for the Arntl/Clock activator complex (4.8 times higher than positive control; Fig. S5A) than “high-scoring” sequences or the positive control, whereas the “low-scoring” E-box sequence showed approximately the same affinity to the Bhlhb2 repressor.
Fig. 4.
Defining the relationship between affinity and amplitude. (A) Binding affinity between synthetic elements and DNA binding activators or repressors was detected by competitive DNA binding assays. The binding between labeled oligonucleotides of positive control elements (10 pmol) and transcription factors were competed by each of unlabeled oligonucleotides (0, 1, 3, 10, 30, and 100 pmol) for positive control elements (blue), high-scoring elements (red), low-scoring elements (green), or negative control elements (black). Known clock-controlled elements (E-box of Per1, D-box of Per3, or RRE of Arntl) and mutated clock-controlled elements are the positive and negative controls, respectively. Arntl/Clock, Dbp, Rora were used as DNA binding activator of E-box, D-box, and RRE, respectively. Bhlhb2, Nfil3, Nr1d1, were used as DNA binding repressors of E-box, D-box, and RRE, respectively. The relative signal without competitor is 1.0. Error bars indicate SEM determined from independent experimental duplicates. (B and C) In silico analysis of affinity to amplitude mechanism. Gene expression of idealized transcriptional activators (blue dotted line) and repressors (gray dotted line) and the normalized output of different strengths of activator binding affinity (weaker affinity 1/Ka = 4 is red line and stronger affinity 1/Ka = 20 is green line) are indicated (B). The relative amplitude of oscillation of the output plotted against the strength of activator binding affinity (C). Amplitude was normalized so that the maximum value is set to 100%.
To assist in interpreting these results, we used in silico modeling (Fig. 4 B and C) and treated affinity of activators and repressors as parameters and amplitude as the output of the model. Interestingly, this analysis showed that a high affinity activator complex coupled with a normal affinity repressor complex capitulated lower amplitude rhythms (Fig. 4B), suggesting that the enhanced retention of an activator alone promotes its saturation on a promoter and consequently dampens amplitude in competition-based models. Further, in silico analysis showed not only affinity strength of the activator and repressor (Fig. S5B) but also the appropriate affinity balance of activators and repressors is necessary for high-amplitude circadian oscillations (Fig. 4C; See also SI Appendix Discussions for in silico modeling).
Supporting this notion, a new clock gene, clockwork orange (cwo, a fly ortholog of mammalian Bhlhb2 and Bhlhb3) was recently reported to directly suppress gene expression of several clock genes through E-box elements (37–39). Quantitative and qualitative impairment of cwo revealed an important role of this transcriptional repressor for high-amplitude oscillation of the Drosophila circadian clock. The findings in this report, along with the studies of the cwo gene, collectively show that a competitive balance between direct activator(s) and direct repressor(s) for the E-box element is important for driving high-amplitude oscillations of circadian output genes. In addition to this in vivo biological evidence in the fly, we listed the evolutionary conserved “low-scoring” E-boxes in the mammalian genome (predicted low-amplitude) as candidates for unbalanced affinities. Interestingly, this list includes E-boxes on core clock genes (Cry2, Bhlhb3, Nr1d1, and Rorc) and some low-amplitude clock-controlled genes such as Id2, and to promote follow-up, is available at http://promoter.cdb.riken.jp/circadian.html.
What could explain the discrepancy between E-box motifs and other circadian response elements? We hypothesized that these differences might be encoded at the protein sequence level of the DNA-binding domains of activators and repressors. Interestingly, the DNA binding domains of transactivators and transrepressors of the RRE and D-box are more similar to each other (65% identity and 81% homology for RRE regulators, and 44% identity and 69% homology for D-box regulators) than those of E-box transactivators and transrepressor (22∼30% identity and 55∼57% homology) (Table S4). Based on these findings, we speculate that the DNA-binding domains for transactivators and transrepressors of the RRE and D-box have evolved similar affinities. In contrast, the evolutionarily and structurally divergent regulators of E-boxes, 108 bHLH proteins including several families of activators and repressors, as well as the unrelated period and cryptochrome gene families, may have required the co-evolution of specific DNA-binding domains and E-box sequences with specific flanking regions to generate higher amplitude rhythmicity.
Conclusion
In summary, we have applied a comparative genomics strategy to the understanding of a dynamic transcriptional regulatory system, the mammalian circadian clock. Our informatics strategy employs a model-based search with excellent statistical properties, the evolutionary conservation of putative transcriptional regulatory elements across mouse and human non-coding regions, and statistical evaluation of false discovery rates in each prediction. Experimental validation of this strategy in vitro and in vivo using real-time monitoring of transcriptional activity and quantitative PCR assay has led to the identification of dozens of novel clock-controlled genes and the elements that likely dictate their rhythmicity. High-scoring conserved E-boxes (mean HMM-score = 16.15) had a 40% rate of validation, while low-scoring conserved E-boxes (mean HMM-score = 2.5143) had a 7.1% probability of generating high-amplitude rhythmicity in reporter assays. Linear interpolation from these two numbers generates an estimate of approximately 347 novel conserved E-boxes that likely confer circadian rhythmicity (see also SI Appendix). Moreover, to demonstrate their predictive nature, we have taken these in silico models and designed synthetic elements that exhibit high-amplitude transcriptional rhythmicity as well as the best canonical regulatory elements. Furthermore, experimental measurement and in silico analysis of affinity of regulators to synthetic elements revealed the importance of the appropriate affinity balance between activators and repressors for high-amplitude rhythmicity. Surprisingly, for E-box sequences, lower affinity DNA element generates higher amplitude rhythms. The experimental, analytical, and synthetic approaches discussed here are especially timely as genomics tools are increasingly uncovering the complexity and flexibility of transcriptional regulatory circuits. We predict the general themes and resources reported here will enhance understanding of the biology mediated by complex and dynamic transcriptional regulation including the mammalian circadian clock.
Materials and Methods
Detailed information on the construction of the mammalian promoter/enhancer database, determination of distance from TSSs for natural and randomly positioned elements, calculation of FDR for putative elements, animals, genome sequences, oligonucleotide sequences, plasmid constructions, quantitative PCR, rhythmicity analysis of real-time bioluminescence data, amplitude analysis of real-time bioluminescence data, rhythmicity analysis of quantitative PCR data, over representation analysis of clock-controlled genes, estimation of the number of high-amplitude E-boxes, microarray expression data analysis of genes with predicted clock-controlled elements, affinity analysis of competitive DNA binding data, and in silico analysis of affinity to amplitude mechanism are available in SI Appendix.
Real-Time Circadian Reporter Assays.
Real-time circadian assays were performed as previously described (40) with the following modifications. NIH 3T3 cells (American Type Culture Collection) were grown in DMEM (Invitrogen) supplemented with 10% FBS (JRH Biosciences) and antibiotics (25 units ml−1 penicillin, 25 μg ml−1 streptomycin; Invitrogen). Cells were plated at 5 × 104 cells per well in 24-well plates 24 h before transfection. Cells were transfected with 0.32 μg of plasmids in total (0.13 μg reporter plasmid and 0.19 μg empty plasmid) per well using FuGENE6 (Roche Applied Science) according to the manufacturer's instructions. After 72 h, medium in each well was replaced with 500 μl of culture medium (DMEM/10% FBS) supplemented with 10 mM Hepes (pH 7.2), 0.1 mM luciferin (Promega), antibiotics and 0.01 μM forskolin (nacalai tesque). Bioluminescence was measured with photomultiplier tube (PMT) detector assemblies (Hamamatsu Photonics). The modules and cultures were maintained in a darkroom at 30 °C and interfaced with computers for continuous data acquisition until 96 h after forskolin stimulation. Photons were counted 2 min at 24-min intervals.
Construction, Search, and Design of Putative cis-Acting Elements.
A HMM is a statistical model in which the target system is assumed to be a Markov process with unknown parameters. A HMM describes a probability distribution over input training sequences, i.e., probabilities of the state transition and emission. The extracted model can be used to find the probability of query sequence that is a product of all transition and emission probabilities at training sequences. Nucleotide sequences for known functional clock-controlled elements, 12 E-boxes (18 bp), 10 D-boxes (24 bp), and 15 RREs (23 bp), experimentally verified in previous (21) and current studies (Table S1 and Fig. S2), were used as a training dataset to construct HMMs. We also attempted to construct an HMM for the E'-box, but were unable (i.e., positive controls exhibited poor scores) because of the small number of experimentally validated E'-box (only three: Per2, Bhlhb3, and Cry1) and the relatively short core consensus sequence of the E-box. Thus, we did not use an E'-box HMM in this study. The lengths of these known functional elements were based on our previous experiments (21) and these were sufficient to produce circadian transcriptional activity in circadian reporter assays. These sites were aligned without gaps according to the direction of consensus sequences (TTATG[T/C]AA for the D-box; ref. 22), [A/T]A[A/T]NT[A/G]GGTCA for the RRE; ref. 24). Because the consensus sequence for E-box is palindromic (CACGTG; ref. 19), we generated all possible alignments by changing sequence directions (forward and reverse) and selected one alignment as described below. These alignments were used to build HMMs using hmmt program in the HMMER 1.8.4 software package (31) with default parameters (using sim annealing, starting kT for sim annealing run as 5.0, and multiplier for sim annealing as 0.95). We used the older version 1.8.4 package (the current version is 2.3.2) in this study because the version 2 series was optimized for analysis of protein sequences. Following construction, models were used to search genomic regions for putative clock-controlled elements using the hmmls program with default parameters (by using threshold matches score to report as 0) except use ‘-c’ option only for bidirectionally search. The average score was used in the search for the conserved elements between human and mouse. To select only one alignment for each E-box, we constructed 2048 HMMs of all possible alignments, and calculated match scores of 12 known E-box sequences in directional HMMER search. We selected the alignment that generated the highest average match score for further work.
To design the “high-scoring” and “low-scoring” sequence of clock-controlled elements, bidirectional HMMER searches were performed against all possible sequences of the same lengths as training dataset (18 bp for E-box, 24 bp for D-box, and 23 bp for RRE) that contain ordinary consensus sequence at the center (CACGTG for E-box; ref. 19; TTATGTAA for D-box; ref. 22, [A/T]A[A/T]NT[A/G]GGTCA for RRE; ref. 24), then filtered out those that naturally exist in either the human or mouse genome. The sequence of the highest and lowest score was selected as the “high-scoring” and “low-scoring” sequences, respectively. All HMMER searches, except the directional search in the selection of E-box alignments, were performed bidirectionally. The higher score was adopted if match scores were obtained for both directions at the same position. The training data are available in Table S2. The HMMs are publicly available on the circadian section of the mammalian promoter/enhancer database: http://promoter.cdb.riken.jp/circadian.html.
Competitive DNA Binding Assays.
In vitro transcription/translation of Flag-tagged mouse protein from pMU2-Arntl, pMU2-Clock, pMU2-Bhlhb2, pMU2-Dbp, pMU2-Nfil3, pMU2-Nr1d1, and pMU2-Rora were performed with TNT T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's specifications. In vitro transcribed/translated Arntl and Clock proteins were mixed in equal volume. The complementary oligonucleotides of three tandem repeats sequence of designed and control cis-acting elements, which were labeled with biotin on 5′-end or non-labeled (for competitor) (Hokkaido System Science), were annealed to generate probes. Competitive DNA binding assays were performed with NoShift Transcription Factor Assay Kit (Novagen) according to the manufacturer's specifications with the following modifications. Ten pmol biotinylated annealed oligonucleotides were incubated with competitor oligonucleotides (final concentration were 0, 1, 3, 10, 30, and 100 pmol) and 5 μl of in vitro transcribed/translated reticulocyte lysates in the binding mixture. After the samples bound to streptavidin-coated microassay plate, the wells were washed, and Anti-Flag M2 Monoclonal Antibody-Peroxidase Conjugate (SIGMA) was applied into the each well. The wells were washed, and TMB substrate was added to each sample to develop a colorimetric signal, which was subsequently read on a spectrophotometer at 450 nm (Power Wave XS, BioTek). Nr1d1 and Rora proteins were used with additional modifications. Binding reactions were performed with their own binding buffer (8 mM Tris-HCl, pH 7.5, 40 mM NaCl, 0.4 mM EDTA, 1.6 mM MgCl2, 3.2% Glycerol, 0.4 mM DTT, 0.4 mg/ml BSA, and 0.5 μM poly dI;dC); 1 μM ZnSO4 is further added into the binding buffer for Rora proteins and were incubated for 90 min at room temperature. And NoShift Wash Buffer and NoShift Antibody Dilution Buffer were diluted up to 0.5 × solution using water in dilution for a working solution.
Supplementary Material
Acknowledgments.
We thank Hajime Tei and Yoshiyuki Sakaki for Arntl and Clock expression vectors, Yutaka Suzuki, Sumio Sugano, and Seiichi Hashimoto for advice on the mammalian promoter/enhancer database, Takao Kondo for high-throughput monitoring systems, Hideki Ukai, Ryotaku Kito, and Kazuhiro Yagita for real-time circadian reporter assay, Rikuhiro G. Yamada, Tetsuya J. Kobayashi, and Tekeya Kasukawa for statistical analysis, and Michael Royle for critical reading. This work was supported by RIKEN Research Collaborations with Industry Program (H.R.U.), and in part, by RIKEN Center for Developmental Biology (CDB) internal grant (H.R.U.), New Energy and Industrial Technology Development Organization (NEDO) Scientific Project grant (H.R.U.) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Genome Network Project to H.R.U.). J.B.H. is supported by grants from the National Institute of Mental Health and National Institute of Neurological Disorders and Stroke of the National Institutes of Health (P50MH074924–01, Joseph S. Takahashi, P. I, NIMH; 1R01NS054794–01A2, J.B.H, P. I, NINDS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802636105/DCSupplemental.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 4.Ota T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 5.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 6.Gerhard DS, et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura K, et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: Database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–D101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 10.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuldiner O, Shor S, Benvenisty N. A computerized database-scan to identify c-MYC targets. Gene. 2002;292:91–99. doi: 10.1016/s0378-1119(02)00668-6. [DOI] [PubMed] [Google Scholar]
- 14.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 16.Conkright MD, et al. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 17.Hoh J, et al. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallikas O, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 21.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 22.Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 23.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding HP, Lazar MA. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol Cell Biol. 1993;13:3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 26.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 27.Paquet ER, Rey G, Naef F. Modeling an evolutionary conserved circadian cis-element. PLoS Comput Biol. 2008;4:e38. doi: 10.1371/journal.pcbi.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakahata Y, et al. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharov AA, Dudekula DB, Ko MS. CisView: A browser and database of cis-regulatory modules predicted in the mouse genome. DNA Res. 2006;13:123–134. doi: 10.1093/dnares/dsl005. [DOI] [PubMed] [Google Scholar]
- 30.Loots G, Ovcharenko I. ECRbase: Database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics. 2007;23:122–124. doi: 10.1093/bioinformatics/btl546. [DOI] [PubMed] [Google Scholar]
- 31.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 32.Rozenberg JM, et al. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 34.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 35.Munoz E, Brewer M, Baler R. Circadian Transcription. Thinking outside the E-Box. J Biol Chem. 2002;277:36009–36017. doi: 10.1074/jbc.M203909200. [DOI] [PubMed] [Google Scholar]
- 36.Reinke H, et al. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto A, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim C, et al. clockwork orange Encodes a Transcriptional Repressor Important for Circadian-Clock Amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato TK, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.