Abstract
Human DNA topoisomerase I (hTop1p) catalyzes the relaxation of supercoiled DNA and constitutes the cellular target of the antitumor drug camptothecin (CPT). The X-ray crystal structure of the enzyme covalently joined to DNA and bound to the CPT analog Topotecan suggests that there are two classes of mutations that can produce a CPT-resistant enzyme. The first class includes changes in residues that directly interact with the drug, whereas a second class alters interactions with the DNA and thereby destabilizes the drug binding site. The Thr729Ala, that is part of a hydrophobic pocket in the enzyme C-terminal domain, belongs to a third group of mutations that confer CPT resistance, but do not interact directly with the drug or the DNA. To understand the contribution of this residue in drug resistance, we have studied the effect on hTop1p catalysis and CPT sensitivity of four different substitutions in the Thr729 position (Thr729Ala, Thr729Glu, Thr729Lys and Thr729Pro). Tht729Glu and Thr729Lys mutants show severe CPT resistance and furthermore, Thr729Glu shows a remarkable defect in DNA binding. We postulate that the maintenance of the hydrophobic pocket integrity, where Thr729 is positioned, is crucial for drug sensitivity and DNA binding.
INTRODUCTION
Human DNA topoisomerase I is a monomeric highly conserved enzyme composed by 765 amino acids organized in four mains domains: the N-terminal domain (residues 1–214), highly disorganized, contains the nuclear localization signals and is the only region of the protein not yet crystallized; the core domain (residues 215–635) that can be divided in three subdomains, subdomain I (residues 215–232 and 320–433), II (residues 233–319) and III (residues 434–635); the linker domain (residues 636–712) composed by two protruding coiled coil α-helices, one of them engages direct contact with the DNA molecule; the C-terminal domain (residues 713–765) contains the Tyr723 that performs a nucleophilic attack to the 3′phosphate of the DNA, forming a transient covalent protein–DNA complex that is resolved by a second transesterification mediated by the 5′-OH of the scissile DNA strand (1–3). The topoisomerase I is a bi-lobed protein that clamps completely around duplex DNA through protein–DNA phosphate interaction. One lobe is constituted by the C-terminal catalytic domain and core subdomain III, the other one (called ‘cap’) by core subdomains I and II. Human DNA topoisomerase I has a medical relevance because it is the natural target of plant alkaloid camptothecin (CPT) whose derivatives are frequently used as anticancer drug (4–7). CPT interacts reversibly with the enzyme blocking the DNA–protein complex after the cleavage, slowing the religation and causing severs damages to the DNA. Furthermore, replication forks progression can generate double-strand breaks inducing apoptotic response. A number of mutations that renders the enzyme resistant to the CPT have been described (8). Some of them are located near to the active site and in any case around the CPT binding site or contacting directly the DNA molecule, few other mutations are located far from the active site (Ala653Pro, Glu418Lys) (9,10), influencing the dynamic of the protein–drug interaction and modifying the intradomain interactions.
The X-ray crystal structure of human topoisomerase I covalently joined to double-stranded DNA and bound to the CPT analog Topotecan (TPT) does not explain all mutations that affect CPT sensitivity (11).
The Thr729Ala mutation has been firstly identified to impart drug resistance on human topoisomerase I in the CPT-11 (Irinotecan, a CPT analog) resistant human lung cancer cell line, PC-7/CPT (12) but our data reveal that the equivalent effect with CPT can not be observed if the enzyme, harboring the same mutation, is expressed in the yeast Saccaromyces cerevisiae. Interestingly, looking at the protein structure, Thr729 seems to be too distant to contact the drug directly and it is not clear how the structure or stability of the intercalation drug-binding pocket is affected by the mutation. The Thr729 resides in the hydrophobic core of the C-terminal domain 12.4 Å from the catalytic tyrosine and 13.1 Å from Asn722 that establishes a water-mediated contact directly to the drug. Changing the Asn722 to Ser, shortening the side chain, is sufficient to impart CPT resistance to the protein (13,14). According to these data, Redinbo and co-authors (15) assumed that the basis of the Thr729 mutants CPT resistance come from the destabilization of the C-terminal region that could lead to the specific shift in position of Asn722, consequentially inducing the breakage of the water-mediated contact with the drug.
In order to confirm these hypotheses, we analyzed the effect of Thr729 substitutions to Lys, Pro and Glu on the in vivo and in vitro catalytic activity and drug sensitivity of the human DNA topoisomerase I. These three substitutions were chosen for their positive or negative charge, Lys and Glu, respectively, and for their capability in distorting the α-helix as in the case of Pro. Our results show that the 729 position is a key point in maintaining the correct geometry of the hydrophobic pocket of the C-terminal domain. In fact, even if the enzyme keeps on its catalytic properties and its sensitivity to CPT in the presence of a Thr729Ala mutation, a dramatic CPT resistance effect could be observed when the Thr729 was mutated to Lys or Glu, and a minor consequence could be seen in the presence of a Pro. Furthermore, the Thr729Glu mutant shows a remarkable defect in the DNA binding indicating that the integrity of the C-terminal geometry is essential for the preservation of the correct interactions between the enzyme and its substrate during the progression of the catalytic cycle. Dynamic simulation experiments described in the accompanying paper, propose a structural and dynamical interpretation for the role played by residue 729 in these long-range protein–DNA communications (Chillemi et al.)
MATERIALS AND METHODS
Yeast strains and plasmids
Camptothecin (Sigma-Aldrich, Europe) was dissolved in Me2SO to a final concentration of 4 mg/ml and stored at −20°C. Anti-FLAG M2 affinity gel, 3xFLAG peptide and M2 monoclonal antibody were purchased from Sigma. Saccharomyces cerevisiae strain EKY3 (ura3-52, his3Δ;200, leu2Δ;1, trp1Δ;63, top1::TRP1, MATα) were previously described (16,17). Plasmid YCpGAL1-hTOP1 in which the human topoisomerase I gene is expressed under the galactose inducible promoter was described previously (18,19). The htop1Thr729Ala, htop1Thr729Pro, htop1Thr729Lys and htop1Thr729Glu were generated by oligonucleotide-directed mutagenesis of the hTop1YCpGAL1 (Stratagene, USA-QuickChange XL-Site-Directed Mutagenesis Kit). All the constructs contain the N-terminal sequence FLAG which specify for the DYKDDDY octapeptide, recognized by the M2 monoclonal antibody (20).
Drug sensitivity assay
Yeast EKY3 strains were transformed with YCp50, YCpGAL1-hTOP1, YCpGAL1-htop1Thr729Ala, YCpGAL1-htop1Thr729Pro, YCpGAL1-htop1Thr729Lys, YCpGAL1-htop1Thr729Glu and YCpGAL1-htop1Ala653Pro-Thr718Ala vectors by LiOAc treatment (21) and selected on synthetic complete (SC)-uracil medium supplemented with 2% dextrose. Transformants were grown to an A595 = 0.3 and 5 µl aliquots of serial 10-fold dilutions were spotted onto SC-uracil plates plus 2% dextrose or 2% galactose, with or without the indicated concentrations of CPT.
Purification of DNA topoisomerase I
Epitope-tagged hTop1 proteins were partially purified from galactose-induced EKY3 cells transformed with YCpGAL1-hTOP1, YCpGAL1-htop1Thr729Ala, YCpGAL1-htop1Thr729Pro, YCpGAL1-htop1Thr729Lys or YCpGAL1-htop1Thr729Glu by phosphocellulose chromatography as previously described (22,23). hTop1 proteins eluted in TEEG [50 mM Tris (pH 7.4) 1 mM EDTA, 1 mM EGTA, 10% glycerol] plus 0.8 M KCl were diluted to a final 150 mM KCl and applied to an anti-Flag M2 affinity gel column. The column was washed with TBS [50 mM Tris (pH 7.4), 150 mM KCl] and the hTop1 proteins were eluted with a solution of 100 μg/ml 3xFlag peptide in TBS. The active fractions were dialyzed using a Spectra/Por Float-A-Lyzer 50 000 MWCO to eliminate the competitor Flag peptide. Proteins were quantified using Bradford assay and immunoblot densitometry analysis. The fractions were adjusted to a final concentration of 40% glycerol and stored at −20°C.
hTop1p activity in vitro
hTop1p activity was assayed in a DNA relaxation assay (18,22). Equal concentrations of hTop1 proteins were incubated in 30 µl reactions containing 0.5 μg of negatively supercoiled pHC624 plasmid DNA in 20 mM Tris (pH 7.5), 0.1 mM Na2EDTA, 10 mM MgCl2, 50 μg/ml acetylated BSA and the indicated concentration of KCl. Reactions were incubated at 37°C and terminated with a final concentration of 0.5% SDS. Two sets of time course relaxation experiments were conducted. In the first set, the assay was performed with an excess of DNA, 400 fmol, relative to an enzyme concentration of 130 fmol, (DNA/enzyme ratio 3:1). The second set contained an excess of enzyme, 1200 fmol, compared to DNA (DNA/enzyme ratio 1:3). Both the experiments were performed in the presence of KCl 150 mM and 37°C of temperature for the indicated time and stopped by addition of SDS with a final concentration of 0.5% (w/v) (24). Reaction products were resolved in 1% agarose gels, stained with ethidium bromide and visualized with a BioRad Gel doc system.
To quantify relative levels of supercoiled and relaxed DNA topoisomers, the stained gels were first exposed to UV light to induce photo-nicking of the DNA, then restained with ethidium bromide. These steps obviated any contribution of DNA topology to ethidium bromide binding and consequently, relative band intensity.
DNA cleavage assays
CPT-induced stabilization of hTop1-DNA covalent complexes was assessed in DNA cleavage assays as described (16,22,23). Briefly, equal concentrations or equal activities of purified hTop1 proteins were incubated in 50 µl reactions with a single 3′ 32P-end-labeled 900-bp DNA fragment, which contains a high affinity hTop1p cleavage site (25) and was excised from plasmid pBlueAK3–1 (23). Where indicated, reactions were supplemented with 10 µM CPT. Me2SO was added to the no drug controls. Following incubation at 37°C for 30 min, reactions were terminated by the addition of 1% SDS and the cleavage products were resolved in a 7 M urea/8% polyacrylamide gel and visualized by Phosphorimager analysis (26).
Kinetics of religation using oligonucleotide substrate
To assess enzyme catalyzed DNA cleavage and religation, a truncated suicide oligonucleotide substrate was used. A 14-nt oligo CL14 (5′-GAAAAAAGACTTAG-3′), which contains a hTop1p high affinity cleavage site, was 5′-end-labeled with [γ32P] ATP and T4 polynucleotide kinase. A 25-nt CP25 (5′-TAAAAATTTTTCTAAGTCTTTTTTC-3′) complementary strand, which was 5′phosphorylated with unlabeled ATP and a 11-nt R11 (5′-AGAAAAATTTT-3′) religation oligonucleotide. The three strands were annealed at a molar ratio of 2 : 3 : 1 of CP25: CL14: R11 in 100 mM NaCl, 1 mM EDTA and 1 mM DTT by heating to 95°C for 5 min, followed by slow cooling to room temperature. The resulting DNA substrate (20 nM) was incubated with equal concentrations of hTop1 proteins in 20 mM Tris (pH 7.5), 0.1 mM EDTA, 10 mM MgCl2, 50 µg/ml acetylated BSA and 150 mM KCl at 37°C. At the times indicated, 5 µl aliquots were quenched with 0.5% SDS and heating to 75°C. The reaction products were trypsinized and resolved in 7 M urea/20% acrylamide gels. The percentage of religation was determined by PhosphorImager and ImageQuant software and normalized on the total amount of radioactivity in each lane.
DNA-binding assay
To assess enzyme DNA binding a 100-nt DNA oligonucleotide (5′-GCTATCGAATTCGCTATAATTCATATGATAGCGGATCCAAAAAAGACTTAGAAAAAAAAAAAGCTTAAGC AACATATGGTATCCGGAATTCAAT-3′) containing the Tetrahimena termofila high affinity DNA topoisomerase I binding site was 5′-end-labeled with [γ32P] ATP and T4 polynucleotide kinase. The complementary strand was 5′ phosphorylated with unlabeled ATP. The two strands were annealed at a molar ratio of 1:1 in 100 mM NaCl, 1 mM EDTA and 1 mM DTT by heating to 95°C for 5 min, followed by slow cooling to room temperature. DNA binding of hTop1 was assayed by nitrocellulose filter binding as previously described (24,27). Briefly, hTop1 and mutant proteins (0–5 pmol) were incubated in 100 μl of binding buffer [20 mM Tris (pH 7.5), 10 mM MgCl2, 0.1 mM EDTA, 50 µg/ml gelatin and 1 mM β-mercaptoethanol], 150 mM KCl, containing 20 000 c.p.m. (50–100 fmol) of 5′-end-labeled DNA substrate for 5 min at room temperature. Cellulose acetate microcentrifuge spin columns (Micron Separations Inc., USA) with 6-mm diameter nitrocellulose filter (0.45 µm) were pre-washed with 100 μl of salmon sperm solution 100 μg/ml to reduce the DNA non-specific binding, subsequently the samples were applied on the filter and the unbound DNA was eluted by centrifugation at 6000 r.p.m. (2800g). After a subsequent wash with 100 µl of binding buffer 200 mM KCl, the amounts of labeled DNA, eluted and filter retained, were counted using a Beckman LS801 liquid scintillation counter.
The percentage of DNA binding was calculated as (filter-bound c.p.m.)/(filter-bound c.p.m.) + (combined eluent c.p.m.). The obtained data were corrected for the non-specific binding of free DNA, which was generally 1–2% of the total DNA. Protein-filter binding was verified by immunoblot.
RESULTS
An altered CPT sensitivity was shown by Thr729 mutants when expressed in yeast
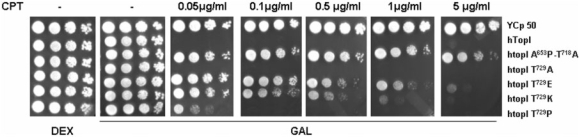
To assess the in vivo consequences of htop1Thr729 mutant expression, the viability and CPT sensitivity of top1Δ; cells transformed with GAL1-top1 constructs were assayed (Figure 1). The top1Δ; cells expressing human TOP1 exhibit a severe drop in viability in the presence of CPT, whereas cells expressing Thr729 mutants displayed different level of CPT sensitivity.
Figure 1.
An altered CPT sensitivity was shown by T729 mutants when expressed in yeast. Exponential cultures of EKY3 (top1Δ) cells transformed with vector control YCp50, YCpGAL1-hTOP1, YCpGAL1-htop1Ala653Pro,Thr718Ala [a known CPT resistant mutant (29)], YCpGAL1-htop1Thr729Ala, YCpGAL1-htop1Thr729Pro, YCpGAL1-htop1Thr729Lys and YCpGAL1-htop1Thr729Glu were serially 10-fold diluted and spotted onto SC-uracil plates supplemented with dextrose (Dex) or galactose (Gal) and, as indicated, CPT. Cell viability was assessed following incubation at 30°C.
Thr729Lys and Thr729Glu mutations render the enzyme clearly resistant to CPT being viable in the presence of the drug. However, a difference in the CPT concentration that renders the cells able to grow in Gal media was observed: Thr729Glu mutant produces viable colonies until 1 μg/ml of CPT while Thr729Lys mutant is less resistant, growing in the presence of 0.5 μg/ml of drug. Even though the introduction of a Pro normally disturbs the α-helix folding, the Thr to Pro substitution does not dramatically interfere with the hTop1p sensitivity to CPT. In fact, cells expressing htopIThr729Pro protein display only a minor capability to grow in the presence of 0.05 μg/ml of CPT. A correlation between the point mutation htopIThr729Ala and the alteration in the sensitivity to hTopIp inhibitor irinotecan, was previously described in the CPT11 resistant human lung cancer cell lines, PC-7/CPT. Nevertheless, our results indicate that the expression of the same mutation in Saccaromyces cerevisiae does not show resistance to CPT.
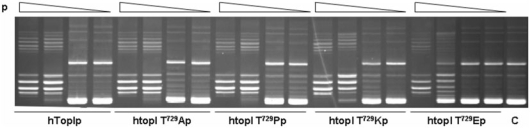
Thr729 mutants are catalytically active in vitro
To assess whether the imposed substitutions in the 729 position could alter the relaxation activity of the enzyme, the mutants were purified from galactose-induced yeast top1Δ cells, as described in the Materials and methods section and tested for their specific activity. Serial dilutions of equal protein concentrations were incubated with negatively supercoiled plasmid DNA and the reaction products resolved in agarose gels. As illustrated in Figure 2, at optimum salt condition for the wild-type enzyme (i.e. 150 mM), the specific activity of htopIThr729Glu was significantly reduced, while htop1Thr729Ala, htop1Thr729Pro and htop1Thr729Lys show an overall activity quite comparable to that of hTop1p. The observed reduction of htop1Thr729Glu specific activity could result from several alterations in enzyme catalytic cycle, including DNA association, cleavage, strand rotation and ligation. To discriminate between these possibilities, we analyzed step by step the catalytic cycle of the mutants and compared it to that of the wild-type enzyme.
Figure 2.
Thr729 mutants are catalytically active in vitro. A total of 120 pmol of purified hTop1p, hTop1Thr729Ala, hTop1Thr729Pro, hTop1Thr729Lys and hTop1Thr729Glu were serially 10-fold diluted and incubated in DNA relaxation assays with negatively supercoiled plasmid DNA. Following incubation at 37°C for 30 min, the reaction products were resolved in agarose gels and visualized after staining with ethidium bromide. C indicates no enzyme control.
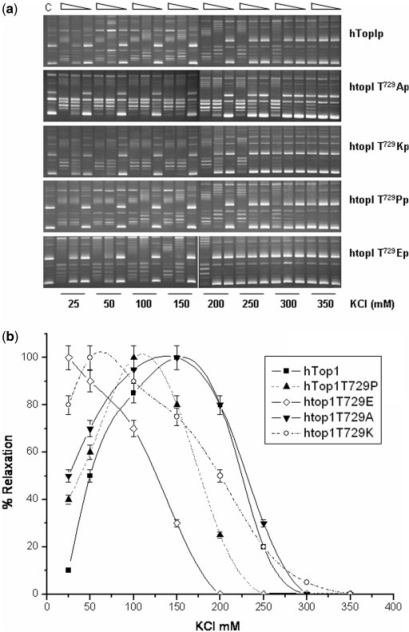
We first assessed the effects of ionic strength on mutant enzyme catalysis. hTop1p activity exhibits a salt optimum around 150 mM. Deviations from this value have previously been shown to correspond to increased or decreased DNA binding, for salt optimum more or less than 150 mM, respectively. Limiting amounts of hTop1p or htop1 mutants were incubated in plasmid relaxation assays containing increasing concentrations of KCl (Figure 3A). The percentage of relaxed plasmid DNA topoisomers produced ([relaxed DNA topoisomers]/[relaxed + supercoiled DNA topoisomers]) following incubation with the enzymes was quantified and plotted as a function of salt concentration (Figure 3B). Thr729 mutants display significant differences in their requirements for optimum of KCl concentration. In fact, while htopIThr729Ala shows a maximum in the catalytic activity comparable to that of the wild-type enzyme (150 mM KCl), DNA relaxation catalyzed by htop1Thr729Pro and htop1Thr729Lys was enhanced at lower salt concentrations (100 mM KCl). The minimum KCl concentration is shown by the Thr729Glu mutant where the optimum is reached at 25 mM KCl. These results are consistent with a general lower affinity effect of htop1Thr729 mutants for DNA than wild-type enzyme that becomes maximum in the case of the htop1Thr729Glu protein.
Figure 3.
The htop1Thr729Glu and htop1Thr729Lys mutant enzymes exhibit a lower salt optimum than wild-type hTop1p in plasmid DNA relaxation assays. (A) Limiting amounts of purified hTop1p, htop1Thr729Ala, htop1Thr729Pro, htop1Thr729Lys or htop1Thr729Glu were serially 10-fold diluted and incubated with plasmid DNA at 37°C, in the presence of the indicated concentration of KCl. The reaction products were resolved in agarose gels as described in Figure 2. (B) The percentage of relaxed plasmid DNA topoisomers obtained with hTop1p (filled square), htop1Thr729Ala (inverted filled triangle), htop1Thr729Pro (filled triangle), htop1Thr729Lys (open circle) and htop1The729Glu (open diamond) at the indicated concentration of KCl were determined in a minimum of three independent assays. Error bars indicate SDs from the mean.
Thr729 mutant sensitivity to CPT in vitro
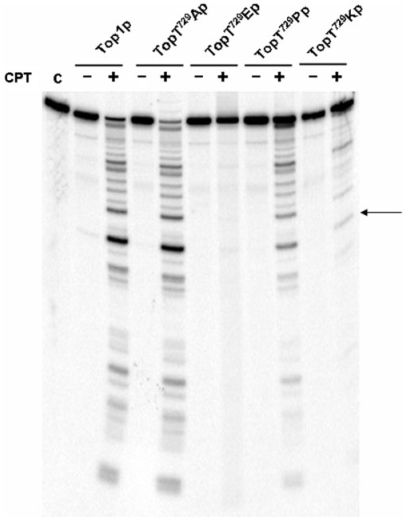
To assess the mechanism of Thr729 mutant resistance to CPT, the ability of CPT to stabilize the covalent DNA–enzyme complex was assessed in a DNA cleavage assay.
Equal concentrations of hTop1p and mutants proteins were incubated with a 900-bp 3′-end-labeled DNA fragment. Following incubation at 37°C for 30 min, the cleavable complexes were trapped by SDS and the cleaved DNA fragments were resolved in a denaturing polyacrylamide gel, Figure 4. Under these conditions, the relative intensity and distribution of cleaved DNA fragments can be used to assess mutation and/or CPT-induced alterations in the steady-state levels of covalent enzyme–DNA intermediates.
Figure 4.
Cleavage–religation equilibrium. Equal concentrations of hTop1p, htop1Thr729Al, htop1Thr729Pro, htop1Thr729Lys or htop1Thr729Glu were incubated with a single 32P-end-labeled DNA fragment in absence or presence of CPT as described in Materials and methods section. After 30 min the reactions were stopped with 0.5% SDS and the cleavage products resolved by urea/PAGE. C indicates no enzyme control. The arrow indicates the position of a high affinity cleavage site.
When wild-type protein is incubated without CPT, no cleavage of the labeled DNA strand is detected but when it is exposed to the drug, a dramatic increase in cleaved DNA fragments is observed, indicating that CPT stabilize the covalent enzyme–DNA intermediate. The same result is observed in the case of htop1Thr729Ala showing that this mutant is not resistant to CPT in vitro. A different behavior is observed with htop1Thr729Glu, htop1Thr729Pro and htop1Thr729Lys mutants. The htop1T729Glu does not trap any significant covalent DNA–enzyme intermediate, while cleavage occurs in the presence of CPT for the htop1Thr729Pro and htop1Thr729Lys, providing a direct evidence for drug binding to the mutant–DNA complexes. The extent of cleavage correlate with the in vivo drug resistance of the two mutants, showing a reduction of cleavable complex formation that is most evident in the case of htop1Thr729Lys.
The same effect can be detected using equal activity of proteins (Supplementary Figure 4S).
Taking together these data strongly confirm the differential drug resistance produced by the htop1Thr729 mutants in the in vivo assays and suggest that the Thr729 could have a key role in maintaining the correct geometry during the protein binding with the drug. To address such dynamic interactions, we used an oligonucleotide-based DNA substrate to uncouple hTop1p catalyzed DNA cleavage from religation.
Analysis of the religation rate using a suicide substrate
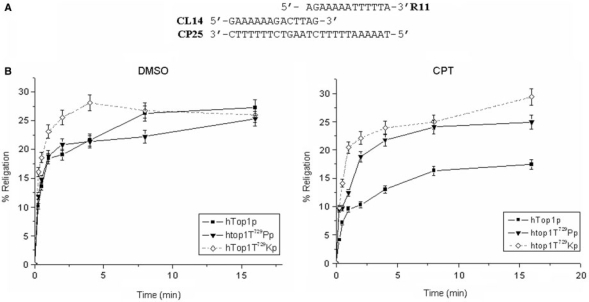
A common strategy to uncouple DNA cleavage from religation is to use an oligonucleotide-based substrate that contains a truncated scissile strand. For example, as depicted in Figure 5A, hTop1p cleavage of a high affinity site within a suicide DNA substrate liberates a dinucleotide and traps the covalent hTop1p–DNA complexes. In the presence of hTop1p, religate DNA molecules accumulate very rapidly in the absence of CPT, whereas the rate of DNA religation is dramatically reduced in the presence of CPT (Figure 5B). In contrast, the rate of DNA religation was substantially enhanced in the case of the htop1Thr729Pro or htop1Thr729Lys. We also tested the htop1Thr729Glu kinetic of religation using the described suicide DNA substrate and other substrates that differ for length and nucleotide sequence. We were unable to detect any cleavage of suicide DNA substrates in the absence and in the presence of an annealed religation strand. These data are consistent with a decrease in mutant enzyme binding to DNA.
Figure 5.
Analysis of the religation rate using a suicide substrate. (A) In the oligonucleotide-based suicide substrate, cleavage of the 5′-radiolabeled scissile strand (14 nt) liberates an AG dinucleotide to trap the covalent DNA–hTop1p complex. (B) The percentage of DNA relegation, in the presence and in the absence of CPT, was determined by ImageQuant software and normalized to the total amount of radioactivity in each lane: hTop1p (filled square), htop1Thr729Pro (filled traingle), htop1Thr729Lys.
The substitution of Thr729 involves a mechanism of decreased DNA binding by DNA topoisomerase
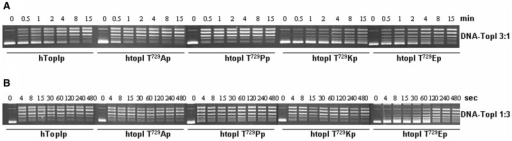
The failure in the recognition of a suicide DNA substrate, the absence of any trapped DNA cleaved products in the presence of CPT and the reduced capability to relax the DNA in physiologic ionic strength conditions by the htop1Thr729Glu enzyme, are strictly related to one of the steps involved in the catalytic cycle of the enzyme: the equilibrium DNA binding/DNA dissociation. To address this point, we analyzed the kinetics of DNA relaxation of the three CPT-resistant mutants htop1Thr729Pro, htop1Thr729Lys and htop1Thr729Glu, compared to that of the wild-type enzyme. The experiments were conducted in the presence of 150 mM KCl, using equal protein amounts with an excess or a deficit of negatively supercoiled plasmid DNA compared to the enzyme (DNA/enzyme ratio 3 : 1 or 1 : 3) (Figure 6). In the presence of excess DNA, all the enzyme molecules are assumed to interact with the substrate. In these conditions, the speed of the relaxation activity can be expressed as a function of the enzyme Km. As shown in Figure 6, both the htop1Thr729Lys and htop1Thr729Glu enzymes relax the DNA more slowly than wild-type accomplishing the same final relaxation level.
Figure 6.
Time course of plasmid DNA relaxation catalyzed by hTop1p, htop1Thr729Pro, htop1Thr729Lys and htop1Thr729Glu. Equal concentrations of hTop1p, htop1Thr729Pro, htop1Thr729Lys and htop1Thr729Glu were incubated with supercoiled plasmid DNA in the presence of 150 mM KCl in the presence of an excess of DNA (A) or protein (B). At the times indicated, aliquots were treated with 0.5% SDS and the reaction products were resolved in agarose gels as in Figure 2.
In the presence of excess protein versus the DNA substrate, hTopIp relaxes DNA in a processive manner. The same effect cannot be obtained in the case of the Thr729Glu mutant, which continues to be distributive in relaxing negatively supercoiled DNA.
These results suggest that the replacement of the Thr729 to Glu alters significantly the tertiary structure of the C-terminal hTop1p domain compromising the enzyme affinity to the DNA.
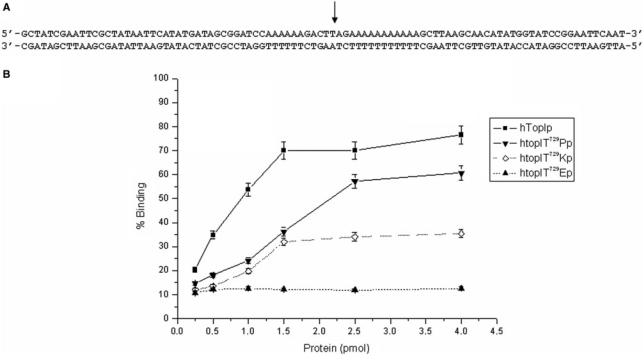
In order to quantify the difference in DNA binding by the three obtained CPT-resistant enzymes, a filter binding assay was performed. Increasing amounts of wild-type enzyme and mutants were incubated with a 100-bp labeled DNA fragment, Figure 7A, containing the specific hTop1p recognition sequence as described in the Material and methods section. The reaction mixtures were applied to a nitrocellulose filter and the amount of DNA retained was counted. Figure 7B shows the percent of bound DNA as a function of the enzyme concentration. As it can be appreciated, the amount of DNA bound by the three mutant proteins was significantly reduced relative to the wild-type. Coherently with the reduced catalytic activity observed in the relaxation assays and in the DNA cleavage assay, the DNA binding profile obtained with the single mutant htop1Thr729Lys and htop1Thr729Glu proves that changes in the hydrophobic properties of the C-terminal region of the enzyme radically affect the DNA binding/DNA dissociation equilibrium by hTop1p.
Figure 7.
DNA-binding assay. Increasing amounts of hTop1p, htop1Thr729Pro, htop1Thr729Lys and htop1Thr729Glu were incubated with an end-labeled 100-nt double-strand DNA fragment as described in Materials and methods section. The percentage of DNA binding as a function of enzyme concentrated is illustrated.
DISCUSSION
Mutations that impart resistance to CPT have been identified in several regions of human topoisomerase I. Few of them involve residues making direct contacts with the drug, many others cluster in the Lips region between the CAP (subdomain I and II) and CAT (subdomain III and C-terminal domain) regions. Some others are located in the CAT domain (2,15,28). Some of these mutations alter the enzyme catalytic cycle giving important clues on enzyme activity.
The Thr729 to Ala substitution that has been isolated in a CPT11 resistant tumor cell line (12) is located in the C-terminal domain and its side chain forms a 2.6-Å hydrogen bond with the hydroxyl group of Tyr619. This interaction stabilize the contacts between the C-terminal and core subdomain III regions of the CAT, that extends from the top half of the molecule downward by a couple of long helices (8 and 9) functioning as an hinge that opens and closes the enzyme around DNA. Thr729 and Tyr619 are positioned respectively in α-helix 21 and α-helix 17 that together with helix 16 constitute a hydrophobic pocket (11). This three-helix structure bridges the linker, the active site region and the hinge. In addition, as discussed by Chrencik et al. (15), the Thr729 residue could have an important role in the geometry of the enzyme C-terminal region. In the X-ray structure, in fact, Thr729 forms a water-mediated contact with the backbone carbonyl oxygen atom of Asp725 and is positioned on the same α-helix of the Asn 722 residue, which interacts with the CPT through a water-mediated bond.
Our analyses of Thr729 mutations in human Top1p, in vitro and in vivo, support this model but suggest that the charge and geometry of amino acid side chains at this position, directly impact the active site architecture and the sensitivity of hTop1p to CPT.
Surprisingly the substitution Thr729Ala, when tested in the S. cerevisiae yeast system, did not show any level of drug resistance as in the tumor cell line.
Our results clearly indicate that the introduction of a basic or an acidic side chain (Thr729Lys or Thr729Glu) changes the quality of DNA binding, rendering in this case the enzyme CPT resistant. In fact, the Thr729Glu mutation completely abolish DNA binding, while the Thr729Lys substitution can still bind to DNA but with a reduced affinity if compared to the wild-type enzyme.
In contrast, when the Thr729 was substituted by a hydrophobic residue as Ala or Pro, the same effect was not observed. The Thr729Ala mutation shows CPT sensitivity comparable to that of the wild-type without any observable alterations in the specific catalytic activity, both in the plasmid DNA relaxation and in cleavage/religation assays.
The replacement of Thr729 by Pro, a residue that could distort the α-helix, reduces the enzyme sensitivity to CPT with a less extent than the two polar substitutions Thr729Lys and Thr729Glu. Taken together, our data support the idea that changing the hydrophobic interactions between helices 16, 17 and 21, by the addition of a charged residue, must have a dramatic effect in enzyme stability. It has been previously demonstrated that topo58, a C-terminal deletion of topo70, but comprising the core subdomain III, is still able to bind DNA with the same affinity than the topo70 enzyme (29). It is quite surprising that a single hydrophilic substitution in the C-terminal may have such a dramatic effect on DNA binding.
The results reported in the accompanying paper reinforce our data showing a different dynamic behavior for the Thr729Pro and the Thr729Lys mutants, if compared to the wild-type protein. In fact, while in the presence of the Thr729Lys mutation some high fluctuations in the region comprising helices 16 and 17 has been observed, the same region shows a reduction of mobility in the Thr729Pro mutant.
Moreover, the evidence that CPT has little effect on the rate of DNA religation by Thr729Pro and Thr729Lys mutants, suggests that their alterations in protein–DNA interaction can also affect drug binding to the covalent complex.
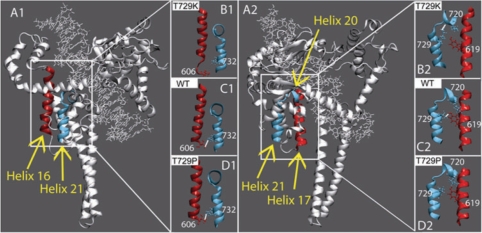
As shown in Figure 8, panel C2, in the wild-type protein a direct hydrogen bond between helix 21 and helix 17 is formed by the Thr729 and Tyr619. The results obtained by the dynamic simulation described in the accompanying paper have shown that in the case of the Thr729Pro mutant the direct hydrogen bond became lost without altering the relative positions of the two helices (Figure 8, panel D2). On the contrary, the Thr729Lys mutation provokes a more dramatic effect. As it can be seen in Figure 8, panel B2, the loss of the direct hydrogen bond is followed by the moving away of helix 21 from helix 17 with a direct effect on the drug-binding pocket.
Figure 8.
Structure modifications induced by the Thr729 mutations. Panel A1: helix 16, in core domain, and helix 21, in the C-terminal domain are highlighted in red and light blue colours, respectively. Panel B1–C1–D1: only the helices are shown in representative snapshots of the htopIThr729Lys, with hTopIp and htopIThr729Pro simulations, respectively. The side chains of Thr606 and Trp732, in helixes 16 and 17, respectively, are shown in ball and stick. Stable direct hydrogen bonds, when present, are indicated with a white line. Panel A2: helix 17, in core domain, and helices 20 and 21, in the C-terminal domain are highlighted in red and light blue colours, respectively. Panel B2–C2–D2: only the helices are shown in representative snapshots of the htopIThr729Lys, hTopIp and htopIThr729Pro simulations, respectively. The side chains of Lys720, in helix 20, of the mutated residue 729, in helix 21, and of Tyr619, in helix 17 are shown in ball and stick. Stable direct hydrogen bonds, when present, are indicated with a white line.
Moreover the two mutations exert a different effect on DNA religation as it can be seen in Figure 5. In fact, while htopIThr729Lys shows an increase in the rate of DNA religation in the absence of CPT, the activity of htopIThr729Pro is comparable to that of the wild-type enzyme in the same conditions. The addition of CPT dramatically reduces the religation rate for the wild-type protein, but does not show any effect in the case of htopIThr729Lys and a mild reduction for the htop1Thr729Pro.
In addition, topIThr729Glu was unable to cleave any suicide substrate used in our assays supporting the hypothesis that the drug resistance is due to the reduced DNA binding.
The kinetics of DNA relaxation in the presence of an excess of substrate, displays a distributive mode of action both for htopIThr729Lys and htopIThr729Glu. The same results cannot be observed in the presence of an excess of protein. In these conditions, only htopIThr729Glu relaxed DNA distributively while htopIThr729Lys restored its processivity. This data are strongly supported by the binding assay, where htop1Thr729Glu was unable to bind the given substrate independently by the protein concentration.
We previously demonstrated that the flexibility of the linker domain affects the geometry of the active site influencing the DNA cleavage/religation equilibrium of the enzyme and the CPT sensitivity through long-range conformational contacts (9,30).
In addition, a dynamic coupling between the linker domain flexibility and the active site of hTop1p may also be inferred from X-ray structures of Topo70 in the presence and the absence of the CPT analog TPT, where the flexibility of the linker domain prevents its structural determination. However, in the ternary Topo70–DNA–TPT complex, the structure of the linker domain could be resolved (11).
A detailed comparison of the ternary structure of the Topo70–DNA covalent complex in the presence and absence of TPT shows that helix 16 is half a turn shorter in the absence of the drug. In the presence of TPT Thr606 and Trp732, sited on the helices 16 and 21, respectively, forms an hydrogen bond between Trp732 and Thr606 (Figure 8B) that is lost in the absence of the drug, where Thr606 is positioned in the misfolded part of the helix 16. The data described in the accompanying paper show that this interaction is similarly lost in the htop1Thr729Lys dynamic simulation (Figure 8, panels B1, C1 and D1) where a partial destructuration of the helix 16, has been observed.
It is interesting to observe that the linker domain is directly connected to the hydrophobic pocket formed by helices 16, 17 and 21 by two long flexible loops that extend from the two coiled-coil towards helix 17 directly and helix 21 via the catalytic Tyr region. This coordinated structure could be strongly influenced by the radical change imposed by the introduction of charged residues. The dynamic cross-correlation map of the htop1Thr729Pro and htop1Thr729Lys mutants compared to that of the wild-type, described in the accompanying paper, show a dramatic change in the protein dynamics. In fact, while in the wild-type protein the linker and the C-terminal domains move in an anticorrelated way, the Thr729Lys mutation abolishes this motions and the Thr729Pro provokes an increased rigidity in the same domains.
Taken together, the experimental and the simulation data indicate that a single change in a crucial residue may alter dramatically the behavior of DNA topoisomerase IB, giving important clues on its dynamic mode of action.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministero dell’Istruzione, dell’Università e della Ricerca, Cofinanziamento 2005–2007; Ministero della Salute. Funding for open access charge: Miur COFIN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mary-Ann Bjornsti, Alessandro Desideri and Ivan Micetic for helpful discussions.C.L. is recipient of an Adriano Buzzati-Traverso Fellowship.
REFERENCES
- 1.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 2.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 3.Mo YY, Wang C, Beck WT. A novel nuclear localization signal in human DNA topoisomerase I. J. Biol. Chem. 2000;275:41107–41111. doi: 10.1074/jbc.M003135200. [DOI] [PubMed] [Google Scholar]
- 4.Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Galindo C, Radomski K, Stewart CF, Furman W, Santana VM, Houghton PJ. Clinical use of topoisomerase I inhibitors in anticancer treatment. Med. Pediatr. Oncol. 2000;35:385–402. doi: 10.1002/1096-911x(20001001)35:4<385::aid-mpo1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco GS. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resist. Update. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 9.Fiorani P, Bruselles A, Falconi M, Chillemi G, Desideri A, Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- 10.Fiorani P, Chillemi G, Losasso C, Castelli S, Desideri A. The different cleavage DNA sequence specificity explains the camptothecin resistance of the human topoisomerase I Glu418Lys mutant. Nucleic Acids Res. 2006;34:5093–5100. doi: 10.1093/nar/gkl670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staker BL, Hjerrild K, Feese MD, Behnke CA, Burgin AB, Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota N, Kanzawa F, Nishio K, Takeda Y, Ohmori T, Fujiwara Y, Terashima Y, Saijo N. Detection of topoisomerase I gene point mutation in CPT-11 resistant lung cancer cell line. Biochem. Biophys. Res. Commun. 1992;188:571–577. doi: 10.1016/0006-291x(92)91094-7. [DOI] [PubMed] [Google Scholar]
- 13.Fertala J, Vance JR, Pourquier P, Pommier Y, Bjornsti MA. Substitutions of Asn-726 in the active site of yeast DNA topoisomerase I define novel mechanisms of stabilizing the covalent enzyme-DNA intermediate. J. Biol. Chem. 2000;275:15246–15253. doi: 10.1074/jbc.275.20.15246. [DOI] [PubMed] [Google Scholar]
- 14.Fujimori A, Harker WG, Kohlhagen G, Hoki Y, Pommier Y. Mutation at the catalytic site of topoisomerase I in CEM/C2, a human leukemia cell line resistant to camptothecin. Cancer Res. 1995;55:1339–1346. [PubMed] [Google Scholar]
- 15.Chrencik JE, Staker BL, Burgin AB, Pourquier P, Pommier Y, Stewart L, Redinbo MR. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 16.Megonigal MD, Fertala J, Bjornsti MA. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 17.Kauh EA, Bjornsti MA. SCT1 mutants suppress the camptothecin sensitivity of yeast cells expressing wild-type DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1995;92:6299–6303. doi: 10.1073/pnas.92.14.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjornsti MA, Benedetti P, Viglianti GA, Wang JC. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 19.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 20.Fiorani P, Amatruda JF, Silvestri A, Butler RH, Bjornsti MA, Benedetti P. Domain interactions affecting human DNA topoisomerase I catalysis and camptothecin sensitivity. Mol. Pharlmacol. 1999;56:1105–1115. doi: 10.1124/mol.56.6.1105. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A. Lithium Acetate Yeast Transformation. New York: Cold Spring Harbor Laboratory Press; 1994. pp. 133–134. [Google Scholar]
- 22.Benedetti P, Fiorani P, Capuani L, Wang JC. Camptothecin resistance from a single mutation changing glycine 363 of human DNA topoisomerase I to cysteine. Cancer Res. 1993;53:4343–4348. [PubMed] [Google Scholar]
- 23.Knab AM, Fertala J, Bjornsti MA. A camptothecin-resistant DNA topoisomerase I mutant exhibits altered sensitivities to other DNA topoisomerase poisons. J. Biol. Chem. 1995;270:6141–6148. doi: 10.1074/jbc.270.11.6141. [DOI] [PubMed] [Google Scholar]
- 24.Frøhlich RF, Andersen FF, Westergaard O, Andersen AH, Knudsen BR. Regions within the N-terminal domain of human topoisomerase I exert important functions during strand rotation and DNA binding. J. Mol. Biol. 2004;336:93–103. doi: 10.1016/j.jmb.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Bonven BJ, Gocke E, Westergaard O. high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985;41:541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- 26.Fiorani P, Hann CL, Benedetti P, Bjornsti MA. Drug-induced stabilization of covalent DNA topoisomerase I-DNA intermediates. DNA cleavage assays. Methods Mol. Biol. 2001;95:291–301. doi: 10.1385/1-59259-057-8:291. [DOI] [PubMed] [Google Scholar]
- 27.Hann CL, Carlberg AL, Bjornsti MA. Intragenic suppressors of mutant DNA topoisomerase I-induced lethality diminish enzyme binding of DNA. J. Biol. Chem. 1998;273:31519–31527. doi: 10.1074/jbc.273.47.31519. [DOI] [PubMed] [Google Scholar]
- 28.van der Merwe M, Bjornsti MA. Mutation of Gly721 alters DNA topoisomerase I active site architecture and sensitivity to camptothecin. J. Biol. Chem. 2008;283:3305–3315. doi: 10.1074/jbc.M705781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Champoux JJ. Reconstitution of enzymatic activity by the association of the cap and catalytic domains of human topoisomerase I. J. Biol. Chem. 2002;277:30815–30823. doi: 10.1074/jbc.M205302200. [DOI] [PubMed] [Google Scholar]
- 30.Losasso C, Cretaio E, Palle K, Pattarello L, Bjornsti MA, Benedetti P. Alterations in linker flexibility suppress DNA topoisomerase I mutant-induced cell lethality. J. Biol. Chem. 2007;282:9855–9864. doi: 10.1074/jbc.M608200200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.