Abstract
The microarray analysis of total cellular RNA is a common method used in the evaluation of radiation-induced gene expression. However, profiling the cellular transcriptome does not take into account posttranscriptional processes that affect gene expression. To better define the genes whose expression is influenced by ionizing radiation, we used polysome-bound RNA to generate gene translation profiles for a series of tumor and normal cell lines. Cell lines were exposed to 2 Gy, polysome-bound RNA isolated 6 hours later, and then subjected to microarray analysis. To identify the genes whose translation was affected by radiation, the polysome-bound RNA profiles were compared with their corresponding controls using significance analysis of microarrays (<1% false discovery rate). From the statistically significant genes identified for each cell line, hierarchical clustering was performed by average linkage measurement and Pearson’s correlation metric. Ingenuity Pathway Analysis was used for distributing genes into biological networks and for evaluation of functional significance. Radiation-induced gene translation profiles clustered according to tissue of origin; the cell lines corresponding to each tissue type contained a significant number of commonly affected genes. Network analyses suggested that the biological functions associated with the genes whose translation was affected by radiation were tumor type–specific. There was also a set of genes/networks that were unique to tumor or normal cells. These results indicate that radiation-induced gene translation profiles provide a unique data set for the analysis of cellular radioresponse and suggest a framework for identifying and targeting differences in the regulation of tumor and normal cell radiosensitivity.
Introduction
Radiotherapy continues to be a primary treatment modality for most solid tumors. Recent approaches aimed at improving the efficacy of radiation involve the development and application of molecularly targeted agents, a strategy that requires a thorough understanding of the fundamental processes comprising cellular radioresponse. Towards this end, the modulation of gene expression has long been assumed to play a regulatory role in a cell’s response to radiation, and thus, to provide a source of potential targets for radiation modifiers. Over the last several years, the microarray analysis of total cellular RNA has been applied extensively as a means of providing a genome-wide perspective of radiation-induced changes in gene expression. Although such analyses have been reported for a variety of irradiated tumor and normal cell lines, the profiles generated have revealed few commonly affected genes, even among cell lines originating from the same histology (1–4). Moreover, although these microarray analyses accurately reflect changes at the mRNA level, there has been an overall lack of data correlating radiation-induced changes in mRNAs with their corresponding proteins. Although there are exceptions involving individual genes (5), the vast majority of mRNA changes detected after irradiation have not been extended to the protein level. Along these lines, in a direct comparison of radiation-induced proteins with their corresponding mRNAs, Szkanderová et al. reported no correlation for the 10 proteins evaluated (6). Given that protein is the operational end product of gene expression, the lack of correlation between mRNA and protein changes combined with the heterogeneity among cell lines has made it difficult to assign a functional significance to radiation-induced gene expression.
Profiling the cellular transcriptome, the traditional microarray approach, does not take into account posttranscriptional processes that contribute to the regulation of gene expression. It is well established that posttranscriptional events such as mRNA splicing, nuclear export, stabilization, and translation initiation provide an infrastructure for regulating gene expression, a process that can function independent of transcription. For example, in eukaryotic cells exposed to a number of types of stress, changes in mRNA levels do not correlate with protein production (7–10). Accounting for the discrepancy between the transcriptome and proteome is translational control (11–13), which has also been shown to play a significant role in regulating gene expression during such fundamental processes as embryogenesis (14), gliomagenesis (15), and T-cell activation (16). It should also be noted that in a recent analysis of the NCI-60 cell lines, at best, a 65% agreement was found between transcript and protein expression profiles under basal growth conditions (17).
Thus, to better understand the effects of radiation on gene expression, it will be necessary to take into account posttranscriptional regulation, i.e., translational control. Because one of the final steps in gene translation is the loading of a mRNA onto polysomes, we have begun to focus on the microarray analysis of polysome-bound RNA (3). In contrast with total cellular RNA or mRNA (traditional microarray analysis), this procedure bypasses the posttranscriptional infrastructure to generate profiles of mRNAs that are undergoing translation, which significantly reduces the discrepancy between the transcriptome and the proteome (16, 18, 19). We initially applied this microarray approach to human glioma cell lines irradiated with 7 Gy (3). The data generated showed that the number of genes whose translational activity was modified by radiation was approximately 10-fold greater than those whose transcription was affected. Moreover, this study showed that, in contrast with the radiation-induced transcriptome, there is a correlation between the genes whose translational activity was affected and the expression of their corresponding proteins. These results suggested that defining gene translation profiles might provide a novel perspective of radiation-induced regulation of gene expression. Therefore, to extend our initial findings to other cell types and to a clinically relevant dose, we have now used polysome-bound RNA to generate gene translation profiles for 18 human cell lines after exposure to 2 Gy.
Materials and Methods
Cell lines and treatment
The human tumor cell lines used in this study included five gliomas (U87, U251, SF126, SF539, SF295), four pancreatic carcinomas (ASPC1, BxPC3, MiaPaca, PANC1), three breast carcinomas (MDA-MB-231, MCF7, T47D), and two non–small cell lung carcinomas (HOP62, A549). The normal human cell lines evaluated were BJ (skin fibroblast), MRC5 and MRC9 (lung fibroblasts), and MEC (mammary epithelial cells). Cell lines were obtained from the National Cancer Institute, Developmental Therapeutics Program repository or American Type Culture Collection with the exception of the mammary epithelial cell line, which was purchased from Cambrex BioScience, Inc. Cells were grown in either DMEM or RPMI medium as suggested from the source, supplemented with 10% fetal bovine serum and glutamate (5 mmol/L). Cell cultures were maintained at 37°C and 5% CO2. Cultures were irradiated using a Pantak X-ray source at a dose rate of 1.55 Gy/min. Cell lines were exposed to 2 Gy or sham-irradiated, polysome-bound RNA was isolated 6 h later and subjected to microarray analysis. Each cell line was evaluated in biological replicates.
Polysome RNA preparation and microarray hybridization
Polysome preparation and probe labeling were performed as described (3). Polysome-bound samples from sucrose-gradient fractions were pooled and subjected to microarray analysis. Experimental RNA was labeled with Cy3-dUTP and reference RNA (Stratagene Universal Reference) was labeled with Cy5-dUTP. Each microarray chip contained 7,680 human cDNA clones (National Cancer Institute, ROSP 8K Human Array), and methods for microarray hybridization and washing were done as previously described (20). Hybridized arrays were scanned with GenePix 4000A scanner (Axon Instruments, Inc.) at wavelengths of 635 and 532 nm for Cy5- and Cy3-labeled probes, respectively. The resulting TIFF images were analyzed with GenePix Pro 4.0 software (Axon Instruments). The images and raw intensity data were stored at mAdb tools (Center for Information Technology, NIH).6 The microarray data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE10547).7
Statistical analysis filtering and clustering
Preliminary filtering of raw data was performed using mAdb tools with the following setting. All flagged spots were treated as missing values and the data were subjected to spot quality filter with signal to background ratios of >2, a minimum background-corrected signal of 250 counts, and 60% of pixels in the spots with an intensity greater than a SD plus background. Further data analysis was carried out using R software package (R Development Core Team, 2005)8 and custom-written scripts. Genes with missing values in >70% of arrays were removed from the analysis and the rest of the missing values were imputed using K-means nearest neighbor method. Lowess normalization (21) was performed on log2-transformed data. The normalized signal intensity was divided by the control channel intensity and the resulting log ratios from duplicate hybridizations were averaged. After the above processing, 6,227 probes remained for further analysis. To show biological replicate similarity and cell grouping based on gene expression patterns, hierarchical clustering was performed using average linkage and Pearson’s correlation metric. As expected, biological replicates clustered with the smallest distance metric reflecting a high degree of similarity (data not shown).
Tissue-specific gene expression profiling
To obtain a list of potential radiation-responsive genes according to the tissue of origin, data were divided into five groups containing cell lines corresponding to each of the four tumor types (breast carcinoma, glioma, lung carcinoma, and pancreas carcinoma) and normal tissue. Genes that were significantly different between untreated and irradiated samples were identified using the two-class paired response parameter of the significance analysis of microarrays (SAM) algorithm with 100 permutations (22). The resulting delta values from SAM analysis for each data set were adjusted to obtain the largest gene list that gave a <1% false discovery rate (FDR). From the statistically significant genes identified by SAM, Venn diagrams were constructed to identify genes common to any two cancer categories from the four cancer types tested, and between genes common to any two cancer categories set and normal cell lines.
Network and pathway analysis
Ingenuity pathway analysis (IPA; Ingenuity Systems)9 was used for evaluating the functional significance of radiation-induced gene profiles. Specified lists of genes identified by SAM as being affected by radiation were used for network generation and pathway analyses implemented in IPA tools. HUGO official gene names for the selected gene lists were uploaded into the IPA, which was then mapped to the Ingenuity Pathway Knowledge Base. The so-called focus genes were then used for generating biological networks. A score was generated for each network according to the fit of the original set of significant genes. This score reflects the negative logarithm of the P value, which indicates the likelihood of the focus genes in a network being found together due to random chance. Using a 99% confidence level, scores of ≥2 were considered significant. Significances for biological functions were then assigned to each network by determining a P value for the enrichment of the genes in the network for such functions compared with the whole Ingenuity Pathway Knowledge Base as a references set.
Results
The goal of this study was to compare the radiation-induced gene translation profiles generated from human tumor cell lines that corresponded to tumors that are typically treated with radiotherapy, a panel that included five gliomas, four pancreatic carcinomas, three breast carcinomas, and two non–small cell lung carcinomas. In addition, radiation-induced gene translation profiles were generated for four normal human cell lines: a skin fibroblast (BJ), two lung fibroblasts (MRC5, MRC9), and mammary epithelial cells. All cell lines used in this study were capable of forming colonies in monolayer culture and were evaluated in log-phase growth. Specifically, cell lines were exposed to 2 Gy or sham-irradiated, polysome-bound RNA was isolated 6 hours later and subjected to microarray analysis. Each cell line was evaluated in biological replicates.
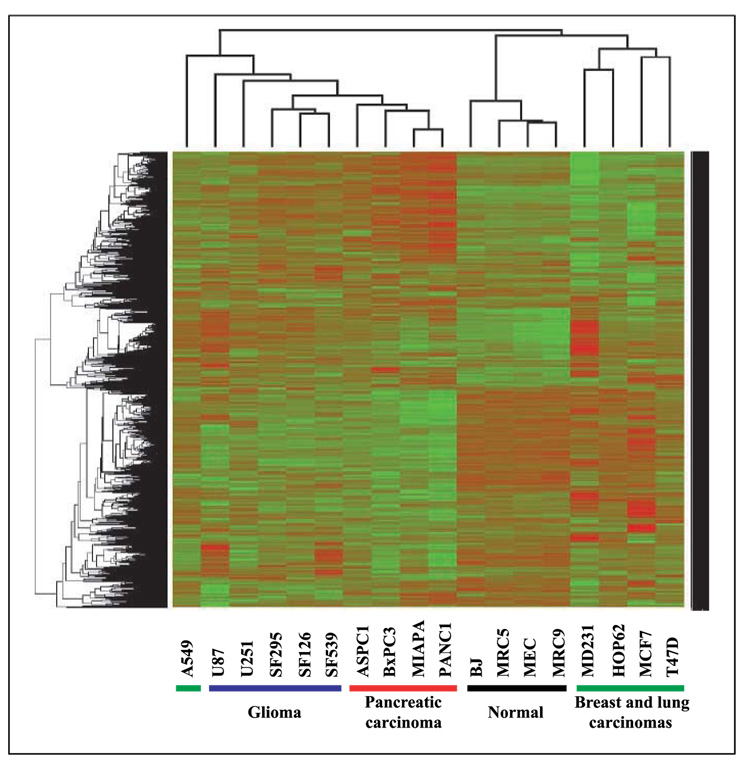
To identify the genes whose translation was either up-regulated or down-regulated by radiation, biological replicates were averaged for each cell line and compared with the polysome-bound RNA profiles generated from their corresponding control (unirradiated) cells using SAM (<1% FDR). The cell lines were then compared in terms of the genes whose translational activity was affected by radiation using a two-way heat map with average linkage distance and Pearson’s correlation metric (Fig. 1). The glioma and pancreatic carcinoma cell lines clustered according to tumor type; the breast and lung tumor cell lines were considerably more heterogeneous, being interspersed among each other. In contrast, the normal cell lines, which included skin fibroblasts, lung fibroblasts, and mammary epithelial cells were strikingly homogeneous, forming a definitive cluster separate from the tumor cell lines. This cluster analysis suggests similarities among cell lines within the specific histologies with respect to radiation-induced translational gene expression profiles. Accordingly, we then determined the specific genes that were commonly affected in the cell lines corresponding to each tumor type and normal tissue (Table 1). Each of the tumor types as well as the normal cells contained a significant number of commonly affected genes as determined by SAM. The glioma and pancreatic carcinoma cell lines each had a greater number of common genes than in the breast and lung carcinomas, which is consistent with greater degree of heterogeneity among the breast and lung tumor cell lines as illustrated in Fig. 1. The four normal cell lines also contained a relatively large number of commonly affected genes. Thus, these data indicate that although there were cell line–specific responses, among cell lines initiated from the same histology, there was a common subset of genes whose translational activity was affected by radiation.
Figure 1.
Two-way heat map comparing the genes whose translational activities were affected by radiation in each cell line. Cell lines were irradiated (2 Gy) and polysome-bound RNA collected 6 h later. To identify genes whose translation was either up-regulated or down-regulated by radiation, biological replicates were averaged for each cell line and compared with the polysome-bound RNA profiles generated from their corresponding control (unirradiated) cells using SAM (<1% FDR). Those genes were then used to derive unsupervised cluster maps using average linkage distance measurement and Pearson’s correlation metric. Up-regulation (red) and down-regulation (green) in irradiated cells.
Table 1.
Number of genes whose translation state was affected by radiation in cell lines as a function of the tissue of origin
| Tissue of origin | Down-regulated | Up-regulated |
|---|---|---|
| Breast tumors | 796 | 711 |
| Lung tumors | 397 | 382 |
| Glioma | 1,398 | 1,566 |
| Pancreas tumors | 2,279 | 1,844 |
| Normal | 1,385 | 1,974 |
NOTE: Genes that were significantly different between untreated and radiation-treated samples were identified using the two-class paired response parameter SAM (<1% FDR) for the cell lines corresponding to each tissue of origin.
To provide insight into the potential functional implications of tumor type specificity in the radiation-induced gene translation profiles, the genes common to each of the tumor histologies shown in Table 1 were placed in the context of the known interactome using IPA. The five most significant networks and their corresponding top functions for each tumor type are shown in Table 2. Given the degree of enrichment in the networks, which is reflected by the significance score and the number of focus molecules (out of a possible 35), these results indicate that the genes subject to radiation-induced translational control do not simply comprise a random list and may be of potential functional consequence. As expected for data derived from tumor cell lines, cancer and cell cycle were the most represented functions in each of the histologies. However, there were also a number of functions that seemed histology-specific. For example, the top network for breast, lung, glioma, and pancreas was associated with gene expression, viral function, embryonic development and cellular assembly and organization, respectively.
Table 2.
Functions associated with the top five networks for genes whose translation state was affected by radiation for each tumor type
| Score | Focus molecules | Top functions | |
|---|---|---|---|
| Breast | |||
| 1 | 40 | 34 | Gene expression, cell cycle, DNA replication, recombination, and repair |
| 2 | 40 | 34 | Cellular development, cancer, cell cycle |
| 3 | 35 | 32 | Cell-to-cell signaling and interaction, nervous system development and function, cancer |
| 4 | 33 | 31 | Small molecule biochemistry, cell morphology, cancer |
| 5 | 33 | 31 | Cell cycle, cellular assembly and organization, cellular movement |
| Lung | |||
| 1 | 53 | 35 | Viral function, cancer, hematologic disease |
| 2 | 38 | 29 | Cardiovascular system development and function, cellular growth and proliferation, cellular movement |
| 3 | 38 | 29 | Cancer, cell death, hematologic disease |
| 4 | 38 | 29 | Neurologic disease, cancer, reproductive system disease |
| 5 | 38 | 29 | Cancer, endocrine system disorders, cell cycle |
| Glioma | |||
| 1 | 32 | 35 | Cancer, embryonic development, organ development |
| 2 | 32 | 35 | Cell death, cancer, cell morphology |
| 3 | 32 | 35 | Cancer, protein trafficking, cell cycle |
| 4 | 32 | 35 | Gene expression, cellular growth and proliferation, hematologic system development and function |
| 5 | 30 | 34 | Cellular assembly and organization, cellular compromise, lipid metabolism |
| Pancreas | |||
| 1 | 27 | 35 | Cellular assembly and organization, cellular compromise, lipid metabolism |
| 2 | 27 | 35 | Carbohydrate metabolism, drug metabolism, small molecule biochemistry |
| 3 | 27 | 35 | Cellular assembly and organization, cancer, cell cycle |
| 4 | 27 | 35 | Cell cycle, DNA replication, recombination, and repair, cell signaling |
| 5 | 27 | 35 | RNA posttranscriptional modification, cellular assembly and organization, cellular compromise |
NOTE: Genes whose translation was affected by radiation in each tumor type as defined by SAM (<1% FDR) were subjected to IPA, the top five networks with their top three associated function categories are shown. Score refers to statistical significance (negative logarithm of the P value); focus molecules refers to the number of genes (out of a possible 35) in each network.
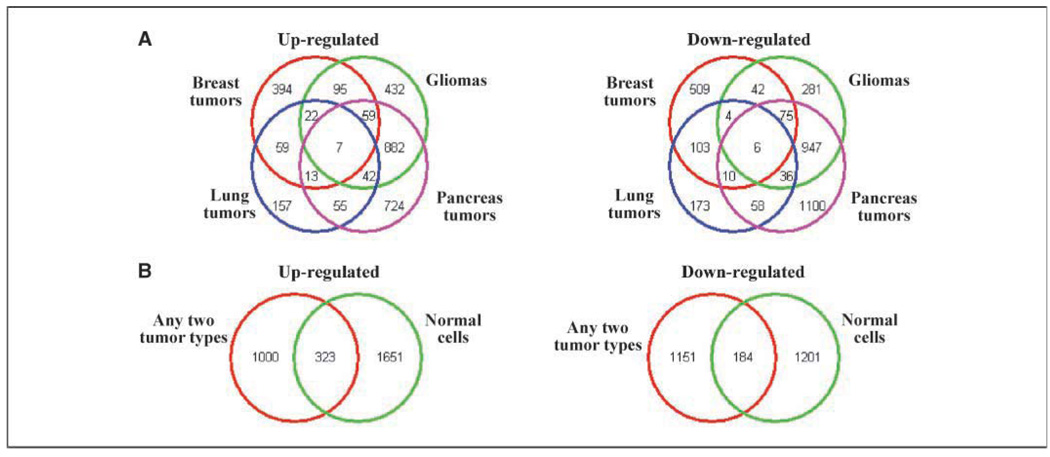
The next step was to compare the gene subsets comprising the radiation-induced translational profiles of tumor and normal cells. Towards this end, we initially identified genes that were commonly affected across the four tumor histologies. As shown by the Venn diagrams in Fig. 2A, although there were few genes in common among the four tumor types, there were a substantial number of genes in common between any two cancer types. Therefore, to account for tumor cell line heterogeneity and to generate a list of tumor-derived radiation-affected genes, the genes common to any two tumor types (2,658 genes: 1,323 increased and 1,335 decreased) were selected for further analysis (23). This subset of tumor genes was then compared with those affected in the normal cells (as shown in Table 1). The Venn diagram (Fig. 2B) indicates that a set of 507 (increased plus decreased) genes was detected in the radiation-induced translational profiles of both tumor and normal cell lines. However, importantly, there was also a set of genes that were unique to either tumor cells or normal cells.
Figure 2.
Comparison of tissue types regarding genes whose translation states were affected by radiation. A, Venn diagrams depicting the number of genes that were commonly up-regulated (left) or down-regulated (right) by radiation in four tumor types (pancreas, gliomas, lung, and breast). For each tumor type, the genes were those defined by SAM (<1% FDR) using the cell lines comprising each histology as shown in Table 1. B, Venn diagrams depicting the number of genes affected in the normal cells as defined in Table 1 using SAM (<1% FDR) and genes that were commonly affected in any two of the tumor types determined in A. Up-regulated (left) and down-regulated (right) by radiation.
The gene sets unique to tumor or normal cells as defined in Fig. 2 were then distributed into networks of known biological pathways using IPA, with the biological functions associated with the top 10 networks identified for tumor and normal cells shown in Table 3. Although there was some overlap in functions, there were also clear distinctions between tumor and normal cells. The number 1 ranked network from tumor cells was associated with cancer, cell cycle and skeletal and muscular disorders, whereas for normal cells, the top network was associated with cell to cell signaling and interaction, tissue development, and organismal development. For each cell type, there was a significant enrichment of genes associated with functions typically associated with radioresponse such as cell cycle, cell signaling, and DNA replication, recombination, and repair. In addition, both normal and cancer cells were enriched in genes involved in RNA posttranscriptional modifications, which has only recently been associated with radioresponse (3).
Table 3.
Functions associated with the top 10 networks for genes whose translation state was affected by radiation in cancer and normal cell lines
| Score | Focus molecules | Top functions | |
|---|---|---|---|
| Cancer | |||
| 1 | 37 | 35 | Cancer, cell cycle, skeletal and muscular disorders |
| 2 | 35 | 34 | Connective tissue development and function, tissue development, amino acid metabolism |
| 3 | 35 | 34 | Cell cycle, cell signaling, DNA replication, recombination, and repair |
| 4 | 35 | 34 | RNA posttranscriptional modification, cell signaling, carbohydrate metabolism |
| 5 | 35 | 34 | RNA posttranscriptional modification, cell death, connective tissue development and function |
| 6 | 32 | 33 | Cellular assembly and organization, gene expression, cell cycle |
| 7 | 32 | 33 | Cancer, hematologic disease, gastrointestinal disease |
| 8 | 30 | 32 | Cell death, posttranslational modification, cancer |
| 9 | 30 | 32 | Cancer, cellular movement, protein trafficking |
| 10 | 30 | 32 | Cell signaling, DNA replication, recombination, and repair, gene expression |
| Normal | |||
| 1 | 33 | 35 | Cell-to-cell signaling and interaction, tissue development, organismal development |
| 2 | 30 | 34 | Cell cycle, cellular assembly and organization, DNA replication, recombination, and repair |
| 3 | 30 | 34 | Cellular assembly and organization, cancer, cell cycle |
| 4 | 30 | 34 | RNA posttranscriptional modification, gene expression, cellular assembly and organization |
| 5 | 30 | 34 | Amino acid metabolism, posttranslational modification, small molecule biochemistry |
| 6 | 30 | 34 | Hematologic disease, gene expression, lipid metabolism |
| 7 | 30 | 34 | Inflammatory disease, immune response, cellular development |
| 8 | 30 | 34 | Gene expression, cell signaling, lipid metabolism |
| 9 | 30 | 34 | DNA replication, recombination, and repair, cell cycle, cancer |
| 10 | 28 | 33 | Cell cycle, embryonic development, cellular assembly and organization |
NOTE: Genes whose translation was affected by radiation in normal or cancer cell lines as defined by SAM (<1% FDR) were subjected to IPA, the top 10 networks with their top 3 associated function categories are shown. Score refers to statistical significance (negative logarithm of the P value); focus molecules refers to the number of genes (out of a possible 35) in each network.
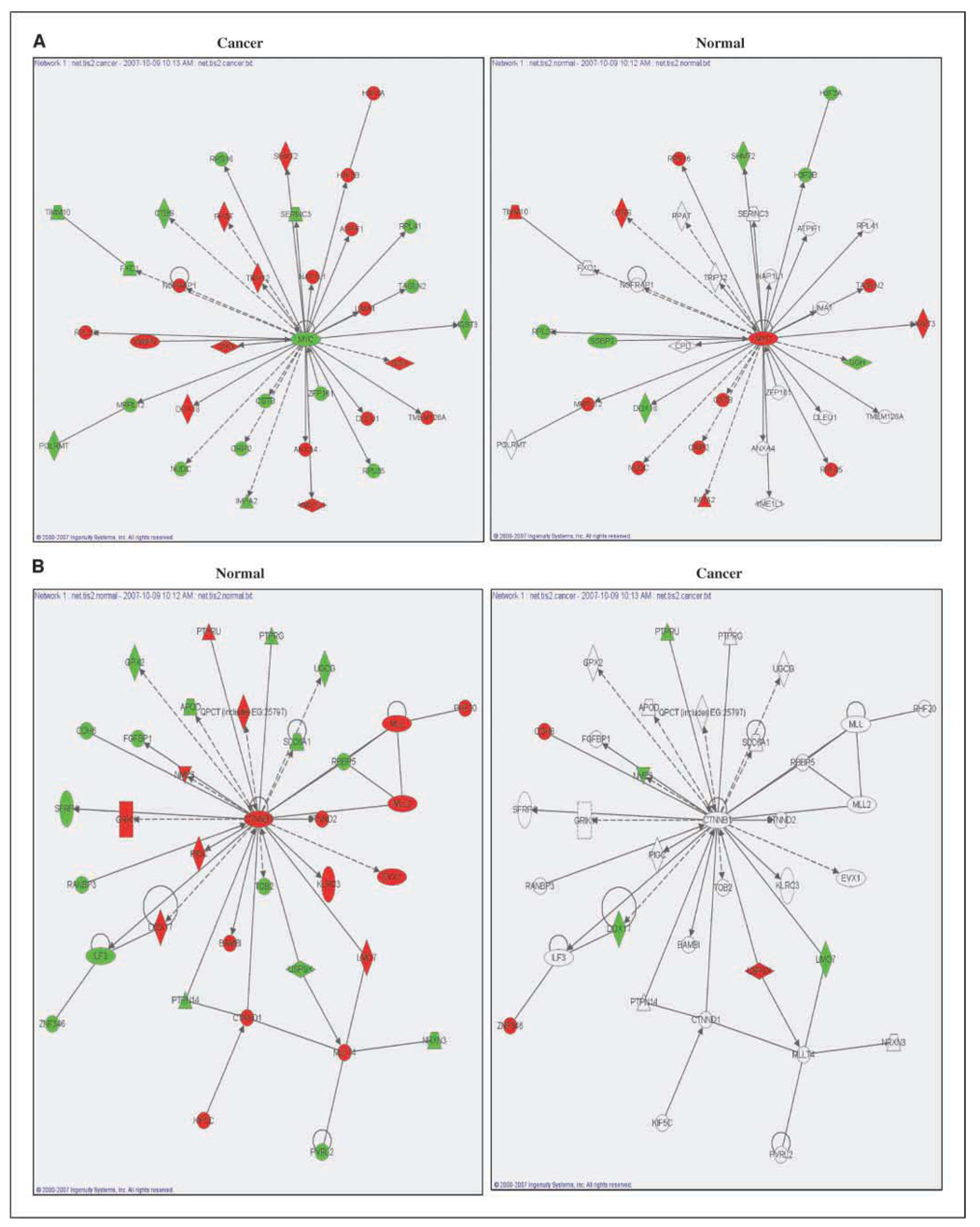
To better illustrate the differences in radiation-induced translational profiles between tumor and normal cells, the genes comprising the top-ranked networks for cancer cells and normal cells are shown in Fig. 3. The top network derived from the genes unique to the cancer cell lines centered on a decrease in Myc, with the other genes in this network either increased or decreased in response to radiation (Fig. 3A, left). Evaluation of this same network overlaying the gene values from the normal cell gene subset revealed fewer genes affected by radiation, and for those that were, the effect of radiation was in the opposite direction as compared with the cancer cells, including Myc (Fig. 3A, right). The hub of the top network derived from the genes unique to normal cells was β-catenin (CTNNB1; Fig. 3B, left). In the normal gene enrichment, β-catenin was up-regulated and was surrounded by 34 interacting genes whose translation was affected by radiation. Analysis of this network overlaying the gene values from the cancer cell gene subset revealed far fewer genes affected by radiation (Fig. 3B, right). Thus, the top networks with Myc and β-catenin as hubs are indicative of differences between cancer and normal cells in terms of radiation-induced gene translational profiles.
Figure 3.
The top-ranked network for cancer and normal cells as defined by IPA. A, top network from cancer cells (left); the same network overlaid on normal cells (right). B, network from normal cells (left); the same network overlaid on cancer cells (right). Up-regulation (red) and down-regulation (green) in irradiated cells.
Discussion
Whereas there are a number of reports defining the cellular transcriptome after irradiation, the goal of the current study was to generate a whole genome perspective of radiation-induced changes in gene translation for a series of cell lines commonly used in cancer research. In previous work directly comparing the microarray analyses of total RNA and polysome-bound RNA after the irradiation of three glioma cell lines, few, if any genes were found to be commonly affected in both procedures (3). These results suggested that the radiation-induced changes in transcription and translation are not coordinated, with each proceeding through different regulatory mechanisms. This is consistent with a similar role for translational control of gene expression observed after other forms of cell stress (9, 10). The independence of these two events was further supported by data indicating that the radiation-induced changes in translation, as detected in the microarray analysis of polysome-bound RNA, occur through the recruitment of existing mRNAs to and away from polysomes (3). Thus, these results, combined with those for other types of stress, indicate that as compared with transcriptional changes, analysis of the radiation-induced changes in gene translation provides a unique data set.
Along these lines, analyses of radiation-induced gene expression using total cellular RNA (i.e., traditional microarray analysis) have revealed few commonly affected genes among cell lines initiated from the same tissue type (1–4). Accordingly, based on the analysis of the cellular transcriptome, radiation-induced gene expression is considered to be independent of the tissue of origin and highly dependent on the individual cell genotype (1, 4). However, when evaluated at the level of translation, as shown here, radiation-induced gene expression exhibits a significant degree of tissue dependence. For the 18 cell lines evaluated, radiation-induced translational profiles, for the most part, clustered according to tissue of origin, with each histologic category containing a significant number of commonly affected genes. A tissue type dependency for radiation-induced gene expression would have a number of potential implications. With respect to cancer treatment, if gene expression influences tumor cell radiosensitivity, then the optimal preclinical development of targets for radiation modifiers would take into consideration tumor type. In addition, independent of whether the induced changes in gene expression directly contribute to radiosensitivity, its tissue type dependency may provide a source of biomarkers indicative of radiation exposure in treatment as well as environmental settings.
A long sought goal in the use of radiation in cancer treatment has been to identify exploitable differences in the radioresponse of tumor and normal tissue. Comparisons of the radiation-induced gene translation profiles obtained from the tumor and normal cell lines revealed clearly different gene subsets as well as an overall greater degree of homogeneity among the four normal cell lines. Radiosensitivity or the mode of cell death is unlikely to account for the difference between these normal and tumor cells in that, in contrast with cells of hematopoietic or lymphatic origin, the normal cells in this study die through mitotic catastrophe, as do the solid tumor cell lines. A more likely explanation pertains to translation in general. It is becoming increasing well recognized that abnormal translation is a fundamental characteristic of tumor cells and a potential target for cancer treatment (24). This abnormality has been attributed to signaling pathways involved in translation such as those mediated by Ras, phosphoinositide-3-kinase, or mTOR (15, 24), which have also been implicated in radioresponse, as well as by overexpression of components of the general translational machinery such as eIF4E (25). Whether these molecules play a role in the radiation-induced regulation of gene translation remains to be investigated. However, the disparity between the radiation-induced gene translation profiles generated for cancer and normal cell lines are consistent with the abnormal translation in tumor cells.
In addition to general differences between tumor and normal cells, because of the strong correlation between translational control and the cell proteome, gene translation profiles may serve as high-throughput approach for identifying the specific proteins that selectively participate in the radioresponse of each cell type. For example, the top functional network derived from the genes unique to the cancer cell lines had Myc at its primary hub, the expression of which was decreased by radiation. Whereas the effects of radiation on all the proteins in this network remained to be defined, the decrease in Myc protein is consistent with that previously reported for human tumor cell lines (26). Myc is well established to play a role in transformation and cell cycle control; although it has been implicated in radioresponse, the specific processes have yet to be clearly defined (27). For normal cells, the primary hub protein in the top network was β-catenin, which is consistent with recent reports showing that irradiation of normal mammary progenitor cells results in increased β-catenin protein (28, 29). Moreover, β-catenin protects against radiation-induced death in these normal cells (28, 29), although the specific processes have not been defined. Thus, whereas further investigations validating specific protein changes are clearly required, the study presented here suggests that radiation-induced translational gene expression profiles provide a framework for identifying and targeting differences in the regulation of tumor and normal cell radiosensitivity.
Acknowledgments
Grant support: NIH grant from the National Cancer Institute, CA126943 (P.J. Tofilon).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 2.Camphausen K, Purow B, Sproull M, et al. Orthotopic growth of human glioma cells quantitatively and qualitatively influences radiation-induced changes in gene expression. Cancer Res. 2005;65:10389–10393. doi: 10.1158/0008-5472.CAN-05-1904. [DOI] [PubMed] [Google Scholar]
- 3.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–1061. doi: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 4.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 5.Azzam EI, de Toledo SM, Little JB. Expression of CONNEXIN43 is highly sensitive to ionizing radiation and other environmental stresses. Cancer Res. 2003;63:7128–7135. [PubMed] [Google Scholar]
- 6.Szkanderová S, Port M, Stulik J, et al. Comparison of the abundance of 10 radiation-induced proteins with their differential gene expression in L929 cells. Int J Radiat Biol. 2003;79:623–633. doi: 10.1080/09553000310001606821. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ideker T, Thorsson V, Ranish J, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 9.Shenton D, Smirnova JB, Selley JN, et al. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 10.Smirnova JB, Selley JN, Sanchez-Cabo F, et al. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol Cell Biol. 2005;25:9340–9349. doi: 10.1128/MCB.25.21.9340-9349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 12.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 13.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 14.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 15.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 16.Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- 17.Shankavaram UT, Reinhold WC, Nishizuka S, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 18.Jechlinger M, Grunert S, Tamir IH, et al. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- 19.Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci U S A. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang YY, Chen Y, Gadisetti Chandramouli VR, et al. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246–6254. [PubMed] [Google Scholar]
- 21.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zembutsu H, Ohnishi Y, Tsunoda T, et al. Genome-wide cDNA microarray screening to correlate gene expression profiles with sensitivity of 85 human cancer xenografts to anticancer drugs. Cancer Res. 2002;62:518–527. [PubMed] [Google Scholar]
- 24.Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007;26:5973–5990. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- 25.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E-from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 26.Demasters G, Di X, Newsham I, Shiu R, Gewirtz DA. Potentiation of radiation sensitivity in breast tumor cells by the vitamin D3 analogue, EB 1089, through promotion of autophagy and interference with proliferative recovery. Mol Cancer Ther. 2006;5:2786–2797. doi: 10.1158/1535-7163.MCT-06-0316. [DOI] [PubMed] [Google Scholar]
- 27.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Chen MS, Woodward WA, Behbod F, et al. Wnt/β-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 29.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/β-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]