Abstract
Single-molecule force spectroscopy studies and steered molecular dynamics simulations have revealed that protein topology and pulling geometry play important roles in determining the mechanical stability of proteins. Most studies have focused on local interactions that are associated with the force-bearing β-strands. Interactions mediated by neighboring strands are often overlooked. Here we use Top7 and barstar as model systems to illustrate the critical importance of the stabilization effect provided by neighboring β-strands on the mechanical stability. Using single-molecule atomic force microscopy, we showed that Top7 and barstar, which have similar topology in their force-bearing region, exhibit vastly different mechanical-stability characteristics. Top7 is mechanically stable and unfolds at ∼150 pN, whereas barstar is mechanically labile and unfolds largely below 50 pN. Steered molecular dynamics simulations revealed that stretching force peels one force-bearing strand away from barstar to trigger unfolding, whereas Top7 unfolds via a substructure-sliding mechanism. This previously overlooked stabilization effect from neighboring β-strands is likely to be a general mechanism in protein mechanics and can serve as a guideline for the de novo design of proteins with significant mechanical stability and novel protein topology.
INTRODUCTION
The mechanical properties of proteins play important roles in a variety of biological processes and also make proteins attractive potential building blocks for nanomechanical applications (1–3). Detailed studies of the mechanical properties of proteins using single-molecule atomic force microscopy (AFM) and steered molecular dynamics (SMD) simulations have revealed valuable insights into the design of mechanically stable proteins (4–11). It is recognized that both protein topology (4–8,12) and pulling geometry (13–15) play important roles in determining the mechanical stability of proteins.
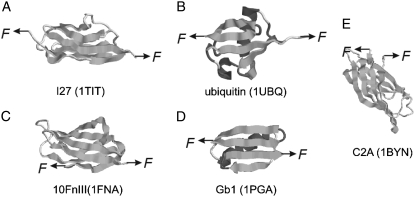
It has been observed that the vast majority of the mechanically stable proteins identified so far share a common structural feature that is characterized by the topology of force-bearing terminal β-strands, where the two terminal β-strands are arranged in parallel and constitute a shear topology. Intensively studied examples include titin I27, ubiquitin, FnIII, and GB1 (Fig. 1) (13,16–21). Upon stretching, the two terminal β-strands shear against each other to provide mechanical resistance to protein unfolding. This structural feature has been considered as a general criterion for proteins to be mechanically stable and has been used as an important condition to screen proteins with significant mechanical stability (17–19).
FIGURE 1.
Shear topology is a common feature of mechanically stable proteins. Images A–D show the three-dimensional structures of representative proteins that are mechanically stable and possess shear topology: (A) I27, (B) ubiquitin, (C) the 10th FnIII domains of fibronectin, and (D) GB1. For comparison, the three-dimensional structure of C2A, which is mechanically weak and possesses a typical unzipping topology, is shown in E.
However, most of the studies to date have focused on force-bearing terminal strands and their associated interactions. Interactions mediated by neighboring β-strands are often overlooked. Here, we use two proteins—Top7 and barstar (which have similar protein topologies)—as model systems to illustrate the critical importance of stabilization provided by neighboring β-strands in defining the mechanical stability of proteins.
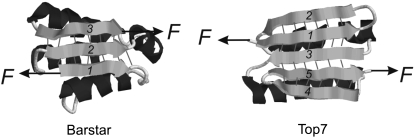
Both proteins are not naturally occurring mechanical proteins. Top7 is a computationally designed protein of 92 residues (22), and barstar is a small protein of 89 residues and is a natural intracellular inhibitor of extracellular ribonuclease barnase (23). From the protein topology perspective, and by looking at the β-sheet of Top7 and barstar alone (Fig. 2), the arrangements of terminal force-bearing β-strands in both proteins appear to be very similar. The two force-bearing strands are pointing to opposite directions and are not directly connected but spaced by a third β-strand (22). All three β-strands are interconnected by backbone hydrogen bonds. This arrangement constitutes a shear topology, which is typical for mechanically stable proteins. The major structural difference in the β-sheet between Top7 and barstar is that there are two additional β-strands flanking the force-bearing strands in the β-sheet of Top7 such that the two force-bearing strands of Top7 are further stabilized from both sides.
FIGURE 2.
Three-dimensional structures of barstar (PDB code: 1BTA) and Top7 (PDB code: 1QYS). The two force-bearing β-strands of barstar and Top7 are arranged in a similar fashion: force-bearing β-strands are arranged in parallel and point in opposite directions. They are spaced by a third β-strand and do not interact with each other directly. Backbone hydrogen bonds (thin bars) in the β-sheet connect all three β-strands together. The difference between the two proteins is that the force-bearing β-strands of barstar are located at the edge of the β-sheet, whereas the force-bearing β-strands of Top7 are further protected by two additional β-strands.
A previous study has shown that Top7 is mechanically stable (17). Since the topology and pulling geometry of Top7 and barstar are very similar, barstar is predicted to be mechanically stable if the protein topology of force-bearing strands is the predominant factor in determining the mechanical stability of proteins. To test this prediction, we carried out single-molecule AFM experiments to directly measure the mechanical stability of barstar, and then applied SMD to illustrate the unfolding mechanism.
The rest of the article is designed as follows: In the Materials and Methods section we give detailed descriptions of the protein engineering of the barstar polyprotein and AFM setup. Also presented are the computer simulation details. In the Results section we present the force measurement of barstar from AFM and the SMD unfolding comparison of barstar and Top7. In the Discussion we address the question of whether shear topology between the pulling strands is a sufficient condition to provide strong mechanical resistance.
MATERIALS AND METHODS
Protein engineering
Plasmid containing barstar was a kind gift from Dr. Jayant Udgaonkar. DNA encoding barstar was PCR-amplified with restriction sites BamHI at the 5′ end and BglII followed by two stop codons and KpnI at the 3′end, respectively. The PCR-amplified product was digested with restriction enzymes BamHI and KpnI and subcloned into plasmid pUC19 digested with similar enzymes. The gene encoding barstar was confirmed using DNA sequencing. pUC19-barstar was digested with BamHI and KpnI and subcloned into plasmid pUC19-GB1 digested with BglII and KpnI to obtain pUC19-(GB1-barstar). To obtain pUC19-(GB1-barstar)2, plasmid pUC19-(GB1-barstar) was digested with BamHI and KpnI and insert released was subcloned into plasmid pUC19-(GB1-barstar) digested with BglII and KpnI. For protein expression, (GB1-barstar)2 was digested with restriction enzymes BamHI and KpnI and subcloned into plasmid pQE80L digested with similar enzymes. pUC19-(GB1-barstar)4 was obtained from pQE80-(GB1-barstar)2 using a strategy similar to that described above for obtaining pUC19-(GB1-barstar)2.
The expression vector pQE80L contains an N-terminal 6 residue histidine tag to facilitate purification of expressed proteins. The polyprotein was expressed in DH5α strain. For protein purification, cells were lysed by incubation with lysozyme (1 mg/mL). However, protein was found to be in the inclusion bodies. To purify the polyprotein, the inclusion bodies were resuspended in 6 M guanidine hydrochloride (GdnCl) in PBS for 4 h at room temperature. The solution was centrifuged at 9000 × g for 30 min. The supernatant was then incubated with Ni-NTA beads. The beads were washed with 6 MGdnCl containing 10 mM imidazole. They were further washed extensively with PBS containing 10 mM imidazole. Protein was eluted with PBS containing 300 mM imidazole at room temperature. The eluted sample was extensively dialyzed against PBS at 4°C. The dialyzed sample was spun at 9000 × g for 30 min at 4°C. The supernatant was collected and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). The protein was found to be >95% pure.
Barstar* was obtained by extending the 5′ and 3′ end of barstar with DNA encoding four amino acid linkers at each end. To extend the termini of arstar, it was PCR-amplified with forward and reverse primers that contain DNA encoding amino acids SGAG and GSAG at the 3′ and 5′ ends, respectively. Amplified PCR product was subsequently subcloned into plasmid pUC19 to obtain pUC19-(barstar*).
For AFM experiments, polyprotein (GB1)4-barstar*-(GB1)4 was constructed instead of (GB1-barstar*)4 to improve protein solubility. pUC19-(barstar*) was then digested with enzymes BamHI and kpnI and subcloned into pQE80-(GB1)4 digested with BglII and KpnI to generate pUC19-(GB1)4-(barstar*). (GB1)4 flanked with BamHI and KpnI was further subcloned into pQE80L-(GB1)4-barstar* digested with BglII and KpnI.
Protein was expressed in E. coli strain DH5α. Cell lysis was carried out as described above. Constructing (GB1)4-barstar*-(GB1)4 greatly improved the solubility of the expressed protein. Approximately 40% of the expressed polyprotein (GB1)4-barstar*-(GB1)4 was found to be in the soluble fraction. Protein was purified from the soluble fraction using Ni-NTA affinity chromatography.
Circular dichroism spectroscopy measurements
Far-UV CD measurements were carried out on a Jasco-J810 spectropolarimeter flushed with nitrogen gas. The spectra were recorded in a cuvette with a path length of 0.2 cm at a scan rate of 20 nm min−1. For each protein sample an average of three scans were reported. CD spectra of polyprotein (GB1)8 and (GB1)4-(barstar)2-(GB1)4, which was designed according to the procedure described above for (GB1)4-(barstar*)2-(GB1)4, were measured. The molar ellipticity of (GB1)8 and (GB1)4-(barstar)2-(GB1)4 was calculated according to the following equation: [θM] = (100 × [θobs])/(d × C), where [θM] and [θobs] are the molar and observed ellipticity, respectively; d is the pathlength (in centimeters); and C is the molar protein concentration. [θM] for (GB1)8 was subtracted from that of (GB1)4-(barstar)2-(GB1)4 to obtain the θM value for (barstar)2. The mean residue ellipticity ([θM.R.E.]) for barstar was obtained from the molar ellipticity according to the following equation: [θM.R.E.] = [θM]/(n − 1), where n is the number of amino acids present in (barstar)2 in (GB1)4-(barstar)2-(GB1)4.
Single-molecule AFM
Single-molecule AFM experiments were carried out on a custom-built atomic force microscope that was constructed as described previously (24) The spring constant of each individual cantilever (Si3N4 cantilevers from Vecco, with a typical spring constant of 40 pN nm−1) was calibrated in solution using the equipartition theorem before and after each experiment (25,26). All of the force-extension measurements were carried out in PBS buffer. About 1 μL of the polyprotein sample (∼500 ng) was added onto a clean glass coverslip covered by PBS buffer and was allowed to absorb for 5 min before proceeding to the AFM measurements.
During the unfolding experiment, the AFM tip was brought into contact with the substrate with a typical contact force of several nanonewtons and then pulled away. Occasionally, molecules adsorbed onto the AFM tip, allowing them to be stretched between the AFM tip and the glass substrate.
SMD simulations
Barstar was subjected to a simulated equilibration, constant velocity, and constant force stretching in SMD. The aqueous environment was modeled using explicit water representation, i.e., protein was solvated in a water box with periodic boundary conditions. The water box was large enough for the equilibration and for the first 50 Å of stretching (length 124 Å, width 64 Å, height 50 Å). The whole protein-water system contained ∼36,000 atoms. The SMD simulations were performed in constant velocity (pulling velocity ranges from 1 m/s to 10 m/s) as well as constant force (at 800 pN) modes. The model preparation and data analysis were performed with visual MD (VMD) (27) and an MD simulation with nanoscale MD (NAMD) (28). During the 1-ns equilibration, barstar was reasonably stable from the initial Protein Data Bank (PDB) structure 1BTA, with the root mean-square deviation (RMSD) in the range of 2 Å. This final equilibrated structure was the starting point used in the pulling SMD. The SMD simulations were also performed on Top7 using the same setup as described previously (17).
RESULTS
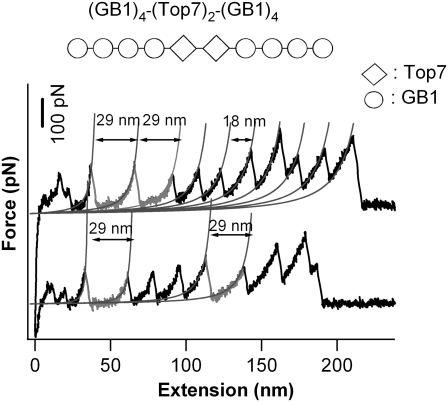
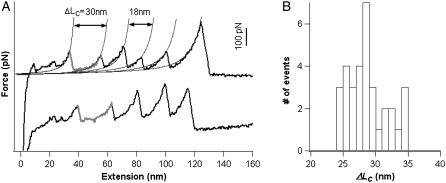
The model proteins that were compared here are Top7 and barstar.The mechanical unfolding of Top7 has been investigated in detail using single-molecule AFM and SMD (17). As shown in Fig. 3, stretching Top7 results in very clear unfolding force peaks with ΔLc of ∼29 nm, corresponding to the unfolding of fully folded Top7 domains. The average unfolding force of Top7 is ∼150 pN at a pulling speed of 400 nm/s. These results indicate that Top7 is mechanically stable.
FIGURE 3.
Single-molecule AFM measurements show that Top7 is mechanically stable. Force-extension relationships of (GB1)4-(Top7)2-(GB1)4 polyprotein chimera. The top diagram shows the schematic of the engineered polyprotein chimera; diamonds stand for Top7, and circles represent GB1 domains. The unfolding events of Top7 (gray) are characterized by an unfolding force of 150 pN and ΔL of ∼29 nm. The thin lines correspond to the WLC fits to the consecutive unfolding events. All of the single-molecule AFM experiments were carried out as described previously (19).
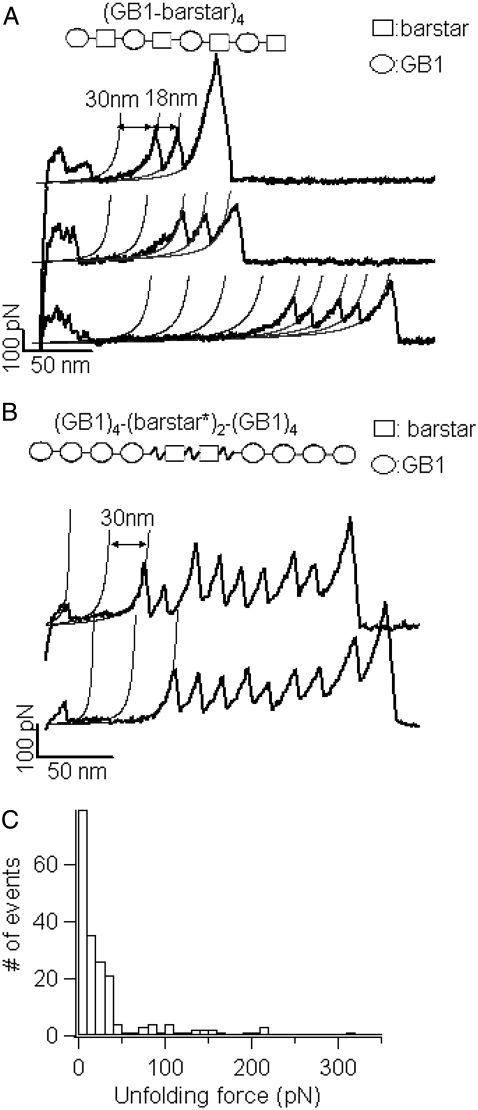
Using polyprotein engineering techniques (16), we constructed a heteropolyprotein, (GB1-barstar)4, in which barstar alternated with GB1, for single-molecule AFM experiments (Fig. 4 A). Here the well-characterized GB1 domains served as an internal marker, enabling us to identify single-molecule stretching events and discern the fingerprint of the mechanical unfolding of barstar in force-extension curves of the polyprotein chimera (19,29). Stretching the polyprotein (GB1-barstar)4 resulted in force-extension curves with characteristic sawtooth patterns (for examples see Fig. 4 A, black curves). These force-extension curves are characterized by a long featureless “spacer” followed by up to four GB1 unfolding events, which are characterized by the unfolding force of ∼180 pN and contour length increment (ΔLc) of ∼18 nm as measured by fitting the worm-like chain model (30) of polymer elasticity to the consecutive unfolding force peaks (19).
FIGURE 4.
Single-molecule AFM measurements show that barstar is mechanically labile. (A) Force-extension relationships of (GB1-barstar)4 polyprotein chimera. The force-extension curves of (GB1-barstar)4 are characterized by the long featureless spacer preceding the unfolding events of GB1 domains, which are characterized by ΔL of ∼18 nm and unfolding forces of ∼180 pN. The featureless spacers result from the unraveling of barstar domains at low forces and the subsequent extension of the unfolded barstar domains. (B) Force extension relationships of (GB1)4-(barstar*)2-(GB1)4 polyprotein chimera. In both A and B, thick lines correspond to WLC fits of ΔL of ∼18 nm, whereas thin lines were generated using the WLC model with ΔL of ∼30 nm, which indicate the locations where the mechanical unfolding of barstar should occur. As evident from the force-extension curves, barstar predominantly unfolds at forces below the detection limit of our AFM. (C) Unfolding force histogram of barstar indicates that barstar is mechanically labile. There is no well-defined unfolding force peak present, and barstar largely unfolds at forces below 50 pN.
Barstar contains 89 amino acid residues and is ∼32.0 nm long (89 aa × 0.36 nm/aa) when unfolded and fully extended. Since the N- and C-termini of barstar are 1.8 nm apart (31), the complete mechanical unraveling of barstar would result in unfolding events of contour length increment ΔLc of ∼30 nm (89*0.36 nm − 1.8 nm = 30.2 nm) if barstar has significant mechanical stability. If barstar is mechanically more stable than GB1, the unfolding events of barstar will occur at higher forces and appear after the unfolding events of GB1 in force-extension curves. However, we did not observe any unfolding event of ΔLc of ∼30 nm after the GB1 unfolding events. The last peaks in force-extension curves, which generally correspond to the detachment of the fully unfolded polyprotein chains from either the AFM tip or substrate, can be as high as 1 nN, effectively excluding the possibility that the unfolding events of barstar were not observed because the stretching force was not high enough to trigger the unfolding of barstar. Therefore, the mechanical stability of barstar should be lower than that of GB1 domains. The thin lines in Fig. 4 A, which were generated based on the expected ΔLc of 30 nm for barstar, indicate the locations where the mechanical unfolding of barstar should have occurred. However, despite the regular appearance of GB1 unfolding events in the force-extension curves of (GB1-barstar)4, no apparent barstar unfolding events were present in the majority of the force-extension curves. Instead, a featureless long spacer preceding the first GB1 unfolding event is the dominant feature in the majority of the force extension curves. Furthermore, the initial length of the spacer roughly correlates with the number of GB1 unfolding events in the force extension curve: the more GB1 unfolding events occur, the longer the spacer is. Since barstar alternates with GB1 in the heteropolyprotein, it is certain that at least one barstar has been stretched if we observe two unfolding events of GB1 in a given force-extension curve of (GB1-barstar)4 (32). Therefore, the featureless spacer preceding the GB1 unfolding events must correspond to the mechanical unraveling and stretching of barstar domains. These results strongly indicate that, in contrast to the prediction based on shear topology and the structural similarity with Top7, barstar is mechanically labile and unfolds at very low forces despite its thermodynamic stability (33) and shear topology (31).
To avoid any potential steric constraint for barstar from the neighboring domains in the polyprotein, we also engineered an extended variant of barstar (denoted as barstar*), in which the barstar was extended on both termini by four amino-acid flexible linkers (SGAG for N-terminal extension and GSAG for C-terminal extension, respectively). We then engineered polyprotein construct (GB1)4-(Barstar*)2-(GB1)4 for single molecule AFM experiments (Fig. 4 B), where GB1 domains served as the fingerprint for identifying single molecule stretching event. If we observe five or more GB1 unfolding events in a given force-extension curve, we can make sure that the two barstar domains have been stretched. Hence, the force-extension curve of (GB1)4-(Barstar*)2-(GB1)4 that contains five or more GB1 unfolding events must contain features due to the stretching and unfolding of the two barstar domains. Fig. 4 B shows the force-extension curves of stretching (GB1)4-(Barstar*)2-(GB1)4. Similar to those observed for (GB1-barstar)4, in most of the force-extension curves we did not observe unfolding event of ΔL of 30 nm, which would correspond to the mechanical unfolding of barstar. Instead, we observed a featureless spacer of length approximately corresponding to mechanical unraveling of two barstar domains before the first GB1 unfolding event.
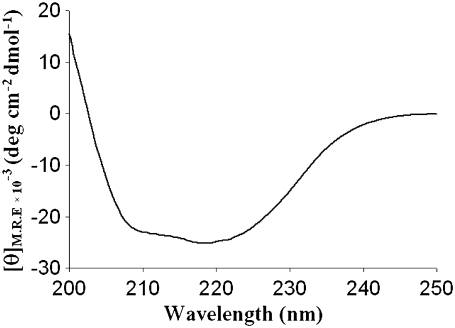
The folded state of barstar was investigated using far-ultraviolet circular dichroism (far-UV CD) spectroscopy for (GB1)4-(barstar)2-(GB1)4. Far-UV CD has been used extensively to examine the secondary structural content in proteins. The far-UV CD spectra of barstar present in the designed heteropolyprotein were calculated from the CD spectra of the heteropolyprotein (GB1)4-(barstar)2-(GB1)4 and polyprotein (GB1)8 according to the procedure described in Materials and Methods. As shown in Fig. 5, CD spectra showed two minima at 208 nm and 222 nm consistent with the natively folded state of barstar. Similar results were obtained using (GB1)4-(barstar*)2-(GB1)4, indicating that the designed polyprotein maintains the native folded state of barstar.
FIGURE 5.
Far-UV CD indicates the natively folded state of barstar in the designed (GB1)4-(barstar)2-(GB1)4 polyprotein. CD spectra for barstar in the designed poplyprotein exhibit two minima at 208 and 222 nm, consistent with the α/β structure of barstar.
In a small percentage of force-extension curves, we observed some putative unfolding events of barstar of ΔL of ∼30 nm (Fig. 6), which is consistent with the expected contour length increment of the unfolding of barstar. Fig. 4 C shows the unfolding force histogram for barstar compiled from all the force-extension curves, including the featureless ones as well as the putative ones. For those force extension curves that did not show any unfolding event for barstar, the unfolding force is taken as zero, which means that barstar domains unfold at forces below the resolution of our instrument (∼20 pN). It is evident that the majority of barstar domains unfold at forces below 50 pN, corroborating that barstar is mechanically weak.
FIGURE 6.
A small percentage of barstar molecules exhibit putative unfolding events. (A) Representative force-extension curves of (GB1)4-barstar-(GB1)4 show putative unfolding events of barstar. In these force-extension curves, unfolding events of ΔL of ∼30 nm were observed (colored in gray), which coincide with the ΔL expected from complete unfolding of barstar. Thin lines show the WLC fits of ΔL of ∼18 nm and∼30 nm, respectively. (B) Histogram of contour length increment of putative barstar unfolding events. It is important to note that such putative unfolding events of barstar are a small percentage of the events observed in our single-molecule AFM experiments.
The AFM data indicate that Top7 and barstar, two proteins with similar shear topology arrangements for the force-bearing β-strands as well as similar pulling geometries, have very different mechanical-stability characteristics. These results suggest that protein mechanical stability cannot be predicted solely based on the apparent static topology picture. Other important yet subtle factors may make important contributions to determine the mechanical stability of proteins. To understand the molecular mechanism underlying the dramatic difference in mechanical stability for Top7 and barstar, we carried out SMD simulations to directly compare the unfolding processes and their associated molecular events.
SMD has been used extensively to reveal the molecular mechanism underlying mechanical unfolding processes and the mechanical stability of proteins (5–7,10). To directly compare the mechanical stability of the two proteins, we carried out SMD simulations for Top7 and barstar in constant force mode as well as constant velocity mode.
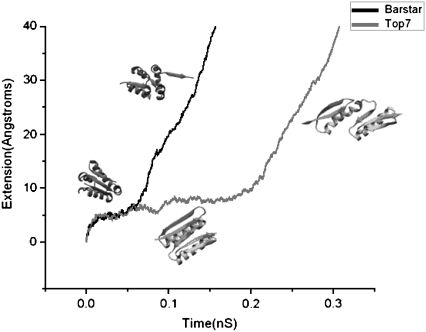
As shown in Fig. 7, under a constant pulling force of 800 pN, it takes ∼50 ps for barstar to cross the unfolding barrier. The unfolding process of barstar is characterized by breaking the backbone hydrogen bonds connecting β-strands 2 and 3 (or sometimes between 1 and 2) one by one, resulting in the gradual peeling of the C-terminal (or sometimes the N-terminal) β-strand away from the remaining structure of barstar. This peeling mechanism is in sharp contrast to the apparent shear topology of the two force-bearing strands of barstar, as well as the cooperative (concurrent) breaking of the backbone hydrogen bonds connecting the terminal force-bearing β-strands, which is generally observed for the mechanically stable proteins of shear topology, such as I1 and I27 (5,34–36) and ubiquitin (13).
FIGURE 7.
Constant-force SMD simulation trajectories of the mechanical unfolding of barstar (black) and Top7 (gray) at a stretching force of 800 pN. SMD simulations reveal that the mechanical unfolding of barstar is initiated by the peeling of the C-terminal force-bearing β-strand from the remainder of barstar, whereas Top7 unfolds via a substructure-sliding mechanism. Top7 is mechanically more stable than barstar: it takes barstar ∼50 ps to cross the unfolding barrier, whereas it takes considerably longer (∼200 ps) for Top7 to cross the barrier.
In contrast to barstar, constant-force SMD for Top7 revealed quite different pictures in the unfolding processes. Compared to the unfolding of barstar, it takes considerably longer (∼200 ps) for Top7 to cross the unfolding barrier under the same stretching force. Similar to what constant-velocity SMD revealed (17), constant-force SMD showed that the main unfolding barrier for Top7 corresponds to simultaneous rupture of the backbone hydrogen bonds connecting β-strands 1 and 3, resulting in a substructure-sliding mechanism for unfolding of Top7. After crossing the main energy barrier, β-strands 3, 4, and 5 remained together, whereas strand 1 stayed together with strand 2, suggesting that the cohesive interactions in these two regions are strong enough to hold them together.
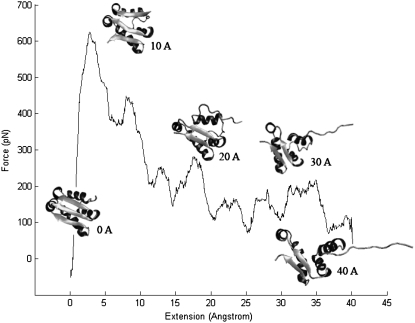
Constant-velocity SMD of barstar (one of the simulations presented in Fig. 8) showed that with 1 m/s pulling speed, the unfolding force is around 600–650 pN, much lower than the Top7 unfolding force of 900–1100 pN under the same pulling velocity (17). The unfolding events of barstar are similar in constant velocity pulling and constant force pulling simulations, i.e., the end strand peels off during unfolding.
FIGURE 8.
Force-extension curves of the mechanical unfolding of barstar obtained from constant-velocity SMD simulations at 1 m/s. The trajectories reveal that stretching barstar from its N- and C-termini results in the peeling, instead of shearing, of the C-terminal β-strand. Snapshots of the structure of barstar along the mechanical unfolding pathway are shown at different extensions.
Comparing the trajectories of the unfolding of barstar with Top7, it becomes evident that the presence of two additional β-strands 2 and 4 in Top7, which form β-hairpins with strands 1 and 5, provided necessary stabilization. The presence of β-strands 2 and 4 on the edge of Top7 makes it difficult to bend the force-bearing β-strands 1 and 5, and effectively prevents peeling of the terminal β-strands from the remaining structure of the protein. In contrast, the lack of structural elements on the edge of barstar to protect the force-bearing β-strands makes it easy to peel terminal β-strands from barstar to trigger its mechanical unfolding. This effect explains the observed sharp difference in mechanical stability between barstar and Top7.
DISCUSSIONS
In this work we have directly compared two proteins, barstar and Top7, that have similar topology in the force-bearing strands yet differ in topology outside those central segments. From this comparison, it becomes evident that the stabilization effect of the neighboring strands, which previously has been overlooked, also plays important roles in determining the overall mechanical stability of proteins. Although mechanical stability is considered to be a local property of proteins and largely determined by the local topology and local interactions in the critical region of proteins, our results demonstrate that the actual unfolding pathway (shearing versus peeling) cannot be predicted solely on the basis of the apparent static topology picture of proteins, i.e., the arrangement of force-bearing β-strands and the array of hydrogen bonds connecting β-strands with respect to the force vector. The delicate stabilization provided by neighboring β-strands seems to be a general feature in protein mechanics. A survey of the proteins studied by single-molecule force spectroscopy revealed that all of the mechanically stable proteins studied to date, including I27 (16), ubiquitin (13), and GB1 (19), exhibit the feature of neighboring β-strand stabilization. The terminal force-bearing strands in all of these proteins are protected by neighboring β-strands. And in many cases, the protecting neighboring β-strands form β hairpins with the force-bearing β-strands. These observations corroborate the notion that the stabilization effect provided by neighboring β-strands is a common feature in the design of mechanically stable proteins. This neighboring-strands stabilization effect can also help to elucidate why some proteins that have a typical unzipping topology (which is indicative of weak mechanical stability) exhibit reasonable mechanical stability. The C2A domain is a good example in this regard. The C2A domain has a characteristic unzipping topology (Fig. 1 E) yet unfolds at a force of 60 pN in AFM (4), which is weaker than many typical mechanical proteins, but stronger than barstar.
Overall, our results reveal that protein local topology is not the only determining factor in the mechanical stability of proteins. The stabilization effect provided by neighboring strands plays important roles in determining the mechanical stability as well as the actual unfolding pathways of proteins. Such a stabilization effect seems to be a general feature in protein mechanics and can serve as an important consideration in identifying novel proteins of significant mechanical stability, as well as in the de novo design of proteins with novel topology for mechanical applications. More theoretical analyses on the spectrum of the states of the protein energy landscape may also provide a better understanding of the mechanical resistance of proteins (37).
Acknowledgments
We thank Prof. Jayant B. Udgaonkar and Prof. David Baker for providing the plasmids encoding the gene of barstar and Top7, respectively.
This work was supported by the Natural Sciences and Engineering Research Council of Canada, Canada Research Chairs program, and Canada Foundation for Innovation (H. Li) and National Institutes of Health grant P01AI060915 (to H. Lu).
Editor: Peter Hinterdorfer.
References
- 1.Balzani, V. V., A. Credi, F. M. Raymo, and J. F. Stoddart. 2000. Artificial molecular machines. Angew. Chem. Int. Ed. Engl. 39:3348–3391. [DOI] [PubMed] [Google Scholar]
- 2.Bao, G., and S. Suresh. 2003. Cell and molecular mechanics of biological materials. Nat. Mater. 2:715–725. [DOI] [PubMed] [Google Scholar]
- 3.Tatham, A. S., and P. R. Shewry. 2000. Elastomeric proteins: biological roles, structures and mechanisms. Trends Biochem. Sci. 25:567–571. [DOI] [PubMed] [Google Scholar]
- 4.Carrion-Vazquez, M., A. F. Oberhauser, T. E. Fisher, P. E. Marszalek, H. Li, and J. M. Fernandez. 2000. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 74:63–91. [DOI] [PubMed] [Google Scholar]
- 5.Lu, H., B. Isralewitz, A. Krammer, V. Vogel, and K. Schulten. 1998. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 75:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu, H., and K. Schulten. 2000. The key event in force-induced unfolding of Titin's immunoglobulin domains. Biophys. J. 79:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paci, E., and M. Karplus. 2000. Unfolding proteins by external forces and temperature: the importance of topology and energetics. Proc. Natl. Acad. Sci. USA. 97:6521–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klimov, D. K., and D. Thirumalai. 2000. Native topology determines force-induced unfolding pathways in globular proteins. Proc. Natl. Acad. Sci. USA. 97:7254–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockwell, D. J. 2007. Force denaturation of proteins—an unfolding story. Current Nanoscience. 3:3–15. [Google Scholar]
- 10.Sotomayor, M., and K. Schulten. 2007. Single-molecule experiments in vitro and in silico. Science. 316:1144–1148. [DOI] [PubMed] [Google Scholar]
- 11.Vogel, V. 2006. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 35:459–488. [DOI] [PubMed] [Google Scholar]
- 12.Li, P. C., and D. E. Makarov. 2004. Ubiquitin-like protein domains show high resistance to mechanical unfolding similar to that of the 127 domain in titin: evidence from simulations. J. Phys. Chem. B. 108:745–749. [Google Scholar]
- 13.Carrion-Vazquez, M., H. Li, H. Lu, P. E. Marszalek, A. F. Oberhauser, and J. M. Fernandez. 2003. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10:738–743. [DOI] [PubMed] [Google Scholar]
- 14.Brockwell, D. J., E. Paci, R. C. Zinober, G. S. Beddard, P. D. Olmsted, D. A. Smith, R. N. Perham, and S. E. Radford. 2003. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Biol. 10:731–737. [DOI] [PubMed] [Google Scholar]
- 15.Dietz, H., F. Berkemeier, M. Bertz, and M. Rief. 2006. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 103:12724–12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 96:3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma, D., O. Perisic, Q. Peng, Y. Cao, C. Lam, H. Lu, and H. Li. 2007. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc. Natl. Acad. Sci. USA. 104:9278–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockwell, D. J., G. S. Beddard, E. Paci, D. K. West, P. D. Olmsted, D. A. Smith, and S. E. Radford. 2005. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 89:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao, Y., C. Lam, M. Wang, and H. Li. 2006. Nonmechanical protein can have significant mechanical stability. Angew. Chem. Int. Ed. Engl. 45:642–645. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., H. H. Huang, C. L. Badilla, and J. M. Fernandez. 2005. Mechanical unfolding intermediates observed by single-molecule force spectroscopy in a fibronectin type III module. J. Mol. Biol. 345:817–826. [DOI] [PubMed] [Google Scholar]
- 21.Lu, H., and K. Schulten. 1999. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins. 35:453–463. [PubMed] [Google Scholar]
- 22.Kuhlman, B., G. Dantas, G. C. Ireton, G. Varani, B. L. Stoddard, and D. Baker. 2003. Design of a novel globular protein fold with atomic-level accuracy. Science. 302:1364–1368. [DOI] [PubMed] [Google Scholar]
- 23.Guillet, V., A. Lapthorn, R. W. Hartley, and Y. Mauguen. 1993. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure. 1:165–176. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez, J. M., and H. Li. 2004. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 303:1674–1678. [DOI] [PubMed] [Google Scholar]
- 25.Florin, E. L., M. Rief, H. Lehmann, M. Ludwig, C. Dornmair, V. T. Moy, and H. E. Gaub. 1995. Sensing specific molecular-interactions with the atomic-force microscope. Biosens. Bioelectron. 10:895–901. [Google Scholar]
- 26.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 27.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38, 27–38. [DOI] [PubMed]
- 28.Kale, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151:283–312. [Google Scholar]
- 29.Cao, Y., and H. Li. 2007. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat. Mater. 6:109–114. [DOI] [PubMed] [Google Scholar]
- 30.Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- 31.Lubienski, M. J., M. Bycroft, S. M. Freund, and A. R. Fersht. 1994. Three-dimensional solution structure and 13C assignments of barstar using nuclear magnetic resonance spectroscopy. Biochemistry. 33:8866–8877. [PubMed] [Google Scholar]
- 32.Li, H., A. F. Oberhauser, S. D. Redick, M. Carrion-Vazquez, H. P. Erickson, and J. M. Fernandez. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA. 98:10682–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khurana, R., and J. B. Udgaonkar. 1994. Equilibrium unfolding studies of barstar: evidence for an alternative conformation which resembles a molten globule. Biochemistry. 33:106–115. [DOI] [PubMed] [Google Scholar]
- 34.Marszalek, P. E., H. Lu, H. Li, M. Carrion-Vazquez, A. F. Oberhauser, K. Schulten, and J. M. Fernandez. 1999. Mechanical unfolding intermediates in titin modules. Nature. 402:100–103. [DOI] [PubMed] [Google Scholar]
- 35.Gao, M., H. Lu, and K. Schulten. 2002. Unfolding of titin domains studied by molecular dynamics simulations. J. Muscle Res. Cell Motil. 23:513–521. [DOI] [PubMed] [Google Scholar]
- 36.Lu, H., and K. Schulten. 1999. Steered molecular dynamics simulation of conformational changes of immunoglobulin domain I27 interpret atomic force microscopy observations. Chem. Phys. 247:141–153. [Google Scholar]
- 37.Klimov, D. K. T. 1998. Linking rates of folding in lattice models of proteins with underlying thermodynamic characteristics. J. Chem. Phys. 109:4119–4125. [Google Scholar]