Abstract
State transitions, or the redistribution of light-harvesting complex II (LHCII) proteins between photosystem I (PSI) and photosystem II (PSII), balance the light-harvesting capacity of the two photosystems to optimize the efficiency of photosynthesis. Studies on the migration of LHCII proteins have focused primarily on their reassociation with PSI, but the molecular details on their dissociation from PSII have not been clear. Here, we compare the polypeptide composition, supramolecular organization, and phosphorylation of PSII complexes under PSI- and PSII-favoring conditions (State 1 and State 2, respectively). Three PSII fractions, a PSII core complex, a PSII supercomplex, and a multimer of PSII supercomplex or PSII megacomplex, were obtained from a transformant of the green alga Chlamydomonas reinhardtii carrying a His-tagged CP47. Gel filtration and single particles on electron micrographs showed that the megacomplex was predominant in State 1, whereas the core complex was predominant in State 2, indicating that LHCIIs are dissociated from PSII upon state transition. Moreover, in State 2, strongly phosphorylated LHCII type I was found in the supercomplex but not in the megacomplex. Phosphorylated minor LHCIIs (CP26 and CP29) were found only in the unbound form. The PSII subunits were most phosphorylated in the core complex. Based on these observations, we propose a model for PSII remodeling during state transitions, which involves division of the megacomplex into supercomplexes, triggered by phosphorylation of LHCII type I, followed by LHCII undocking from the supercomplex, triggered by phosphorylation of minor LHCIIs and PSII core subunits.
INTRODUCTION
Photosystem II (PSII) is a large multisubunit pigment protein complex that uses light energy to oxidize water and reduce plastoquinone. In PSII, light energy is captured by the peripheral antenna and is transferred to the core complex, where it is trapped. In green plants, the peripheral antennas are composed of major trimeric and minor monomeric light-harvesting complex II (LHCII) proteins (Dekker and Boekema, 2005). In the vascular plant Arabidopsis thaliana, there are three major trimeric LHCII proteins. Types I and II are encoded by five and four duplicated genes (Lhcb1.1-1.5 and Lhcb2.1-2.4), respectively, while type III is encoded by a single gene (Lhcb3.1; Jansson, 1999). In the green alga Chlamydomonas reinhardtii, there are four major LHCII proteins (types I to IV) encoded by five, one, two, and one genes (LhcbM3, -4, -6, -8, and -9; LhcbM5; LhcbM2 and -7; and LhcbM1), respectively (Minagawa and Takahashi, 2004). Three minor monomeric LHCII polypeptides, CP29, CP26, and CP24, are encoded by the genes Lhcb4, -5, and -6, respectively, in Arabidopsis (Jansson, 1999), whereas C. reinhardtii only contains the first two (Teramoto et al., 2001). Single-particle image analysis of electron micrographs revealed that these peripheral antenna proteins are bound to both sides of the PSII core, each of which consists of one LHCII trimer and two LHCII monomers in spinach (Spinacia oleracea) (Boekema et al., 1995) and C. reinhardtii (Nield et al., 2000). This protein complex organization is called the C2S2 supercomplex, where “C” and “S” refer to the PSII core complex and strongly bound LHCII trimer, respectively (Dekker and Boekema, 2005). In spinach, another supercomplex, C2S2M2 supercomplex, has been reported, where two moderately bound LHC II trimers along with CP24 are associated with the C2S2 supercomplex (Boekema et al., 1998).
Photosystem I (PSI) has a broad absorption peak in the far-red region as well as peaks in the blue and red regions, whereas PSII has peaks in the blue and red but not in the far-red region. Thus, an imbalance of energy distribution between the two photosystems tends to occur in natural environments, where light quality and quantity fluctuate with time (Bellafiore et al., 2005; Tikkanen et al., 2006). Under such conditions, state transitions occur to balance the light-harvesting capacity of the two photosystems (Bonaventura and Myers, 1969; Murata, 1969). When the plastoquinone pool gets reduced (Allen et al., 1981), a protein kinase is activated through the cytochrome bf complex (Wollman and Lemaire, 1988; Vener et al., 1997). This kinase phosphorylates the LHCII that is bound to PSII in the appressed region of the thylakoids (Bennett, 1977). The phosphorylation of LHCII then leads to the lateral migration of LHCII to PSI in the unappressed region (Andersson et al., 1982), where it acts as the peripheral antenna for PSI (State 2). Oxidation of the plastoquinone pool induces the opposite effect, regenerating State 1 (Bennett, 1980).
The association of LHCII proteins with PSI has been observed indirectly as changes in the quantum yield of PSI and PSII during state transitions (Samson and Bruce, 1995). Phosphorylated LHCII polypeptides were detected in the unappressed region of thylakoid membranes where PSI is preferentially located (Bassi et al., 1988). However, only recently have several lines of direct biochemical evidence shown the binding of LHCII polyepeptides to PSI (Pesaresi et al., 2002; Zhang and Scheller, 2004; Takahashi et al., 2006). Such an association was first observed in an Arabidopsis psae1-1 mutant where a fraction of LHCII was associated with PSI when the mutant plants were exposed to low-light (State 2 favoring) conditions, giving rise to a high molecular mass protein-pigment complex (Pesaresi et al., 2002). This large complex, however, seemed to be an aggregated product since the mutant did not show state transitions, likely due to a low level of PsaH. The next attempt was through cross-linking of the mobile LHCII proteins with the PSI-LHCI supercomplex in Arabidopsis (Zhang and Scheller, 2004). More major LHCII proteins, including Lhcb1 and -2, were cross-linked to PsaH, I, and L subunits in State 2 than in State 1. A further report was provided from a study on C. reinhardtii, where the PSI-LHCI/II supercomplex isolated from State 2 cells contained three LHCII polypeptides, CP26, CP29, and LhcbM5, suggesting a pivotal role for the minor monomeric LHCII in state transitions (Takahashi et al., 2006). Mobile LHCII proteins were also observed in single particle images of the PSI-LHCI/II supercomplexes. Boekema and colleagues (Kouřil et al., 2005) reported a large density along the side of PsaH/L/A/K in electron micrographs of Arabidopisis PSI, which they assigned to the LHCII trimer, and Barber and colleagues located a smaller density near PsaH in C. reinhardtii, which they assigned to CP29 (Kargul et al., 2005).
Although much has been learned about the molecular details of state transitions on the PSI side, those on the PSII side remain elusive. Which LHCII polypeptides are dissociated from PSII? What is the dissociation order? Does the dissociation cause any effects on PSII macroorganization? Notably, the roles of phosphorylated LHCII have been obscure. It was originally proposed that the phosphorylation of LHCII induces conformational changes, leading to its dissociation from PSII (Nilsson et al., 1997), but an Arabidopsis mutant lacking the H subunit of PSI shows that phosphorylated LHCIIs remain attached to PSII (Lunde et al., 2000). It was thus proposed that phosphorylation of LHCII increases its affinity for PSI over PSII and results in its migration from PSII to PSI (Allen and Forsberg, 2001). While many LHCII polypeptides, including minor LHCII, are phosphorylated during the transition from State 1 to 2 (Turkina et al., 2006), whether all of these phosphorylations simply exert the same effect, namely, increasing affinity for PSI, or each of those plays a unique role in the progress of state transitions has remained largely unknown.
In this study, we introduced two new approaches to provide insights on the fate of LHCII polypeptides associated with PSII during the course of state transitions. One uses the detergent tridecyl-β-d-maltoside (TM), which has a one-unit-longer aliphatic tail than the more commonly used mild detergent dodecyl-β-d-maltoside (β-DM). We found this detergent more suitable for solubilizing larger membrane protein complexes in C. reinhardtii. The other approach uses nickel affinity chromatography, which has been successfully employed for the discrete and rapid purification of multisubunit membrane protein complexes, including the PSII core complex from C. reinhardtii (Sugiura et al., 1998). We used a C. reinhardtii mutant carrying a His-tagged CP47 (Suzuki et al., 2003) to isolate the various PSII complexes retaining peripheral antennas under both State 1 and 2 conditions. Using these approaches, we characterized the polypeptide composition, supramolecular organization, and phosphorylation of the PSII complexes both in State 1 and 2. The results indicate that with the undocking of LHCIIs, the PSII-LHCII supercomplex and its multimer (PSII-LHCII megacomplex) were transformed into PSII core complexes, leading to the transition from State 1 to 2. The undocking process was accompanied by a multistep phosphorylation of LHCIIs and PSII core subunits. Based on the obtained results, we present a new model of the PSII complex remodeling during state transitions, which involves divisions of the megacomplex into supercomplexes and LHCII undocking from the supercomplex, triggered by the phosphorylation of LHCII type I, minor LHCIIs, and PSII core subunits.
RESULTS
Isolation of PSII Complexes in States 1 and 2
Since relatively few attempts have been reported for purifying the Chlamydomonas PSII-LHCII supercomplex, we sought to establish a streamlined protocol to obtain discrete and intact PSII complexes retaining peripheral antennas. Thylakoids prepared from a genetically modified C. reinhardtii strain, PsbB-His, in which a 6x His-tag was fused to the C terminus of CP47 (Suzuki et al., 2003), were subjected to nickel affinity chromatography. A highly active PSII core complex with no attached LHCIIs was previously purified from this strain using β-DM as a detergent (Suzuki et al., 2003). To solubilize PSII particles that retain peripheral LHCIIs, we first tested a β-DM epimer n-dodecyl-α-d-maltoside, which has been reported to partially solubilize spinach thylakoids and allow the purification of PSII-LHCII supercomplexes (Boekema et al., 1998). However, n-dodecyl-α-d-maltoside yielded similar results to β-DM in C. reinhardtii (data not shown). We then tested another mild detergent, TM, which has the same head group as β-DM but with one unit longer aliphatic chain than β-DM.
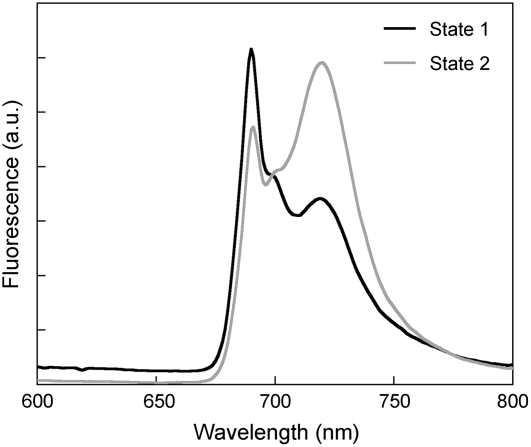
C. reinhardtii cells were grown under low-light conditions (∼20 μmol photons m−2 s−1) until the early exponential phase, and the culture was split into two: one for preparing thylakoids in State 1 and another for preparing them in State 2. Induction of state transitions was done by manipulating the redox state of the plastoquinone pool as described previously (Bulté et al., 1990). Cells were treated with a PSII inhibitor DCMU for 20 min under low light to oxidize the plastoquinones, which locked them in State 1. State 2–locked cells were obtained by treatment with the uncoupler carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP) for 20 min under low light to reduce the plastoquinone pool. Fluorescence emission spectra at 77K of cells in State 1 showed that the peak at 689 nm (F689), originating from PSII, was higher than the peak at 718 nm (F718) from PSI (Figure 1, State 1). On the other hand, cells locked in State 2 exhibited the opposite trend, as F718 was higher than F689 (Figure 1, State 2). These results confirmed that the energy distribution between the two photosystems was altered and the cells were indeed locked in the designated state. The results of these treatments are consistent with previous reports (Bulté et al., 1990; Finazzi et al., 1999).
Figure 1.
The 77K Steady State Fluorescence Emission Spectra of C. reinhardtii Cells Locked in States 1 and 2.
C. reinhardtii cells grown under 20 μmol photons m−2 s−1 were treated with 10 μM DCMU or 5 μM FCCP for 20 min to induce to State 1 or State 2, respectively. Excitation was given at 440 nm. Spectra were normalized to the 699-nm (CP47) emission peak (Lin and Knox, 1991).
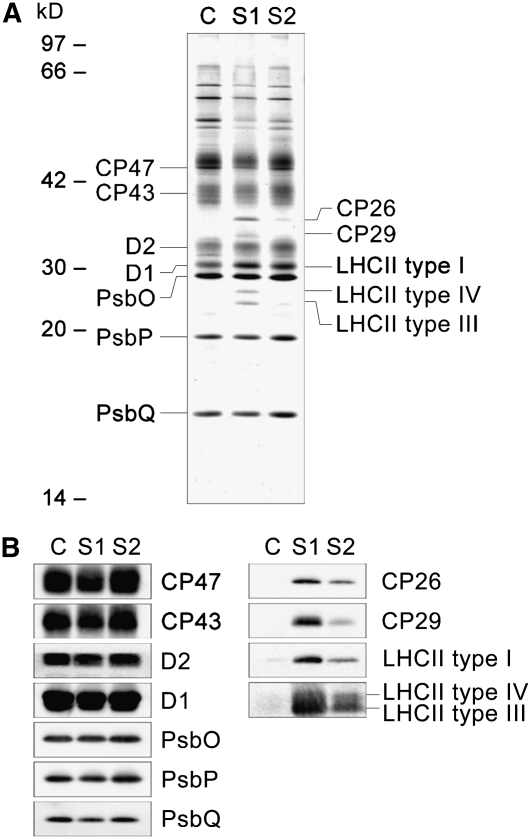
Thylakoid membranes isolated from States 1 and 2 locked cells were solubilized by TM and loaded onto a nickel affinity column. For comparison, we also isolated PSII cores using β-DM solubilization according to the procedure described by Suzuki et al. (2003). Protein complexes carrying His-tagged CP47 were bound to the column and eluted with 250 mM imidazole. Immunoblot analysis confirmed that PSI and PSII subunits were clearly separated in the flow-through and the eluate fractions, respectively (see Supplemental Figure 1 online), indicating that nickel affinity chromatography could be used to purify PSII particles. Each eluate was analyzed by SDS-PAGE and immunoblotting (Figures 2A and 2B) to confirm that the eluted complexes harbor PSII core subunits, including D1, D2, CP43, CP47, PsbO, PsbP, and PsbQ. In addition, the TM-solubilized protein complexes eluted from the column contained minor LHCIIs (CP26 and CP29) and major LHCIIs type I, III, and IV, suggesting that these eluates contain PSII complexes retaining the peripheral antennas (Figures 2A and 2B). The retained LHCIIs were estimated to represent 43 and 16% of LHCIIs in the State 1 and 2 samples, respectively, by immunoblotting using an anti-LHCII type I antibody. The LHCIIs recovered in the flow-through are most likely not, or only marginally, bound to PSII.
Figure 2.
Polypeptide Composition of Affinity-Purified PSII Complexes in States 1 and 2.
Polypeptides in PSII complexes, isolated using nickel affinity chromatography, were analyzed by SDS-PAGE stained with Coomassie blue (A) and immunoblotting using antibodies specific for each protein as labeled (B). C, isolated PSII cores; S1, PSII complexes isolated from State 1 thylakoid membranes; S2, PSII complexes isolated from State 2 thylakoid membranes. One microgram of chlorophyll was loaded in each lane.
The Three PSII Fractions and Their Relative Abundance in States 1 and 2
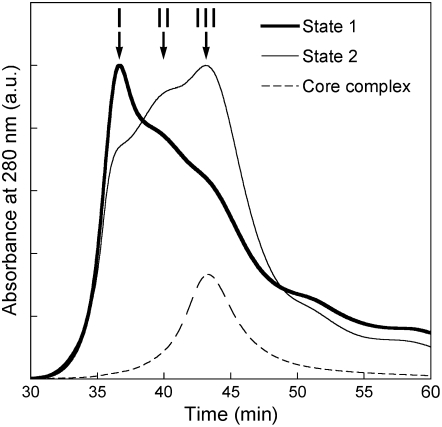
Comparing the amount of bound LHCIIs, it is clear that the PSII complexes in State 1 bind more LHCIIs than in State 2 (Figure 2B). This is consistent with the existing model, where mobile LHCIIs migrate from PSII to PSI in the transition from State 1 to 2, thereby decreasing the PSII antenna size (see, for instance, Allen and Forsberg, 2001). The decreased amount of LHCIIs associated with PSII complexes in State 2 (Figures 2A and 2B) suggests that the PSII-LHCII supercomplex has a decreased molecular mass in State 2 compared with State 1. We thus performed gel filtration to analyze the molecular mass of the protein complexes isolated by nickel affinity chromatography. The gel filtration chromatogram showed at least three peaks (or shoulders) at 36.6 (Fraction I), 39.7 (Fraction II), and 43.5 min (Fraction III) retention time, which corresponds to a molecular mass of ≥2000, 900 to 1000, and 300 to 400 kD, respectively (Figure 3). SDS-PAGE and immunoblot analysis confirmed that all three fractions contained PSII core subunits, including D1, D2, CP43, CP47, PsbO, PsbP, and PsbQ (see Supplemental Figures 2 and 3 online). In State 1, the PSII subunits (D1, CP47, and PsbO) were more abundant in Fraction I, whereas in State 2 they were more abundant in Fraction III (see Supplemental Table 1 online). Since a control PSII core sample isolated from β-DM–solubilized membranes peaked at ∼43.5 min, Fraction III was assigned as PSII core (Figure 3).
Figure 3.
Gel Filtration Elution Profile of PSII Particles in States 1 and 2 Isolated with Nickel Affinity Chromatography.
PSII complexes isolated with nickel affinity chromatography were separated by gel filtration on a Superdex 200 PC 3.2/30 column. Three major fractions are designated as I to III. The elution profile of the purified PSII cores is shown as a dashed line. Traces are shown for 280-nm absorption. Thick line, State 1; thin line, State 2.
The LHCII association with the PSII fractions was tested by their chlorophyll and carotenoid compositions using HPLC (Table 1). The ratio of chlorophyll a to b, which is inversely correlated to the abundance of LHC proteins, was similar in Fractions I and II but markedly increased in Fraction III, where the ratio was approximately the same as that of PSII core, confirming that Fraction III contains PSII core and Fractions I and II contain LHCII proteins in addition to PSII core. The ratio of neoxanthin, which is bound only to LHCII, to β-carotene, which is bound only to PSII core proteins, in Fraction I was nearly the same as that in Fraction II but significantly larger than that in Fraction III, which indicates that more LHCIIs are associated with the PSII particles in Fractions I and II than in Fraction III (Table 1). The functional coupling of the bound LHCIIs to PSII core was determined by measuring the functional absorption cross section of PSII (σPSII), which indicated that the bound LHCIIs in Fraction I and II increased σPSII from 163 Å2/quanta in Fraction III to 366 and 350 Å2/quanta in Fractions I and II, respectively (Table 1). These findings were further supported by the SDS-PAGE gel, indicating that Fractions I and II contain significant amounts of LHCII proteins, while Fraction III does not (see Supplemental Figure 2 online).
Table 1.
Characterization of the Three PSII Fractions Obtained by Ni-Affinity Chromatography and Gel Filtration
| PSII Fractionsa | I | II | III | Core |
|---|---|---|---|---|
| Chlorophyll a/b (w/w)b | 3.22 ± 0.37 | 3.36 ± 0.24 | 4.30 ± 0.75 | 4.27 ± 0.13 |
| Neoxanthin/β-carotene (mol/mol) | 0.26 ± 0.13 | 0.25 ± 0.13 | 0.13 ± 0.03 | <0.001 |
| σPSII (Å2/quanta) | 366 ± 20 | 350 ± 26 | 163 ± 88 | ND |
PSII fractions were isolated from State 2–locked samples. Values shown are means of more than three measurements ± sd.
Chlorophyll concentrations were determined as described by Niyogi et al. (1997).
The relative abundance of Fractions I to III was dramatically changed during state transitions. In State 1, Fraction I was predominant, whereas in State 2, Fraction III was more pronounced (Figure 3). It is thus inferred from these results that there is more LHCII-retaining PSII (Fractions I and II) in State 1 and more LHCII-less PSII (Fraction III) in State 2, which is most likely due to the removal of LHCIIs from the PSII-LHCII supercomplex upon transition from States 1 to 2.
LHCII Polypeptide Composition in the Three PSII Fractions
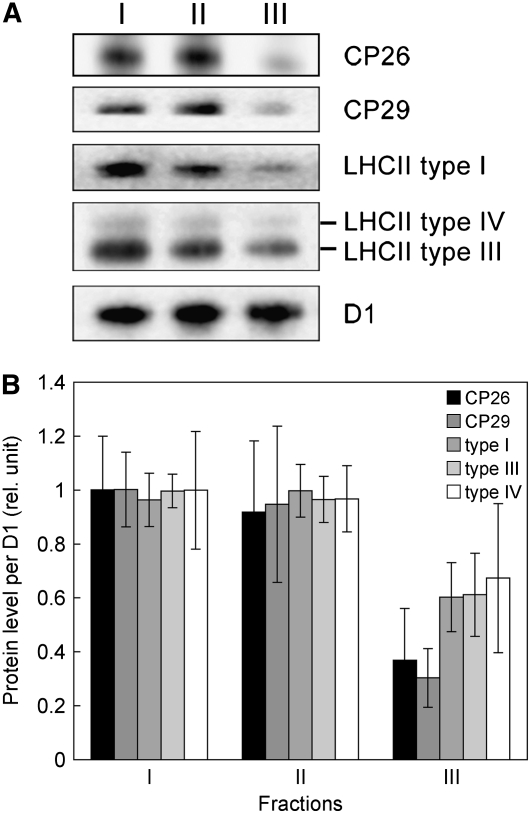
The composition of LHCII polypeptides in each gel filtration fraction was determined using immunoblotting. Both Fractions I and II contained all of the minor and major LHCIIs except for LHCII type II (Figure 4A). The relative amount of each LHCII to the PSII core (D1 protein) was essentially the same in the two fractions (Figure 4B). In Fraction III, however, the levels of bound LHCIIs were significantly reduced, confirming that this fraction is mainly composed of PSII complexes that lost most of their LHCII (Figures 4A and 4B).
Figure 4.
Relative Abundance of LHCII Polypeptides in the Gel Filtration Fractions I to III.
(A) Minor LHCIIs (CP26 and CP29) and major LHCIIs (types I, III, and IV) in the three PSII fractions were detected using immunoblotting with specific antibodies as indicated. I, II, and III indicate the corresponding fractions isolated from a State 1–locked sample using gel filtration (Figure 3). Each protein sample was normalized to the quantity of D1 protein. Three independent experiments were performed, and a representative result is shown.
(B) Protein levels detected by immunoblotting (A) were quantified using ImageJ software. Values are means of three measurements ± sd normalized to the highest value in the three fractions.
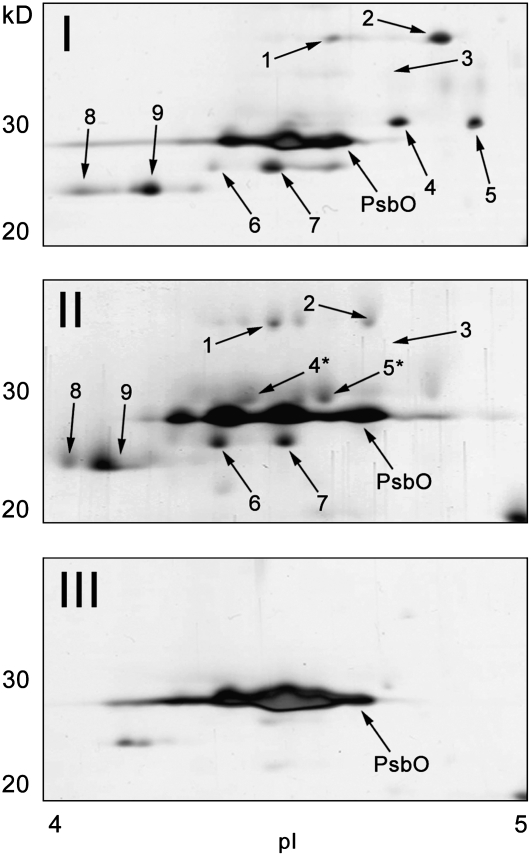
The major LHCII type I and III in C. reinhardtii are encoded by five (LhcbM3, -M4, -M6, -M8, and -M9) and two (LhcbM2 and -M7) duplicated genes, respectively, while both LHCII types II and IV are encoded by single genes, LhcbM5 and -M1, respectively (Minagawa and Takahashi, 2004). To further determine which LHCII gene products are associated with PSII in these fractions, we conducted two-dimensional gel electrophoresis (2-DE). The first dimension was isoelectric focusing, which uncovered distinct LHCII spots because hydrophobic PSII core subunits did not penetrate the gel. Several spots that have been previously reported as LHCIIs (Stauber et al., 2003) appeared in the region between 20 and 42 kD and pH 4.0 and 5.0 on the 2-DE gels (Figure 5). Polypeptides in these spots were then identified by tandem mass spectrometry (MS/MS) analysis after in-gel trypsin digestion (see Supplemental Table 2 online). All of the minor and major LHCIIs except for LhcbM5 and -M8 were detected in Fraction I (Figure 5). Spot 3, corresponding to CP29, was hardly visualized by silver staining in Figure 5 but was clearly detected by immunoblotting (see Supplemental Figure 5 online) and was confirmed by MS/MS (see Supplemental Table 2 online). The same LHCII polypeptides were identified in Fraction II, but they were markedly shifted toward the acidic side (see spots 4 and 5, corresponding to LhcbM3/4/6 and LhcbM4/6/9, respectively in Figure 5, panel II). These shifts could be due to the phosphorylation of these proteins as indicated previously (Hippler et al., 2001). In Fraction III, only a trace amount of LHCII type III (LhcbM2/7) was detected in the 2-DE gel (Figure 5, panel III). These results further support the conclusion that Fractions I and II contain PSII-LHCII supercomplexes with similar polypeptide compositions, and Fraction III contains PSII cores and a small amount of PSII core with LHCII type III. It is of note that among the four types of major LHCIIs in C. reinhardtii, LHCII type II was barely detected in any of the PSII-LHCII supercomplexes in either State 1 or 2; however, it has been reported to be associated with PSI in State 2 (Takahashi et al., 2006). LHCII type II is thus more likely either to be bound at the outer ridge of the peripheral antenna for PSII in State 1, such that it is removed by TM solubilization, or to exist as an unbound form in State 1 and associate with PSI in State 2.
Figure 5.
Two-Dimensional Separation of LHCII Polypeptides in the Gel Filtration Fractions I to III.
Fractions I to III isolated from a State 2–locked sample were subjected to 2-DE. Shown are the regions between 20 and 40 kD and pH 4.0 and 5.0 on the silver-stained 2-DE gels. Each numbered spot was identified by MS/MS analysis (see Supplemental Table 2 online) after in-gel trypsin digestion. Asterisks indicate putatively phosphorylated polypeptides (see corresponding spots 13 and 14 in Figure 8).
Supramolecular Organization of the Three PSII Particles
According to single-particle image analysis of PSII-LHCII supercomplexes in higher plants, the C2S2M2 supercomplex is the most predominant (Dekker and Boekema, 2005). However, the largest PSII-LHCII supercomplex in C. reinhardtii has been reported to be the C2S2 supercomplex, which might be due to the lack of CP24 in this organism (Nield et al., 2004; Dekker and Boekema, 2005). The PSII-LHCII supercomplex in Fraction II is likely a C2S2 supercomplex, based on the estimated molecular mass and the polypeptide composition (Boekema et al., 1995): two major LHCII trimers, two copies of CP26 and CP29, and one PSII core dimer. The largest PSII particle with molecular mass of ≥2000 kD in Fraction I could be a multimer of the C2S2 supercomplex, since the polypeptide composition (Figure 5), relative amount of LHCIIs to D1 (Figure 4B), chlorophyll a/b ratio, neoxanthin/β-carotene ratio, and functional absorption cross section (Table 1) of PSII particles in Fraction I are all similar to those in Fraction II.
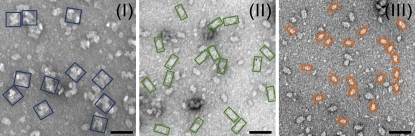
To investigate the supramolecular organization of the PSII particles in the three gel filtration fractions, negatively stained single particles were observed via electron microscopy. Figure 6 shows a distribution of the top-down views of PSII particles in Fractions I to III. Large (33.0 × 33.0 nm, blue), medium (16.5 × 33.0 nm, green), and small particles (11.0 × 20.5 nm, orange) are predominant in Fractions I, II, and III, respectively (Figure 6; see Supplemental Figure 4 online). These sizes correspond to two C2S2 supercomplexes (Nield et al., 2000), one C2S2 supercomplex (Nield et al., 2000), and one PSII core dimer (Barber, 2006), respectively. Dekker and Boekema (2005) showed that several types of dimerized PSII-LHCII supercomplexes, so-called megacomplexes, are formed through the lateral association of supercomplexes. Although most of the megacomplexes reported so far in higher plants are dimers of C2S2M2 supercomplexes, the dimerization of C2S2 supercomplexes was also suggested in C. reinhardtii (Dekker and Boekema, 2005). Based on these results, it is thus inferred that the predominant PSII particles with molecular masses of ≥2000 kD, 800 to 900 kD, and 300 to 400 kD in Fractions I, II, and III are (C2S2)2 PSII-LHCII megacomplexes, C2S2 PSII-LHCII supercomplexes, and PSII core dimers, respectively.
Figure 6.
Electron Micrographs of Single-Particle PSII Complexes in the Gel Filtration Fractions I to III.
Fractions I to III isolated from State 2–locked cells were negatively stained with uranyl acetate. Projections of particles inferred to be PSII-LHCII megacomplexes, PSII-LHCII supercomplexes, and PSII core complexes are indicated by blue, green, and orange boxes, respectively. Bars = 50 nm.
Phosphorylations Involved in the Detachment of LHCII
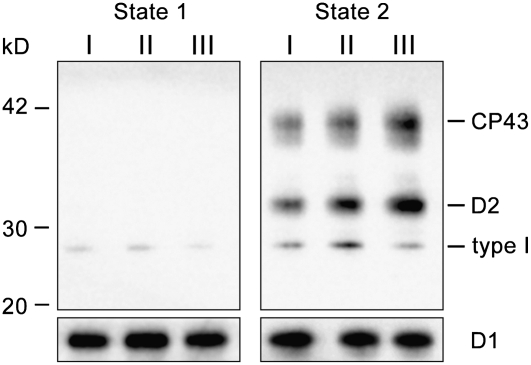
Phosphorylated polypeptides in the three gel filtration fractions were determined by immunoblotting using an antiphosphothreonine antibody (Figure 7). The obtained results demonstrate that PSII proteins in State 1 were mostly not phosphorylated, whereas PSII-bound LHCII type I and the core subunits CP43 and D2 were strongly phosphorylated in State 2. This clearly indicates that phosphorylation by itself, at least of LHCII type I, does not readily induce detachment of LHCIIs from PSII. Intriguingly, LHCII type I was much more phosphorylated in Fraction II than in Fraction I in State 2 (Figure 7, Table 2). This implies a possibility that the phosphorylation of LHCII type I triggers the division of the PSII-LHCII megacomplex in Fraction I into the PSII-LHCII supercomplexes in Fraction II. The levels of phosphorylation in the PSII core subunits, CP43 and D2, were highest in the core complex and lowest in the megacomplex (Table 2). Thus, the phosphorylation of the PSII core subunits occurs in a different manner from that of LHCII type I, suggesting a distinct mechanism for the regulation of the kinases for LHCII polypeptides and PSII core subunits.
Figure 7.
Phosphorylated Polypeptides in the Gel Filtration Fractions I to III from State 1– and State 2–Locked Samples.
Phosphorylated proteins were detected using immunoblotting with an antiphosphothreonine antibody. I, II, and III indicate the corresponding fractions isolated using gel filtration (Figure 3). Each protein sample was normalized to the quantity of D1 protein.
Table 2.
Relative Phosphorylation Levels of LHCII Type I, CP43, and D2 in the PSII Fractions during State 2
| PSII Fractionsa | I | II | III |
|---|---|---|---|
| LHCII type I (%) | 100 | 203 ± 33 | 97 ± 30 |
| CP43 (%) | 100 | 142 ± 19 | 160 ± 32 |
| D2 (%) | 100 | 159 ± 35 | 172 ± 21 |
Phosphorylation levels are normalized to the quantity of D1 protein and quantified by ImageJ. Values shown are means of three measurements ± sd normalized to the value for Fraction I.
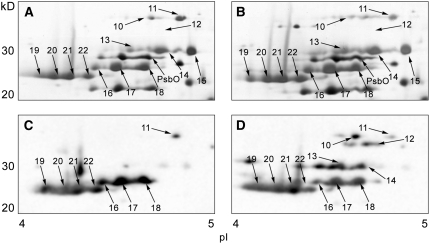
We also characterized the unbound form of LHCII polypeptides to get further insights on their dissociation from PSII. Free LHCIIs in flow-through fractions obtained after nickel affinity chromatography were subjected to 2-DE analysis (Figure 8). The compositions of unbound LHCIIs were nearly the same in State 1 and 2 (Figures 8A and 8B; see Supplemental Figure 5 and Supplemental Table 3 online); however, there were clear differences in their levels of phosphorylation. Spots corresponding to free CP26, CP29, and LHCII type I (LhcbM3, -4, and -6) were more phosphorylated in State 2 (Figures 8C and 8D). Although the free CP26 was also phosphorylated in State 1 (spot 11), possibly more amino acid residues in CP26 were phosphorylated in State 2 (spot 10). In Figure 7, CP26 and CP29 associated with PSII were shown to be nonphosphorylated. Therefore, phosphorylations of CP26 and CP29 most likely lead to their dissociation from PSII. Considering the location of the minor LHCIIs bordering major LHCII trimers and the PSII core (Harrer et al., 1998), we infer that the phosphorylation of minor LHCIIs induces the detachment of all LHCIIs, including major LHCIIs, from PSII. Interestingly, LHCII type III (LhcbM2 and -7) and type IV (LhcbM1) in the flow-through fractions were phosphorylated irrespective of the state (Figures 8C and 8D), whereas those associated with PSII were not (Figure 7). Therefore, unlike LHCII type I, once LHCII type III and IV are phosphorylated they will be undocked from PSII no matter the state.
Figure 8.
Two-Dimensional Separation and Phosphorylation Analysis of Unbound LHCII Polypeptides from the State 1– and State 2–Locked Samples.
Nickel affinity chromatography flow-through from the State 1 ([A] and [C]) and State 2 ([B] and [D]) samples was subjected to 2-DE. Shown are the regions between 20 and 40 kD and pH 4.0 and 5.0 on the 2-DE gels. Protein spots were silver stained ([A] and [B]), and the numbered spots were identified by MS/MS analysis (see Supplemental Table 3 online). Phosphorylated proteins were detected using immunoblotting with an antiphosphothreonine antibody ([C] and [D]). Loaded protein amounts were approximately the same in (A) and (B), and in (C) and (D).
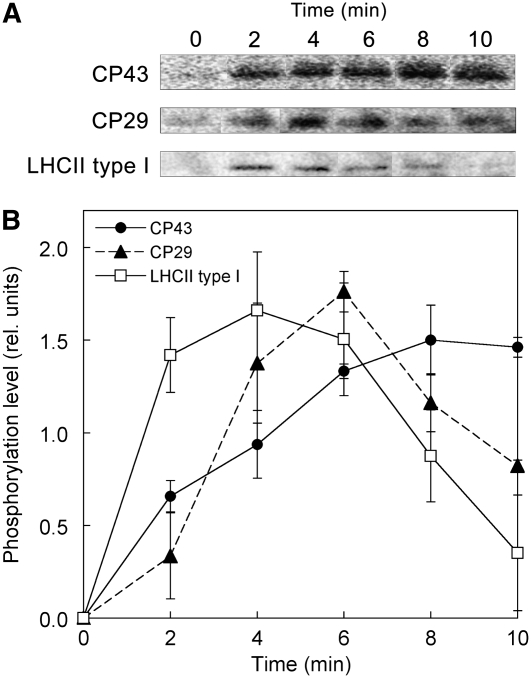
Lastly, we examined the kinetics of phosphorylation for the most conspicuously phosphorylated proteins of LHCII and PSII in the course of transition from State 1 to 2. Whole cell samples, taken every 2 min under blue light illumination (light 2), were subjected to immunoblotting using an antiphosphothreonine antibody (Figure 9). While the phosphorylation level of LHCII type I was immediately elevated (followed by a slow decline), that of CP29 was gradually increased, but similarly dephosphorylated after 6 min. The phosphorylation of the core subunit CP43 was even slower and reached a plateau after 6 min. The distinct phosphorylation kinetics of the three polypeptides suggests their distinct roles in state transitions. Since Fraction II is the main fraction harboring the phosphorylated LHCII type I (Figure 7), its population could be increased in the initial phase of transition from State 1 to 2. This in turn suggests that the division of the megacomplex occurs first with the LHCII type I phosphorylation and then the other phosphorylations trigger the undocking of LHCIIs from PSII, until just the PSII core complex remains.
Figure 9.
Phosphorylation Kinetics in the Course of Transition from State 1 to State 2.
C. reinhardtii cells settled in State 1 under weak far-red light (720 nm) for 15 min were illuminated by weak blue light (467 nm) to induce a transition to State 2. Phosphorylated polypeptides in cells sampled at every 2 min were detected by immunoblotting with an antiphosphothreonine antibody (A), and kinetics of the relative phosphorylation levels are shown (B). Data represent an average of three independent measurements and are expressed as mean ± sd.
DISCUSSION
The Two-Step Transition from State 1 to State 2: Megacomplex to Supercomplex, and Supercomplex to Core Complex
In this study, PSII particles from State 1– and State 2–locked thylakoids were characterized biochemically and structurally. The profile of the gel filtration and single particle images of the electron micrographs demonstrated that megacomplexes, which are presumably multimers of PSII-LHCII supercomplexes, are predominant in State 1, whereas PSII core complexes with much less LHCII are predominant in State 2. We also identified an intermediate state of transition, the PSII-LHCII supercomplex monomer, which has the same polypeptide and pigment compositions as the megacomplex. We propose that the transition from State 1 to 2 proceeds in two steps on the PSII side: first, the PSII megacomplex is divided into supercomplexes and then LHCIIs (both major and minor) dissociate from these supercomplexes.
Role of Phosphorylated LHCII
It is well accepted that LHCII phosphorylation is crucial for the migration of LHCIIs from PSII to PSI (Depége et al., 2003). To give insights into the molecular details of the undocking process of LHCIIs from PSII, phosphorylation of the PSII core subunit and of LHCII polypeptides both bound and unbound to PSII was thoroughly characterized. The results suggest distinct roles for each phosphorylated protein: (1) LHCII type I (LhcbM3, -4, -6, and -9), (2) minor LHCII (CP26 and CP29), (3) PSII core subunits (CP43 and D2), and (4) LHCII type III (LhcbM2 and -7) and type IV (LhcbM1).
(1) LHCII Type I (LhcbM3, -4, -6, and -9)
It has been hypothesized that phosphorylated major LHCIIs do not necessarily dissociate themselves from PSII (Lunde et al., 2000). It is clear that LHCII type I remains associated with PSII upon phosphorylation. Surprisingly, the phosphorylation of LHCII type I is induced earlier in the course of state transition, and it is mostly localized in the supercomplex, not in the megacomplex. This suggests that the phosphorylation of LHCII type I likely causes division of the megacomplex. As has been previously described by Dekker and Boekema (2005), the two C2S2 supercomplexes constituting the megacomplex are bridged by an LHCII trimer in C. reinhardtii. Therefore, the phosphorylation of the most abundant trimeric LHCII, type I, may induce the division of the megacomplex due to its altered conformation (Sprang et al., 1988; Nilsson et al., 1997).
(2) Minor Monomeric LHCII (CP26 and CP29)
Our previous work indicates that the minor monomeric LHCIIs are shuttled to PSI, thereby acting as a linker between PSI and major trimeric LHCII during a transition to State 2 (Takahashi et al., 2006). The minor LHCIIs, especially CP29, are indeed strongly phosphorylated in State 2 (Kargul et al., 2005; Takahashi et al., 2006; Turkina et al., 2006). This study provides further evidence for the pivotal role of the minor LHCIIs. While both CP26 and CP29 associated with the megacomplex and supercomplex were not phosphorylated, those of the free form were phosphorylated (Figure 8D). Thus, only phosphorylated minor LHCIIs are dissociated from PSII. Since the minor monomeric LHCIIs are bordering major LHCII trimers and the PSII core (Harrer et al., 1998; Yakushevska et al., 2003), any LHCII proteins associated with PSII will be forced to leave when the minor LHCIIs are dissociated. Thus, it is inferred that phosphorylation of CP26 and CP29 triggers undocking of the entire peripheral antenna during state 2 transition.
Recently, Turkina et al. (2006) reported the exact phosphorylated amino acid residues on CP29 in C. reinhardtii cells under State 2 as well as under high-light conditions. Transfer of the cells from State 1 to 2 conditions increases the number of phosphorylated residues in CP29 from two to four, whereas high-light treatment causes seven phosphorylated residues. Based on the mapping of those phosphorylated residues at the interface of PSII core and the peripheral antenna proteins, they proposed that the hyperphosphorylation of CP29 causes uncoupling of itself and other LHCII proteins from PSII in cells under State 2 or high-light stress. Our proposal, based on this study, agrees with their report.
(3) PSII Core Subunits (CP43 and D2 Protein)
We demonstrated that CP43 and D2 protein are phosphorylated under State 2 conditions, but the distribution of phosphorylated core proteins among the megacomplex, supercomplex, and core complex, as well as the phosphorylation kinetics, is completely different from that of LHCII. These results are compatible with previous findings that the phosphorylation of PSII core subunits and LHCIIs were both under the redox control by the plastquinone pool and that only LHCII phosphorylation depends on cytochrome bf complex (Bennett, 1991). Two distinct kinases, STN7 and STN8, were assigned as the kinases for LHCIIs and PSII core subunits, respectively (Bellafiore et al., 2005; Bonardi et al., 2005).
Since the two phosphorylation events (PSII core subunits and LHCIIs) occur simultaneously during the transition to State 2, we must emphasize that the two processes are potentially interlinked. Although it is controversial (Andronis et al., 1998, Fleischmann and Rochaix, 1999, Bonardi et al., 2005), PSII core subunit phosphorylation has been suggested to act as a protection mechanism from proteolytic degradation of photodamaged PSII complex under high-light conditions (Koivuniemi et al., 1995; Kruse et al., 1997; Turkina et al., 2006). Since cells were grown in low light in this study (20 μmol photons m−2 s−1), the core subunits are not under the high-light stress. However, the bare PSII core complex (after dissociation of LHCIIs during state transition) is potentially more susceptible to photodamage (Finazzi et al., 2001), so that the core phosphorylation could also be effective to protect cores after the dissociation of LHCII upon state transition.
Alternatively, core subunit phosphorylation might be specifically required for undocking of the peripheral antenna. Our results revealed that the strongest phosphorylation of PSII subunits was found in Fraction III (Figure 7). It is thus possible that the simultaneous phosphorylation of both minor LHCIIs and PSII core subunits facilitates dissociation of peripheral antenna via coulombic repulsion. A similar model has been proposed previously by Turkina et al. (2006). This possibility might not be the case, if land plants and green algae share a common mechanism for state transitions, as an stn8 mutant of Arabidopsis does perform state transitions normally, although PSII core phosphorylation was suppressed (Bonardi et al., 2005). Further studies incorporating the kinase mutants are expected to clarify these issues.
(4) LHCII Type III (LhcbM2 and -7) and Type IV (LhcbM1)
In contrast with LHCII type I, LHCII type III (LhcbM2 and -7) and IV (LhcbM1) were not associated with PSII when they were phosphorylated (Figures 7 and 8). These unbound phosphorylated polypeptides were not only in the State 2 samples but also in the State 1 samples. These results may imply that the phosphorylation-induced undocking of LHCII type III and IV occurs irrespective of the state and plays a unique role in photosynthesis other than state transitions. Care should be taken, however, since the quantity of phosphorylated polypeptides cannot be directly compared between State 1 and 2 gels, as approximately the same amount of proteins were resolved in the two 2-DE gels although the amounts of unbound LHCIIs in State 1 cells should be significantly lower than those in State 2 cells (Figures 8C and 8D). It is unclear whether LHCII type III and IV are more phosphorylated under State 2. Simply put, their behavior upon phosphorylation is different from that of LHCII type I; they leave PSII upon phosphorylation.
It should be noted that a mutant of the LhcbM1 gene in C. reinhardtii (npq5) has been shown to undergo normal state transitions (Elrad et al., 2002). Interestingly, the npq5 mutant was originally isolated as exhibiting little energy-dependent quenching (qE quenching) (Niyogi et al., 1997). This implies that the phosphorylated (dissociated) LhcbM1 and -2 may, in fact, be involved in qE quenching in addition to their roles in state transitions. Considering that C. reinhardtii displays little to no expression of the essential protein for qE quenching, PsbS (Niyogi et al., 2005), and only has a limited capacity of qE quenching when grown under normal light (Finazzi et al., 2006), the underlying mechanism for qE quenching in this green alga is not necessarily the same as that elucidated in higher plants (see reviews in Horton and Ruban, 2005; Niyogi et al., 2005). Further investigation on the significance of phosphorylation of LhcbM1 and -2 will clarify this point.
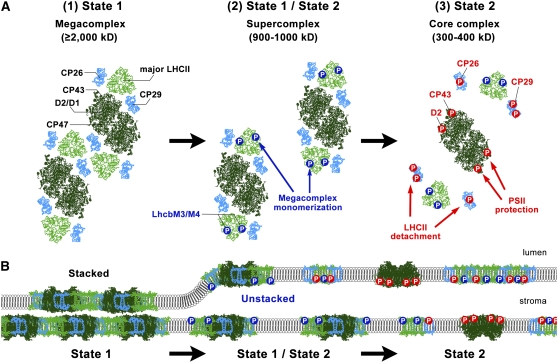
Based on the above observations, we propose the following model for the State 1 to 2 transition in C. reinhardtii: (1) unphosphorylated LHCIIs stabilize the megacomplex (State 1); (2) the phosphorylation of LHCII type I in the major LHCII trimer triggers the division of the megacomplex, resulting in individual C2S2 supercomplexes; (3) the phosphorylation of CP26 and CP29 as well as of PSII core subunits D2 and CP43 induces the undocking of all LHCIIs from PSII; and (4) free LHCIIs reassociate with the PSI-LHCI supercomplex yielding State 2 (illustrated in Figure 10A).
Figure 10.
A Proposed Model for the Remodeling of PSII during the Transition from State 1 to State 2.
(A) A top-down view from the stromal side of the membrane showing the division of the PSII megacomplex and detachment of peripheral antenna: (1) unphosphorylated LHCIIs stabilize the megacomplex; (2) the phosphorylation of LHCII type I in the major LHCII trimer triggers the division of the megacomplex, resulting in discrete C2S2 supercomplexes; (3) the phosphorylation of CP26 and CP29 as well as PSII core subunits D2 and CP43 induces the undocking of most LHCIIs from the PSII core complex. The crystal structure of the PSII core complex (dark green) is from 2AXT.pdb, and the LHCII monomer (light blue) and trimer (light green) structures are from 2NHW.pdb. Structural organizations of the megacomplex and supercomplex refer to Dekker and Boekema (2005) and Nield et al. (2004), respectively. The first- and second-step phosphorylations are indicated by blue and red circles with a letter “P,” respectively.
(B) A side-view of the membrane plane showing alterations in the thylakoid ultrastructure: (1) thylakoids are more stacked in State 1 (left); (2) after membrane unstacking, which is likely caused by the phosphorylation of LHCII type I, more LHCII proteins, including CP26 and CP29 and the core proteins CP43 and D2 protein, are phosphorylated (middle), leading to the undocking of most peripheral antenna proteins from PSII (right).
Ultrastructure of Thylakoid Membranes in State Transitions
It has been inferred that state transitions and the ultrastructure of thylakoid membranes are closely related (Allen and Forsberg, 2001). The functional absorption cross section of PSII is reported to be large in grana and small in stroma lamella (Garab and Mustárdy, 1999; Danielsson et al., 2006; Veerman et al., 2007), indicating that PSII in the stacked region of the thylakoid membrane tends to harbor more antenna proteins. In fact, a highly ordered paracrystalline arrangement of supercomplexes, from which the megacomplexes are thought to be solubilized, was observed only in the highly stacked grana core in Arabidopsis (Boekema et al., 2000). In contrast with the grana/stroma lamellae-type structure in higher plants, thylakoid membranes in Chlamydomonas are organized such that long bands of two to four thylakoid membranes are extensively but loosely appressed (Bertos and Gibbs, 1998). If the megacomplexes described above are localized in the appressed region, the thylakoid membranes in C. reinhardtii need to have a larger appressed region in State 1 than in State 2 to accommodate the larger number of megacomplexes. Indeed, an electron micrograph of C. reinhardtii thin sections from State 1 cells show a higher degree of membrane stacking, whereas those in State 2 were relatively unstacked (see Supplemental Figure 6 online). Currently, it is unclear whether state transitions affect the ultrastructure of thylakoid membranes or vice versa.
Figure 10B shows a model of state transitions that incorporates stacking/unstacking of thylakoid membranes. Since PSII megacomplexes are localized in the appressed region (Boekema et al., 2000), the diffusion of LHCII proteins is restricted (Kirchhoff et al., 2004) under State 1 conditions. After the membranes unstack, which may be caused by the phosphorylation of LHCII type I, more LHCII proteins, including CP26, CP29, and the core proteins CP43 and D2, are phosphorylated, leading to the undocking of all peripheral antenna proteins from PSII, which is partly reflected in the phosphorylation kinetics (Figure 9).
Further studies will be needed to characterize the significance of our other findings, including the phosphorylation of LHCII type III and IV in State 1, the redox-dependent phosphorylation of PSII core subunits, and possible changes in the thylakoid ultrastructure during state transitions. Such studies will provide new perspectives on the mutually related mechanisms of photosynthetic acclimation, including state transitions, qE quenching, and repair of photodamaged PSII centers.
METHODS
Strain and Growth Conditions
A genetically modified Chlamydomonas reinhardtii strain PsbB-His, carrying a 6x His-tag at the C terminus of the psbB gene (Suzuki et al., 2003), was grown in Tris-acetate-phosphate medium (Gorman and Levine, 1965) under a light intensity of 20 μmol photons m−2 s−1 aerated with ambient air at 23°C.
Induction of State Transitions
PsbB-His cells were treated as described previously (Bulté et al., 1990) to lock State 1 and 2. Briefly, the cells were incubated with 10 μM DCMU or 5 μM FCCP for 20 min under the same conditions as their growth to induce State 1 or State 2, respectively.
Isolation of Thylakoid Membranes and PSII Particles
Cells were harvested at mid-log phase (5 × 106 cells/mL), and thylakoid membranes were isolated similar to methods described by Chua and Bennoun (1975) with minor modifications. The buffer used for preparing thylakoid membranes contained 25 mM MES, 0.33 M sucrose, and 1.5 mM NaCl, pH 6.5. Disruption of cells was done using a BioNeb system (Glas-Col) at 10 kgf/cm2. Thylakoid membranes were adjusted to 1 mg chlorophyll/mL (for isolating PSII-LHCII supercomplexes) in a buffer containing 25 mM MES, 100 mM NaCl, and 12.5% (w/v) glycerol, pH 6.5, and solubilized with 1.0 to 1.2% (w/v) TM (Anatrace) at 4°C for 30 min in the dark. Solubilized thylakoid membranes were applied to a column with ProBond Resin (Invitrogen), which was preequilibrated with the same buffer supplemented with 15 mM imidazole and 0.03% (w/v) TM. Chromatography was performed as described previously (Suzuki et al., 2003). The eluate was diluted 10 times with a buffer containing 25 mM MES, pH 6.5, and 0.03% TM and concentrated to 1 mg chlorophyll/mL using an Amicon Ultra 100 KMWCO filter. Chlorophyll concentrations were determined as described by Porra et al. (1989) unless otherwise indicated.
Gel Filtration
PSII complexes isolated by nickel affinity chromatography (20 μg chlorophyll) were separated on a SMART System with a Superdex 200 PC 3.2/30 column (GE Healthcare), which was preequilibrated with 25 mM MES and 0.03% TM, pH 6.5. Gel filtration was conducted at 10°C with a flow rate of 25 μL/min. Protein concentrations were detected from their absorbance at 280 nm. Molecular masses of the protein complexes were estimated with a standard curve drawn with blue dextran (molecular mass of 2000 kD), thyroglobulin (molecular mass of 669 kD), and apoferritin (molecular mass of 443 kD) from GE Healthcare.
SDS-PAGE, 2-DE, Immunoblotting, and MS/MS
Polypeptides were separated by SDS-PAGE as described previously (Ikeuchi and Inoue, 1988). The first dimension of 2-DE, isoelectric focusing, was performed according to the procedure described by Hippler et al. (2001) with an Immobiline DryStrip (pH 4 to 7) on a Multiphor II system (GE Healthcare). The second dimension, SDS-PAGE, was conducted as described above. Polypeptide bands were stained with Quick CBB (Wako) or silver stain MS kit (Wako).
Immunoblotting was performed as described previously (Takahashi et al., 2006). Antibodies against D1, D2, CP43, CP47, PsbO, PsbP, and PsbQ (1/5000 dilution) were kind gifts from M. Ikeuchi at the University of Tokyo, those against LhcbM6 for detecting LHCII type I and P25K for detecting LHCII type III and IV (1/10,000 dilution) were provided by M. Hippler at the University of Münster, and those against CP26, CP29, LhcbM5 (1/10,000 dilution), and PsaA/B (1/5000 dilution) were described by Takahashi et al. (2006). Phosphorylated proteins were detected with an antiphosphothreonine (42H4) monoclonal antibody (1/5000 dilution) from New England Biolabs. Anti-rabbit secondary antibody was used for all but antiphosphothreonine monoclonal antibody for which anti-mouse secondary antibody was used. To identify phosphorylated proteins, the duplicated blots were checked either with antibodies specific to each polypeptide (SDS-PAGE) or MS/MS analysis (2-DE). Quantification of protein levels was done with ImageJ software (Abramoff et al., 2004).
In-gel trypsin digestion of polypeptides was performed as described by Shevchenko et al. (1996). The digested samples were applied to a Paradigm MS4 dual solvent system (Michrom BioResources). Solutions of 2 and 90% (v/v) acetonitrile with 0.1% (v/v) formic acid were used as the mobile phases A and B, respectively. The flow rate was 2 μL/min with a gradient as follows: 5% of B from 0 to 40 min, 95% from 40 to 50 min, and 5% from 50 to 60 min. Mass spectra were measured with a Finningan LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific). Obtained mass spectra were analyzed by SEQUEST software (revision 3.3; Thermo Fisher Scientific) using a genomic database for C. reinhardtii (version 3.0 assembly). The obtained peptide information was filtered based on the following criteria: number of top matches = 1, probability < 0.001, and Sf-final score > 0.85.
Fluorescence Analysis
Steady state fluorescence spectra at 77K were measured as described previously (Takahashi et al., 2006). Functional absorption cross section of PSII (σPSII) was determined according to Kolber et al. (1998) with a fluorescence induction and relaxation (FIRe) fluorimeter (Satlantic). Samples were dark-adapted for 10 min, and a 100-μs single-turnover flash was applied by a blue LED (455 nm) to cumulatively saturate PSII. The absolute σPSII values were calculated according to the calibration factor provided by the manufacturer.
Phosphorylation Kinetics from State 1 to State 2
PsbB-His cells grown as described above were illuminated with far-red light through a band-pass filter (720 ± 10 nm) for 15 min to induce State 1, which was followed by 10 min illumination with blue light through a band-pass filter (467 ± 10 nm) to induce State 2. Cells were sampled every 2 min in the course of transition to State 2. Polypeptides were isolated from the SDS-solubilized cells by precipitating with chloroform/methanol (Wessel and Flügge, 1984) and subjected to SDS-PAGE. Phosphorylation of polypeptides was detected by immunoblotting using an antiphosphothreonine antibody as described above. Three independent experiments were performed.
HPLC Analysis
Pigments of protein samples were extracted with 80% (v/v) acetone. HPLC was performed using a 4.6 × 250- mm VP-ODS C18 column (Shimadzu). Separation of pigments was done by a linear gradient of acetone/water as described previously (Niyogi et al., 1997). Molar extinction coefficients of 36,620 and 122,510 M−1 cm−1 were used for neoxanthin and β-carotene, respectively.
Transmission Electron Microscopy
Observation of single-particle PSII complexes was done according to Ortega et al. (2000). An H-7650 ZeroA electron microscope was used, and micrographs were digitally captured at a magnification of ×30,000. For observation of thylakoid membranes, cells induced to State 1 or State 2 were fixed in 1% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, and 1% osmium tetraoxide, dehydrated, and embedded in epoxy resin. Ultrathin sections were stained with 2% uranyl acetate and 0.2% lead citrate. A JEM 1200EX electron microscope (JEOL) was used, and samples were imaged at a magnification of ×50,000.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specificity of Nickel Affinity Chromatography.
Supplemental Figure 2. Polypeptide Composition in Gel Filtration Fractions I to III during States 1 and 2.
Supplemental Figure 3. Relative Abundance of the PSII Core Subunits in the Gel Filtration Fractions from State 1– and State 2–Locked Samples.
Supplemental Figure 4. Distribution of PSII Particle Projections on Electron Micrographs.
Supplemental Figure 5. Evidence for the Correspondence between Spot #12 in Figure 8D and CP29.
Supplemental Figure 6. Structure of Thylakoid Membranes in State 1 and State 2.
Supplemental Table 1. Relative Protein Levels of the PSII Core Subunits in the Three PSII Fractions.
Supplemental Table 2. MS/MS Analysis of the LHCII Proteins Bound to the PSII Complexes.
Supplemental Table 3. MS/MS Analysis of the Free LHCII Proteins in the Nickel Affinity Chromatography Flow-Through.
Supplementary Material
Acknowledgments
We thank Wesley Swingley for critical comments on the manuscript. We thank Shin-ichiro Ozawa at Okayama University for the help with MS/MS. We also thank Masahiko Ikeuchi and Michael Hippler for the kind gift of the antibodies. Our work was supported by the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (18004526 to M.I.), Grants-in-Aid for Scientific Research for Plant Graduate Student from the Nara Institute of Science and Technology (to M.I.), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (18GS0318 to Y.T. and J.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jun Minagawa (minagawa@lowtem.hokudai.ac.jp).
Online version contains Web-only data.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image Processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Allen, J.F., Bennett, J., Steinback, K.E., and Arntzen, C.J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291 21–25. [Google Scholar]
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6 317–326. [DOI] [PubMed] [Google Scholar]
- Andersson, B., Åkerlund, H.-E., Jergil, B., and Larsson, C. (1982). Differential phosphorylation of the light-harvesting chlorophyll–protein complex in appressed and non-appressed regions of the thylakoid membrane. FEBS Lett. 149 181–185. [Google Scholar]
- Andronis, C., Kruse, O., Deak, Z., Vass, I., Diner, B.A., and Nixon, P.J. (1998). Mutation of residue threonine-2 of the D2 polypeptide and its effect on photosystem II function in Chlamydomonas reinhardtii. Plant Physiol. 117 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, R., Giacometti, G.M., and Simpson, D.J. (1988). Changes in the organization of stroma membranes induced by in vivo state 1-state 2 transition. Biochim. Biophys. Acta 935 152–165. [Google Scholar]
- Barber, J. (2006). Photosystem II: An enzyme of global significance. Biochem. Soc. Trans. 34 619–631. [DOI] [PubMed] [Google Scholar]
- Bellafiore, S., Barneche, F., Peltier, G., and Rochaix, J.-D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433 892–895. [DOI] [PubMed] [Google Scholar]
- Bennett, J. (1977). Phosphorylation of chloroplast membrane polypeptides. Nature 269 344–346. [Google Scholar]
- Bennett, J. (1980). Chloroplast phosphoproteins. Evidence for a thylakoid-bound phosphoprotein phosphatase. Eur. J. Biochem. 104 85–89. [DOI] [PubMed] [Google Scholar]
- Bennett, J. (1991). Protein phosphorylation in green plant chloroplasts. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 281–311. [Google Scholar]
- Bertos, N.R., and Gibbs, S.P. (1998). Evidence for a lack of photosystem segregation in Chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 34 1009–1016. [Google Scholar]
- Bulté, L., Gans, P., Rebéillé, F., and Wollman, F.-A. (1990). ATP control on state transitions in vivo in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1020 72–80. [Google Scholar]
- Boekema, E.J., Hankamer, B., Bald, D., Kruip, J., Nield, J., Boonstra, A.F., Barber, J., and Rögner, M. (1995). Supramolecular structure of the photosystem II complex from green plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema, E.J., van Breemen, J.F.L., Van Roon, H., and Dekker, J.P. (2000). Arrangement of photosystem II supercomplexes in crystalline macrodomains within the thylakoid membrane of green plant chloroplasts. J. Mol. Biol. 301 1123–1133. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., van Roon, H., and Dekker, J.P. (1998). Specific association of photosystem II and light-harvesting complex II in partially solubilized photosystem II membranes. FEBS Lett. 424 95–99. [DOI] [PubMed] [Google Scholar]
- Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., Jahns, P., and Leister, D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437 1179–1182. [DOI] [PubMed] [Google Scholar]
- Bonaventura, C., and Myers, J. (1969). Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189 366–383. [DOI] [PubMed] [Google Scholar]
- Chua, N.-H., and Bennoun, P. (1975). Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. USA 72 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson, R., Suorsa, M., Paakkarinen, V., Albertsson, P.A., Styring, S., Aro, E.M., and Mamedov, F. (2006). Dimeric and monomeric organization of photosystem II. Distribution of five distinct complexes in the different domains of the thylakoid membrane. J. Biol. Chem. 281 14241–14249. [DOI] [PubMed] [Google Scholar]
- Dekker, J.P., and Boekema, E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706 12–39. [DOI] [PubMed] [Google Scholar]
- Depége, N., Bellafiore, S., and Rochaix, J.-D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299 1572–1575. [DOI] [PubMed] [Google Scholar]
- Elrad, D., Niyogi, K.K., and Grossman, A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi, G., Barbagallo, R.P., Bergo, E., Barbato, R., and Forti, G. (2001). Photoinhibition of Chlamydomonas reinhardtii in state 1 and state 2. Damages to the photosynthetic apparatus under linear and cyclic electron flow. J. Biol. Chem. 276 22251–22257. [DOI] [PubMed] [Google Scholar]
- Finazzi, G., Furia, A., Barbagallo, R.P., and Forti, G. (1999). State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1413 117–129. [DOI] [PubMed] [Google Scholar]
- Finazzi, G., Johnson, G.N., Dall'Osto, L., Zito, F., Bonente, G., Bassi, R., and Wollman, F.-A. (2006). Nonphotochemical quenching of chlorophyll fluorescence in Chlamydomonas reinhardtii. Biochemistry 45 1490–1498. [DOI] [PubMed] [Google Scholar]
- Fleischmann, M.M., and Rochaix, J.-D. (1999). Characterization of mutants with alterations of the phosphorylation site in the D2 photosystem II polypeptide of Chlamydomonas reinhardtii. Plant Physiol. 119 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garab, G., and Mustárdy, L. (1999). Role of LHCII-containing macrodomains in the structure, function and dynamics of grana. Aust. J. Plant Physiol. 26 649–658. [Google Scholar]
- Gorman, D.S., and Levine, R.P. (1965). Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer, R., Bassi, R., Testi, M.G., and Schäfer, C. (1998). Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L. (liverwort), which contains reaction center and antenna proteins. Eur. J. Biochem. 255 196–205. [DOI] [PubMed] [Google Scholar]
- Hippler, M., Klein, J., Fink, A., Allinger, T., and Hoerth, P. (2001). Towards functional proteomics of membrane protein complexes: Analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28 595–606. [DOI] [PubMed] [Google Scholar]
- Horton, P., and Ruban, A. (2005). Molecular design of the photosystem II light-harvesting antenna: photosynthesis and photoprotection. J. Exp. Bot. 56 365–373. [DOI] [PubMed] [Google Scholar]
- Ikeuchi, M., and Inoue, Y. (1988). A new 4.8-kDa polypeptide intrinsic to the PS II reaction center, as revealed by modified SDS-PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 29 1233–1239. [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4 236–240. [DOI] [PubMed] [Google Scholar]
- Kargul, J., Turkina, M.V., Nield, J., Benson, S., Vener, A.V., and Barber, J. (2005). Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J. 272 4797–4806. [DOI] [PubMed] [Google Scholar]
- Kirchhoff, H., Tremmel, I., Haase, W., and Kubitscheck, U. (2004). Supramolecular photosystem II organization in grana thylakoid membranes: Evidence for a structured arrangement. Biochemistry 43 9204–9213. [DOI] [PubMed] [Google Scholar]
- Koivuniemi, A., Aro, E.-M., and Andersson, B. (1995). Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34 16022–16029. [DOI] [PubMed] [Google Scholar]
- Kolber, Z.S., Prášil, O., and Falkowski, P.G. (1998). Measurements of variable chlorophyll fluorescence using fast repetition rate techniques: Defining methodology and experimental protocols. Biochim. Biophys. Acta 1367 88–106. [DOI] [PubMed] [Google Scholar]
- Kouřil, R., Zygadlo, A., Arteni, A.A., de Wit, C.D., Dekker, J.P., Jensen, P.E., Scheller, H.V., and Boekema, E.J. (2005). Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44 10935–10940. [DOI] [PubMed] [Google Scholar]
- Kruse, O., Zheleva, D., and Barber, J. (1997). Stabilization of photosystem two dimers by phosphorylation: Implication for the regulation of the turnover of D1 protein. FEBS Lett. 408 276–280. [DOI] [PubMed] [Google Scholar]
- Lin, S., and Knox, R.S. (1991). Studies of excitation energy transfer within the green alga Chlamydomonas reinhardtii and its mutants at 77K. Photosynth. Res. 27 157–168. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., and Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408 613–615. [DOI] [PubMed] [Google Scholar]
- Minagawa, J., and Takahashi, Y. (2004). Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82 241–263. [DOI] [PubMed] [Google Scholar]
- Murata, N. (1969). Control of excitation transfer in photosynthesis I. Light-induced change of chlorophyll a fluoresence in Porphyridium cruentum. Biochim. Biophys. Acta 172 242–251. [DOI] [PubMed] [Google Scholar]
- Nield, J., Orlova, E.V., Morris, E.P., Gowen, B., van Heel, M., and Barber, J. (2000). 3D map of the plant photosystem II supercomplex obtained by cryoelectron microscopy and single particle analysis. Nat. Struct. Biol. 7 44–47. [DOI] [PubMed] [Google Scholar]
- Nield, J., Redding, K., and Hippler, M. (2004). Remodeling of light-harvesting complexes in Chlamydomonas in response to environmental changes. Eukaryot. Cell 3 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, A., Stys, D., Drakenberg, T., Spangfort, M.D., Forsén, S., and Allen, J.F. (1997). Phosphorylation controls the three-dimensional structure of plant light harvesting complex II. J. Biol. Chem. 272 18350–18357. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Björkman, O., and Grossman, A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Li, X.-P., Rosenberg, V., and Jung, H.-S. (2005). Is PsbS the site of non-photochemical quenching in photosynthesis? J. Exp. Bot. 56 375–382. [DOI] [PubMed] [Google Scholar]
- Ortega, J., Singh, S.K., Ishikawa, T., Maurizi, M.R., and Steven, A.C. (2000). Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell 6 1515–1521. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Lunde, C., Jahns, P., Tarantino, D., Meurer, J., Varotto, C., Hirtz, R.-D., Soave, C., Scheller, H.V., Salamini, F., and Leister, D. (2002). A stable LHCII–PSI aggregate and suppression of photosynthetic state transitions in the psae1-1 mutant of Arabidopsis thaliana. Planta 215 940–948. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Samson, G., and Bruce, D. (1995). Complementary changes in absorption cross-sections of photosystems I and II due to phosphorylation and Mg2+-depletion in spinach thylakoids. Biochim. Biophys. Acta 1232 21–26. [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Sprang, S.R., Acharya, K.R., Goldsmith, E.J., Stuart, D.I., Varvill, K., Fletterick, R.J., Madsen, N.B., and Johnson, L.N. (1988). Structural changes in glycogen phosphorylase induced by phosphorylation. Nature 336 215–221. [DOI] [PubMed] [Google Scholar]
- Stauber, E.J., Fink, A., Markert, C., Kruse, O., Johanningmeier, U., and Hippler, M. (2003). Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2 978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M., Inoue, Y., and Minagawa, J. (1998). Rapid and discrete isolation of oxygen-evolving His-tagged photosystem II core complex from Chlamydomonas reinhardtii by Ni2+ affinity column chromatography. FEBS Lett. 426 140–144. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Minagawa, J., Tomo, T., Sonoike, K., Ohta, H., and Enami, I. (2003). Binding and functional properties of the extrinsic proteins in oxygen-evolving photosystem II particle from a green alga, Chlamydomonas reinhardtii having His-tagged CP47. Plant Cell Physiol. 44 76–84. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Iwai, M., Takahashi, Y., and Minagawa, J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto, H., Ono, T.-A., and Minagawa, J. (2001). Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42 849–856. [DOI] [PubMed] [Google Scholar]
- Tikkanen, M., Piippo, M., Suorsa, M., Sirpio, S., Mulo, P., Vainonen, J., Vener, A.V., Allahverdiyeva, Y., and Aro, E.M. (2006). State transitions revisited - a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62 779–793. [DOI] [PubMed] [Google Scholar]
- Turkina, M.V., Kargul, J., Blanco-Rivero, A., Villarejo, A., Barber, J., and Vener, A.V. (2006). Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 5 1412–1425. [DOI] [PubMed] [Google Scholar]
- Veerman, J., McConnel, M.D., Vasil'ev, S., Mamedov, F., Styring, S., and Bruce, D. (2007). Functional heterogeneity of photosystem II in domain specific regions of the thylakoid membrane of spinach (Spinacia oleracea L.). Biochemistry 46 3443–3453. [DOI] [PubMed] [Google Scholar]
- Vener, A.V., van Kan, P.J., Rich, P.R., Ohad, I.I., and Andersson, B. (1997). Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: Thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl. Acad. Sci. USA 94 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, D., and Flügge, U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138 141–143. [DOI] [PubMed] [Google Scholar]
- Wollman, F.-A., and Lemaire, C. (1988). Studies on kinase-controlled state transitions in Photosystem II and b6f mutants from Chlamydomonas reinhardtii which lack quinone-binding proteins. Biochim. Biophys. Acta 933 85–94. [Google Scholar]
- Yakushevska, A.E., Keegstra, W., Boekema, E.J., Dekker, J.P., Andersson, J., Jansson, S., Ruban, A.V., and Horton, P. (2003). The structure of photosystem II in Arabidopsis: Localization of the CP26 and CP29 antenna complexes. Biochemistry 42 608–613. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Scheller, H.V. (2004). Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 279 3180–3187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.