Abstract
Clerocidin (CL), a microbial diterpenoid, reacts with DNA via its epoxide group and stimulates DNA cleavage by type II DNA topoisomerases. The molecular basis of CL action is poorly understood. We establish by genetic means that CL targets DNA gyrase in the Gram-positive bacterium Streptococcus pneumoniae, and promotes gyrase-dependent single- and double-stranded DNA cleavage in vitro. CL-stimulated DNA breakage exhibited a strong preference for guanine preceding the scission site (−1 position). Mutagenesis of −1 guanines to A, C or T abrogated CL cleavage at a strong pBR322 site. Surprisingly, for double-strand breaks, scission on one strand consistently involved a modified (piperidine-labile) guanine and was not reversed by heat, salt or EDTA, whereas complementary strand scission occurred at a piperidine-stable −1 nt and was reversed by EDTA. CL did not induce cleavage by a mutant gyrase (GyrA G79A) identified here in CL-resistant pneumococci. Indeed, mutations at G79 and at the neighbouring S81 residue in the GyrA breakage-reunion domain discriminated poisoning by CL from that of antibacterial quinolones. The results suggest a novel mechanism of enzyme inhibition in which the −1 nt at the gyrase-DNA gate exhibit different CL reactivities to produce both irreversible and reversible DNA damage.
INTRODUCTION
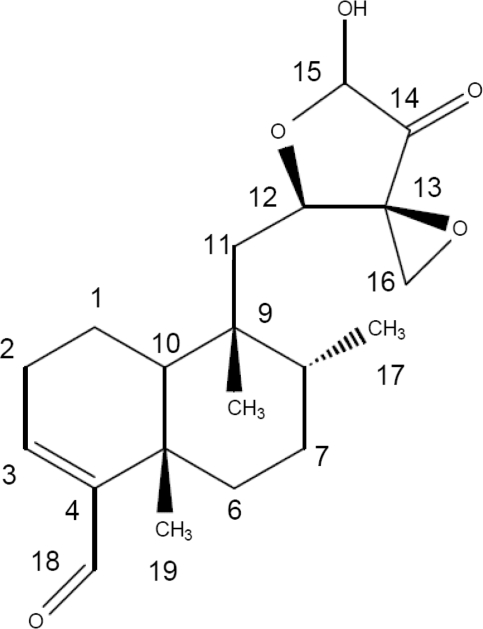
Clerocidin (CL) (Figure 1) is a cytotoxic diterpenoid natural product (1) that inhibits bacterial DNA gyrase and mammalian DNA topoisomerase II (2,3). Unlike other topoisomerase poisons, CL induces an irreversible DNA break when guanine (G) is present at the −1 position from the cleavage site (3–5). In the absence of topoisomerases, CL itself exhibits an intrinsic DNA alkylating activity against guanines that are exposed in single-stranded DNA regions, promoting spontaneous depurination and subsequent DNA scission at the abasic site (6). These observations and other data suggest that mammalian type II topoisomerases potentiate CL attack at guanines that have in some way been ‘processed’ in the enzyme-DNA complex (5,6). Thus, although toxicity has precluded clinical use, CL is of interest as a probe of the normally transient cleavage complex formed in the reaction cycle of type II topoisomerases.
Figure 1.
Chemical structure of CL.
Recent work has focused on the structural features of CL and DNA necessary for intrinsic- and mammalian topo II-mediated DNA cleavage. Both reactions require the highly strained epoxide moiety in the C12–C16 region (6,7) (Figure 1): opening of the epoxide ring by prolonged incubation with ethanol abolishes drug activity. CL reacts very slowly with duplex DNA (requires 24–48 h) but more rapidly with single-stranded DNA producing irreversible cleavage at G and C (7,8). The epoxide ring reacts covalently with the N-7 position of guanine resulting in facile release of the modified base (7). The aldehyde linked to C4 is not essential as related epoxy antibiotics such as terpentecin and UCT4B lack the moiety yet retain activity (3,9). The diterpenoid ring system is dispensable for intrinsic alkylating activity and can be replaced by naphthalene (7). Unlike the intrinsic reaction with duplex DNA, topo II-mediated cleavage induced by CL is rapid (complete in minutes) and occurs irreversibly at guanines but with one-third of DNA scission sites occurring reversibly at cytosines (5). It is known that the reactive NH2 and N3 sites of cytosine are involved in base pairing and are thereby protected from CL alkylation. Therefore, the C preference presumably arises from non-covalent interaction of CL with the enzyme–DNA complex in a manner akin to classical topo II poisons (8). These features reinforce the idea that the conformation of bound DNA in the topoisomerase complex has a major influence on CL action. Indeed, substantial distortion of nicked duplex DNA bound to yeast topo II has recently been observed by X-ray crystallography (10).
CL is potentially an interesting probe of bacterial type II topoisomerases. Thus, although gyrase and topo IV share structural and mechanistic similarities, they manipulate DNA in distinct ways to produce different DNA products (11–15). Gyrase, an GyrA2GyrB2 complex specified by gyrA and gyrB genes, introduces negative supercoils into DNA and has important roles in maintaining bacterial chromosome supercoiling and in the initiation and elongation phases of DNA replication. Topo IV, a ParC2ParE2 tetramer, encoded by parC and parE genes, preferentially relaxes positively supercoiled DNA and unlinks pre-catenanes and catenated chromosomes formed during DNA replication. Both enzymes act by passing a duplex DNA segment through a transient enzyme–DNA break formed by the covalent cleavage complex (14,16,17). Whereas for topo IV, DNA scission occurs in a 34-bp DNA ‘gate’ segment (18), gyrase wraps a 150-bp DNA segment that contains both the ‘gate’ and ‘transported’ DNA segments (19–21). It is not known whether DNA wrapping by gyrase (and its absence in the topo IV complex) affects base potentiation and CL action.
Although CL studies have concentrated on mammalian topo II, the drug does have antibiotic activity against Escherichia coli and particular potency against Gram-positive species such as Staphylococcus aureus (2,3). Initial work reported that CL inhibits gyrase (from E. coli) (2), which is also the target of antibacterial quinolone poisons in Gram-negative bacteria. Recently, we showed that CL inhibits purified recombinant S. pneumoniae topo IV to produce irreversible DNA scission at −1G and on that basis proposed that topo IV is the CL target in Gram-positive bacteria (22), as it is for many quinolones (23). In the present study, we have determined the target of CL in S. pneumoniae, a Gram-positive system amenable to genetic manipulation through its ability to undergo DNA transformation. Contrary to our previous suggestion (22), we find that gyrase is the primary target. In complementary biochemical experiments, we show that CL traps gyrase cleavage complexes on DNA, and we explore fully the unusual chemical, DNA and protein requirements for this reaction. This study is the first to demonstrate that CL differentially modifies each half of a topoisomerase-DNA gate, and new data reported here illuminates key differences in CL action versus antibacterial quinolone poisons both in vivo and in vitro.
MATERIALS AND METHODS
Bacterial strains, reagents and plasmids
Streptococcus pneumoniae 7785 is a ciprofloxacin-susceptible clinical strain isolated at St George's Hospital Medical School (24). The strain and its defined quinolone-resistant mutants (25) were grown either on brain heart infusion-agar plates containing 10% horse blood or in liquid culture in Tryptic Soy Broth (TSB) (Difco). Streptococcus pneumoniae R6, a standard laboratory strain competent for genetic transformation, and E. coli strain BL21(λDE3)pLysS were from our laboratory collection. TOPO10 competent cells were from Invitrogen. CL, a gift from Leo Pharmaceutical Products (Ballerup, Denmark), was prepared as a 5-mg/ml stock solution in DMSO, stored frozen at −80°C and used within 4 weeks. Competence stimulating peptide was synthesized by Activotec, Cambridge, UK. Supercoiled and relaxed pBR322 were purchased from New England Biolabs and John Innes Enterprises, respectively. [γ-33P]ATP (3000 Ci/mmol) was from GE Healthcare.
Selection of CL-resistant S. pneumoniae mutants
A culture of S. pneumoniae 7785 was grown in TSB at 37°C to an OD600 of 0.8 (about 5 × 108 cfu/ml). Bacteria from one millilitre of culture were spun down, plated on CL-BHI blood plates and incubated aerobically at 37°C for 20 h. Drug challenge at 0.8, 1.6 and 3 μg/ml allowed recovery of mutants CL6, CL7, CL8 and CL9, mutant CL10, and mutants CL11, CL12 and CL13, respectively. The minimum inhibitory concentrations (MICs) of the mutants for CL were determined by the 2-fold dilution method (25). Briefly, ∼105 colony forming units of bacteria was spotted on agar plates made up with brain heart infusion medium supplemented with 10% horse blood and containing serial 2-fold dilutions of drug. Plates were examined for growth after overnight aerobic incubation at 37°C. The known ciprofloxacin and gemifloxacin MICs of strain 7785 were determined in parallel as a control (26). The eight mutants all exhibited a CL MIC of 12 μg/ml and a ciprofloxacin MIC of 1 μg/ml. For parent strain 7785, the MICs for CL, ciprofloxacin and gemifloxacin were 0.2, 1 and 0.06 μg/ml, respectively.
DNA sequence analysis of topoisomerase genes in resistant mutants
To obtain S. pneumoniae genomic DNA, strain 7785 and its drug-resistant mutants were grown in 10 ml of TSB broth to an OD600 of 0.6 and harvested by centrifugation. The bacterial pellet was re-suspended in 2 ml of 50 mM Tris–HCl, pH 7.5, 10 mM EDTA. SDS and proteinase K were added to final concentrations of 1% and 100 μg/ml, respectively and the suspension was incubated at 42°C for 1 h. Phenol extraction was performed twice, the DNA was recovered by ethanol precipitation and redissolved in Tris–HCl, pH 7.5, 1 mM EDTA. Genomic DNA from strain CL12 was used as a template to amplify the gyrase and topo IV genes using Taq DNA polymerase. PCR was performed for 30 cycles: denaturation at 93°C for 45 s, annealing at 52°C for 45 s and amplification at 74°C for 3 min. Full-length genes were amplified using the following primer pairs: VGA35/VGA36 for gyrA, P7164/P7165/for gyrB, N6894/VPC2 for parC and N7043/7044 for parE (27). Automated sequence analysis of each PCR product was done by Lark Technologies, Inc employing a series of nested primers complementary to the amplified DNA. The four gene sequences from CL12 (totalling nearly 9 kb) were compared with those of the parent S. pneumoniae 7785 (27). The gyrase and topo IV genes of three other randomly chosen mutants CL10, CL11 and CL13 were also sequenced.
Genetic transformation of S. pneumoniae
The 2.4-kb gyrA gene was amplified by PCR from parent strain S. pneumoniae 7785 and CL-resistant mutant CL12 using primers VGA35/VGA36 (27) and used to transform S. pneumoniae strain R6 following the approach of Pozzi et al. (28). Briefly, S. pneumoniae R6 was grown in tryptic soy broth (TSB, Difco) and 30 min before onset of stationary phase, cultures were frozen in 10% glycerol. Competence medium (TSB pH 8.0, 10% glycerol, 0.16% bovine serum albumin, 0.01% CaCl2) (10 ml) was mixed with 0.5 ml of thawed R6 cell stock. Competence stimulating peptide was then added to 100 ng/ml along with transforming DNA (final concentration 1 μg/ml). The cell mix was incubated at 37°C for 2.5 h and then plated on TSB-agar plates containing CL at 0.8, 1.6 and 3 μg/ml, respectively. Plates were incubated aerobically at 37°C for 48 h. CL-resistant colonies were obtained by transformation with CL12 gyrA DNA but not with the 7785 gyrA DNA. The gyrA genes of five randomly chosen CL-resistant R6 transformants were sequenced in full and the antibiotic susceptibiliy of the strains was determined by the 2-fold dilution method. All five mutants exhibited a CL MIC of 12–24 μg/ml, compared with an MIC 0.2–0.4 μg/ml for the recipient R6 strain. The ciprofloxacin MIC of the transformants was the same as for R6, i.e. 0.5 μg/ml.
Recombinant gyrase and topo IV subunits
Streptococcus pneumoniae GyrA, GyrB, ParC, ParE and GyrA S81F were expressed as His-tagged proteins in E. coli from plasmids pXP10, pXP9, pXP13, pXP14 and pXP15, respectively, and were purified to >95% homogeneity as described previously (27,29). GyrA G79A and GyrA G79D were prepared by expression from gyrA plasmid pXP10 whose gyrA codon 79 had been altered using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's protocol. The mutagenic primers used for PCR to obtain gyrA (G79A) were 5′GGT-AAA-TAT-CAC-CCA-CAC-GCG-GAT-TCC-TCT-ATT-TAT-GAA (nucleotide positions 217–255, altered nucleotide 237 is underlined) and its complement, 5′TTC-ATA-AAT-AGA-GGA-ATC-CGC-GTG-TGG-GTG-ATG-TTT-ACC. For gyrA (G79D), the same forward primer was made except nucleotides C237 and G238 were replaced with A and C, respectively. The appropriate complementary changes were introduced into the reverse primer. The mutant gyrA genes were sequenced in full to confirm the presence of the single desired mutation at codon 79. The mutant GyrA proteins were expressed in E. coli and purified by Ni–agarose chromatography as described previously for wild-type GyrA (27). When induced and purified under identical conditions, GyrA, GyrA S81F and GyrA G79A showed similar specific activities in a standard DNA supercoiling assay (27) of ∼5 × 104 U/mg: the specific activity of the GyrA G79D protein was ∼2.5 × 104 U/mg.
Mutagenesis of specific DNA cleavage sites
Our mapping studies (see below) identified a strong site of gyrase cleavage site at 1073 in pBR322 whose sequence is known in full (30). Mutant sites bearing alterations at 1073 and at 1078 on the complementary strand were generated from pBR322 using complementary primers in the QuikChange Mutagenesis Kit according the manufacturer's instructions. Three forward primers were made 5′CTGTCCAGGCAGGTAGATXACGAYCATCAGGGACAGCTTCAA (nucleotide position 1055–1096) in which positions G1073 and C1078 (marked by X and Y) were pair-wise replaced with A and T, C and G or T and A, respectively. Three reverse primers were designed to complement the altered sequences. Mutated plasmids were recovered and the introduction of site changes was confirmed by DNA sequence analysis.
A similar approach was used to mutate the ‘E’ cleavage site for topo IV found in the S. pneumoniae parE gene (31). This site was shown to be efficiently cleaved by topo IV in the presence of gemifloxacin and therefore ideal for mutational studies (31) Three mutagenic forward primers were created i.e. 5′TACCAAGGTCATGAAXGACTYTGCACGTAAAACAGGTCTTCT-3′ (parE nucleotide positions 882 to 923) in which T897 and A902 (denoted by X and Y) were substituted pair-wise with G and C, C and G and A and T. Three complementary (reverse) primers were made accordingly. Plasmid pXP1 carrying the E site was used as template for PCR mutagenesis. Mutations at the E site were confirmed by DNA sequence analysis of recovered plasmids.
5′-end labelling of primers and DNA cleavage substrates
Oligonucleotide primers (30 pmol) were labelled at their 5′ end by incubation with [γ-33P] ATP (30 pmol) and T4 polynucleotide kinase. The mixture was incubated at 37°C for 30 min followed by heat inactivation at 90°C for 3 min. The labelled primers were used in PCR to generate cycle sequencing chain termination ladders and to amplify DNA fragments specifically labelled at one or the other 5′ end. A 290-bp product, the ‘S fragment’ internal to S. pneumoniae parE (nucleotides 1179–1468) was amplified from plasmid pXP1 using primers S6398 and S6399 as described earlier (31). Similarly, 295-bp (nucleotides 905–1200) and 234-bp (966–1200) fragments were amplified from pBR322 using primers 5′Gyra (pBR905) and Apbr1200 (pBR1200), and pBR966 and pBR1200, respectively (31). A 256-bp E fragment from S. pneumoniae parE (containing the strong E cleavage site for topo IV) was amplified from pXP1 using primers EFOR and EREV (31). To label one or other strand, PCR was set up with 10 pmol of 5′-end 33P-labelled forward primer plus an equal amount of the unlabelled reverse primer. DNA fragments were purified with the QIAquick PCR purification kit (Qiagen, CA). The same procedures were used to generate PCR products carrying mutated versions of the strong pBR322 site and of the E site.
DNA supercoiling and DNA cleavage assays
DNA gyrase activity was examined in the standard DNA supercoiling assay (27) using a reaction buffer containing 35 mM Tris–HCl, pH 7.5, 6 mM MgCl2, 1.8 mM spermidine, 24 mM KCl, 5 mM DTT, relaxed pBR322 (0.4 μg), 36 μg/ml BSA, 6.5% glycerol (w/v), 1.4 mM ATP. Wild-type and mutant gyrase activities were reconstituted from appropriate GyrA and GyrB subunits. Subunit specific activity was determined by serial dilution and assay in the presence of a 5-fold excess of the complementing subunit (to activate all the available limiting subunit).
For cleavage of supercoiled DNA, gyrase (0.45 μg GyrA and 1.7 μg GyrB) was incubated with supercoiled pBR322 DNA (0.4 μg) in the absence or presence of CL or ciprofloxacin in gyrase supercoiling buffer from which ATP had been omitted. After 1-h incubation at 37°C, SDS and proteinase K were added to final concentrations of 1% and 100 μg/ml, respectively, and incubation was continued for another hour at 42°C. Loading buffer was added to the samples and DNA products were separated by electrophoresis in 1% agarose in TBE buffer (90 mM Tris base, 90 mM boric acid, 2.5 mM EDTA) prior to ethidium bromide staining and photography under UV light using an Alpha Innotech digital camera. The same protocol was used to study cleavage of short unlabelled PCR fragments (200–300 bp) except inclusion of ATP was required for efficient breakage and reaction products were separated on 2.5% low-gelling temperature agarose in TBE buffer.
Cleavage of end-labelled DNA substrates was carried out in DNA supercoiling assay buffer. The mixture (20 μl) contained 300 fmol end-labelled DNA substrate, 2 μg of GyrA and 4 μg of GyrB proteins, 1 mM ATP, and 200 μM CL or 100 μM gemifloxacin. After incubation for 1 h at 37°C, samples were treated with SDS and proteinase K as described for non-radioactive substrates, divided into two equal aliquots and precipitated with ethanol. One dried aliquot of each sample was taken up in 100 μl of 1-M piperidine, heated at 90°C for 30 min (to release any modified bases) and the piperidine was removed under vacuum. DNA samples (treated and untreated with piperidine) were taken up in formamide gel loading buffer (32% formamide, 3.3 mM NaOH, 0.085% bromophenol blue, 0.085% xylene cyanol) and heated at 90°C for 2–4 min. The DNA products were separated by electrophoresis in a denaturing 6% polyacrylamide gel alongside an appropriate chain termination sequencing ladder (obtained with the Fmol DNA cycle sequencing kit) and/or a G+A ladder generated by the Maxam–Gilbert method (32). Gels were dried under vacuum on to Whatmann 3MM filter paper, exposed to a PhosphorImager screen, and visualized using software from Amersham Biosciences and ImageQuant.
RESULTS
DNA gyrase is the target of CL in S. pneumoniae
As a first step in understanding CL action, we examined its activity against wild-type S. pneumoniae strain 7785 alongside clinically important quinolones. CL was a potent inhibitor with a minimum inhibitory concentration of 0.2 μg/ml (0.6 μM), which compares favourably with values of 0.06 (0.12 μM) and 1–2 μg/ml (3–6 μM) for gemifloxacin and ciprofloxacin, respectively (26). Mutants of S. pneumoniae 7785 resistant to CL were obtained in a single step by plating on agar containing various inhibitory levels of the drug. Mutant CL12 was 60-fold less susceptible to CL but remained fully susceptible to ciprofloxacin. DNA sequence analysis revealed a novel gyrA mutation (GGG to GCG, G79A) with no other changes in gyrA or gyrB, or in the topo IV parC and parE genes. The same gyrA mutation was present in three other randomly chosen mutants selected independently at different drug levels. Repeated recovery of the same gyrA mutation in several single-step CL-resistant mutants implicates gyrase in CL action in vivo.
To show conclusively that gyrase is the primary target of CL in S. pneumoniae, we used genetic transformation to reintroduce the gyrA (G79A) mutation into a CL-susceptible S. pneumoniae strain and examined the CL susceptibility of the transformants. For these experiments, we amplified just the gyrA(G79A) gene of mutant CL12 as a 2.4-kb DNA fragment and used it to transform S. pneumoniae strain R6, which (unlike 7785) is competent to undergo genetic transformation by uptake of donor DNA and chromosomal integration by homologous recombination (28). Using the gyrA (G79A) gene as donor DNA, CL-resistant transformants were readily recovered on CL-agar plates: about 0.02% of the recipient R6 cells became transformed, i.e. very much higher than the observed frequency of spontaneous mutants (∼10–8). No transformants were obtained using as a control the wild-type gyrA gene from the parental 7785 strain. The gyrA genes of five randomly chosen transformants were amplified by PCR and sequenced in full. Each carried the G79A codon change (G to C at position 236). Moreover, the gyrA genes exhibited mosaic polymorphism for silent nucleotide changes indicative of uptake/integration of donor CL12 gyrA, i.e. at position 225 (C to T, 5 of 5 transformants), 264 (C to T, 4 of 5 transformants) and at 141 (C to T, one of five transformants). Therefore, the resistant colonies were genuine transformants and not spontaneous mutants. Crucially, all five transformants exhibited a CL MIC of 12–24 μg/ml (the same as strain CL12) versus 0.2–0.4 μg/ml for the recipient R6 strain and (like CL12) all remained susceptible to ciprofloxacin. It appears that the gyrA (G79A) mutation alone can account for the 60-fold increase in CL resistance (seen for CL12) and that gyrase is the primary target of the drug in S. pneumoniae.
CL promotes gyrase cleavage of supercoiled DNA
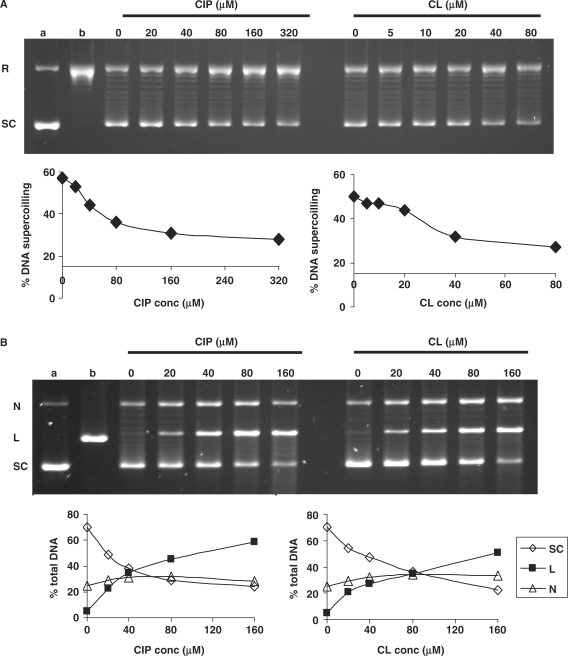
Given the outcome of the genetic experiments, we tested the effects of CL on S. pneumoniae gyrase in catalytic and DNA cleavage assays (Figure 2). As a control, we employed ciprofloxacin, a fluoroquinolone often used as a comparator for inhibition/cleavage studies. In the absence of drug (Figure 2A, lanes 0), gyrase converted relaxed plasmid DNA (lane b) into the supercoiled form in an ATP-dependent reaction. CL inhibited this activity in a dose-dependent fashion with 50% inhibition (IC50) at 40 μM, i.e. comparable to the ciprofloxacin IC50 of ∼80 μM (Figure 2A, compare left and right lanes).
Figure 2.
CL inhibits DNA gyrase and stimulates enzyme-mediated DNA cleavage. (A) Inhibition of DNA supercoiling by DNA gyrase. Relaxed pBR322 DNA was incubated with S. pneumoniae gyrase (1U) and 1.4 mM ATP in the absence or presence of CL or the quinolone ciprofloxacin (CIP) at the concentrations indicated on the figure. Reaction mixtures were loaded on a 1% agarose gel and DNA bands were visualized by staining with ethidium bromide and uv light. Lane a, supercoiled pBR322 (SC); lane b, relaxed pBR322 (R). (B) Stimulation of gyrase-mediated DNA breakage. Supercoiled pBR322 was incubated at 37°C for 1 h with gyrase (in the absence of ATP) with CL or CIP at the concentrations indicated on the figure. After addition of SDS and incubation with proteinase K, DNA products were separated and displayed by electrophoresis in 1% agarose. (Lane a) is the input supercoiled pBR322, (lane b) is pBR322 linearized with EcoRI. N, L and SC denote nicked, linear and supercoiled plasmid DNA, respectively. In (A) and (B), gel images were captured using an Alpha Innotech digital camera and DNA bands were quantitated and analysed using associated software.
For the DNA cleavage assay, supercoiled pBR322 was incubated with gyrase in the absence or presence of CL or ciprofloxacin at concentrations up to 160 μM. DNA cleavage was induced by addition of SDS and, after proteinase K treatment, the DNA was separated and displayed by agarose gel electrophoresis (Figure 2B). There was little or no enzyme-induced DNA breakage in the absence of drug (lanes 0) but addition of CL (or ciprofloxacin) caused a dose-dependent production of both nicked and linear DNA corresponding to the capture of single-stranded and double-stranded DNA breaks, respectively (Figure 2B). Digital quantitation of DNA bands showed that CL induced 25% linearization of input DNA (CC25) at 20–40 μM compared with a value of 20 μM for ciprofloxacin (Figure 2B).
ATP stimulation and sequence specificity of DNA breakage
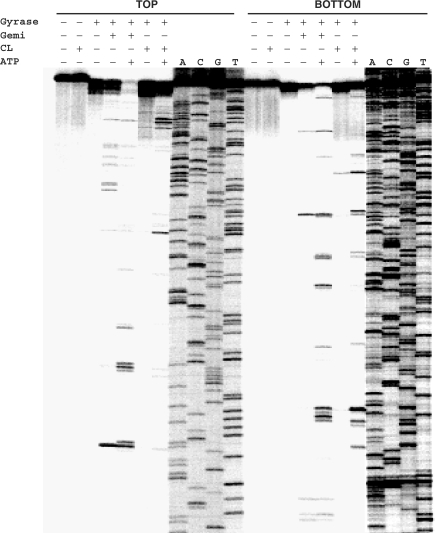
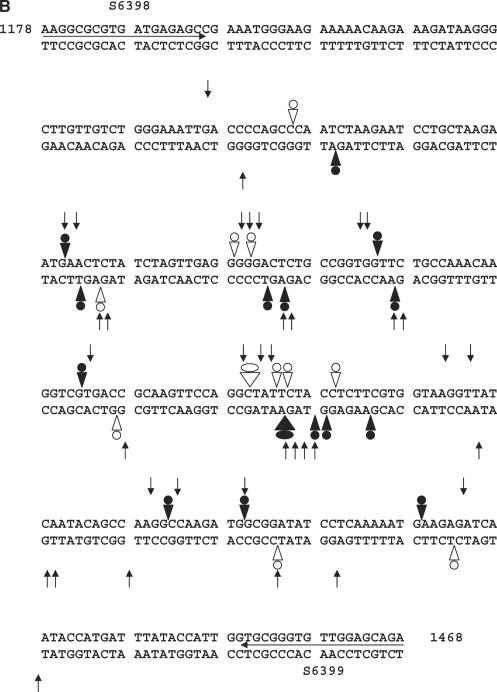
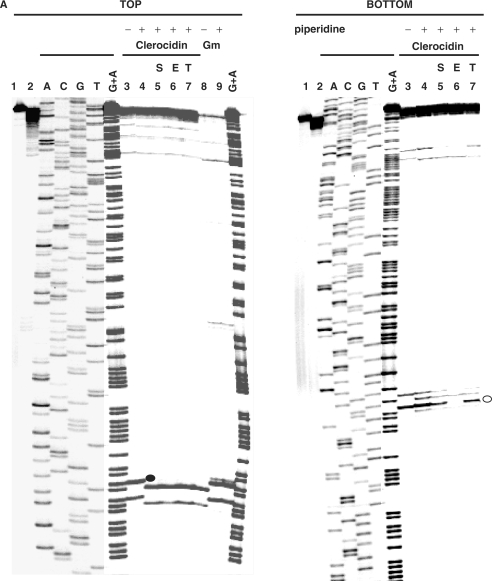
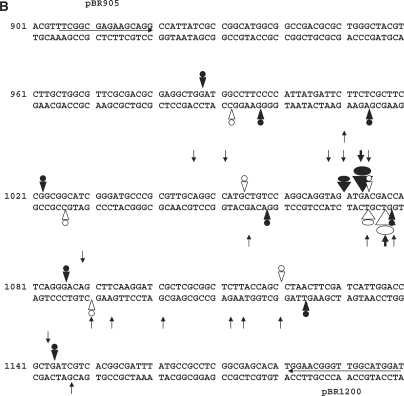
The specificity of gyrase-mediated DNA breakage promoted by CL was examined using the 5′-end 33P-labelled S fragment from the S. pneumoniae parE gene and compared to that of gemifloxacin, an anti-pneumococcal quinolone whose induction of sequence-specific cleavage has been studied in detail for pneumococcal gyrase and topo IV (31). Cleavage of TOP and BOTTOM DNA strands by both drugs was markedly enhanced by the inclusion of ATP and required the presence of gyrase: no breakage was seen with CL alone consistent with the known poor reactivity of CL toward duplex DNA (Figure 3, Introduction section). CL stimulated a unique repertoire of cuts, though some sites were also cleaved with gemifloxacin (Figure 3). Quinolones are known to stabilize DNA cleavage fragments that possess 3′ OH ends and hence co-migrate with chain termination DNA sequencing products (21,31). CL-generated fragments also co-migrated with dideoxy sequencing products (and some quinolone-stimulated fragments) (Figure 3 and later Figures 4–6) allowing their mapping at the sequence level. CL-induced gyrase scission occurred either at single sites or more frequently in pairs on complementary strands showing the characteristic 4-bp stagger expected of double-stranded DNA breakage by gyrase (31). A compilation of sites mapped in the pneumococcal S fragment or plasmid pBR322 (see below) showed a strong preference for guanine at the −1 position or the equivalent C at +5 (Table 1). Ten sites of double-stranded DNA cleavage had both −1G and +5C, i.e. guanine at both −1 positions. This is the first sequence analysis of CL-promoted cleavage by gyrase.
Figure 3.
CL-promoted DNA cleavage mediated by gyrase occurs at specific sites and is stimulated by ATP. The 290-bp S fragment (from the S. pneumoniae parE gene) 33P-labelled at the 5′ end of the TOP or BOTTOM strand was cleaved by S. pneumoniae gyrase in the absence or presence of 200 μM CL or 100 μM gemifloxacin (Gemi) and 1 mM ATP. These drug concentrations were chosen so as to achieve comparable cleavage levels of the linear DNA substrate. DNA products were separated by electrophoresis in a denaturing 6% polyacrylamide gel alongside DNA sequencing products (ACGT) obtained by the chain termination method using the same 5′-labelled oligonucleotide primers employed in PCR.
Figure 4.
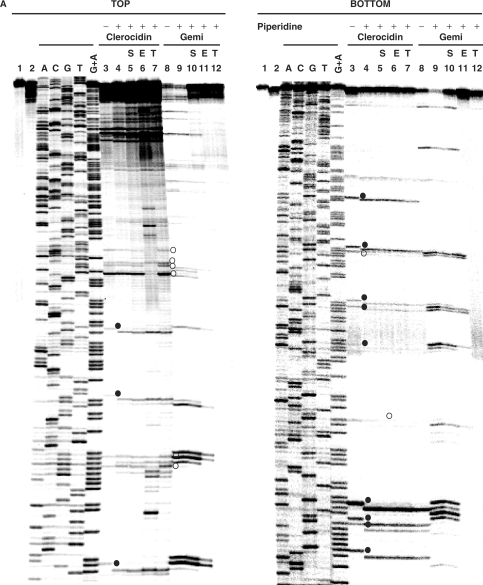
CL induces DNA modification and irreversible/reversible DNA breakage at gyrase cleavage sites. (A) Probing CL modification and cleavage reversibility. The S fragment, 33P-labelled at the 5′ end of the TOP or BOTTOM strand, was incubated with S. pneumoniae gyrase and 1 mM ATP (lanes 2–12) in the absence (lane 2) or presence of 200 μM CL (lanes 3–7) or 100 μM gemifloxacin (lanes 8–12). Samples in lanes 5 and 10, 6 and 11 and 7 and 12 were further incubated with 0.6 M NaCl for 15 min (denoted by S), with 35 mM EDTA for 15 min (E), or heating to 65°C for 15 min (T), respectively. After cleavage induction with SDS and proteinase K, DNA was precipitated with ethanol. Cleavage products in lanes 4–7 and 9–12 were further incubated with 1-M piperidine at 90°C for 30 min and then lyophilized. Reaction products were loaded and run on 6% denaturing polyacrylamide gels alongside the G + A Maxam–Gilbert chemical sequencing products, and the ACGT dideoxy sequencing products obtained using the same 5′ end-labelled oligonucleotide primers. Lane 1 is untreated substrate DNA. Lane 2 is a control for gyrase-processed DNA. Lanes 3 and 8 are cleavage products that were not treated with piperidine. DNA cleavage products that undergo shortening after hot piperidine are denoted by filled circles; those unaffected by piperidine are labelled with open circles. Gemifloxacin cleavage products were uniformly unaffected by hot piperidine (compare lanes 8 and 9) and were formed reversibly (lanes 10–12). Several new bands appearing after EDTA treatment (TOP strand, lane 6) may arise from sealing of sites proximal to the 5′ end revealing distal cleavage sites. (B) Mapping CL-gyrase cleavage sites in the S fragment. Arrowheads and arrows show cleavage sites stimulated by CL and gemifloxacin, respectively. Filled and open arrowheads denote sites of irreversible and reversible cleavage, respectively. Filled and open circles denote CL sites at which the 3′ nucleotide was released by hot piperidine or was wholly (or partially) unaffected by piperidine treatment, respectively. The large arrowheads and circles denote the most efficient cleavage site. Different to the 4-bp gyrase stagger, two sites with an apparent 2-bp overhang likely arise from closely spaced single cuts. Numbers refer to nucleotide positions in the S. pneumoniae parE gene (31) counting from the first nucleotide of the initiation codon ATG. Horizontal arrows denote the sequences of forward (S6398) and reverse oligonucleotides (S6399) used in PCR.
Figure 5.
CL-stimulated gyrase cleavage at the pBR322 ‘1073’ site. (A) DNA modification and EDTA reversal. A 234-bp pBR322 fragment (nucleotides 966–1200), labelled with 33P at the 5′ end of the TOP or BOTTOM strand, was incubated with gyrase and 1 mM ATP in the absence (lane 2) or presence of 200 μM CL (lanes 3–7) or 100 μM gemifloxacin (Gm) (lanes 8 and 9). Samples in lanes 5–7 were further incubated with NaCl (S), EDTA (E) or at 65°C (T) as described in the Figure 4 legend. After treatment with SDS and proteinase K, reaction products were precipitated with ethanol. Samples in lanes 4–7 and 9 were incubated with hot piperidine. Samples were lyophilized and run on 6% denaturing polyacrylamide gels alongside G+A chemical sequencing and ACGT chain termination sequencing products. Lanes 1 and 2 are controls for untreated DNA and gyrase processed DNA, respectively. Lanes 3 and 8 are non-piperidine treated cleavage products. The main cleavage site at 1073 produced CL-stabilized fragments indicated by a filled circle (TOP strand, piperidine-shifted and irreversible) and by an unfilled circle (BOTTOM strand, largely unshifted by piperidine and EDTA reversible). (B) Location of DNA gyrase cleavage sites in the pBR322 fragment. Symbols are used as described in the Figure 4B legend. Large arrowheads denote the ‘1073’ site; smaller arrowheads indicate weaker sites. Nucleotide sequence from ref 30.
Figure 6.
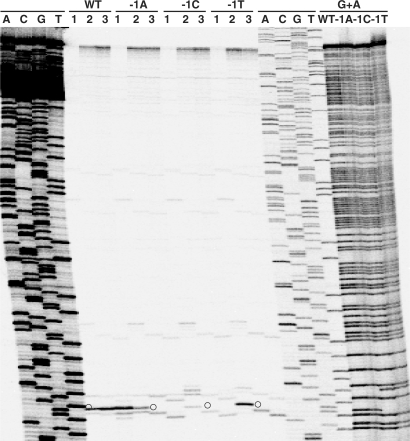
Substitution of −1 guanines at the pBR322 ‘1073 site’ with adenine, cytosine or thymine abolishes gyrase DNA cleavage stimulated by CL but not by gemifloxacin. The 295-bp pBR322 fragment 5′ 33P-labelled on the bottom strand was amplified by PCR from pBR322 plasmids carrying the wild-type site (G at both −1 positions, indicated by WT) or mutant sites bearing A, C or T at both −1 positions (denoted by −1A, −1C and −1T). The fragments were used in a DNA cleavage assay with gyrase and 1 mM ATP and either 100 μM gemifloxacin (lanes 1) or 200 μM CL (lanes 2 and 3). DNA was recovered by ethanol precipitation and run on an 6% denaturing polyacrylamide gel alongside ACGT chain termination DNA sequencing products obtained from a wild-type DNA template and G+A Maxam–Gilbert chemical sequencing products generated for each of the four substrates. Prior to electrophoresis, one set of CL cleavage products was treated with hot piperidine (lanes 3). Open circle denotes cleavage product derived from the major cleavage site.
Table 1.
Alignment of DNA cleavage sites for S. pneumoniae gyrase induced by clerocidin
| Sites involving cleavage on complementary DNA strands | ||
| −1 +5 | ||
|---|---|---|
| CCCCAGCC CAAT CTAAGAAT | Site 1 | S fragment |
| TAAGAATG AACT CTATCTAG | 2 | S fragment |
| AGTTGAGG GGGA CTCTGCCG | 3 | S fragment |
| TTGAGGGG GACT CTGCCGGT | 4 | S fragment |
| CAAGGTCG TGAC CGCAAGTT | 5 | S fragment |
| TTCCAGGC TATT CTACCTCT | 6 | S fragment |
| CAGGCTAT TCTA CCTCTTCG | 7 | S fragment |
| AGGCTATT CTAC CTCTTCGT | 8 | S fragment |
| ATTCTACC TCTT CGTGGTAA | 9 | S fragment |
| CCAAGATG GCGG ATATCCTC | 10 | S fragment |
| CAAAAATG AAGA GATCAATA | 11 | S fragment |
| GGTAGATG ACGA CCATCAGG | Site 1 | pBR322-1073 |
| GCAGGTAG ATGA CGACCATC | 2 | pBR322-1070 |
| GTAGATGA CGAC CATCAGGG | 3 | pBR322 |
| GAGGCTGG ATGG CCTTCCCC | 4 | pBR322 |
| CGCTTCCG GCGG CATCGGGA | 5 | pBR322 |
| AGGCCATG CTGT CCAGGCAG | 6 | pBR322 |
| CATCAGGG ACAG CTTCAAGG | 7 | pBR322 |
| TTACCAGC CTAA CTTCGATC | 8 | pBR322 |
| Sites with cleavage on one DNA strand | ||
| AGATAGAG TTCA TTCTTAGC | Site 12 | S fragment |
| GCCGGTGG TTCT GCCAAACA | 13 | S fragment |
| TTTGGCAG AACC ACCGGCAG | 14 | S fragment |
| AGCCAAGG CCAA GATGGCGG | 15 | S fragment |
| GACCGCTG ATCG TCACGGCG | 9 | pBR322 |
| CGGAAGCG AGAA GAATCATA | 10 | pBR322 |
| ATAATGGG GAAG GCCATCCA | 11 | pBR322 |
DNA sequences surrounding the breakage point are shown for one strand of each double-stranded breakage site or for sites of single-stranded breakage promoted by clerocidin in the parE S fragment (31) and in pBR322 (905–1200 nt region). The central four nucleotides (+1 to +4) correspond to those in the 4-bp overhang produced by DNA breakage by gyrase. Preferred guanines at –1 and cytosines at +5 (indicating guanine at –1 on the complementary strand) are shown in boldface.
Selective DNA modification and cleavage irreversibility at the DNA gate
In the absence of topoisomerases, it is known that the epoxide moiety of CL reacts at N-7 of unpaired guanines (6). Incubation over extended times (24–48 h) leads to loss of the N-7 alkylated base and strand scission at the abasic site, a process used in the Maxam–Gilbert DNA sequencing protocol and facilitated by hot piperidine (32). To determine whether CL modifies bases exposed in the gyrase cleavage complex, we compared the mobility of cleavage products from the TOP and BOTTOM strands of the S fragment (Figure 4A) before (lanes 3) and after hot piperidine (lanes 4). A number of fragments (denoted by filled circles on Figure 4A) suffered a 1-nt shift in mobility after hot piperidine consistent with the loss of a modified nucleotide at their extreme 3′ end, i.e. the nucleotide at −1. After hot piperidine, these fragments co-migrated with the Maxam–Gilbert ladder which have 3′ phosphate termini, Shifted fragments invariably arose from scission at −1G and their production could not be reversed with salt, EDTA or high temperature (Figure 4A, lanes 5–7). However, the mobility of other cleavage fragments (denoted by open circles in Figure 4A) was not affected by hot piperidine (lanes 3 and 4). These fragments arose from cleavage at sites with A, C, T or even G at −1, and invariably their production could be at least partially reversed with EDTA (lanes 6), though not with salt or heat (lanes 5 and 7). Control experiments showed that gemifloxacin-promoted cleavage products were universally unaffected by piperidine consistent with the known non-covalent interaction of quinolones with the cleavage complex (lanes 8 and 9). Moreover, cleavage by gemifloxacin (lane 8), a high-affinity quinolone (26), was partially reversed with high salt (lane 10) and fully reversed with EDTA and heat (lanes 11 and 12), consistent with sealing of unmodified 3′ OH breaks. Thus, in contrast to quinolones, it appears that CL stimulates both irreversible and reversible gyrase cleavage of the S fragment with evidence of covalent modification at some guanine sites.
Reversible and irreversible DNA scission by CL with covalent modification at some sites was also seen using a different DNA substrate, namely a PCR product amplified from plasmid pBR322 (nucleotides 966–1200) and 5′-33P end-labelled in the TOP or BOTTOM strands (Figure 5A).
Access to data from both strands of the S fragment and of the pBR322 fragment, enabled us to examine the link between guanine modification and irreversibity/reversibility at both −1 positions for a number of double-stranded gyrase breakage sites. The results are summarized for each fragment in Figures 4B and 5B, which bring out a novel and previously unreported feature. In fact, all the sites show evidence of differential CL attack, namely complete guanine modification and irreversible cleavage on one strand (filled arrowhead and circle) is invariably accompanied by reversible cleavage at a wholly (or partially) unmodified nucleotide on the complementary strand (open arrowhead and circle). When the second −1 nt was A, C or T, there was no detectable piperidine-sensitive modification, and DNA scission was wholly reversible at this position. However, sites with two −1G nucleotides (e.g. the major pBR322 site in Figure 5) showed complete guanine modification on one strand accompanied on the second strand by partial guanine modification (0–60% shift to smaller fragment with hot piperidine depending on the site) and at least partial reversibility. Evidently, CL reacts differentially at the two −1 positions of the gyrase–DNA gate, even when both nucleotides are guanines.
Mutagenesis establishes CL preference for −1G
The role of the −1 nt in CL action was further investigated using directed mutagenesis of the major pBR322 gyrase site at 1073, a site bearing G at both −1 positions (Figure 5B) and which was cleaved some 10× more efficiently than other sites, e.g. in the S fragment. Given that sites exhibiting a single −1G can be cleaved (Table 1, Figures 4B and 5B), we constructed mutants of the pBR322 site in which both −1 positions (G1073 and complementary strand G1078) were altered to either A, C or T nucleotides. The sites, contained in end-labelled 295-bp PCR products, were used as cleavage substrates for gyrase in the presence of gemifloxacin or CL, and the products were displayed on a denaturing polyacrylamide gel (Figure 6). Gemifloxacin promoted efficient double-stranded DNA cleavage at the wild-type (WT) site and its −1A, −1C and −1T mutants (lanes 1) producing the expected fragment (circles in Figure 6), thereby showing that the mutant sites are still recognized by gyrase. CL induced cleavage at the WT site to produce a DNA fragment that co-migrated with the gemifloxacin-stabilized fragment and with the appropriate dideoxy DNA sequencing product (WT, lanes 1 and 2). However, this fragment was absent for CL cleavage of the mutant −1A, −1C and −1T substrates (lanes 2). Instead, enhanced cleavage was seen at nearby sites i.e. 3′ of G1079 (smaller fragment) and G1075 (larger fragment) (Figure 6, −1A, −1C and −1T, lanes 2) and both fragments exhibited a one-base shortening on piperidine treatment (lanes 3). These mutagenesis data provide the first direct evidence that CL-induced gyrase cleavage of DNA is prevented when G is absent from both −1 positions.
Protein requirements for poisoning: role of GyrA G79 and S81
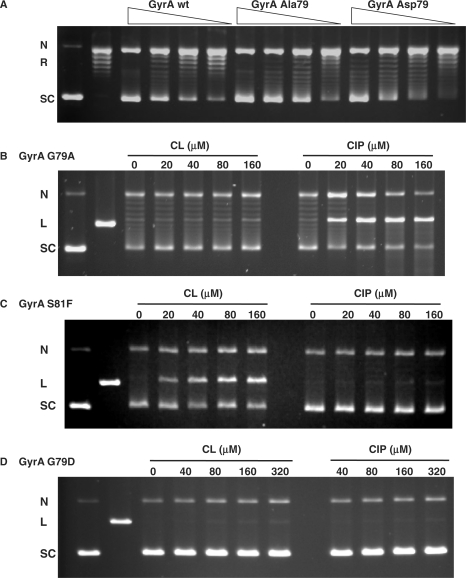
In contrast to the DNA substrate, very little is known about the enzyme interactions that govern CL activity. The identification of a novel gyrA mutation (G79A) conferring CL-resistance in S. pneumoniae is therefore of considerable interest. The mutation affects a conserved glycine residue that lies in the CAP-like helix–turn–helix motif of GyrA thought to bind DNA (33,34). A different mutation of the equivalent glycine residue (to aspartate or cysteine) has been associated (albeit rarely) with clinical resistance to quinolones in E. coli (35,36). Moreover, in S. pneumoniae, adjacent mutations in GyrA of S81 to F or Y commonly cause resistance to quinolones (25,29,37) implying that a localized region of GyrA is implicated in binding different gyrase poisons. To understand better how CL interacts to poison gyrase, we expressed the S. pneumoniae G79A, S81F and G79D mutant GyrA subunits in E. coli and purified the proteins to homogeneity. In a supercoiling assay, the three mutant proteins reconstituted a gyrase activity comparable to that of the wild-type enzyme (Figure 7A, see ref. 29 for GyrA S81F data).
Figure 7.
Mutations at G79 and S81 in GyrA can discriminate gyrase poisoning by CL from that of quinolones. (A) Recombinant GyrA A79 and GyrA D79 proteins reconstitute a DNA supercoiling activity comparable to that of wild-type gyrase (wt). Relaxed pBR322 DNA was incubated with increasing amounts of wild-type or mutant S. pneumoniae GyrA subunits in the presence of 1 mM ATP and a fixed amount of wild type GyrB. DNA products were separated by electrophoresis in 1% agarose. (B, C and D) Differential poisoning of mutant gyrases by CL and CIP. Supercoiled pBR322 DNA was incubated with mutant gyrases (GyrA G79A, S81F or A79D) in the absence of ATP in a DNA cleavage assay containing CL or ciprofloxacin (CIP) at the concentrations indicated on the figure. In the four panels, N, R, L and SC denote nicked, relaxed, linear (EcoRI-cut) and supercoiled pBR322 DNA, respectively.
Unlike the equipotent DNA cleavage promoted by CL and ciprofloxacin with WT gyrase (Figure 2B), enzyme reconstituted with GyrA G79A induced cleavage of supercoiled pBR322 strongly with ciprofloxacin but only weakly with CL (Figure 7B). These biochemical data agree with our genetic studies showing that the GyrA G79A alteration accounts for the CL-resistant but ciprofloxacin-sensitive phenotype of the S. pneumoniae CL12 mutant. Conversely, gyrase reconstituted with GyrA S81F promoted DNA cleavage with CL but not with ciprofloxacin (Figure 7C). This result is in accord with data showing that quinolone-resistance mutations in gyrase or topo IV conferred no bacterial cross resistance to CL (GyrA S81F, ParC S79F) or mild (2- to 4-fold) hypersensitivity (GyrA S81Y, ParC S79Y) (Pan,X.S. and Fisher,L.M., unpublished data). Finally, even though very active in DNA supercoiling (Figure 7A), the GyrA G79D complex did not induce DNA cleavage with either CL or ciprofloxacin even at 320 μM (Figure 7D). The responses of the mutant gyrases in ATP-independent plasmid cleavage were reproduced in ATP-dependent cleavage of end-labelled DNA (Pan,X.S. and Fisher,L.M., unpublished data). It appears that the sidechains of residues 79 and 81 in GyrA are a critical determinant of whether gyrase can be poisoned by CL and quinolones.
DISCUSSION
We have shown that CL targets gyrase in the Gram-positive bacterium S. pneumoniae and induces single- and double-stranded DNA breakage by the enzyme in vitro. Using a variety of biochemical approaches, we demonstrate that gyrase-mediated DNA scission induced by CL has unusual characteristics including a preference for guanine bases, differential nucleotide modification, elements of irreversibility and reversibility and differences in protein requirements that distinguish it from enzyme poisoning by antibacterial quinolones. It appears that CL has a unique antibiotic mechanism that involves selective chemical inactivation of gyrase-DNA complexes.
Previously, we showed that CL acts on topo IV to cause irreversible DNA cleavage at −1G leading to the reasonable conclusion that topo IV is the intracellular drug target (22). However, the genetic data presented here demonstrates that CL primarily targets gyrase in vivo. Presumably, CL also reacts with topo IV in vivo but the enzyme is a secondary target. In the earlier work, we did not have access to genetic evidence and did not know that gyrase is also potently (and irreversibly) inhibited by the drug with IC50 and CC25 values (for DNA supercoiling inhibition and DNA cleavage) of 40 and 20–40 μM (Figure 2). Ideally, both genetic and biochemical approaches should be applied in establishing whether gyrase or topo IV is a cellular target.
Biochemical analysis showed that CL-induced DNA cleavage by gyrase occurred at specific sites that partially overlapped the cleavage repertoire seen for the quinolone gemifloxacin (Figures 3–5). Scrutiny of many CL sites revealed a very strong preference for G at −1 on one, or both, strands (Table 1). Mutagenesis of the strong pBR322 1073 site showed that substitution of both −1G positions with A, C or T abolished CL-promoted DNA breakage (Figure 6). We also conducted parallel mutagenesis on the well-characterized E site for S. pneumoniae topo IV (31) and examined double-stranded cleavage of 256-bp E-site PCR products on native agarose gels. We found that replacement of the two WT −1T nucleotides with G induced strong double-stranded CL-topo IV cleavage, which was absent for −1A, −1C and −1T (Pan, X.S. and Fisher,L.M., unpublished data). Thus, CL requires at least one −1 guanine to induce DNA breakage by gyrase and topo IV.
Perhaps the most surprising and novel finding of the present study was that CL reacts differently with the two halves of the gyrase-DNA gate (Figures 4 and 5). Previous studies have reported irreversible and reversible CL-mediated single-strand cleavage by mammalian topo II at G and C, respectively, and have alluded to DNA cleavage products with abnormal electrophoretic mobility (4). However, the link between DNA modification and (ir)reversibility has not been explored. Our analysis of 19 gyrase sites revealed the same consistent feature of quantitative guanine modification and irreversible cleavage on one strand, accompanied on the complementary strand by wholly reversible cleavage at an apparently unmodified −1 A, C or T nucleotide, and variable but site-reproducible modification-reversibility at −1 G (Figures 4–6). These results are the first to show differential reaction of CL at the two −1 nt of a topoisomerase-DNA complex.
We have reported that CL-induced DNA damage by S. pneumoniae topo IV could not be reversed by heat or salt (i.e. similar to gyrase, Figures 4 and 5) nor by EDTA (unlike partial EDTA reversal seen for gyrase) implying that CL induces exclusively irreversible DNA breaks with topo IV (22). In light of the unusual pattern of CL action at gyrase sites, we have re-examined and repeated the earlier EDTA reversal work. In those experiments, supercoiled pBR322 DNA was reacted with 50 μM CL and topo IV producing nicked circles (∼30% of total DNA), full-length linear DNA (one double-strand break per plasmid) (40%) and the remainder a smear of smaller linear DNA species (arising from two or more double-stranded breaks per plasmid). EDTA reversal caused no detectable recovery of supercoiled DNA that would arise from sealing of nicked circles or full sealing of a double-stranded DNA break (hence our conclusion of irreversible DNA damage). However, EDTA did promote an increase in nicked DNA that on closer scrutiny arises from depletion of the smear of linear DNA species. This latter aspect was missed originally and can only be explained by irreversible and reversible scission at each topo IV site such that EDTA converts double-stranded DNA breaks into DNA nicks allowing recovery of linear and multiply nicked circular DNA, but not the supercoiled DNA substrate. (The same behaviour was also seen for EDTA reversal of CL-induced cleavage of pBR322 by gyrase.) Unfortunately, the assay with plasmid DNA does not allow examination of particular topo IV sites. However, by using end-labelled S- and pBR322-fragments as substrates with product analysis on sequencing gels, we have found that CL-induced topo IV cleavage does generate piperidine-shifted and -unshifted DNA fragments, which would correspond to irreversible and reversible sites (Pan,X.S. and Fisher,L.M., unpublished data). Evidently, differential reactivity of the two halves of the topoisomerase-DNA gate also occurs with topo IV and thus does not require DNA wrapping in the enzyme complex.
Our results showing covalent modification of gyrase (and topo IV) cleavage complexes support and extend the earlier model of CL action against topo II (6). We suggest that CL first binds preferentially and reversibly to gyrase–DNA complexes in which at least one −1 position is occupied by guanine. After transient gate opening by gyrase to form the cleavage complex, the guanine is potentiated for attack (10). CL is not able to alkylate the canonical B form of double-stranded DNA for steric reasons (6) and thus the DNA must be significantly distorted or cleaved to allow CL attack from the major groove. N-7 modification of the −1G is then sufficient to block religation perhaps by mis-positioning the 3′OH end. Resealing of the complementary DNA strand might also be impeded by this guanine modification (‘one-drug’ mechanism) or might arise from non-covalent interactions with a second bound CL molecule (‘two-drug’ mechanism).
It seems likely that the alkylated guanine is stably retained in the gyrase complex as suggested by the exact co-migration of CL-stimulated DNA fragments with their quinolone- or dideoxy-generated counterparts (Figures 3–6). Alternatively, the modified guanine may have been lost (prior to or post DNA-gate opening) (as seen for CL attack on free 2′ deoxyguanosine) (7) producing DNA fragments with an abasic site at the 3′ end, and which are reported to have retarded electrophoretic mobility (38). It is known that an engineered abasic site at the −1 position (replacing G) induces E. coli topo IV-mediated cleavage and that salt, but not EDTA, can reverse abasic site-induced topo IV cleavage complexes (39). However, arguing against abasic sites, we found that neither salt, EDTA, nor heat was able to reverse gyrase cleavage at the CL-modified −1 position (Figures 4–6).
Several observations bear on the model. First, simple epoxides do not induce DNA cleavage by gyrase (Pan,X.S. and Fisher,L.M., unpublished data) suggesting that capture of gyrase–DNA complexes by CL involves specific binding interactions, e.g. with the diterpenoid ring as shown for topo IV (22). Second, CL was inactive against a mutant topo IV unable to form a cleavage complex implying that gate opening is required for drug action (22). Indeed, increased access to the enzyme–DNA gate could underlie ATP-independent cleavage of supercoiled plasmid (Figure 2) but ATP-dependent breakage of linear DNA (Figure 3) involving either enhanced enzyme–DNA affinity or ATP-driven cycling of the enzyme complex. Third, like CL, quinolones such as gemifloxacin also exhibit a preference (among several determinants) for −1G (31,40) explaining the partial overlap in gyrase cleavage sites for the two drug classes. Fourth, CL modification of one −1G and (for some sites) low-level modification of the other −1G (Figures 4–6) could imply a two-drug mechanism. Attack at one guanine to generate a bulky adduct may inhibit reaction with the other, or could reflect sequence-dependent differences in guanine reactivities (38).
The novel gyrA (G79A) alteration blocks CL-stimulated DNA cleavage (Figure 7B) and is the first mutation to be identified with CL resistance in vivo, though earlier work had indirectly linked gyrA with CL resistance in E. coli (2). G79 is highly conserved throughout type II topoisomerases and is located between the α3 and α4 helices of a CAP-like motif that forms the GyrA DNA breakage-reunion domain (31). We suggest that G79 of GyrA, the −1 guanine at the DNA gate, and the reactive epoxide moiety of CL lie together in the ternary complex, and that mutation of G79 blocks CL binding.
Interestingly, the phenotypes of the various mutants involving GyrA G79/S81 and ParC S79 (Figure 7 and Results section) may explain earlier work reporting cross-resistance of CL and quinolones in E. coli yet CL activity against quinolone-resistant S. aureus (2). Mutation of the conserved E. coli GyrA G81 (equivalent to S. pneumoniae GyrA G79) to aspartate (GGT to GAT) could be obtained in a single-step to produce the observed cross-resistance (Figure 7D). However, the same S. pneumoniae GyrA alteration (G79D, GGG to GAT or GAC) requires 2 nt changes and would not be recovered in our single-step mutant selection. Similarly, S. pneumoniae GyrA S81 (and ParC S79) are conserved in S. aureus (as GyrA S84 and GrlA S80) and their mutation is often implicated in quinolone resistance of both species (25,29,37,41). That the common quinolone-resistance mutations at these residues do not reduce CL activity (see Results section, Figure 7 and ref. 22) explains the lack of cross-resistance in S. aureus. An attractive rationale binding together these various findings is that different gyrase poisons occupy different, perhaps overlapping sites at the DNA–enzyme interface that are impacted differently by neighbouring side-chain mutations.
In summary, we have shown that CL is an unusual inhibitor of bacterial type II topoisomerases that exerts its potent anti-Gram-positive antibacterial activity by differentially modifying the two halves of the gyrase–DNA gate. That CL reacts with similar −1G specificity and selectivity against gyrase and topo IV suggests that both enzymes bind DNA in a distorted conformation. Precise understanding of these enzyme–DNA interactions awaits successful X-ray structure studies.
FUNDING
Biotechnology and Biological Sciences Research Council, UK (BBD01882X1 to L.M.F.). Funding for open access charge: St George's, University of London.
Conflict of interest statement. None declared.
REFERENCES
- 1.Andersen N, Lork H, Rassmussen P. The relative and absolute configuration of clerocidin and its co-metabolites. Tetrahedron Lett. 1984;25:469–472. [Google Scholar]
- 2.McCullough JE, Muller MT, Howells AJ, Maxwell A, O'Sullivan J, Summerill RS, Parker WL, Wells JS, Bonner DP, Fernandes PB. Clerocidin, a terpenoid antibiotic, inhibits bacterial DNA gyrase. J. Antibiot. 1993;46:526–530. doi: 10.7164/antibiotics.46.526. [DOI] [PubMed] [Google Scholar]
- 3.Kawada S, Yamashita Y, Fujii N, Nakano H. Induction of a heat-stable topoisomerase II-DNA cleavable complex by non-intercalative terpenoids, terpentecin and clerocidin. Cancer Res. 1991;51:2922–2925. [PubMed] [Google Scholar]
- 4.Binaschi M, Zagotto G, Palumbo M, Zunino F, Farinosi R, Capranico G. Irreversible and reversible topoisomerase II cleavage stimulated by clerocidin: sequence specificity and structural drug determinants. Cancer Res. 1997;57:1710–1716. [PubMed] [Google Scholar]
- 5.Borgnetto ME, Tinelli S, Carminati L, Capranico G. Genomic sites of topoisomerase II activity determined by comparing DNA breakage enhanced by three distinct DNA poisons. J. Mol. Biol. 1999;285:545–554. doi: 10.1006/jmbi.1998.2330. [DOI] [PubMed] [Google Scholar]
- 6.Gatto B, Richter S, Moro S, Capranico G, Palumbo M. The topoisomerase II poison alkylates non-paired guanines of DNA: implications for irreversible stimulation of DNA cleavage. Nucleic Acids Res. 2001;29:4224–4230. doi: 10.1093/nar/29.20.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter S, Gatto B, Fabris D, Takao K, Kobayashi S, Palumbo M. Clerocidin alkylates DNA through its epoxide function: evidence for a fine-tuned mechanism of action. Nucleic Acid Res. 2003;31:5149–5156. doi: 10.1093/nar/gkg696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter S, Menegazzo I, Fabris D, Palumbo M. Concerted bis-alkylating activity of clerocidin towards unpaired cytosine residues in DNA. Nucleic Acids Res. 2004;32:5658–5667. doi: 10.1093/nar/gkh898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawada SZ, Yamashita Y, Uosaki Y, Gomi K, Iwasaki T, Takiguchi T, Nakano H. UCT4B, a new antitumor antibiotic with topoisomerase II-mediated DNA cleavage activity, from Streptomyces sp. J. Antiot. 1992;45:1182–1184. doi: 10.7164/antibiotics.45.1182. [DOI] [PubMed] [Google Scholar]
- 10.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 11.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 13.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 14.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Zechiedrich EL, Cozzarelli NR. Role of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 16.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 17.Mizuuchi K, Fisher LM, O’Dea MH, Gellert M. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc. Natl Acad. Sci. USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng H, Marians KJ. The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 19.Fisher LM, Mizuuchi K, O’Dea MH, Ohmori H, Gellert M. Site-specific interactions of DNA gyrase with DNA. Proc. Natl Acad. Sci. USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 21.Morrison A, Cozzarelli NR. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad. Sci. USA. 1981;78:1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter SN, Leo E, Giaretta G, Gatto B, Fisher LM, Palumbo M. Clerocidin interacts with the cleavage complex of Streptococcus pneumoniae topoisomerase IV to induce selective irreversible DNA damage. Nucleic Acids Res. 2006;34:1982–1991. doi: 10.1093/nar/gkl127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins PG, Fluit PG, Schmitz FJ. Fluoroquinolones:structure and target sites. Curr. Drug Targets. 2003;4:181–190. doi: 10.2174/1389450033346920. [DOI] [PubMed] [Google Scholar]
- 24.Pan XS, Fisher LM. Cloning and characterisation of the parC and parE genes of Streptococcus pneumoniae encoding topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan XS, Fisher LM. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yague G, Morris JE, Pan XS, Gould KA, Fisher LM. Cleavable–complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other quinolones. Antimicrob. Agents Chemother. 2002;46:413–419. doi: 10.1128/AAC.46.2.413-419.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan XS, Fisher LM. Streptococcus pneumoniae DNA gyrase and topoisomerase IV:overexpression, purification, and differential inhibition by quinolones. Antimicrob. Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pozzi G, Masala L, Ianelli F, Manganelli R, Havarstain LS, Piccoli L, Simon D, Morrison DA. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan XS, Yague G, Fisher LM. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 2001;45:3140–3147. doi: 10.1128/AAC.45.11.3140-3147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peden KW. Revised sequence of the tetracycline-resistance gene of pBR322. Gene. 1983;22:277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- 31.Leo E, Gould KA, Pan XS, Capranico G, Sanderson MR, Palumbo M, Fisher LM. Novel symmetric and asymmetric DNA scission determinants for Streptococcus pneumoniae topoisomerase IV and gyrase are clustered at the DNA breakage site. J. Biol. Chem. 2005;280:14252–14263. doi: 10.1074/jbc.M500156200. [DOI] [PubMed] [Google Scholar]
- 32.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth. Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 33.Morais-Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 34.Laponogov I, Veselkov DA, Sohi MK, Pan XS, Achari A, Yang C, Ferrara JD, Fisher LM, Sanderson MR. Breakage-reunion domain of Streptococcus pneumoniae topoisomerase IV: crystal structure of a gram-positive quinolone target. PLoS ONE. 2007;2:e301. doi: 10.1371/journal.pone.0000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cambau E, Bordon F, Collatz E, Gutmann L. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not nalidixic acid. Antimicrob. Agents Chemother. 1993;37:1247–1252. doi: 10.1128/aac.37.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone-resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan XS, Ambler J, Mehtar S, Fisher LM. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun D, Clement JJ, Hurley LH. Structure of the altromycin B (N-7-guanine)-DNA adduct. A proposed prototypic DNA adduct structure for the pluramycin antitumor antibiotics. Biochemistry. 1993;32:8068–8074. doi: 10.1021/bi00083a003. [DOI] [PubMed] [Google Scholar]
- 39.Shea ME, Hiasa H. Replicative helicases can translocate through abasic site-induced covalent topoisomerase IV-DNA complexes. Nucleic Acids Res. 2001;29:614–621. doi: 10.1093/nar/29.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter S, Giaretta G, Comuzzi V, Leo E, Mitchenall LA, Fisher LM, Maxwell A, Palumbo M. Hot-spot consensus of luoroquinolone-mediated DNA cleavage by Gram-negative and Gram-positive type II DNA topoisomerases. Nucleic Acid Res. 2007;35:6075–6085. doi: 10.1093/nar/gkm653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrero L, Cameron B, Crouzet J. Analysis of stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]