Abstract
Proper assembly of RNA into catalytically active three-dimensional structures requires multiple tertiary binding interactions, individual characterization of which is crucial to a detailed understanding of global RNA folding. This work focuses on single-molecule fluorescence studies of freely diffusing RNA constructs that isolate the GAAA tetraloop-receptor tertiary interaction. Freely diffusing conformational dynamics are explored as a function of Mg2+ and Na+ concentration, both of which promote facile docking, but with 500-fold different affinities. Systematic shifts in mean fluorescence resonance energy transfer efficiency values and line widths with increasing [Na+] are observed for the undocked species and can be interpreted with a Debye model in terms of electrostatic relaxation and increased flexibility in the RNA. Furthermore, we identify a 34 ± 2% fraction of freely diffusing RNA constructs remaining undocked even at saturating [Mg2+] levels, which agrees quantitatively with the 32 ± 1% fraction previously reported for immobilized constructs. This verifies that the kinetic heterogeneity observed in the docking rates is not the result of surface tethering. Finally, the KD value and Hill coefficient for [Mg2+]-dependent docking decrease significantly for [Na+] = 25 mM vs. 125 mM, indicating Mg2+ and Na+ synergy in the RNA folding process.
INTRODUCTION
RNA catalytic and biological functionality, such as translation and self-splicing, requires that RNA molecules fold into specific three-dimensional forms (1). Central to achieving and maintaining a correctly folded RNA structure are tertiary contacts (2,3), although the contributions of individual tertiary interactions to the RNA folding pathway and conformational dynamics are not well understood (4,5). Proper RNA folding also requires counterions to minimize repulsions of the negatively charged phosphate backbone through site-specific coordination to the RNA and/or nonspecific (delocalized) electrostatic screening (2,6–15). However, tertiary interactions and counterions do not always lead to the desired result; they can stabilize misfolded or kinetically trapped states, thereby preventing RNA from rapidly achieving a catalytically competent structure (2,16–19). These kinetically trapped states often result in long-lived intermediate conformations and heterogeneous folding rates (20–22). This complicated folding behavior is not limited to large ribozymes with multiple domains, with similarly complex behavior noted even for simpler RNA systems such as the hairpin ribozyme (23,24). Clearly any predictive understanding of RNA folding dynamics will require addressing not only global issues of topological structure, but also individual tertiary interactions and their dependence on cationic environment.
In response to this need, we have been systematically investigating RNA tertiary binding motifs at the single-molecule level to characterize isolated interactions as a function of solution environment. The work presented here focuses on cation-dependent RNA folding due to the ubiquitous GNRA-hairpin tetraloop-receptor interaction (25), whereby a GAAA tetraloop docks into an 11-nucleotide internal receptor loop (3,8). These GAAA tetraloop and receptor structures, both free and receptor-bound, have been extensively studied by NMR and x-ray crystallography (8,26–28). Thermodynamic and kinetic contributions of the tetraloop-receptor interaction to RNA folding have been investigated at the ensemble level in a variety of cationic environments in isolation (29,30), in P4-P6 domains (4,31) and in group II introns (32). In bulk studies, however, there is no synchronization between folding/unfolding events for different molecules. Therefore, one must either perturbatively induce a short-lived synchronization (e.g., laser-induced temperature or pH jump methods and rapid mixing) to initiate observable folding dynamics, or be restricted to observing static equilibrium properties of the system. Conformational heterogeneity, for example, due to misfolding events, is largely obscured because of the limited resolution of different states by traditional ensemble methods such as gel electrophoresis.
Studies at the single-molecule level allow direct observation of conformational populations, providing a powerful tool for investigating structural fluctuations in nucleic acids (23,24,33–38). In particular, time-resolved, single-molecule fluorescence resonance energy transfer (smFRET) methods allow thorough characterization of RNA-folding dynamics under both equilibrium and nonequilibrium conditions. Folding and unfolding rate constants are directly measurable, intermediate or misfolded conformations are explicitly identifiable, and subpopulations that exhibit different rate constants and conformations (i.e., kinetic and static heterogeneity) are readily distinguishable (23,24,33–36,38–42). Such measurements often profit from the longer observation times afforded by immobilization of the molecule, e.g., tethering to a cover glass. Immobilization raises concerns that surface proximity and/or tethering methods may influence the folding dynamics and/or of functionality of biomolecules (43,44). Seminal studies of the Tetrahymena thermophila and hairpin ribozymes showed that catalysis was unaffected by immobilization (23,38), which suggests that RNA functionality is uninfluenced by surface tethering. Furthermore, Ha and co-workers showed that the kinetic heterogeneity identified for surface-immobilized hairpin ribozymes is also present when the molecules are encapsulated in liposome vesicles (45), where interactions with vesicular walls are thought to be minimal (46,47). Single-molecule studies of freely diffusing species complement tethered-molecule investigations by providing the capability to interrogate and resolve conformational populations in solution without potential for surface interaction. By way of example, Deniz and co-workers have revealed many insights into the [Mg2+]-dependent folding of the hairpin ribozyme through a systematic conformational study utilizing freely diffusing smFRET (37). Additionally, the Mg2+-binding parameters observed for immobilized hairpin ribozymes (KD and Hill coefficient) were found to be quantitatively replicated in freely diffusing RNA (37).
The focus of this work is smFRET studies of freely diffusing RNA containing a single tertiary binding motif, specifically the GAAA tetraloop-receptor interaction (Fig. 1). Folding of this RNA construct occurs when the tetraloop docks into the receptor domain via a flexible, single-stranded A7 arm (Fig. 1) (30,48). In particular, we explore the equilibrium effect of divalent (Mg2+) and monovalent (Na+) ions on the tetraloop-receptor docking/undocking in the absence of surface immobilization. Even for this simple, isolated RNA tertiary interaction, the folding dynamics prove quite rich. Both Mg2+ and Na+ lead to proper formation of the tetraloop-receptor contact, though Na+ permits additional electrostatic relaxation of the undocked structure over the concentration range studied. We also find that Mg2+ and Na+ interact synergistically to enable tetraloop-receptor docking. Specifically, at low [Na+] (≈25 mM), Mg2+ displays highly cooperative Hill plots as a function of Mg2+ concentration. Mg2+-induced folding becomes much more efficient (and correspondingly less cooperative) at higher Na+ concentrations (125 mM Na+), emphasizing the influence of electrostatic screening in the tetraloop-receptor construct. These results highlight the role of electrostatic shielding and conformational relaxation in the undocked species, as further demonstrated by a dramatic [Na+]-dependent increase in both peak locations and widths in the FRET distributions.
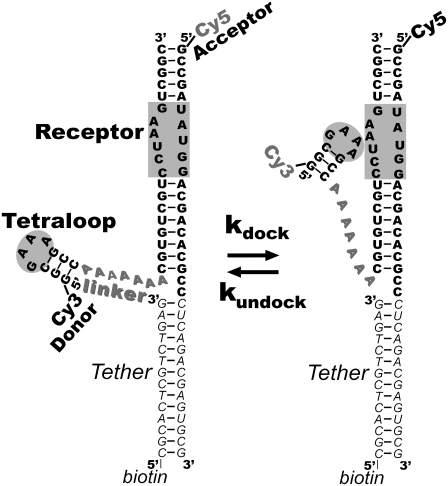
FIGURE 1.
RNA construct for Cy3-Cy5 FRET-monitoring of GAAA tetraloop-receptor docking/undocking. The GAAA tetraloop and receptor are connected by a flexible A7 linker (shadowed gray text) and highlighted in the undocked (left) and docked (right) states. A biotinylated region (italicized) is also retained for quantitative comparison with previous tethered results.
As a secondary thrust, we quantitatively compare our studies of freely diffusing RNA with previous work on the surface-tethered GAAA tetraloop-receptor construct. The previous studies on the immobilized tetraloop-receptor construct revealed significant kinetic heterogeneity, i.e., two populations with distinctly different docking kinetics. These two populations could be characterized as either i), an actively docking population exhibiting multiple docking/undocking transitions during a typical 30 s trajectory before photobleaching and ii), a nondocking population with no transitions evident on a several-minute timescale (30,48). Given the simplicity of this RNA system, such heterogeneity is particularly surprising. The freely diffusing results confirm that the fraction of actively docking versus nondocking populations observed for tethered constructs is native to the RNA. We also find these populations to be independent of salt concentration and therefore not arising from improperly formed secondary structures (i.e., hairpin opening and closing.) Furthermore, quantitative comparison between i), freely diffusing and ii), immobilized conformational populations demonstrate GAAA-tetraloop-receptor folding dynamics to be unaffected by immobilization over a wide range of Mg2+ and Na+ concentrations and cationic environments.
MATERIALS AND METHODS
Preparation of RNA construct
The GAAA tetraloop-receptor tertiary interaction is isolated in an RNA construct (depicted in Fig. 1), where docking and undocking of the tetraloop and receptor domains are coupled by a single-stranded A7 linker. The A7 linker sequence is one of several constructs used to study the effect of the sequence flexibility and length on the dynamics of the tetraloop-receptor interaction (30,48), for which the fraction of actively docking versus nondocking heterogeneity was quantitatively similar. Tetraloop-receptor RNA constructs are prepared as previously described (30,48). Briefly, synthetic amino-modified RNA oligomers (Dharmacon, Lafayette, CO) are labeled with Cy3 and Cy5 N-succinimidyl esters (Amersham Biosciences, Piscataway, NJ) and high performance liquid chromatography purified. (Mention of commercial products is for information only; it does not imply National Institute of Standards and Technology recommendation or endorsement, nor does it imply that products mentioned are necessarily the best available for the purpose.) Annealing of the Cy3- and Cy5-labeled RNA oligomers forms the tetraloop and receptor domains. A DNA oligomer (Integrated DNA Technologies, Coralville, IA) is also hybridized to the complementary extension of one of the RNA oligomers generating the tether domain (Fig. 1). The 14 mer DNA/RNA hybrid tether is biotinylated for immobilization on passivated glass surfaces via biotin-streptavidin chemistry. Though such tethering capabilities are not necessary to perform smFRET experiments on freely diffusing RNA, they enable explicit comparison with our previous studies (30,48). The distances of the Cy3 and Cy5 in the undocked and docked states (Fig. 1) are estimated to be >70 Å and <35 Å, respectively. All solutions for freely diffusing studies of Mg2+-mediated docking are diluted to ∼100 pM RNA in donor (Cy3) strand in a standard buffer containing 50 mM hemisodium HEPES (pH 7.5, Sigma Aldrich, St. Louis, MO), 100 mM NaCl, and 0.1 mM EDTA with varying [MgCl2]. Detailed [Na+]-dependent and monovalent/divalent-synergy studies of the tetraloop-receptor motif are performed in 50 mM hemisodium HEPES (pH 7.5) and 0.1 mM EDTA, with [NaCl] and [MgCl2] varied. Note that 50 mM hemisodium HEPES contains 25 mM Na+ even before added NaCl. All buffers are 0.2 μm sterile filtered and autoclaved before preparing RNA solutions.
Experimental apparatus
To enable smFRET studies of freely diffusing RNA, we have adapted the previously described scanning confocal microscope system (30,48) in a similar manner to Deniz et al. and Rothwell et al. (50,51). Briefly, the fluorescence excitation source is a mode-locked 82 MHz frequency doubled Nd:YAG laser (Model 3800, Spectra Physics, Mountain View, CA), spatially filtered through a single-mode fiber and linear polarizer. Trace infrared light from the Nd:YAG 1064 nm fundamental is removed by a bandpass filter (EX 530/10, Chroma Technology, Rockingham, VT) upon entrance of the excitation laser light into the back port of the microscope. The excitation light is focused through a water immersion objective (Olympus (Center Valley, PA) UPlanApo 60×, 1.2 numerical aperture) to a diffraction-limited spot. Fluorescence emission collected by the microscope objective is isolated from the excitation source with a dichroic beam splitter (550DRLP, Chroma Technology) and spatially filtered with a 50 μm pinhole, limiting detection to the confocal volume. A broad-band-polarizing beam splitter cube divides the fluorescence into vertical and horizontal polarizations with respect to the linearly polarized laser excitation axis. Each polarization is further separated into donor (Cy3) and acceptor (Cy5) channels by a dichroic beam splitter (645DCXR, Chroma Technology), with photon color further defined by transmission through bandpass filters HQ585/70M and HQ700/75M (Chroma Technology), respectively. Photon detection is performed by four avalanche photodiodes (SPCM-AQR-14, Perkin-Elmer Optoelectronics, Fremont, CA). For these studies, however, we focus on unpolarized FRET signals, summing over horizontal/vertical channels. Fluorescence trajectories are acquired for freely diffusing fluorescently labeled RNA constructs using a time-correlated single-photon counting module (SPC-134, Becker & Hickl, Berlin, Germany). A full width half maximum instrument time response of 570 ± 5 ps is measured by prompt Raman scattering signal from H2O.
Sample solutions are studied in a flow-cell sample holder assembled from a fluorinated polymer block (polychlorotrifluoroethylene) by milling a “microchannel” (0.2 mm deep, 10 mm long, 3.0 mm wide) on one of the block faces. Two 0.5 mm diameter holes are drilled in from the opposite side to connect at a 60° angle to the channel ends. A cover glass is attached to the holder with a thin layer of silicone rubber adhesive to form a ∼10 μL volume flow cell. A slot is milled above the channel, leaving only 0.5 mm of polymer above the illumination area to limit back scattering of the excitation source. After flushing in solutions, data are collected under static conditions, with entrance and exit holes covered by tape. All cover glass (22 × 22 mm Corning (Corning, NY) No. 1-1/2) is cleaned by soaking overnight in concentrated nitric acid followed by thorough rinsing with NANOPure water (Barnstead International, Dubuque, IA) and a 45 min treatment with ozone/ultraviolet light (UVO cleaner model 42, Jelight, Irvine, CA). The cover glass surface is passivated by a 120 μL flush and 10 min incubation of bovine serum albumin (10 mg/mL bovine serum albumin), followed by a 1 mL buffer rinse, and then loaded by flowing in 200 μL of the desired RNA solution.
For freely diffusing studies, the laser focus is positioned 15 μm above the bovine serum albumin-passivated cover glass in solution to observe freely diffusing molecule at powers of 50–100 μW. At such powers, photobleaching of freely diffusing molecules is minimal in our experimental apparatus, eliminating the need for an enzymatic oxygen scavenging system (50,52). Labeled RNA diffusing (100 pM) through the confocal excitation region is readily monitored via isolated bursts of fluorescence photons, resulting in a mean occupancy of about λ = 0.03 molecules in a ∼0.5 fL confocal volume. The probability of m molecules in the detection volume is 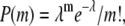 translating into <0.044% probability for events with m ≥ 1.
translating into <0.044% probability for events with m ≥ 1.
FRET analysis of time traces of freely diffusing RNA
Fluorescence photons stored using time-correlated single-photon counting methods are recalled and sorted into 1 ms bins. This is comparable to the mean transit time through the confocal volume, as determined directly from fluorescence correlation spectroscopy (ttransit = 0.38 ± 0.05 ms) (53,54). Based on rate constants obtained from our previous single-molecule studies, this time binning is considerably faster than the conformational dynamics (i.e., 1/kdock or 1/kundock > 5 ms) and therefore predominantly samples single docking/undocking events (48). We can analyze the photon bursts either at the single bin level or with a full-burst algorithm to group photons that most likely arrived from the same molecule (50,53,55). The full-burst-level method, in which several time bins are combined to account for all emission collected from a single molecule's passage through the confocal volume, increases the signal/noise ratio but risks averaging docking/undocking events because of the decreased time resolution (56). For this reason, we choose to analyze bursts at the single-bin level, following the methods proposed by Schultz, Weiss, and co-workers (50,55).
To distinguish RNA emission events from background, a threshold fluorescence signal is determined by a minimum sum of donor and acceptor photons per bin. This criterion requires signal levels (typically 25–35 kHz) to be >10 times the standard deviation of the background, which is sufficient to distinguish labeled RNA constructs with high statistical significance (50). The choice of threshold has been systematically varied and exhibits negligible effects on the results presented here. The FRET efficiency (EFRET),
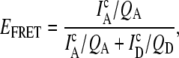 |
(1) |
can be calculated for each event from the corrected donor and acceptor intensities ( and
and  ), with QD and QA as the corresponding quantum yields. The corrected donor and acceptor intensities differ from background-corrected intensities on the detection channels ΔI1 and ΔI2, respectively, because of i), collection efficiencies of donor and acceptor emission on both channels (i.e.,
), with QD and QA as the corresponding quantum yields. The corrected donor and acceptor intensities differ from background-corrected intensities on the detection channels ΔI1 and ΔI2, respectively, because of i), collection efficiencies of donor and acceptor emission on both channels (i.e.,  and ii), nonnegligible direct excitation of the acceptor (i.e., αD versus αA, where αD + αA = 1). Corrections have been formulated for bulk FRET and single-molecule studies, but not including each of the above contributing factors (57,58). Weiss and co-workers have developed elegant methods for implementing such corrections using dual-laser excitation (59). Expressions for these corrections suitable under single-laser conditions are presented below.
and ii), nonnegligible direct excitation of the acceptor (i.e., αD versus αA, where αD + αA = 1). Corrections have been formulated for bulk FRET and single-molecule studies, but not including each of the above contributing factors (57,58). Weiss and co-workers have developed elegant methods for implementing such corrections using dual-laser excitation (59). Expressions for these corrections suitable under single-laser conditions are presented below.
Cross talk correction is implemented by expressing the experimentally observed intensity vector (ΔI1, ΔI2) in terms of the actual donor/acceptor emission vector (ID, IA) via a 2 × 2 matrix equation,
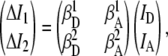 |
(2) |
where absolute collection efficiencies of the donor and acceptor on channels 1 and 2,  (0.00000 ± 0.00003),
(0.00000 ± 0.00003), 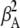 (0.0242 ± 0.0018),
(0.0242 ± 0.0018), 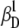 (0.0269 ± 0.0024), and
(0.0269 ± 0.0024), and 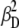 (0.00211 ± 0.00018), are obtained from observed (ΔI1 and ΔI2) signals for known emission rates from donor and acceptor only constructs. The corresponding expressions for ID, IA, and ID/IA are obtained via matrix inversion of Eq. 2:
(0.00211 ± 0.00018), are obtained from observed (ΔI1 and ΔI2) signals for known emission rates from donor and acceptor only constructs. The corresponding expressions for ID, IA, and ID/IA are obtained via matrix inversion of Eq. 2:
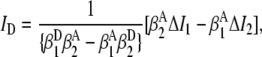 |
(3) |
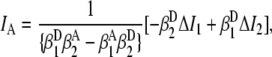 |
(4) |
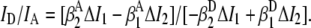 |
(5) |
Correction for direct excitation of the acceptor is most simply incorporated in a modified expression for EFRET:
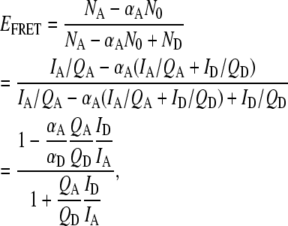 |
(6) |
where N0 = NA + ND = (IA/QA + ID/QD) represents the sum of acceptor and donor excitations and therefore αAN0 is the fractional direct excitation of the acceptor. The quantum yield ratio (QA/QD) = 1.2 ± 0.3 ratios for Cy3 and Cy5 is determined independently via studies of singly labeled constructs. The fraction of acceptor direct excitation (αA = 0.07 ± 0.01, αD = 0.93 ± 0.01) can be obtained from extinction coefficients of singly labeled donor and acceptor at the direct excitation wavelength, 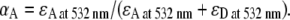 Thus, a corrected EFRET can be readily calculated from the experimental ΔI1 and ΔI2 signals via Eqs. 5 and 6 when collection efficiencies and quantum yields are measured.
Thus, a corrected EFRET can be readily calculated from the experimental ΔI1 and ΔI2 signals via Eqs. 5 and 6 when collection efficiencies and quantum yields are measured.
Note that cross talk and direct excitation corrections in the above expressions treat all experimentally observed intensities equivalently, irrespective of whether photons came from donor-acceptor labeled RNA constructs or donor-only molecules. Consequently, 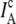 is overcorrected by the removal of acceptor direct excitation when the acceptor is missing or bleached. Thereby, the correction yields negative EFRET values for donor-only molecules, which can be readily identified and isolated in EFRET histograms. As a consistency check, the correction procedure can be tested by removing this direct excitation correction for a sample of donor-only molecules, which, as expected, yields EFRET = 0.01 ± 0.01, centered within uncertainty around zero.
is overcorrected by the removal of acceptor direct excitation when the acceptor is missing or bleached. Thereby, the correction yields negative EFRET values for donor-only molecules, which can be readily identified and isolated in EFRET histograms. As a consistency check, the correction procedure can be tested by removing this direct excitation correction for a sample of donor-only molecules, which, as expected, yields EFRET = 0.01 ± 0.01, centered within uncertainty around zero.
RESULTS AND DISCUSSION
Mg2+-induced tetraloop and receptor docking in freely diffusing RNA
Burst fluorescence methods are used to observe [Mg2+]-dependent folding due to a single GAAA tetraloop-receptor interaction in freely diffusing RNA constructs with Cy3 donor (Cy5 acceptor) fluorescence tags positioned near the tetraloop(receptor), respectively. Docking of the tetraloop and receptor brings the dye pair in closer proximity; from 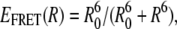 this translates into readily measurable changes in FRET efficiency, where R0 = 53 Å is the calculated Cy3/Cy5 Förster radius for 50% energy transfer probability. Fluorescence time trajectories are obtained as RNA molecules freely diffuse through the confocal detection volume. The left panel in Fig. 2 A shows sample fluorescence traces as a function of time at low and high [Mg2+]. A clear dominance of donor bursts is evident at low [Mg2+] (Fig. 2 A, top), which shifts to predominantly acceptor emission under high [Mg2+] conditions (Fig. 2 A, bottom).
this translates into readily measurable changes in FRET efficiency, where R0 = 53 Å is the calculated Cy3/Cy5 Förster radius for 50% energy transfer probability. Fluorescence time trajectories are obtained as RNA molecules freely diffuse through the confocal detection volume. The left panel in Fig. 2 A shows sample fluorescence traces as a function of time at low and high [Mg2+]. A clear dominance of donor bursts is evident at low [Mg2+] (Fig. 2 A, top), which shifts to predominantly acceptor emission under high [Mg2+] conditions (Fig. 2 A, bottom).
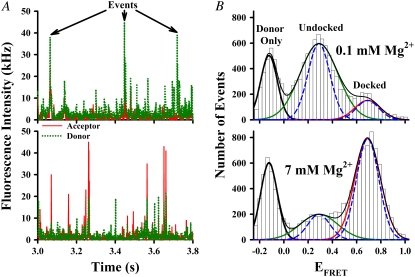
FIGURE 2.
Sample smFRET data analysis of freely diffusing tetraloop-receptor RNA. (A) Sample time traces of donor (dotted green lines) and acceptor (solid red lines) intensities at 0.1 mM Mg2+ (top) and 7 mM Mg2+ (bottom) indicate photon burst events as a molecule traverses the laser focal volume. (B) EFRET histograms generated from events that exceed a 25 kHz threshold at 0.1 mM Mg2+ (top) and 7 mM Mg2+ (bottom) fit to a sum of three Gaussian distributions (black line). The individual Gaussian components reveal distinct populations of donor-only (EFRET < 0, thick black), undocked (green) and docked (red) constructs. Dashed blue lines represent shot-noise limited line-shape predictions.
Analysis of these fluorescence bursts provides statistical information on conformational probabilities, generated from all time bins with burst counts above the intensity threshold and representing ≈104 RNA constructs passing through the detection region. The corresponding EFRET histograms for the tetraloop-receptor constructs at low and high [Mg2+] are shown in Fig. 2 B, top and bottom, respectively. These EFRET distributions reveal three distinct populations for the tetraloop-receptor construct well fit by a sum of Gaussians (55), with the individual components shown in Fig. 2 B (black, green, and red lines). As predicted, the leftmost histogram peak (〈EFRET〉 = −0.124 ± 0.003) arises from donor-only molecules, resulting in EFRET < 0 when corrected for direct excitation of the missing Cy5. Such donor-only bursts arise from incomplete constructs lacking an acceptor-labeled strand, properly assembled RNA constructs lacking the fluorescent acceptor, as well as from free dye and acceptor photobleaching. The integrated number of such donor-only burst events is typically <30% of the total and clearly resolved from each of the physically relevant peaks with EFRET > 0.
In a simple two-state model, the two well-resolved peaks at EFRET > 0 correspond to the conformations of the tetraloop-receptor construct. The peaks centered at a low EFRET (0.28 ± 0.01) and high EFRET (0.687 ± 0.005) reflect the undocked and docked populations, consistent with predictions (EFRET ≈ 0.2, EFRET ≈ 0.8) based on modeling the donor/acceptor separation with helical RNA constraints. Dependence of tetraloop-receptor docking on [Mg2+] is evident in the EFRET histograms; the docked state is far more stable at high [Mg2+], whereas the undocked state is favored at low [Mg2+] (Fig. 2 B).
Closer inspection of the peak widths in Fig. 2 B provides additional insight into the nature of docked versus undocked states for the tetraloop-receptor interaction. The curves (dashed blue lines) in Fig. 2 B represent shot-noise limited peaks, where the width, σSN, is the standard deviation of the Gaussian distribution about a mean EFRET (〈EFRET〉) due to finite photon statistics for donor and acceptor intensities. From standard-error propagation, this shot-noise broadening is σSN = (Em(1 − Em)/T)1/2, where Em is the 〈EFRET〉 and T is the photon threshold for event identification (56). Gaussian fits to the experimental histograms reveal the undocked and docked peak widths to be 1.8 ± 0.2 and 1.1 ± 0.1 times the shot-noise limit, respectively. Deviation from the shot-noise-limited behavior is therefore substantial for the undocked peak, whereas the docked peak indicates no significant broadening beyond shot noise (Fig. 2 B).
There are many possible dynamical sources of such EFRET broadening effects. For example, this broadening can arise from acceptor photobleaching or blinking during passage through the laser focus, as demonstrated in studies by Weiss, Seidel, and co-workers (60,61). However, such broadening is small with respect to what we measure, and would contribute to asymmetric tailing of undocked and docked populations toward the donor-only peak (60,61), which is not evident in the data. Other broadening mechanisms include hindered rotational motion, spectral diffusion, and/or quantum-yield fluctuations of the dye labels. However, each of these have been ruled out as likely broadening mechanisms in Alexa 488-Cy5 FRET pairs by Antonik et al., for which we anticipate behavior similar to Cy3-Cy5 (61). Most relevantly, the aforementioned broadening sources should affect both the undocked and docked states. However, linewidth broadening is evident for only the undocked state, with nearly shot-noise limited predictions for the docked state, and therefore is not likely originating from such photophysical effects. Finally, as will be discussed later, only the undocked peak widths and EFRET values exhibit sensitivity to monovalent (Na+) cation concentration.
A simple model for the observed broadening is that the FRET values reflect a distribution over conformational fluctuations in the nominally two-state picture of docked and undocked states. Docking of the tetraloop with the receptor confines the donor and acceptor to a smaller range of distances and directions, thereby greatly reducing variation in the observed FRET efficiencies. Thus, one would expect negligible dynamical contributions to the docked state widths, in agreement with the nearly shot-noise limited values observed experimentally. Conversely, since flexible motion of the tetraloop linker allows the undocked state to sample a variety of conformations and distances, one would anticipate the undocked EFRET peak to be dynamically broadened beyond the shot-noise limit. Most importantly, this model provides a physical basis for understanding [Na+]-dependent shifts in the mean EFRET values for the undocked state due to ionic strength and Debye shielding effects, which will be addressed later.
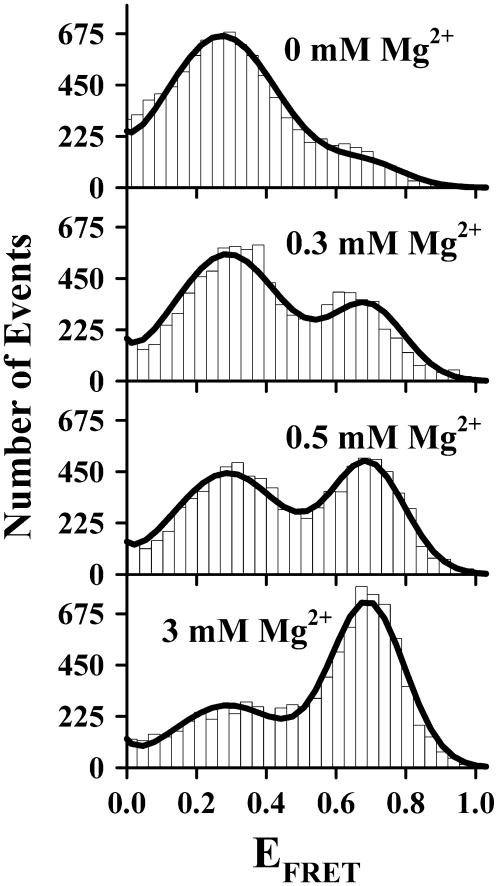
For the moment, however, we focus on [Mg2+]-dependent trends in the tetraloop-receptor docking. Systematic evolution of the docked and undocked populations under standard HEPES buffer conditions (125 mM Na+, pH 7.5) is demonstrated in Fig. 3 for sample concentrations along the [Mg2+] titration by the growth of the high EFRET peak and concomitant reduction of the low EFRET peak, where the EFRET histograms contain the same quantity of total positive EFRET events. Since this occurs over a very small change in total ionic strength, we treat the GAAA tetraloop-receptor interaction as a two-state system, i.e., with distinguishable undocked and docked RNA subpopulations. To determine the fraction of burst events in these subpopulations, the complete set of EFRET histograms has been fit simultaneously to a sum of three Gaussians, with peak widths and centers as adjustable but common parameters for all [Mg2+] conditions. This combined fit procedure permits more accurate characterization of docked versus undocked subpopulations, particularly for the small undocked and docked fractions obtained at the extreme values of [Mg2+]. The fractional populations of undocked and docked species are then readily determined by integration over the respective peaks (50,56).
FIGURE 3.
EFRET population histograms as a function of [Mg2+] with Gaussian fits superimposed. The tetraloop-receptor interaction is promoted by Mg2+, as evidenced by the shift in the relative populations from undocked (low EFRET) to docked (high EFRET) states.
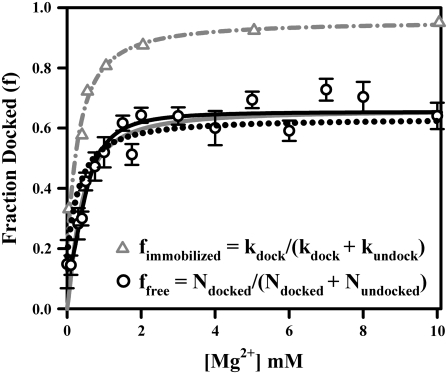
The fractional docked population under freely diffusing conditions, denoted by ffree = Ndocked/(Ndocked + Nundocked), where Ndocked and Nundocked are determined from the integrated Gaussian area of the docked and undocked peaks (37), is plotted versus [Mg2+] as circles in Fig. 4. The data rise smoothly between 0 mM and 1 mM Mg2+ i), from a small but nonzero intercept and ii), reach an asymptotic value less than unity under saturating Mg2+ concentrations. Based on a simple two-state kinetic model summarized in Fig. 5 A, this Mg2+-dependent docked fraction, ffree, can be reasonably well fit to a standard Hill binding equation (37), with the modification that only a fixed fraction, fmax, of the molecules are able to dock,
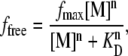 |
(7) |
where [M] is the metal ion concentration. This analysis yields a Mg2+ dissociation constant KD = 0.36 ± 0.6 mM, a Hill coefficient of n = 1.3 ± 0.3 (see Fig. 4, solid gray line) consistent with noncooperative behavior, and an asymptotic docking fraction of fmax = 0.66 ± 0.03. This model is inconsistent with the finite docked fraction (15 ± 8%) experimentally observed at [Mg2+] = 0 mM in Fig. 4, which requires a more complex mechanism (discussed later) involving both divalent Mg2+- and monovalent Na+-mediated docking pathways. Of more immediate relevance, however, the data indicate (1 − fmax) = 0.34 ± 0.03, which would be consistent with 34% of the RNA constructs unable to dock and undock. This nonunity asymptote could in principle suggest that docking proceeds via a Mg2+-bound undocked intermediate, for which the asymptote would reflect the equilibrium of the Mg2+-activated intermediate with the docked state. However, previous studies of surface-immobilized RNA constructs also identified such a subpopulation of nondocking species (48), which is shown below to be quantitatively consistent with the freely diffusing RNA data.
FIGURE 4.
Comparison of Mg2+-dependent fractional docked population for freely diffusing (black circles) and immobilized tetraloop-receptor constructs (gray triangles and dash-dotted line). fimmobilized is calculated from the kinetic rate constants observed in tethered actively docking/undocking constructs, where a nondocking population (32 ± 1%) was previously observed (48). ffree is fit to Eq. 7 (solid gray line), where n = 1.3 ± 0.3, KD = 0.36 ± 0.6 mM, and fmax = 0.66 ± 0.03. Linear scaling of fimmobilized to ffree (Eq. 9) yields 66 ± 2% constructs are actively docking under freely diffusing conditions (dotted black line). ffree is also fit to Eq. 10 (solid black line) derived from the model in Fig. 5 C, which allows for a nonzero docked fraction at 0 mM Mg2+ due to 125 mM Na+.
FIGURE 5.
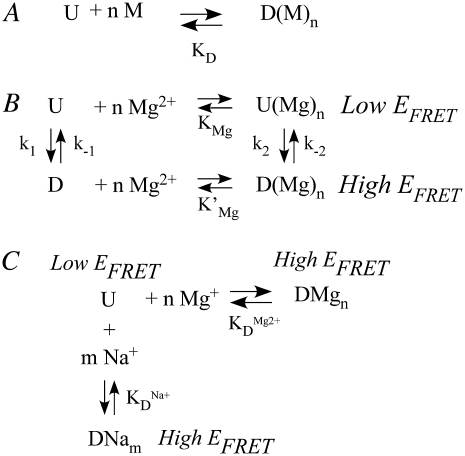
(A) Nominal two-state picture for cooperative binding of metal ions (M) to an undocked state (U), enabling progression to a docked state (D(M)n) with metal ion dissociation constant, KD. (B) Mechanism adapted from Kim et al. (36) to describe docking of the GAAA tetraloop and receptor with and without Mg2+, where KMg and K′Mg are Mg2+ -dissociation constants and the rate constants reflect docking and undocking resolved by FRET. (C) Simplified parallel model to describe [Na+]- and [Mg2+]-dependence for the observed fraction of docked molecules with Mg2+ and Na+ dissociation constants.
Freely diffusing versus immobilized RNA: kinetics and heterogeneity
Single-molecule studies of immobilized tetraloop-receptor constructs indicate that the observed docking (kdock) and undocking (kundock) rate constants increase by 12-fold and decrease by 3-fold, respectively, over a 0–10 mM [Mg2+] range, which motivated a more complex kinetic description than a traditional cooperative-binding model (Fig. 5 A). Of particular relevance to the burst fluorescence results is that these previous tethered studies also provided evidence for kinetic heterogeneity—specifically, a significant fraction (32 ± 1%) of RNA molecules remained permanently undocked on the timescale of photobleaching even under saturating [Mg2+] conditions (48). An important question to consider is whether the observed heterogeneity in kinetic activity is influenced by proximity of immobilized RNA molecules to the protein-passivated glass tethering surface. With burst fluorescence detection, we can now address this question directly by comparing results for freely diffusing RNA with previous data for the immobilized constructs.
To make a quantitative comparison, we invoke the four-state kinetic model described by Kim et al. (36) that includes Mg2+-dependent and independent pathways for the docking of the tetraloop and receptor (Fig. 5 B). According to this model, Mg2+ exchange occurs much faster than subsequent RNA folding/unfolding; thus Mg2+-bound and free states rapidly equilibrate with dissociation constants KMg and K′Mg for undocked and docked states, respectively (Fig. 5 B). Furthermore, the Mg2+-bound and free forms of the undocked and docked states are experimentally indistinguishable by FRET. As a result, the experimental rate constant reflects the combination of k1 and k2 for docking and k−1 and k−2 for undocking, which represent the Mg2+-independent (dependent) pathways, respectively (36,48). Under these conditions, the observed docking and undocking rate constants (Fig. 5 B) are well described by n, k1, k2, k−1, k−2, KMg, and K′Mg, which have already been determined by least-squares analysis of the [Mg2+] dependence of kdock and kundock for the tethered RNA constructs (48).
Due to rapid diffusion, burst methods yield equilibrium population distributions rather than explicit docking and undocking events. An appropriate metric for predicting fractional docked populations from the immobilized studies is therefore
 |
(8) |
where kdock and kundock are experimentally determined and fit as a function of [Mg2+] using the four-state kinetic model (Fig. 5 B), and [U] and [D] are the undocked and docked populations. We can now use data and fits to calculate the equilibrium docking fraction as a continuous function of [Mg2+] under burst conditions. The results are summarized in Fig. 4, where gray triangles represent the docked fraction calculated from rate constants for immobilized species, whereas the gray dash-dotted line represents the corresponding predictions from least-squares fits of kdock and kundock. At high [Mg2+], the freely diffusing data saturate at a docking fraction less than unity, in contrast to the kinetic predictions based on tethered but actively folding/unfolding molecules. More subtly, both the burst data and tethered predictions indicate a finite docking fraction at low [Mg2+], again supporting a Mg2+-independent pathway for folding.
In tethered studies, the rate data are obtained from RNA constructs actively docking and undocking on the millisecond-to-multiple second timescale. Though this correctly reflects the majority RNA population, it does not include the 32% of constructs that exhibit no folding before photobleaching. In the burst studies, all RNAs diffusing through the confocal volume are sampled, including both actively docking/undocking species ([D] and [U]) as well as RNAs not able to dock on the timescale of the experiment ([ND]). Therefore, the freely diffusing and tethered curves in Fig. 4 should be proportional to each other, with a constant scale factor
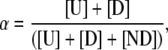 |
(9) |
i.e., ffree = αfimmobilized. We can make this quantitative by least-squares fitting the burst data with a linearly scaled version of the kinetic predictions from the tethered samples, as shown in Fig. 4 (black dotted line) yielding α = 66 ± 2%. This implies a nondocking fraction of 34 ± 2% in the freely diffusing RNA, in agreement with the 32 ± 1% value reported previously for immobilized-RNA constructs. In particular, this confirms that the docking kinetic heterogeneity previously observed is an intrinsic property of the RNA construct and not an artifact of RNA surface immobilization.
Na+-induced docking of the tetraloop and receptor in freely diffusing RNA
The finite intercepts in Fig. 4 indicate the presence of both [Mg2+]-dependent and independent pathways for tetraloop-receptor docking. As a likely source of this [Mg2+]-independent channel, we note that all folding experiments addressed thus far occur in solutions with 125 mM Na+ (100 mM NaCl plus 25 mM Na+ from the 50 mM hemisodium HEPES buffer). Although much weaker, Mg2+ and Na+ have been known to play a role in RNA folding through electrostatic screening and specific binding (2,6,11–13,15,26). For example, Na+-induced folding of the Tetrahymena group I intron requires ∼1700-fold higher concentrations of Na+ than Mg2+ (KD = 460 ± 6 versus 0.270 ± 0.001 mM) (62). Furthermore, at 2500-fold higher concentrations than required for Mg2+, Na+ alone can successfully stabilize the folded conformation of the 16S ribosomal RNA junction (36). In this section, we demonstrate that in the absence of both Na+ and Mg2+, the docking process is effectively blocked by exploiting burst fluorescence time traces for freely diffusing RNA constructs as a function of [Na+].
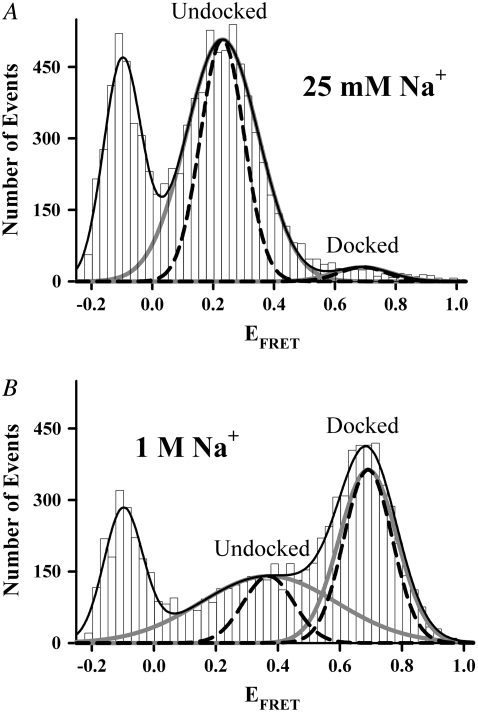
The resulting EFRET distributions from freely diffusing tetraloop-receptor constructs at 0 mM Mg2+ are shown in Fig. 6 for low ([Na+] = 25 mM) and high ([Na+] = 1.0 M) monovalent ion concentrations. At the lower limit of 25 mM Na+ arising from the HEPES buffer, the EFRET distributions are dominated by the undocked conformation (Fig. 6 A), with the docked population visible only very weakly. At [Na+] = 1.0 M, on the other hand, the docked EFRET peak appears (Fig. 6 B) quite prominently, confirming that Na+ can induce docking of the tetraloop and receptor in the absence of Mg2+. However, stabilization of the docked state with Na+ is much weaker than for divalent Mg2+, as indicated by the ≈1000-fold higher range of concentrations required. At the very lowest Na+ concentrations, dissociation of the RNA construct is a possible concern, which would be signaled by an increase in the donor-only population. However, we have tested for this and find no systematic difference in the fractional donor-only population over the range of Mg2+ or Na+ concentrations sampled.
FIGURE 6.
EFRET distributions and Gaussian fits (black lines) showing the donor-only (leftmost peak) population and the individual Gaussian components (gray) and shot-noise predictions (dashed lines) of undocked and docked states at (A) 25 mM Na+ and (B) 1.0 M Na+. Note that the undocked peak shifts to higher center EFRET value and broadens with increasing [Na+] (see text for details).
Relative populations of the docked and undocked states are determined by fitting the Na+-dependent EFRET histograms to Gaussian distributions and calculating fractional populations from integrated areas. Total Gaussian fits (black lines) are shown in Fig. 6, A and B, indicating the undocked and docked components in gray. Shot-noise limited width contributions (black dashed lines) are also shown, again indicating a greater sensitivity of peak widths in the undocked versus docked species. Similar to the Mg2+ results, the docked peak center and width are unaffected by [Na+]. However, in stark contrast with the Mg2+ titration, the undocked peak both shifts and broadens (Fig. 6). More quantitatively, the docked state center EFRET obtained from a global fit of the Na+-titration is 0.691 ± 0.001, in perfect agreement with the EFRET value of 0.687 ± 0.005 obtained from the Mg2+-dependent study. On the other hand, for similar changes in [Na+], the undocked peak shifts by nearly ≈0.200 FRET units and exhibits a ≈2-fold increase in peak width.
The fraction of docked tetraloop-receptor constructs (ffree) is plotted as a function of [Na+] in Fig. 7 A. The data can be well fit by a cooperative-binding model (Eq. 7), yielding an asymptotic saturation fmax = 0.55 ± 0.05, a Hill coefficient 1.3 ± 0.3, and an apparent dissociation constant KD= 180 ± 30 mM. The finite docking fraction at lowest Na+ concentration in Figs. 6 and 7 A is now seen to be completely consistent with residual 25 mM Na+ in the HEPES buffer. However, the KD with respect to Na+ is ∼500-fold larger than that observed for Mg2+, indicating a much lower efficiency for monovalent Na+-mediated docking and comparing quite well with ensemble studies, KD = 220 ± 9 mM (30). Interestingly, fmax is nearly equal for both Na+- and Mg2+-dependent studies, 0.55 ± 0.05 and 0.66 ± 0.03, respectively. This is again consistent with a constant fraction of RNA constructs unable to dock, but now established over an even broader range of both salt concentration and type of cation.
FIGURE 7.
(A) Least-squares fits of fractional docked population (Ndocked/(Ndocked + Nundocked)) versus [Na+] to Eq. 7, resulting in fmax = 0.55 ± 0.05, a Hill coefficient 1.3 ± 0.3, and KD = 180 ± 30 mM. The asymptotic value (fmax) is consistent with Mg2+ studies in Fig. 5, suggesting a ≈32–34% nondocking RNA subpopulation. (B) ffree as a function of [Mg2+] at low [Na+] (25 mM) with a fit to Eq. 10 that also allows for a [Na+] docking pathway (Fig. 5 C), yielding fmax = 0.55 ± 0.04, n = 8 ± 2, and 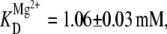 and demonstrating high cooperativity with respect to Mg2+ observed in the presence of minimal Na+.
and demonstrating high cooperativity with respect to Mg2+ observed in the presence of minimal Na+.
As a consistency check, the Na+ titration data can predict the nonzero intercept value previously noted for [Mg2+] = 0 mM with [Na+] = 125 mM. For these purposes, the divalent and monovalent docking processes can be assumed to be independent, as denoted by the kinetic scheme in Fig. 5 C. At equilibrium, the fraction of docked molecules for independent Na+ and Mg2+-induced folding can be easily shown to be
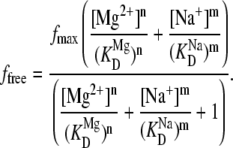 |
(10) |
Based on least-squares fitting of the Na+ titration, the predicted intercept for the fraction docked Mg2+ titration is ffree = 0.2 ± 0.1 at 0 mM Mg2+, 125 mM Na+, which is in excellent agreement with the experimental values fimmobilized = 0.19 ± 0.07 from Eqs. 8 and 9 and ffree = 0.15 ± 0.08. Consequently, the folding in the absence of Mg2+ observed in immobilized and freely diffusing studies can be quantitatively attributed to the presence of 125 mM Na+. Furthermore, this model allows us to describe nonzero intercept of the [Mg2+]-dependent fraction docked (Fig. 4), whereas the simple cooperative binding model (Eq. 7) could not. Using Eq. 10 to fit ffree yields a more precise and physical description of the observed [Mg2+]-dependent docking of the tetraloop-receptor with 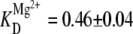 and n = 2.0 ± 0.4 (Fig. 4, solid black line fit to the black circles).
and n = 2.0 ± 0.4 (Fig. 4, solid black line fit to the black circles).
Although Eq. 10 and the model in Fig. 5 C assume independent docking pathways for Na+ and Mg2+, there is evidence that this is not true at low Na+ concentrations. For example, Fig. 7 B displays a titration curve as a function of Mg2+ for low Na+ concentrations (25 mM). As expected from Eq. 10, the intercept nicely matches the value for 25 mM Na+ and 0 mM Mg2+ in Fig. 7 A. Furthermore, at high Mg2+ levels, the docked fraction rises to the typical asymptotic value (fmax ≈ 0.6) seen in other titrations, which again originates from the heterogeneous presence of nondocking RNA constructs. Quite different, however, is the dramatically sigmoidal shape of the titration curve, which implies a much higher level of Mg2+ cooperativity in the absence of Na+. More quantitatively, the data in Fig. 7 B can be fit to the Hill curve of Eq. 10, yielding  and a Hill coefficient of n = 8 ± 2. By way of comparison, when the data at higher Na+ 125 mM are fit to the same model (Fig. 4, solid black line fit to the black circles), these values decrease (i.e., become less cooperative) to
and a Hill coefficient of n = 8 ± 2. By way of comparison, when the data at higher Na+ 125 mM are fit to the same model (Fig. 4, solid black line fit to the black circles), these values decrease (i.e., become less cooperative) to 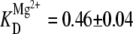 and n = 2.0 ± 0.4. Alternatively stated, the trends in affinities and Hill coefficients indicate that monovalent Na+ greatly diminishes or eliminates docking cooperativity with respect to Mg2+. Examples of high Mg2+ cooperativity at very low Na+ have been noted in previous investigations of the Tetrahymena ribozyme and its P4-P6 domain (62,63). However, the remarkable feature in our study is that such high cooperativities can be exhibited in much simpler RNA constructs docking via a single tertiary interaction. This would also suggest that electrostatic shielding of the phosphate groups by monovalent ions is necessary to facilitate efficient, noncooperative Mg2+-induced docking as discussed in the following two sections.
and n = 2.0 ± 0.4. Alternatively stated, the trends in affinities and Hill coefficients indicate that monovalent Na+ greatly diminishes or eliminates docking cooperativity with respect to Mg2+. Examples of high Mg2+ cooperativity at very low Na+ have been noted in previous investigations of the Tetrahymena ribozyme and its P4-P6 domain (62,63). However, the remarkable feature in our study is that such high cooperativities can be exhibited in much simpler RNA constructs docking via a single tertiary interaction. This would also suggest that electrostatic shielding of the phosphate groups by monovalent ions is necessary to facilitate efficient, noncooperative Mg2+-induced docking as discussed in the following two sections.
Electrostatic relaxation of tetraloop-receptor RNA
The importance of electrostatic screening and relaxation effects in the tetraloop-receptor system is evidenced by a systematic increase in EFRET (and thus decrease in the fluorophore separation) in the undocked state with cation concentration (Fig. 6). To investigate this further, we plot experimental shifts in the undocked 〈EFRET〉 as a function of sodium ion concentration (Fig. 8 A). Upon addition of Na+, the undocked 〈EFRET〉 increases significantly, consistent with partial relaxation of the RNA structure and a statistically closer approach of donor and acceptor. Specifically, EFRET center changes by ≈60%, corresponding to ≈13% (≈7 Å) decrease in Cy3-Cy5 separation. A Hill-type expression commonly used in ensemble FRET studies of two-state systems (30) proves convenient to characterize the [cation]-dependent relaxation of the tetraloop-receptor RNA,
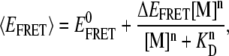 |
(11) |
which for the [Na+]-dependent data yields KD = 180 ± 20 mM, n = 2.1 ± 0.4, 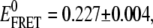 and
and 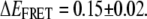 It is worth noting that the KD from fits to Na+-mediated FRET peak shifts is identical within uncertainty (KD = 180 ± 30 mM, Fig. 7 A) to that observed for the undocking/docking transition itself, in support of a common electrostatic origin for both phenomena. Although much smaller in magnitude, a similar Hill-type dependence is observed as a function of Mg2+ for low 25 mM Na+ conditions, as plotted in Fig. 8 B. Once again, the overall highly cooperative shape and affinities (KD = 0.9 ± 0.2 mM) are nearly identical to the corresponding values (KD = 1.06 ± 0.03 mM, Fig. 7 B) obtained for Mg2+-dependent docking at low [Na+]. Interestingly, we do not see appreciable mean EFRET shifts as a function of 0–11 mM Mg2+ under “normal” HEPES buffer, i.e., 125 mM Na+ (see Fig. 2 B). However, from Fig. 8 A, one can easily see that Na+-induced FRET shifts for the undocked peak under these conditions dominate any Mg2+-induced shifts.
It is worth noting that the KD from fits to Na+-mediated FRET peak shifts is identical within uncertainty (KD = 180 ± 30 mM, Fig. 7 A) to that observed for the undocking/docking transition itself, in support of a common electrostatic origin for both phenomena. Although much smaller in magnitude, a similar Hill-type dependence is observed as a function of Mg2+ for low 25 mM Na+ conditions, as plotted in Fig. 8 B. Once again, the overall highly cooperative shape and affinities (KD = 0.9 ± 0.2 mM) are nearly identical to the corresponding values (KD = 1.06 ± 0.03 mM, Fig. 7 B) obtained for Mg2+-dependent docking at low [Na+]. Interestingly, we do not see appreciable mean EFRET shifts as a function of 0–11 mM Mg2+ under “normal” HEPES buffer, i.e., 125 mM Na+ (see Fig. 2 B). However, from Fig. 8 A, one can easily see that Na+-induced FRET shifts for the undocked peak under these conditions dominate any Mg2+-induced shifts.
FIGURE 8.
Evidence for a cation-induced increase in electrostatic compaction and conformational sampling of the undocked state tetraloop-receptor construct. (A and B) Systematic shift in mean EFRET of the undocked peak with increasing [Na+] and [Mg2+], respectively, fit by a Hill-type model (Eq. 11) with  0.227 ± 0.003;
0.227 ± 0.003; 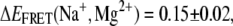 0.07 ± 0.02, n(Na+, Mg2+) = 2.1 ± 0.4, 2.6 ± 0.8, and KD(Na+, Mg2+) = 180 ± 20 mM, 0.9 ± 0.2 mM. (C and D) Systematic shifts in undocked EFRET peak widths as function of [Na+] and [Mg2+], respectively, yielding KD(Na+, Mg2+) = 0.23 M ± 0.02, 1.2 ± 0.4 mM; n(Na+, Mg2+) = 3.6 ± 0.8, 2.7 ± 1.2; and Δσ(Na+, Mg2+) = 0.10 ± 0.01, 0.07 ± 0.03, respectively.
0.07 ± 0.02, n(Na+, Mg2+) = 2.1 ± 0.4, 2.6 ± 0.8, and KD(Na+, Mg2+) = 180 ± 20 mM, 0.9 ± 0.2 mM. (C and D) Systematic shifts in undocked EFRET peak widths as function of [Na+] and [Mg2+], respectively, yielding KD(Na+, Mg2+) = 0.23 M ± 0.02, 1.2 ± 0.4 mM; n(Na+, Mg2+) = 3.6 ± 0.8, 2.7 ± 1.2; and Δσ(Na+, Mg2+) = 0.10 ± 0.01, 0.07 ± 0.03, respectively.
It is worth considering whether these [cation]-dependent peak shifts and broadenings could be induced by changes in donor versus acceptor quantum yields. However, these effects are minor based on both the magnitude and selectivity with which the undocked versus docked peak shift. In the case of Na+, for example, to match ΔEFRET ≈ 0.14 for the undocked peak would require QA/QD to be reduced by >50%, which is observed neither in the donor nor acceptor signal intensities nor analysis of the fluorescence lifetimes. Furthermore, such a reduction would predict a corresponding shift of ΔEFRET ≈ 0.08 in the docked peak, i.e., which is nearly an order of magnitude larger than our experimental uncertainties and not observed. Thus, shifts in the undocked 〈EFRET〉 values must correspond to statistically significant structural changes in average donor-acceptor distances for the RNA construct.
Further confirmation of a cation-mediated structural flexibility in the undocked RNA constructs is also provided by the broadening of the undocked EFRET peak with increased [Na+]. Specifically, the undocked Gaussian peak width, σ, increases from (1.7 ± 0.1)- to (2.7 ± 0.3)-fold excess of shot noise from 25 mM to 1 M Na+ (Fig. 8 C), i.e., greatly exceeding other potential broadening contributions (e.g., triplet formation and photobleaching, as described above) under these experimental conditions. By way of contrast, the docked peak maintains a fixed width near the shot-noise limit (1.2 ± 0.1-fold) over the same range of [Na+]. More quantitatively, the undocked widths (σ) are fit to a Hill-type model (Eq. 11), yielding KD = 230 ± 20 mM, n = 3.6 ± 0.8 and Δσ = 0.10 ± 0.01 (in Fig. 8 C). Once again, this behavior occurs over a comparable range of KD values observed for both i), docking fraction (KD = 180 ± 30 mM) and ii), undocked peak shifts (KD = 180 ± 20 mM), consistent with a common origin of electrostatic screening.
A similar analysis of the undocked peak linewidths as a function of [Mg2+] under low 25 mM Na+ conditions is plotted in Fig. 8 D. The data indicate a systematic increase in linewidths, with an increased KD (1.2 ± 0.4 mM) and a large Hill coefficient (n = 2.7 ± 1.2) that again illustrate high levels of cooperativity with respect to Mg2+. This behavior agrees quantitatively with what was seen previously under low Na+ conditions for Mg2+-dependent i), docking fraction (KD = 1.06 ± 0.03 mM, n = 8 ± 2, Fig. 7 B) and ii), undocked peak shifts (KD = 0.9 ± 0.2 mM, n = 2.6 ± 0.8, Fig. 8 B), underscoring the importance of electrostatic screening in conformational dynamics for the undocked constructs. It is worth reiterating that the undocked peak contains contributions from both actively docking/undocking RNA and nondocking species, as described above, but at high [Na+] the nondocking species is the dominant component. Therefore, electrostatic relaxation of the undocked structure must be responsible for the peak broadening and shifts with increased cation concentration, rather than any fast dynamics due to rapid tetraloop-receptor docking/undocking.
We interpret the origin of the peak shifting and broadening for Na+ as arising from structural relaxation in the undocked RNA constructs, induced by electrostatic screening in an aqueous electrolytic medium. Poisson-Boltzmann or counterion condensation theories may be used to describe the role of monovalent versus divalent ions in structural relaxation of RNA (64). However, due to the polyanionic nature of RNA, a more sophisticated nonlinear Poisson-Boltzmann analysis is sometimes necessary for quantitative determination of the electrostatic potential throughout the entire RNA structure (11,65–71). However, such numerically intensive schemes (69,71,72) often fail to quantitatively model the role of higher order valencies (e.g., Mg2+) in RNA folding (73). Therefore, to provide a qualitative physical interpretation for Mg2+- and Na+-induced conformational changes in the tetraloop-receptor construct, we use a simpler Debye shielding model of point charges screened as a function of ionic strength. Such a Debye treatment slightly underestimates the actual electrostatic screening, though matching all Poisson-Boltzmann trends as a function of ionic strength (74). The relevant parameter in such a model is the 1/e Debye length, 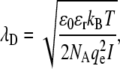 where ɛ0 is the permittivity of free space (8.85 × 10−12 C2/Nm2), ɛr the dielectric constant of water (80.4), qe the elementary charge (1.6 × 10−19 C), kB Boltzmann's constant, T the absolute temperature, NA Avogadro's number, and
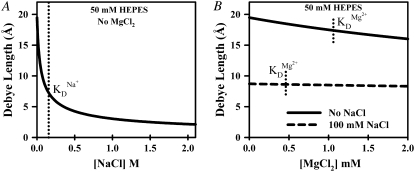
where ɛ0 is the permittivity of free space (8.85 × 10−12 C2/Nm2), ɛr the dielectric constant of water (80.4), qe the elementary charge (1.6 × 10−19 C), kB Boltzmann's constant, T the absolute temperature, NA Avogadro's number, and 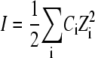 represents ionic strength for a Ci molar concentration of each ion with charge Zi. To illustrate ionic strength effects on charge screening, λD is plotted in Fig. 9 for relevant Na+ and Mg2+ concentrations in 50 mM hemisodium HEPES. The Debye length decreases dramatically from λD ∼ 20 Å to λD < 5 Å (Fig. 9 A) over a comparable range ([Na+] ≈ KD ≈ 180 mM) for which the undocked structure relaxes and broadens (Fig. 8, A and C). By way of contrast, the effect of [Mg2+] on Debye length is completely masked in HEPES buffer with the standard 100 mM NaCl (λD ≈ 8 Å over the entire [Mg2+] range), but becomes much more relevant when NaCl is excluded (Fig. 9 B), consistent with experimental observations in Fig. 7 B and Fig. 8, B and D.
represents ionic strength for a Ci molar concentration of each ion with charge Zi. To illustrate ionic strength effects on charge screening, λD is plotted in Fig. 9 for relevant Na+ and Mg2+ concentrations in 50 mM hemisodium HEPES. The Debye length decreases dramatically from λD ∼ 20 Å to λD < 5 Å (Fig. 9 A) over a comparable range ([Na+] ≈ KD ≈ 180 mM) for which the undocked structure relaxes and broadens (Fig. 8, A and C). By way of contrast, the effect of [Mg2+] on Debye length is completely masked in HEPES buffer with the standard 100 mM NaCl (λD ≈ 8 Å over the entire [Mg2+] range), but becomes much more relevant when NaCl is excluded (Fig. 9 B), consistent with experimental observations in Fig. 7 B and Fig. 8, B and D.
FIGURE 9.
Calculated Debye shielding lengths in the presence of 50 mM hemisodium HEPES buffer with the addition of (A) [NaCl] in the absence MgCl2 and (B) [MgCl2] without and with 100 mM NaCl. Also shown (dotted vertical lines) are the observed KD values for (A) Na+- and (B) Mg2+-facilitated docking.
These monovalent ion effects can be rationalized by efficient screening of phosphate groups in the RNA backbone, leading to reduction in persistence length for the double-stranded and single-stranded regions. This reduction permits greater flexibility and access to more compact RNA conformations, thereby shifting the average EFRET peak for the undocked construct. Likewise, broadening of the EFRET peak can originate from reduced repulsion of the tetraloop and linker from the receptor domain and tether regions of the construct, resulting in a wider distribution of accessible conformations. The observed EFRET peak shifts and broadening effects both occur over a range of [Na+] ∼ 0 −0.5 M, over which the characteristic Debye length decreases by ≈5-fold (Fig. 9 A).
Electrostatic shielding can also be used to interpret the different influences of Mg2+ and Na+ on tetraloop-receptor docking. In the P4-P6 domain of the Tetrahymena ribozyme, both the tetraloop-receptor and adenosine-rich bulge tertiary interactions pack the opposing helical phosphate groups within 8–9 Å (8), requiring tetraloop and receptor helices to be effectively screened for stabilization of the folded structure. The experimentally observed KD for Na+ is consistent with the λD values necessary to allow tetraloop and receptor proximity (see Fig. 9 A). On the other hand, much lower [Mg2+] concentrations are required to achieve the same docked state, with only minor impact on ionic strength and λD (Fig. 9 B). This is consistent with the fact that divalent Mg2+, as a consequence of compact size and high charge density, can intimately localize and coordinate along the RNA at regions of high negative electrostatic potential, whereas monovalent Na+ must rely on the less-efficient mechanism of bulk electrostatic screening (15,67,70,73,75). Similarly, the striking growth in cooperativity for Mg2+-promoted docking can be attributed to the much larger Debye shielding length without NaCl (Fig. 9 B). With insufficient monovalent ionic strength to screen phosphate repulsions, multiple Mg2+ ions must be recruited for relaxing the RNA structure before the more local task of promoting docking, therefore resulting in higher cooperativity and increased KD values. This analysis offers a simple physical model for monovalent-induced structural relaxation of the undocked RNA, consistent with the experimental data, though more rigorous treatment of the electrostatic environment will be necessary to make such comparisons quantitative.
Na+ and Mg2+ synergistically promote tetraloop-receptor docking
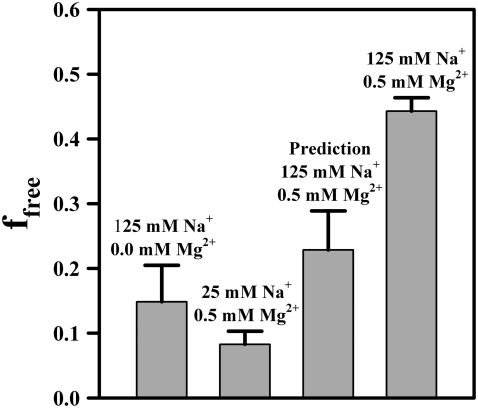
Closer comparison of the cation-dependent population distributions presented in this work also permits one to explore possible competition or synergism between Na+- and Mg2+-induced folding of the tetraloop-receptor constructs. Bokinsky et al. showed that in the presence of 500 mM Na+, the docking transition of the hairpin ribozyme saturates at much higher [Mg2+] than in the absence of Na+, suggesting that Mg2+ and Na+ compete for interaction with the hairpin RNA (33). For the tetraloop and receptor construct, however, we find precisely the opposite scenario: 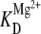 decreases from 1.06 ± 0.03 mM at 25 mM Na+ to 0.46 ± 0.04 mM at 125 mM Na+, whereas at the same time cooperativity with respect to Mg2+ is virtually eliminated (Figs. 7 B vs. 4). This suggests that Na+ enhances the ability of Mg2+ to promote docking, which would imply a more complex description for cation-induced folding than the independent pathways depicted in Fig. 5 C. Synergism between Na+ and Mg2+ in the promotion of the tetraloop-receptor tertiary interaction is best demonstrated by comparing the RNA docking fraction (ffree) for a series of Na+ and Mg2+ concentration pairs. The leftmost two bars in Fig. 10 present data for limits of i), Na+-dominated (125 mM Na+, 0.0 mM Mg2+) and ii), Mg2+-dominated (25 mM Na+, 0.5 mM Mg2+) docking, respectively, with the third bar based simply on the additive Mg2+ and Na+ promoted docking predictions (i + ii). In fact, experimental results (fourth bar) exceed this by nearly twofold, a direct indication that Na+ and Mg2+ interact with positive synergy in promoting the docking event.
decreases from 1.06 ± 0.03 mM at 25 mM Na+ to 0.46 ± 0.04 mM at 125 mM Na+, whereas at the same time cooperativity with respect to Mg2+ is virtually eliminated (Figs. 7 B vs. 4). This suggests that Na+ enhances the ability of Mg2+ to promote docking, which would imply a more complex description for cation-induced folding than the independent pathways depicted in Fig. 5 C. Synergism between Na+ and Mg2+ in the promotion of the tetraloop-receptor tertiary interaction is best demonstrated by comparing the RNA docking fraction (ffree) for a series of Na+ and Mg2+ concentration pairs. The leftmost two bars in Fig. 10 present data for limits of i), Na+-dominated (125 mM Na+, 0.0 mM Mg2+) and ii), Mg2+-dominated (25 mM Na+, 0.5 mM Mg2+) docking, respectively, with the third bar based simply on the additive Mg2+ and Na+ promoted docking predictions (i + ii). In fact, experimental results (fourth bar) exceed this by nearly twofold, a direct indication that Na+ and Mg2+ interact with positive synergy in promoting the docking event.
FIGURE 10.
Evidence for positive Na+ and Mg2+-synergy in promoting tetraloop-receptor docking; ffree for combined Mg2+ and Na+ (rightmost bar) is significantly greater than the prediction (third bar) based on a simple additive model of individual Na+ and Mg2+ results (left two bars).
Electrostatic shielding again provides a simple physical model for positive synergy in the folding of the RNA constructs. At low ionic strengths, additional Mg2+ ions are required to sufficiently relax the RNA before docking, which translates into a large Hill coefficient and increased affinity 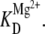 In the presence of only 100 mM NaCl, the Debye length decreases dramatically, cooperativity with respect to Mg2+ is eliminated (as in Fig. 7 B vs. Fig. 4), and
In the presence of only 100 mM NaCl, the Debye length decreases dramatically, cooperativity with respect to Mg2+ is eliminated (as in Fig. 7 B vs. Fig. 4), and 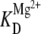 decreases by ≈2-fold because of the increased ionic strength. It would be interesting to test if this synergy changes into competition for docking promotion at even higher Na+ concentrations, as experimentally observed for folding of the hairpin ribozyme (33). Alternatively, the observed Na+-Mg2+ synergy could reflect a specific monovalent binding site that, when filled, facilitates tetraloop-receptor docking. Indeed, Basu et al. have identified a monovalent binding site in the tetraloop-receptor interaction of the P4-P6 domain of the Azoarcus group I intron using x-ray crystallography (26). Increased occupancy of this monovalent site could stabilize tetraloop-receptor binding in a way that complements Mg2+-induced folding and lowers
decreases by ≈2-fold because of the increased ionic strength. It would be interesting to test if this synergy changes into competition for docking promotion at even higher Na+ concentrations, as experimentally observed for folding of the hairpin ribozyme (33). Alternatively, the observed Na+-Mg2+ synergy could reflect a specific monovalent binding site that, when filled, facilitates tetraloop-receptor docking. Indeed, Basu et al. have identified a monovalent binding site in the tetraloop-receptor interaction of the P4-P6 domain of the Azoarcus group I intron using x-ray crystallography (26). Increased occupancy of this monovalent site could stabilize tetraloop-receptor binding in a way that complements Mg2+-induced folding and lowers  Either scenario would be interesting and underscores the importance of studying both the full ribozyme as well as individual tertiary binding interactions at the single-molecule level.
Either scenario would be interesting and underscores the importance of studying both the full ribozyme as well as individual tertiary binding interactions at the single-molecule level.
SUMMARY AND CONCLUSIONS
The [Mg2+]- and [Na+]-dependence of docking for a single GAAA tetraloop-receptor tertiary interaction has been studied under single-molecule fluorescence conditions for freely diffusing RNAs. General expressions are presented that include corrections for cross talk, collection efficiency, quantum yield, and direct excitation of the acceptor for EFRET single-laser excitation. Gaussian fits to the EFRET histograms identify distinct populations corresponding to well-resolved docked versus undocked populations, with EFRET distributions providing a snapshot of equilibrium populations at the single-molecule level.
The fractional docked versus undocked populations have been explored as a function of [Mg2+], which exhibits a nonzero intercept, followed by a rapid increase in folding probability with concentration. The nonzero intercept at Mg2+ = 0.0 M is inconsistent with a two-state Hill analysis and is shown to arise from monovalent Na+-mediated folding in the buffer solution. Independent Na+ titrations indicate efficient folding of the tetraloop-receptor, but with a KD ∼500 times larger than for Mg2+. This shows that both Na+ and Mg2+ lead to equivalent formation of the tetraloop-receptor contact, as supported by identical EFRET values in the docked state. Significant peak shifts (〈EFRET〉) and broadening beyond the shot-noise limit are observed in the undocked but not docked EFRET distributions. This cation-induced peak broadening and shift is interpreted in terms of Debye shielding of the negatively charged RNA backbone, which provides significantly more flexibility to the undocked structures. Furthermore, Mg2+ and Na+ behave synergistically in promoting tetraloop-receptor docking. The presence of even ∼100 mM monovalent Na+ results in a significant decrease in the Mg2+ dissociation constant, as well as eliminating cooperativity as a function of Mg2+. This interdependence of monovalent and divalent ion concentrations raises issues not considered, to our knowledge, in previous kinetic models for tetraloop-receptor docking.
Finally, analysis of the burst studies reveal the existence of a 34 ± 2% subpopulation of nondocking RNA molecules under freely diffusing conditions, in quantitative agreement with smFRET studies of tethered RNA constructs. These results demonstrate that [Mg2+]- and [Na+]-dependent influences on folding kinetics can be studied quantitatively for isolated tetraloop-receptor RNA tertiary interactions under both free and tethered conditions without surface interference. Additional kinetic investigations of tethered RNA should prove invaluable in further testing mechanisms for monovalent/divalent promotion of the tetraloop-receptor interaction.
Acknowledgments
We acknowledge Dr. Arthur Pardi for his contributions to the RNA construct design, as well as for many useful comments in preparation of the manuscript.
Support for this work was provided by the National Science Foundation and by the W. M. Keck Foundation initiative in RNA sciences at the University of Colorado, Boulder. J.L.F. is supported in part by the Optical Science and Engineering Program National Science Foundation-Integrative Graduate Education and Research Traineeship and University of Colorado Biophysics Training (T32 GM-065103) grants. C.D.D was also supported in part by the National Institutes of Health Biophysics Training Grant (GM-065103).
Editor: Samuel Butcher.
References
- 1.Gesteland, R. F., T. R. Cech, and J. F. Atkins. 2006. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Tinoco, I., and C. Bustamante. 1999. How RNA folds. J. Mol. Biol. 293:271–281. [DOI] [PubMed] [Google Scholar]
- 3.Batey, R. T., R. P. Rambo, and J. A. Doudna. 1999. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. 38:2327–2343. [DOI] [PubMed] [Google Scholar]
- 4.Young, B. T., and S. K. Silverman. 2002. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4-P6 RNA domain. Biochemistry. 41:12271–12276. [DOI] [PubMed] [Google Scholar]
- 5.Bartley, L. E., X. W. Zhuang, R. Das, S. Chu, and D. Herschlag. 2003. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 328:1011–1026. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, C. F., and M. T. Record. 1995. Salt nucleic-acid interactions. Annu. Rev. Phys. Chem. 46:657–700. [DOI] [PubMed] [Google Scholar]
- 7.Cate, J. H., and J. A. Doudna. 1996. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 4:1221–1229. [DOI] [PubMed] [Google Scholar]
- 8.Cate, J. H., A. R. Gooding, E. Podell, K. H. Zhou, B. L. Golden, C. E. Kundrot, T. R. Cech, and J. A. Doudna. 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 273:1678–1685. [DOI] [PubMed] [Google Scholar]
- 9.Correll, C. C., B. Freeborn, P. B. Moore, and T. A. Steitz. 1997. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 91:705–712. [DOI] [PubMed] [Google Scholar]
- 10.Pley, H. W., K. M. Flaherty, and D. B. McKay. 1994. 3-dimensional structure of a hammerhead ribozyme. Nature. 372:68–74. [DOI] [PubMed] [Google Scholar]
- 11.Draper, D. E. 2004. A guide to ions and RNA structure. RNA-Publ. RNA Soc. 10:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draper, D. E., D. Grilley, and A. M. Soto. 2005. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34:221–243. [DOI] [PubMed] [Google Scholar]
- 13.Draper, D. E., and V. K. Misra. 1998. RNA shows its metal. Nat. Struct. Biol. 5:927–930. [DOI] [PubMed] [Google Scholar]
- 14.Misra, V. K., and D. E. Draper. 1998. On the role of magnesium ions in RNA stability. Biopolymers. 48:113–135. [DOI] [PubMed] [Google Scholar]
- 15.Woodson, S. A. 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9:104–109. [DOI] [PubMed] [Google Scholar]
- 16.Treiber, D. K., M. S. Rook, P. P. Zarrinkar, and J. R. Williamson. 1998. Kinetic intermediates trapped by native interactions in RNA folding. Science. 279:1943–1946. [DOI] [PubMed] [Google Scholar]
- 17.Treiber, D. K., and J. R. Williamson. 1999. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 9:339–345. [DOI] [PubMed] [Google Scholar]
- 18.Treiber, D. K., and J. R. Williamson. 2001. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 11:309–314. [DOI] [PubMed] [Google Scholar]
- 19.Rook, M. S., D. K. Treiber, and J. R. Williamson. 1999. An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl. Acad. Sci. USA. 96:12471–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thirumalai, D., N. Lee, S. A. Woodson, and D. K. Klimov. 2001. Early events in RNA folding. Annu. Rev. Phys. Chem. 52:751–762. [DOI] [PubMed] [Google Scholar]
- 21.Thirumalai, D., and S. A. Woodson. 1996. Kinetics of folding of proteins and RNA. Acc. Chem. Res. 29:433–439. [Google Scholar]
- 22.Sosnick, T. R., and T. Pan. 2003. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 13:309–316. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang, X. W., H. Kim, M. J. B. Pereira, H. P. Babcock, N. G. Walter, and S. Chu. 2002. Correlating structural dynamics and function in single ribozyme molecules. Science. 296:1473–1476. [DOI] [PubMed] [Google Scholar]
- 24.Tan, E., T. J. Wilson, M. K. Nahas, R. M. Clegg, D. M. J. Lilley, and T. Ha. 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. USA. 100:9308–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa, M., and F. Michel. 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 14:1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu, S., R. P. Rambo, J. Strauss-Soukup, J. H. Cate, A. R. Ferre-D'Amare, S. A. Strobel, and J. A. Doudna. 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 5:986–992. [DOI] [PubMed] [Google Scholar]
- 27.Butcher, S. E., T. Dieckmann, and J. Feigon. 1997. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 16:7490–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jucker, F. M., and A. Pardi. 1995. GNRA tetraloops make a U-turn. RNA. 1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 29.Jaeger, L., and N. B. Leontis. 2000. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed. 39:2521–2524. [DOI] [PubMed] [Google Scholar]
- 30.Downey, C. D., J. L. Fiore, C. D. Stoddard, J. H. Hodak, D. J. Nesbitt, and A. Pardi. 2006. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 45:3664–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, F. L., and T. R. Cech. 1994. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group-I intron domain. J. Mol. Biol. 236:49–63. [DOI] [PubMed] [Google Scholar]
- 32.Abramovitz, D. L., and A. M. Pyle. 1997. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J. Mol. Biol. 266:493–506. [DOI] [PubMed] [Google Scholar]
- 33.Bokinsky, G., D. Rueda, V. K. Misra, M. M. Rhodes, A. Gordus, H. P. Babcock, N. G. Walter, and X. W. Zhuang. 2003. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. USA. 100:9302–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bokinsky, G., and X. W. Zhuang. 2005. Single-molecule RNA folding. Acc. Chem. Res. 38:566–573. [DOI] [PubMed] [Google Scholar]
- 35.Ha, T., X. W. Zhuang, H. D. Kim, J. W. Orr, J. R. Williamson, and S. Chu. 1999. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. USA. 96:9077–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, H. D., G. U. Nienhaus, T. Ha, J. W. Orr, J. R. Williamson, and S. Chu. 2002. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA. 99:4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pljevaljcic, G., D. P. Millar, and A. A. Deniz. 2004. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys. J. 87:457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang, X. W., L. E. Bartley, H. P. Babcock, R. Russell, T. J. Ha, D. Herschlag, and S. Chu. 2000. A single-molecule study of RNA catalysis and folding. Science. 288:2048–2051. [DOI] [PubMed] [Google Scholar]
- 39.Russell, R., X. W. Zhuang, H. P. Babcock, I. S. Millett, S. Doniach, S. Chu, and D. Herschlag. 2002. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA. 99:155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie, S. N. 2001. Single-molecule approach to enzymology. Single Mol. 2:229–236. [Google Scholar]
- 41.Xie, X. S., and H. P. Lu. 1999. Single-molecule enzymology. J. Biol. Chem. 274:15967–15970. [DOI] [PubMed] [Google Scholar]
- 42.Xie, Z., N. Srividya, T. R. Sosnick, T. Pan, and N. F. Scherer. 2004. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc. Natl. Acad. Sci. USA. 101:534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda, R., T. Masaike, K. Adachi, H. Noji, H. Itoh, and K. Kinosita. 2003. The ATP-waiting conformation of rotating F-1-ATPase revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA. 100:9314–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talaga, D. S., W. L. Lau, H. Roder, J. Y. Tang, Y. W. Jia, W. F. DeGrado, and R. M. Hochstrasser. 2000. Dynamics and folding of single two-stranded coiled-coil peptides studied by fluorescent energy transfer confocal microscopy. Proc. Natl. Acad. Sci. USA. 97:13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okumus, B., T. J. Wilson, D. M. J. Lilley, and T. Ha. 2004. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys. J. 87:2798–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boukobza, E., A. Sonnenfeld, and G. Haran. 2001. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J. Phys. Chem. B. 105:12165–12170. [Google Scholar]
- 47.Rhoades, E., E. Gussakovsky, and G. Haran. 2003. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. USA. 100:3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodak, J. H., C. D. Downey, J. L. Fiore, A. Pardi, and D. J. Nesbitt. 2005. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc. Natl. Acad. Sci. USA. 102:10505–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reference deleted in proof.
- 50.Deniz, A. A., M. Dahan, J. R. Grunwell, T. J. Ha, A. E. Faulhaber, D. S. Chemla, S. Weiss, and P. G. Schultz. 1999. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Forster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 96:3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothwell, P. J., S. Berger, O. Kensch, S. Felekyan, M. Antonik, B. M. Wohrl, T. Restle, R. S. Goody, and C. A. M. Seidel. 2003. Multiparameter single-molecule fluorescence spectroscopy reveals heterogeneity of HIV-1 reverse transcriptase: primer/template complexes. Proc. Natl. Acad. Sci. USA. 100:1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grunwell, J. R., J. L. Glass, T. D. Lacoste, A. A. Deniz, D. S. Chemla, and P. G. Schultz. 2001. Monitoring the conformational fluctuations of DNA hairpins using single-pair fluorescence resonance energy transfer. J. Am. Chem. Soc. 123:4295–4303. [DOI] [PubMed] [Google Scholar]
- 53.Fries, J. R., L. Brand, C. Eggeling, M. Kollner, and C. A. M. Seidel. 1998. Quantitative identification of different single molecules by selective time-resolved confocal fluorescence spectroscopy. J. Phys. Chem. A. 102:6601–6613. [Google Scholar]
- 54.Eigen, M., and R. Rigler. 1994. Sorting single molecules: application to diagnostics and evolutionary biotechnology. Proc. Natl. Acad. Sci. USA. 91:5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahan, M., A. A. Deniz, T. J. Ha, D. S. Chemla, P. G. Schultz, and S. Weiss. 1999. Ratiometric measurement and identification of single diffusing molecules. Chem. Phys. 247:85–106. [Google Scholar]
- 56.Deniz, A. A., T. A. Laurence, M. Dahan, D. S. Chemla, P. G. Schultz, and S. Weiss. 2001. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 52:233–253. [DOI] [PubMed] [Google Scholar]
- 57.Jia, Y. W., D. S. Talaga, W. L. Lau, H. S. M. Lu, W. F. DeGrado, and R. M. Hochstrasser. 1999. Folding dynamics of single GCN4 peptides by fluorescence resonant energy transfer confocal microscopy. Chem. Phys. 247:69–83. [Google Scholar]
- 58.Gordon, G. W., G. Berry, X. H. Liang, B. Levine, and B. Herman. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee, N. K., A. N. Kapanidis, Y. Wang, X. Michalet, J. Mukhopadhyay, R. H. Ebright, and S. Weiss. 2005. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88:2939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nir, E., X. Michalet, K. M. Hamadani, T. A. Laurence, D. Neuhauser, Y. Kovchegov, and S. Weiss. 2006. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J. Phys. Chem. B. 110:22103–22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antonik, M., S. Felekyan, A. Gaiduk, and C. A. M. Seidel. 2006. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J. Phys. Chem. B. 110:6970–6978. [DOI] [PubMed] [Google Scholar]
- 62.Heilman-Miller, S. L., D. Thirumalai, and S. A. Woodson. 2001. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol. 306:1157–1166. [DOI] [PubMed] [Google Scholar]
- 63.Deras, M. L., M. Brenowitz, C. Y. Ralston, M. R. Chance, and S. A. Woodson. 2000. Folding mechanism of the Tetrahymena ribozyme P4–P6 domain. Biochemistry. 39:10975–10985. [DOI] [PubMed] [Google Scholar]
- 64.Stigter, D. 1995. Evaluation of the counterion condensation theory of polyelectrolytes. Biophys. J. 69:380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharp, K. A., R. A. Friedman, V. Misra, J. Hecht, and B. Honig. 1995. Salt effects on polyelectrolyte-ligand binding: comparison of Poisson-Boltzmann, and limiting/law counterion binding models. Biopolymers. 36:245–262. [DOI] [PubMed] [Google Scholar]
- 66.Honig, B., and A. Nicholls. 1995. Classical electrostatics in biology and chemistry. Science. 268:1144–1149. [DOI] [PubMed] [Google Scholar]
- 67.Chin, K., K. A. Sharp, B. Honig, and A. M. Pyle. 1999. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 6:1055–1061. [DOI] [PubMed] [Google Scholar]
- 68.Misra, V. K., and D. E. Draper. 2000. Mg2+ binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J. Mol. Biol. 299:813–825. [DOI] [PubMed] [Google Scholar]
- 69.Misra, V. K., and D. E. Draper. 2001. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. USA. 98:12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Misra, V. K., and D. E. Draper. 2002. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 317:507–521. [DOI] [PubMed] [Google Scholar]
- 71.Misra, V. K., R. Shiman, and D. E. Draper. 2003. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 69:118–136. [DOI] [PubMed] [Google Scholar]
- 72.Rocchia, W., E. Alexov, and B. Honig. 2001. Extending the applicability of the nonlinear Poisson-Boltzmann equation: multiple dielectric constants and multivalent ions. J. Phys. Chem. B. 105:6507–6514. [Google Scholar]
- 73.Bai, Y., M. Greenfeld, K. J. Travers, V. B. Chu, J. Lipfert, S. Doniach, and D. Herschlag. 2007. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 129:14981–14988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stigter, D. 1975. Charged colloidal cylinder with a Gouy double-layer. J. Colloid Interface Sci. 53:296–306. [Google Scholar]
- 75.Misra, V. K., and D. E. Draper. 1999. The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J. Mol. Biol. 294:1135–1147. [DOI] [PubMed] [Google Scholar]