Abstract
Protein disulfide isomerase (PDI) oxidizes, reduces, and isomerizes disulfide bonds, modulates redox responses, and chaperones proteins. The Arabidopsis thaliana genome contains 12 PDI genes, but little is known about their subcellular locations and functions. We demonstrate that PDI5 is expressed in endothelial cells about to undergo programmed cell death (PCD) in developing seeds. PDI5 interacts with three different Cys proteases in yeast two-hybrid screens. One of these traffics together with PDI5 from the endoplasmic reticulum through the Golgi to vacuoles, and its recombinant form is functionally inhibited by recombinant PDI5 in vitro. Peak PDI5 expression in endothelial cells precedes PCD, whereas decreasing PDI5 levels coincide with the onset of PCD-related cellular changes, such as enlargement and subsequent collapse of protein storage vacuoles, lytic vacuole shrinkage and degradation, and nuclear condensation and fragmentation. Loss of PDI5 function leads to premature initiation of PCD during embryogenesis and to fewer, often nonviable, seeds. We propose that PDI5 is required for proper seed development and regulates the timing of PCD by chaperoning and inhibiting Cys proteases during their trafficking to vacuoles before PCD of the endothelial cells. During this transitional phase of endothelial cell development, the protein storage vacuoles become the de facto lytic vacuoles that mediate PCD.
INTRODUCTION
In multicellular eukaryotes, the regulated demise of specific sets of cells, termed programmed cell death (PCD), is a physiological paradox of growth and development (Jones and Dangl, 1996; Greenberg, 1996; Lam, 2005). In plants, PCD occurs during anther development, sex determination, tracheary element differentiation, monocot seed germination, nucellus, endothelium, and endosperm degeneration, leaf shape remodeling, leaf, carpel, and petal senescence, and early senescence (Orzaéz and Granell, 1997; Young et al., 1997; Wang et al., 1999; Fath et al., 2000; Fukuda, 2000; Domínguez et al., 2001; Wan et al., 2002; Hiratsuka et al., 2002; Hao et al., 2003; Gunawardena et al., 2004). Besides developmental roles, PCD is also activated in response to biotic and abiotic stresses (Lam, 2005; Yao and Greenberg, 2006), as in the case of the hypersensitive response to pathogen invasion (Heath, 2000; Jones, 2001; Hara-Nishimura et al., 2005).
PCD in plants encompasses at least three major cytological mechanisms (Jones, 2001; Kuriyama and Fukuda, 2002). During senescence, the chloroplasts are degraded initially, followed by the disruption of the vacuoles and the nucleus (Thomas et al., 2003). The second mechanism involves disruption and collapse of the large central vacuole, release of nucleases and proteases, acidification of the cytoplasm, and rapid degradation of nucleic acids and proteins (Obara et al., 2001; Kuriyama and Fukuda, 2002). In cells that undergo apoptosis-like PCD, the nucleus is degraded first, chromatin condenses, and DNA undergoes laddering (Fukuda, 2000). Apoptosis in animals, however, involves phagocytosis during which the DNA is fragmented and Cys proteases, termed caspases, stimulate apoptosis via an activation cascade (Green, 1998; Raff, 1998). In contrast with plants, caspases are constitutively present in most animal cells, residing in the cytosol as a single-chain proenzyme that is converted to an active protease (Martin and Green, 1995; Martins and Earnshaw, 1997). A nonapoptotic pathway of cell death has also been described for animals (Overholtzer et al., 2007).
Although animal apoptosis differs from plant PCD, Cys proteases are also induced in plants undergoing PCD, such as during the differentiation of tracheary elements (Fukuda, 2000), in soybean (Glycine max) cells exposed to stress (Solomon et al., 1999), in gymnosperm embryonic suspensor cells (Suarez et al., 2004), and in senescing organs (Nadeau et al., 1996; Cercos et al., 1999). The Arabidopsis thaliana Cys proteases, SENESCENCE-ASSOCIATED GENE12 and RESPONSIVE TO DEHYDRATION21 (RD21), which are induced during leaf senescence (Gan and Amasino, 1997; Yamada et al., 2001), facilitate nitrogen recycling. During Brassica napus and Ricinus communis seed, flower, leaf, and root development, Cys proteases like the KDEL-tailed Cys endopeptidases are expressed in tissues undergoing PCD, such as nucellar, leaf, and root cap cells (Domínguez and Cejudo, 1998; Linnestad et al., 1998; Xu and Chye, 1999; Wan et al., 2002; Helm et al., 2008), during which nuclear DNA degrades, ricinosomes proliferate, or the extensin scaffolds of the cell wall are digested (Greenwood et al., 2005; Helm et al. 2008). The ricinosomes are derived from the endoplasmic reticulum (ER) and contain ER-resident binding protein (BiP) and protein disulfide isomerase (PDI; EC 5.3.4.1) (Schmid et al., 2001). However, the roles of the ER and PDI in PCD are poorly understood.
The collective observations that PDI occurs within the endomembrane framework connecting the ER, secretory apparatus, the nucleus, and vacuoles prompted us to explore the role of the ER and PDI in PCD. PDI5 retains a classical structure of two thioredoxin motifs and a KDEL sequence, yet is 31 to 91 amino acids shorter than other homologous PDIs (Lu and Christopher, 2008). The more compact nature of PDI5 and the availability of a true gene knockout made PDI5 an intriguing subject of investigation.
Here, we demonstrate that PDI5 is preferentially expressed in floral and developing seed tissues and localizes to the ER and to the protein storage vacuoles (PSVs) and lytic vacuoles (LVs) of endothelial cells. We show that PDI5 interacts with three different Cys proteases, inhibits recombinant Cys protease in vitro, and traffics together with Cys protease RD21 to both PSVs and LVs. Expression of PDI5 in the endothelium precedes major PCD events, such as disruption and collapse of vacuoles, shrinkage and degradation of the cytoplasm, nuclear condensation and fragmentation, and breakdown of the cell wall. Loss of PDI5 leads to premature initiation of PCD in endothelial cells during embryo development and to fewer, often nonviable, seeds. We propose that PDI5 is required for proper embryogenesis and the temporal progression of PCD by chaperoning and inhibiting Cys proteases as they traffic from the ER via the Golgi to vacuoles before the initiation of PCD of endothelial cells.
RESULTS
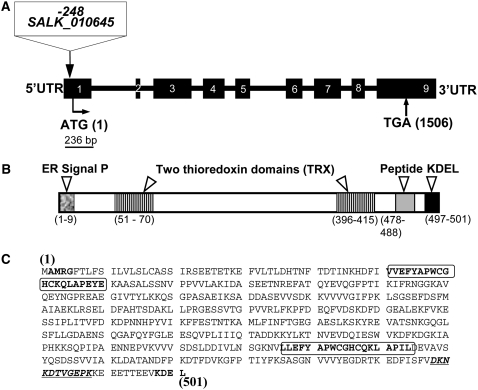
Arabidopsis PDI5 is one of 12 PDI genes found in the Arabidopsis genome (Meiri et al., 2002; Houston et al., 2005). A homozygous T-DNA insertion (SALK_010645; http://signal.salk.edu) within the pdi5 locus was identified using PCR (see Supplemental Figure 1 online), and it is located −248 bp upstream from the initiator ATG codon in the 300-nucleotide 5′ untranslated leader. The PDI5 gene is predicted to encode a spliced sequence of 1506 bp for a deduced polypeptide of 501 amino acids (locus number AAD41430) that contains a putative signal peptide, two thioredoxin catalytic domains (TRX and PWCghC), and an ER KDEL retention motif (Figure 1). We generated a polyclonal rabbit antibody against a peptide in PDI5 (DKNKDTVGEPK, residues 478 to 488; Figures 1B and 1C). The specificity of the antiserum was tested in situ by immunolabeling high-pressure frozen Escherichia coli cells expressing the recombinant PDI5 protein (see Supplemental Figures 2C and 2D online) as well as by immunoblot analysis of protein extracts from these cells (see Supplemental Figures 2A and 2B online).
Figure 1.
Structure and Organization of the Arabidopsis PDI5 Gene and T-DNA Insertion Mutant Site.
(A) Schematic representation of the PDI5 gene map (locus number AAD41430) and T-DNA insertion site location. The T-DNA (SALK_010645) insertion is located in the 300-nucleotide 5′ untranslated region (UTR) of the PDI5 gene at −248 bp upstream from the ATG codon. The gene contains nine exons (black boxes, numbered) separated by eight introns (thick black lines).
(B) Schematic drawing of the fully spliced translated portion of PDI5 cDNA. The white arrowheads indicate the position of various domains, including the ER signal peptide (the bold AMRG at the N terminus in [C]), two thioredoxin catalytic domains (TRX and PWCghC in framed region in [C]), a unique peptide antigen (11 amino acids underlined, bold italics in [C]), and the ER retention motif Lys-Asp-Glu-Leu (KDEL) in the C terminus (bold, unframed in [C]). The numbers correspond to the position and to the numbers of amino acids.
(C) Predicted protein sequence of PDI5 contains 501 amino acids and one stop codon.
PDI5 Is Preferentially Expressed in Flowers and Immature Seeds in Wild-Type Arabidopsis and Is Not Detected in the pdi5Δ T-DNA Insertion Mutant
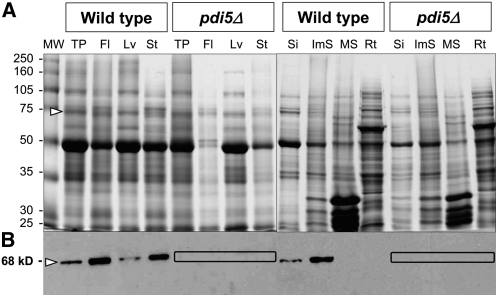
Immunoblot analysis using the anti-PDI5 antiserum on protein extracts of wild-type Arabidopsis plants identified a 68-kD polypeptide (Figures 2A and 2B), which was abundant in flowers, stems, and immature seeds and was less abundant in leaves and siliques (Figure 2B). No immunodetection was seen in any tissues of the homozygous pdi5Δ mutant (T-DNA SALK_010645) (Figure 2B) nor in mature siliques and roots of wild-type plants (Figure 2B). RT-PCR analyses of the three PDI5 gene insertion lines (see Supplemental Figures 1A and 1C online) and the wild type showed that Salk_010645, which is the insertion line used in this study, gave no detectable mRNA. The absence of PDI5 protein and PDI5 mRNA in the mutant confirms that pdi5Δ is indeed a null mutant.
Figure 2.
Immunoblot Analysis of PDI5 in Protein Extracts from Wild-Type and pdi5Δ T-DNA Insertion Mutant Tissues.
(A) SDS-PAGE and Coomassie blue staining of protein extracts from wild-type and pdi5Δ mutant tissues. Open arrowhead points to the 68-kD gel region where the PDI5 protein is expected to migrate. MW, molecular weight standard in kD; TP, total protein extract from whole plants; Fl, flowers; Lv, leaves; St, stems; Si, siliques; ImS, immature seeds; MS, mature seeds; Rt, roots.
(B) Protein gel blot analysis of the gel shown in (A) using anti-PDI5 antibodies. The boxes highlight the lack of PDI5 protein in the T-DNA mutant.
PDI5 Is Expressed in the Gynoecium and in Petals of Florets
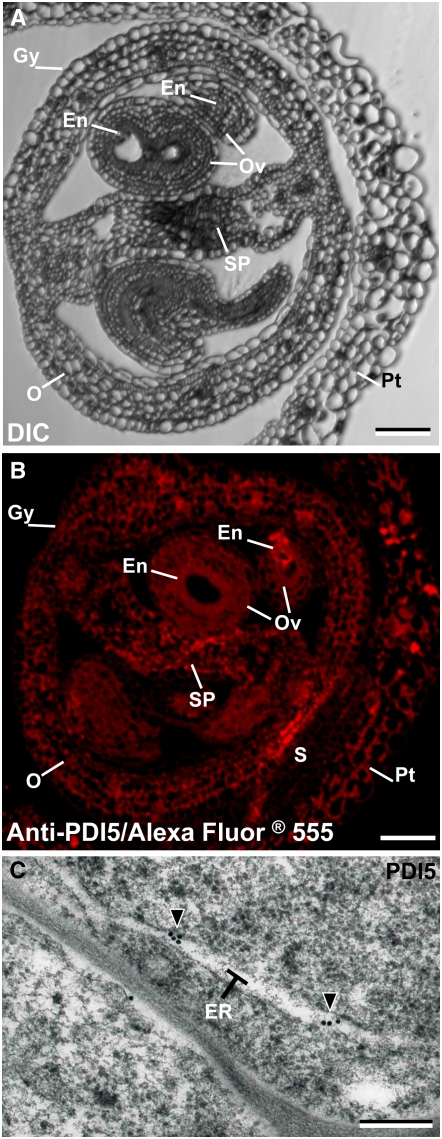
To characterize the expression of PDI5 in young flowers (Figure 2B), we examined the distribution of PDI5 in florets using light, fluorescence, and electron microscopy. Figure 3A illustrates the tissues of a stage 12 Arabidopsis flower (Smyth et al., 1990; Bowman et al., 1991), including the gynoecia organ, which encompasses the developing ovules, the septum, the ovary, and the petals. Immunofluorescence labeling with the polyclonal PDI5 antiserum (Figure 3B) revealed a weak general fluorescence in all gynoecial tissues, including the ovules, the septum, and the ovary. Little labeling was seen in the petals. In electron micrographs of immunolabeled endothelial cells within the ovules, virtually all of the anti-PDI5 labeling was seen over ER cisternae (Figure 3C), consistent with the presence of a predicted KDEL ER retention motif and consistent with the classic localization of PDI in mammals and plants. Immunocytochemistry controls for the specificity of the anti-PDI5 antiserum are shown in Supplemental Figure 3 online. In the absence of the specific antiserum, there was no immunofluorescence or immunogold labeling of endothelial cell sections.
Figure 3.
Localization of PDI5 in Immature Flowers.
(A) to (C) Light, fluorescence, and electron microscopy images of transverse sections of immature florets of the wild-type plant. Gy, gynoecia; O, ovary; En, endothelial cells; Ov, ovule; Pt, petal; SP, septum; S, sepal. Bars = 100 μm in (A) and (B) and 0.2 μm in (C).
(A) Differential interference contrast (DIC) micrograph illustrates the tissues and cellular organization of an early stage 12 Arabidopsis flower. The 12-stage floret contains the gynoecia organ, which encompasses the developing ovules, the septum, and the ovary. Petals surround all gynoecial tissues.
(B) Immunofluorescence labeling of a cross-sectioned floret labeled with anti-PDI5 antibody-Alexa Fluor 555. Petals and the gynoecia organs, such as ovule, endothelium, septum, and ovary, are labeled. Hardly any labeling was seen over the sepals.
(C) Electron micrograph of an ovule gynoecial cell immunolabeled with anti-PDI5 antibody. The gold label is seen over ER cisternae of the endothelial cells of the developing ovule.
During Embryo Development, PDI5 Is Expressed Exclusively in Endothelial Cells before Their PCD
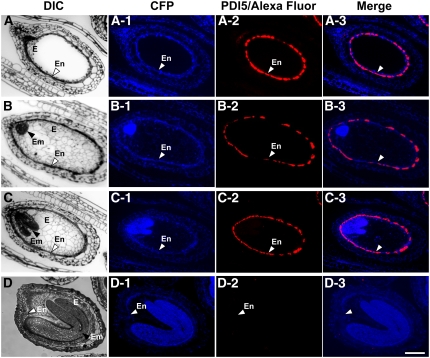
We used light and immunofluorescence microscopy to investigate the cellular distribution of PDI5 in longitudinal sections of developing seeds (silique stages 16 to 19; Smyth et al., 1990) and to correlate the expression of PDI5 in the differentiating cells with the following stages of embryo development: the pre-embryo stage (Figure 4A), the globular stage (Figure 4B), the heart stage (Figure 4C), and the early-bent cotyledon stage (Figure 4D).
Figure 4.
Fluorescent Micrographs Showing the Confinement of PDI5 Expression to the Endothelium in Developing Seeds.
(A) to (D) DIC micrographs of developing seeds. Pre-embryo stage ([A] to [A-3]), globular stage ([B] to [B3]), heart torpedo stage ([C] to [C3]), and early-bent cotyledon stage ([D] to [D3]). The endothelium (En) corresponds to the innermost layer of the integument and defines the interface between endosperm (E) and the future seed coat. In (B) to (D), black arrowheads point to the embryos (Em) at different stages of development. Bar = 100 μm for (A) to (D-3).
(A-1) to (D-1) Autofluorescence micrographs of developing seeds viewed using a cyan fluorescent protein (CFP) filter set.
(A-2) to (D-2) Immunofluorescence micrographs of developing seeds stained with Alexa Fluor 555 and labeled with anti-PDI5 antibodies. White arrowheads denote the endothelial cell layer.
(A-3) to (D-3) Merged images of anti-PDI5 antibody-Alexa Fluor and cyan fluorescent protein.
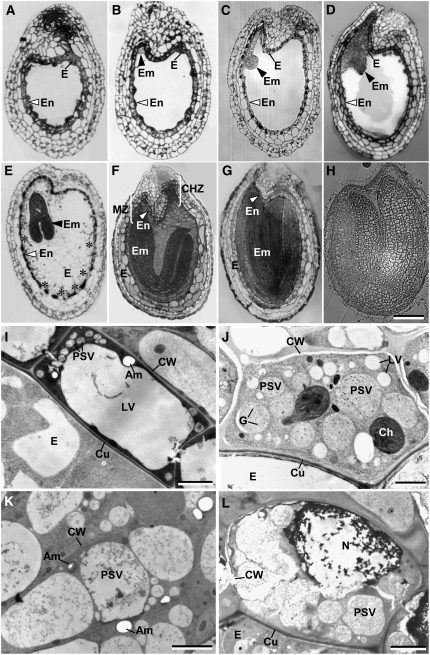
The cell layer that is immunolabeled in Figures 4A to 4C is referred to as the endothelium or endothelial layer in this article. This name has a long history (Esau, 1977) and is used extensively in recent literature (e.g., Schneitz et al., 1995; Otegui and Staehelin, 2000; Beisson et al., 2007). However, it is not universally accepted. Other names given to this cell layer include pigment layer (Bouman, 1975), integumentary tapetum (Kapil and Tiwari, 1978), inner epidermis (Fahn, 1990), endothecium (Kuang et al., 1995), inner integument layer 1 (Beeckman et al., 2000), and L5 ovule integument layer (Beisson et al., 2007). Characteristic features of the endothelial layer include direct contact with the endosperm cells of the embryo sac, a cuticular layer on the cell wall facing the endosperm, and a dense cytoplasm that stains intensely with several histochemical stains (Figures 4 and 5).
Figure 5.
Structural Changes within Endothelial Cells and in the Endothelial Cell Layer during Seed Development.
(A) to (H) Longitudinal sections through developing seeds of wild-type Arabidopsis, illustrating the stages of embryo (Em) and endosperm (E) development and the organization of the endothelial cell layer (En).
(A) Pre-embryo stage. The endothelial cell layer surrounding the embryo sac is completed before embryo growth commences.
(B) Two-celled embryo stage, with a prominent endothelial layer.
(C) and (D) Globular embryo (C) and heart stages (D). The endothelial cell layer begins to thin as the embryo enlarges.
(E) Torpedo embryo stage. The endothelial cell layer starts to break down (asterisks) as the cellular endosperm expands.
(F) and (G) Early (F) and late (G) bent-cotyledon stages. Chalaza (CHZ) and micropilar (MZ) zones are shown.
(H) Coated seed.
(I) to (L) Electron micrographs of longitudinally sectioned endothelial cells at different stages of embryonic development.
(I) Pre-embryo stage endothelial cell (see [A]). The cytoplasm exhibits a normal cellular organization with a large central LV, numerous small PSVs, and an amyloplast (AM) and is surrounded by a cell wall (CW) with a cuticular layer (Cu).
(J) Heart stage endothelial cell (see [D]). Some PSVs are greatly enlarged and contain many electron-dense globoids (G). By contrast, the LVs are much smaller but more numerous than in (I).
(K) Torpedo stage endothelial cell around the micropylar zone (see [E] and [F]). The large PSVs appear to be breaking down.
(L) Bent-cotyledon stage (see [F] and [G]). The process of degradation encompasses the nucleus (N), organelles, and cell wall.
Bars = 100 μm in (A) to (H), 3.00 μm in (I) and (K), 2.00 μm in (J), and 5.00 μm in (L).
The endothelial cell layer is most prominent during the pre-embryo stage and begins to thin thereafter, becoming nearly undetectable in the early-bent cotyledon stage. The autofluorescence of the tissue slices is illustrated in Figures 4A-1 to 4D-1. Accumulation of the PDI5 protein was specific to the endothelial cell layer (Figures 4A-2 to 4C-2), being abundant in the pre-embryo stage ovule (Figure 4A-2) and decreasing thereafter as this layer thinned and underwent PCD (Figures 4B-2 to 4C-2). By the early-bent cotyledon stage, all immunolabeling had disappeared (Figure 4D-2). The merged fluorescence images show the spatial relationship between the immunolabeling pattern of the endothelial layer and the developmental stage of the surrounding tissues (Figures 4A-3 to 4D-3). These findings demonstrated that PDI5 accumulated exclusively in endothelial cells from the pre-embryo up to the heart stage and disappeared when the endothelial cells underwent PCD in the early-bent cotyledon stage. The control experiments involved omission of the primary anti-PDI5 antiserum and/or incubation with the pre-immune serum to test for any cross-reaction of the secondary antibody (AF 555) (see Supplemental Figures 3A to 3D online) and for any nonspecific binding of the primary antibody (see Supplemental Figures 3E to 3H online). All controls consistently produced negative results.
Expression of PDI5 in Endothelial Cells Precedes Vacuolar Changes and the Onset of PCD
To better understand the cellular changes that parallel the decrease in PDI5 expression, we correlated stages of embryo development with modifications in endothelial cell architecture (Figures 5A to 5L). During the pre-embryo and two-cell stages (Figures 5A and 5B), the endothelial cells (Figure 5I) display a typical plant cell architecture with a large central LV and a thin layer of cortical cytoplasm containing many smaller PSVs, amyloplasts, and other cellular organelles. As the embryo develops into the globular and heart stages (Figures 5C and 5D), the PSVs fuse and expand, while the lytic vacuoles become smaller and fragment (Figure 5J). By the time this transformation is complete, the swollen PSVs occupy more than half of the cellular volume (Figure 5K). In the heart-torpedo stage (Figure 5E), the PSVs continue to enlarge and the cytoplasm decreases in size (Figure 5K). Finally, at the bent-cotyledon stage (Figures 5F and 5G), the nucleus, vacuoles, and cytoplasm degenerate and the surrounding tissues envelope the endothelial cells (Figure 5G). As the cellular endosperm expands, the endothelial cell layer degenerates from the central zone (Figure 5E, asterisks) through the chalaza to the micropyle (Figure 5F).
Identification of the PSVs during this transformation involved immunolabeling with anti-δ-TIP antibodies (Figure 6E) and antistorage protein 2S albumin antibodies (see Supplemental Figure 4A online), both of which are markers for PSVs. Unexpectedly, storage protein 2S albumins were also localized in the endothelial cell walls (see Supplemental Figures 4A and 4B online) but not in the walls of surrounding cells or embryonic cells (see Supplemental Figures 4B and 4C online). Control experiments with omission of the primary anti-2S antibody showed no labeling of PSVs (see Supplemental Figures 3J and 3K online) or endothelial cell walls (see Supplemental Figure 4D online). These experiments demonstrate that the endothelial cells exhibit many features of cells undergoing PCD.
Figure 6.
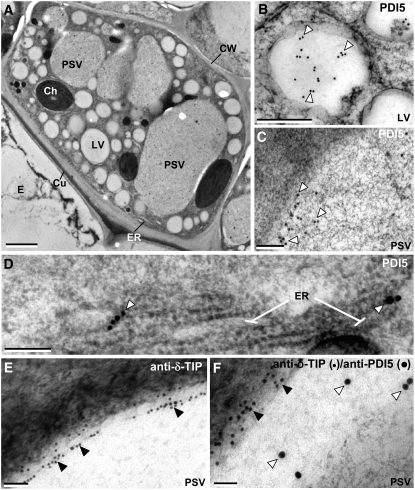
Characterization of Vacuole Types in Senescing Endothelial Cells by Means of Labeling with Anti-PDI5 and Anti-δ-TIP Antibodies.
(A) Electron micrograph of a heart stage endothelial cell. The PSVs are larger than the LVs. Ch, chloroplast; Cu, cuticle; CW, cell wall.
(B) and (C) Immunolocalization of PDI5 (open arrowheads) in an LV (B) and a PSV (C).
(D) Immunolocalization of PDI5 in ER cisternae.
(E) Immunolocalization of δ-TIP (filled arrowheads) in the tonoplast membrane of a PSV.
(F) Double immunolabeling of a PSV with anti-PDI5 (15 nm gold) and anti-δ-TIP (10 nm gold) antibodies.
Bars = 2.00 μm in (A), 0.4 μm in (B), 0.5 μm in (C), 0.2 μm in (D) and (F), and 0.1 μm in (E).
PDI5 Is Found in LVs and PSVs and Preferentially Accumulates in PSVs of Endothelial Cells Undergoing PCD
Sections of developing Arabidopsis seeds were immunolabeled with the PDI5 antiserum. In heart-stage embryos, the endothelial cells exhibited a normal complement of cellular organelles in addition to the swollen PSVs, which were larger than the LVs (Figure 6A). Anti-PDI5 antibody gold labeling was seen in LVs (Figure 6B), PSVs (Figure 6C), and ER cisternae (Figure 6D). To confirm that PDI5 is present in both types of vacuoles, we also labeled sections with the PSV membrane marker, anti-δ-TIP (for Tonoplast Intrinsic Protein) antibody (Jauh et al., 1999), both alone (Figure 6E) and after colabeling with anti-PDI5 antibodies (Figure 6F). The δ-TIP antibodies produced intense labeling over the membrane of PSVs (Figure 6E), and double-labeling experiments demonstrated that those vacuoles also contained PDI5 (Figure 6F). A low level of anti-PDI5 antibody labeling was present over LVs and PSVs of other embryonic cells (see Supplemental Figure 5A online). While the number of LVs is greater than the number of PSVs, the cross-sectional area (μm2) of the PSVs is ∼40-fold greater. The density of anti-PDI5 labeling over endothelial cell vacuoles (LVs and PSVs) was also five times higher than over the vacuoles of embryonic cells. These findings indicate that PDI5 is more highly expressed in heart-stage endothelial cells than in other types of cells in developing seeds and that, within the former, the bulk of PDI5 accumulates in PSVs.
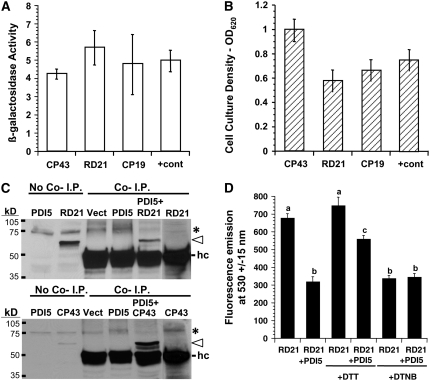
PDI5 Interacts with Three Different Cys Proteases and Inhibits Recombinant RD21 Protease Activity in Vitro
To elucidate the function of PDI5 in endothelial cell vacuoles, we screened two Arabidopsis cDNA libraries for proteins interacting with PDI5 bait using the yeast two-hybrid system. The principal isolates were clones encoding the following three Cys proteases: RD21 (NM_103612), CP43 (NM_123672), and CP19 (NM_112826), which represented 66, 31, and 3.8% of the total recombinants isolated, respectively. Three different validation tests were performed to verify the interaction specificity. β-Galactosidase (β-gal) activities of the reporter gene (Figure 7A), which were based on an equal cell culture density (Figure 7B), represent the degree of interaction between the Cys proteases and PDI5. Cys protease RD21 had the highest level of β-gal activity, but each Cys protease had β-gal levels similar to the positive control. Growth on histidine dropout medium (Kaiser et al., 1994) was also similar to the positive control (see Supplemental Figure 6 online), indicating a solid interaction. Cys protease RD21 has been found in the vacuoles of senescing Arabidopsis leaves (Yamada et al., 2001). We conducted coimmunoprecipitation assays on the two most abundant recombinant Cys proteases (RD21 and CP43) isolated (Figure 7C). The PDI5 antiserum coimmunoprecipitated with RD21 and CP43 only in the presence of recombinant PDI5 (Figure 7C). To determine the nature of the interaction, a Cys protease assay was performed in vitro with recombinant PDI5 and RD21 (Figure 7D). The presence of an equivalent amount of PDI5 inhibits the Cys protease RD21 by >50%. The dithiol reducing agent, DTT, partially disrupts the inhibition, whereas the dithiol oxidizing agent, dithionitrobenzene (DTNB), mimics the inhibition.
Figure 7.
Validation of Protein Interaction with PDI5 Bait in the Yeast Two-Hybrid Assay and of in Vitro Inhibition of RD21 Protease Activity.
(A) Standard β-gal activity measurements (OD570) were conducted according to (Serebriiskii and Golemis, 2000) and normalized to an equal culture density of yeast cells coexpressing PDI5 bait plus one of the interacting Cys proteases (43 [NM_123672], RD21A [NM_103612], or 19 [NM_112826]). Values of two negative controls (coexpression of Cys protease with empty bait vector; PDI5 bait with empty prey vector) were subtracted from the values of Cys proteases coexpressed with PDI5 bait and positive control.
(B) Yeast cell culture densities (OD620) after 2 weeks of growth on His dropout medium with 1 mM 3-AT for each Cys protease coexpressed in the PDI5 bait cell line. The positive control (+cont) consisted of documented interacting partners (Epsin 1 is an interacting partner for the EH domain-containing region of Eps15) as described (Serebriiskii and Golemis, 2000). Values in (A) and (B) represent two experiments done in three replicates (mean ± sd).
(C) Immunoblot of coimmunoprecipitation (Co-I.P.) products (Cys proteases RD21, top panel, or CP43, bottom panel). Open arrowhead indicates coimmunoprecipitated Cys protease (RD21 or CP43) using the anti-PDI5 antiserum. Recombinant RD21 and CP43 were detected with anti-HA tag antiserum. The left two lanes in each panel were controls for no coimmunoprecipiation (no Co-I.P.), in which 40 μg total E. coli protein containing recombinant PDI5, RD21, or CP43 was directly loaded on the gel. The heavy chain (hc) of the anti-PDI5 antisera used in the coimmunoprecipitation is shown in the Co-I.P. lanes. The band in the no Co-I.P. lane labeled RD21 refers to the recombinant RD21 protein that was a positive control for the antiserum used on the immunoblot. No HC protein was detected in this lane. Asterisk denotes a nonspecific background band present in all lanes.
(D) In vitro RD21 Cys protease assay reveals that PDI5 inhibits RD21 activity. Recombinant RD21 activity was measured in the presence and absence of an equivalent amount of PDI5. Addition of 5 mM DTT or DTNB prior to adding PDI5 is noted. The y axis units are fluorescence emission at 530 ± 15 nm. The averages were derived from a sample size of three in six independent experiments (18 total averaged), and the error bars are sd. The a, b, and c refer to significantly different means (P < 0.05) as determined by analysis of variance.
PDI5 Traffics with Cys Proteases from the ER through the Golgi to Vacuoles in Senescing Endothelial Cells
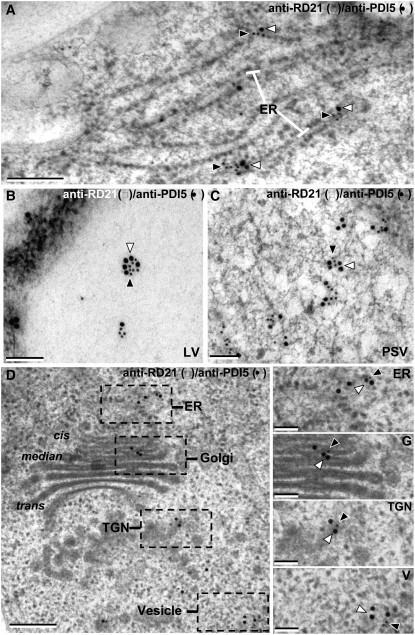
To test the hypothesis that Cys proteases associate with PDI5 during trafficking to vacuoles, we analyzed the subcellular distributions of PDI5 and Cys protease, RD21, in endothelial cells by means of double immunolabeling techniques. PDI5 and RD21 colocalized in ER cisternae (Figures 8A and 8D), LVs (Figure 8B), PSVs (Figure 8C), the Golgi, trans-Golgi network (TGN), and vesicles (Figure 8D) of endothelial cells. Dual immunolocalization showed that PDI5 often surrounded RD21. Quantitative analysis of the colabeling experiments demonstrated that on a per μm2 area of vacuole, the LVs and PSVs contain similar amounts and ratios of anti-PDI5 and anti-RD21 binding sites (Figure 9). Taken together, the yeast two-hybrid, the coimmunoprecipitation, the in vitro Cys protease assay, and the colabeling experiments demonstrate that PDI5 associates with RD21 and that the two proteins traffic together from the ER through the Golgi to LVs and PSVs before the initiation of PCD of endothelial cells. All immunogold-labeling controls involved omission of the primary antibodies and/or substitution with the preimmune serum and gave negative results (see Supplemental Figures 3I to 3M online).
Figure 8.
Coimmunolocalization of Cys Protease RD21 with PDI5 in Senescing Endothelial Cells.
(A) Colocalization of anti-PDI5 (15 nm gold, open arrowheads) and anti-RD21 (10 nm gold, filled arrowheads) within ER-cisternae (ER).
(B) Colocalization of anti-PDI5 (15 nm gold) and anti-RD21 (10 nm gold) antibodies in the LV.
(C) Colocalization of anti-PDI5 (15 nm gold) and anti-RD21 (10 nm gold) antibodies in the PSV.
(D) Colocalization of anti-PDI5 (15 nm gold) and anti-RD21 (10 nm gold) antibodies in the Golgi apparatus and TGN.
Bars = 0.2 μm in (A), 0.1 μm in (B) and (C), 0.2 μm in (D), and 0.15 μm in the (ER), (G), (TGN), and (V) panels.
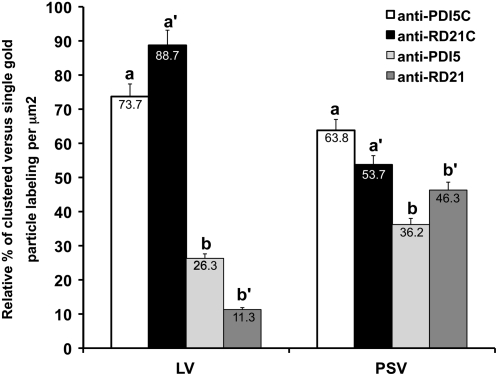
Figure 9.
Quantitative Analysis of the Coimmunogold Labeling of Anti-PDI5 with Anti-RD21 Antibodies in LV and PSV of Senescing Endothelial Cells.
Quantification of PDI5 and RD21 clusters (anti-PDI5C with anti-RD21C) and single (anti-PDI5 and anti-RD21) gold particles in LVs and PSVs of endothelial cells (n = 10 cells). Anti-PDI5- and anti-RD21-gold particles colocalize (a and a′) more frequently in LV than in PSV. The anti-RD21/anti-PDI5 labeling ratio is 1.2 over LVs. The number of gold-labeled clusters of anti-PDI5C with anti-RD21C (a and a′) in endothelial cell vacuoles (LVs and PSVs) was higher than the number of single gold particles in both vacuoles (b and b′).
The pdi5Δ T-DNA Insertion Mutant Affects Seed Set and Seed Viability
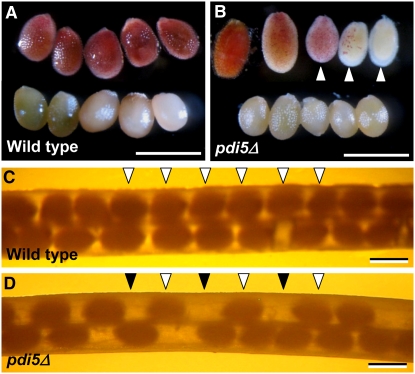
To assess the function of PDI5 on seed development, we used the homozygous pdi5Δ null mutant (Figure 1A) that lacks PDI5 protein (Figure 2B) and PDI5 mRNA (see Supplemental Figure 1C online). The pdi5Δ mutant had siliques with fewer seeds than the wild type, and clear gaps were evident in the seed rows where seed development was aborted (Figures 10C and 10D). The number of aborted seeds varied greatly between siliques. The viability of the developing wild-type and mutant seeds was evaluated using the tetrazolium-based embryo viability test (Boisson et al., 2001). All wild-type seeds (Figure 10A, top) exhibited a uniform red color, indicating viability, whereas the red staining of the mutant seeds varied from a deep orange red color to no staining, indicating that only a fraction of the embryos were viable (Figure 10B, top). These findings suggest that the PDI5 knockout mutation reduces seed set and embryo viability.
Figure 10.
The pdi5Δ Knockout Mutation Affects Seed Viability and Seed Set
(A) and (B) Tetrazolium-based embryo viability test. Bar = 500 μm.
(A) Top row: wild-type embryos show a uniform red color (viable seeds). Bottom row: wild-type embryos boiled for 30 min are light pink (negative control).
(B) Top row: pdi5Δ embryos show variable degrees of red and pink coloring (white arrowheads). Bottom row: boiled mutant embryos are light pink.
(C) and (D) Seed set in wild-type (C) and pdi5Δ mutant (D) siliques. Note the pdi5Δ mutant silique contains fewer seeds (open arrowheads) and gaps (filled arrowheads) (D) than the wild type (open arrowheads). Bars = 1 mm.
DISCUSSION
In developing plant seeds, the endothelial cells have been postulated to serve two functions: transport of nutrients to the embryonic tissues (Kapil and Tiwari, 1978) and, after PCD, participation in the formation of the separation layer between the embryo and the seed coat to allow facile abscission of the coat during germination. Here, we present a mechanism for the regulation of PCD in endothelial cells, whereby PDI5 chaperones and inhibits PCD-related Cys proteases as they traffic from the ER via the Golgi to vacuoles before the proteases are activated.
PDI5 Produced in Endothelial Cells Binds Specifically to PCD-Associated Cys Proteases to Prevent Their Premature Activation during Embryogenesis
A central question of the PCD pathway is how lytic enzymes accumulate without disrupting the biosynthetic apparatus. In plants, Cys proteases are transported to vacuoles and the apoplasm. Some proteases can be processed and converted self-catalytically (and via secondary proteases) from inactive proenzymes to active forms (Martins and Earnshaw, 1997; Beers et al., 2000), or the protease active site can be inhibited by tightly binding cystatin or serpin inhibitors during transport (Solomon et al., 1999; Martínez et al., 2005; Vercammen et al., 2006). In endothelial cells, PDI5 acts as a redox-modifying chaperone that associates with and inhibits Cys proteases to protect Golgi, TGN, and vacuolar membranes from degradation. The abrupt decrease in PDI5 expression and resulting loss of PDI5 coincides with the proteolysis of endothelial cells, in which PDI5 is normally and specifically expressed. Genetically disrupting PDI5 function prematurely hastens the demise of endothelial cells, leading to a diminished embryo viability phenotype (Figures 10B and 10D), which highlights the importance of PDI5-modulated PCD in viable embryogenesis. The partial phenotype in the null pdi5 mutant is readily explained by genetic redundancy; the PDI6 gene, which also resides on chromosome 1, is the most closely related PDI in the Arabidopsis genome. The amino acid sequences of PDI5 and PDI6 are 79% identical to each other (Lu and Christopher, 2008). Therefore, PDI6 could possibly partially, but not completely, compensate for the loss of PDI5.
The association of PDI5 with the Cys proteases CP43 and RD21 was substantial and highly specific. The RD21 and CP43 cDNAs were reproducibly isolated at high frequency by the yeast two-hybrid screen using PDI5 as bait, and their gene products were shown to be associated by means of PDI5 coimmunoprecipitation (Figure 7) and colocalization via immunoelectron microscopy (Figure 8). Numerous other Cys proteases are expressed in the tissues used to make the libraries, but RD21 and CP43 constituted >97% of the clones recovered, passing multiple verification screens. Together, the results support a model for PDI5 specifically binding and quenching the activity of the cognate partner Cys proteases, CP43 and RD21, prior to PCD (Figures 7 and 8).
RD21 and CP43 lack the canonical active sites and conserved features typical of caspases and metacaspases that are involved in PCD in other cells and species (Suarez et al., 2004; Bozhkov et al., 2005). They do contain a redox-sensitive active site, GxCGSCW, comprised of two Cys residues capable of forming a vicinal dithiol bond (Balmer et al., 2003). This site is sensitive to disulfide-specific modifying reagents, such as the reducing agent, DTT, and the oxidizer, DTNB. We provide evidence for a disulfide redox mechanism regulating the in vitro activity of Cys proteases in response to recombinant PDI5; a reducing environment stimulated protease activity in the presence of inhibitory concentrations of PDI5, whereas an oxidizing environment decreased protease activity. In fact, oxidation mimicked the inhibitory effects of recombinant PDI5 on Cys protease activity (Figure 7D). The ability of reducing activity to counteract the oxidizing effect is typical of a mechanism mediated by a thioredoxin-based protein–protein interaction (Balmer et al., 2003).
PDI5 Escorts PCD-Related Cys Proteases to Vacuoles and Regulates PCD Activation
PDI5 is a member of the superfamily of thiodisulfide oxidoreductases. PDIs typically reside in the lumen of the ER, where they catalyze protein folding via the oxidation, reduction, and isomerization of disulfide bonds of most secretory proteins (Freedman et al., 1994; Tu et al., 2000; Wedemeyer et al., 2000; Wilkinson and Gilbert, 2004). PDI5 deviates from this traditional localization. The PDI5 gene is specifically expressed in discrete floral and developing seed tissues (Figure 3B). After synthesis in the ER, PDI5 traffics together with Cys proteases from the ER to the Golgi, TGN, LVs, and, most notably, PSVs even though it possesses a KDEL ER retention signal (Figure 8). As discussed below, these trafficking patterns are neither unique nor incompatible with the role of KDEL mediated ER retention. To demonstrate that the presence of PDI5 and of the Cys protease RD21 in the Golgi, TGN, and vacuole compartments is not simply due to protein overflow from the ER, we mapped the distribution of the ER-resident protein BiP in endothelial cells (see Supplemental Figure 7 online). As seen in that figure, BiP is confined to ER cisternae (81% of gold particles) and to cis-Golgi cisternae (16% of gold particles), from which escaped ER proteins are recycled back to the ER, with only 3% of the label seen over median/trans/TGN cisternae and vacuoles. Thus, the presence of PDI5 and RD21 in Golgi, TGN cisternae, and vacuole compartments appears to reflect a controlled release from the ER and transport through the Golgi to the vacuoles.
In mammals and yeast, PDI isoforms have also been localized to mitochondria and the nucleus (Turano et al., 2002). In plants and algae, PDIs are also present in chloroplasts (Kim and Mayfield, 1997; Levitan et al., 2005; Lu and Christopher, 2006; Shimada et al., 2007). The Chlamydomonas PDI-like protein (RB60) contains a C-terminal signal for ER retention. However, RB60 undergoes dual targeting to the chloroplast and the ER (Levitan et al., 2005). In rat exocrine pancreatic cells, a PDI isoform containing a KDEL signal has also been shown to traffic from the ER through the secretory pathway to the plasma membrane (Yoshimori et al., 1990). Here, we offer one suggestion for why PDI5 is capable of trafficking to PSVs despite its KDEL signal: this signal may help retain noncomplexed PDI5 monomers in the ER, but after binding to a Cys protease (which lacks a KDEL signal), the KDEL signal of PDI5 could become nonfunctional (e.g., masked), thereby enabling the complexed molecules to leave the ER and travel to a PSV. Although it has been well documented in yeast and mammalian cells that PDIs can act as protein folding chaperones, the previous evidence for a chaperone role of PDIs in plants has been limited to the maturation, folding, and packing of seed storage proteins (Shimoni et al., 1995; Li and Larkins 1996; Li et al., 2006). Direct identification of interacting partners and the dual functions of PDI5, both as a chaperone and as a redox-sensitive protease regulator, have not, to our knowledge, been described to date for any member of the PDI family in Arabidopsis.
Vacuolar enlargement and collapse, releasing enzymes into the cytosol that attack organelles and DNA, is a unifying theme of both developmentally controlled and pathogen-induced PCD in plants (Lam, 2005) and in autophagosomes of gymnosperms (Smertenko et al., 2003). Vacuolation of the cytosol as described here differs from apoptotic cell death in animals, which involves phagocytosis, caspase-directed proteolysis, and DNA laddering. The PDI5-induced silencing of noncaspase-type Cys proteases (Figure 7B), until PDI5 levels diminish in the vacuoles (Figures 4D and 8), accentuates the pivotal role that vacuoles play in plant PCD.
In Endothelial Cells, the PSV Becomes the de Facto LV That Mediates PCD
Plants employ three different mechanisms to bring about PCD (Fukuda, 2000; Jones, 2001; Kuriyama and Fukuda, 2002): In leaf and root tissues in which PCD is induced by environmental stress conditions, the nucleus is degraded prior to the breakdown of the cytoplasm and produces DNA laddering in a process that is called apoptosis-like PCD (Ryerson and Heath, 1996; Katsuhara, 1997). During leaf senescence, the chloroplasts are degraded first, followed by nuclear and vacuolar breakdown (Thomas et al., 2003; Otegui et al., 2005). In tracheary cells and during leaf perforation in the lace plant, PCD starts with the breakdown of the large central vacuole, which releases proteases and nucleases to bring about cell degradation (Groover et al., 1997; Obara et al., 2001; Kuriyama and Fukuda, 2002; Gunawardena et al., 2004). As demonstrated in this study, endothelial cells also follow this third pathway (Figures 5I to 5L), but because of differences in cellular architecture, the mechanistic details are somewhat different.
Unlike tracheary cells, which only have a lytic vacuolar system, endothelial cells contain both LVs and PSVs. As demonstrated in Figures 5 and 6, the Cys proteases involved in PCD accumulate in both LVs and PSVs, but more of these enzymes are delivered to the latter even though the LVs initially occupy a larger cell volume than the PSVs. However, shortly after the proteases begin to arrive in the vacuoles, the LVs begin to shrink while the volume of the PSVs is greatly increased. By the time of vacuolar breakdown, the PSV occupies >50% of the cell volume, and the LVs are hardy visible (Figures 5J and 6A).
Plant vacuoles are highly dynamic organelles that can undergo rapid changes in form and volume. The best-studied dynamics relate to the changes in vacuolar architecture associated with the cell cycle (Marty, 1999; Kutsuna et al., 2003; Seguí-Simarro and Staehelin, 2006). For example, in Arabidopsis apical meristem cells, as the cells progress from prometaphase to early telophase, the lytic vacuolar system becomes highly tubular and fragments, and its surface area decreases by ∼50% and its volume by ∼80%. Recovery of the interphase shape and volume occurs in 20 min (Seguí-Simarro and Staehelin, 2006). Thus, the size and shape changes of the vacuoles of endothelial cells preceding PCD are not unusual for plant cells.
What causes the LVs to shrink, and what causes the PSVs to expand during the period leading up to endothelial cell PCD? We postulate that the preferential delivery of PCD-related Cys proteases to PSVs rather than to the LVs (Figures 8 and 9) is responsible for bringing about these vacuolar changes. This biased delivery ensures that all storage proteins are rapidly degraded and the released amino acids can be exported to the surrounding cells prior to cell death. At the same time, the release of amino acids from the storage proteins increases the osmotic potential of the PSVs, thereby drawing water from the LVs to the PSVs and causing protein storage vacuole swelling. Thus, through these processes, the large PSVs become the de facto large LVs of endothelial cells prior to the onset of PCD.
METHODS
Plant Materials, Growth Conditions, and Seed Viability
Seedlings and plants from Arabidopsis thaliana ecotype Columbia (Col-0) wild-type and SALK mutant lines were used for the experiments. T-DNA SALK insertion lines (SALK_010645, SALK_015253, and SALK_136642, http://signal.salk.edu; seeds provided by the ABRC) were identified via PCR of Arabidopsis genomic DNA. The homozygous T-DNA line (SALK_010645) was verified by PCR analysis of genomic DNA of selfed plants using gene-specific PDI5 primers (5′-ATCTTTGAGGCTTTGGGGAAT-3′ and 5′-CTCCGTCTCTTCGCTTCTGAT-3′) and a T-DNA left border primer, LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) as described by Stepanova and Alonso (2003). The accurate insertion positions were determined by sequencing each PCR product. Wild-type and homozygous pdi5Δ mutant seeds were surface-sterilized and then grown on soil (Farfard Super Fine germinating mix; American Clay Works and Supply Company) under 16-h-light/8-h-dark cycles at 19 to 21°C for 4 to 6 weeks and on solid Murashige-Skoog (Gibco) media supplemented with 2% (w/v) sucrose and 0.8% (w/v) phytagar (Gibco BRL).
For seed viability tests, developing embryos were removed from siliques and soaked in 1% 2,3,5-triphenyltetrazolium-chloride solution (Sigma-Aldrich). Samples were incubated for 1 d in the dark at 37°C (Boisson et al., 2001), and photographs of siliques were recorded with a LEICA M212 stereomicroscope.
RT-PCR
Total RNA was treated with RNase-free DNase (Promega) and used to produce the first-strand cDNA with 1.0 units of M-MLV reverse transcriptase and oligo(dT) (Promega). Fifty nanograms of total RNA was used to amplify the transcripts of PDI5 and actin to produce first-strand cDNA. The first-strand cDNAs were mixed with 1× final concentration of reaction buffer (Bio-X-Act Short Mix; Bioline) and 50 ng primers, and PCR was conducted for 30 cycles. The number of cycles to generate a linear range of DNA amplification was previously determined for all PDI genes (Lu and Christopher, 2008). PCR products were visualized by staining with ethidium bromide after agarose gel electrophoresis. The gene-specific primers used in the study corresponded to exon-exon junctions to avoid amplification of genomic DNA. RT-PCR primers for PDI5 were forward, 5′CTCGTGAAGCTGAGGGTATTG-3′, and reverse, 5′-AAGATTGGAGCAAGCTTTTGG-3′. Primers for Actin2 were forward, 5′-TTGCAGGAGATGATGCTCCCAGG-3′, and reverse, 5′-CATTCCCACAAACGAGGGCTGG-3′.
Antibodies, Protein Extraction, SDS-PAGE, and Immunoblot Analysis
Polyclonal antibodies were raised against a unique synthetic peptide (New England Peptide) corresponding to amino acid residues 456 to 468 of PDI5. Affinity-purified antibody specificity was tested by immunoblot analysis and immunogold labeling of a recombinant PDI5 protein expressed in Escherichia coli cells. Flowers, leaves, siliques, and stems were collected from 4- to 6-week-old wild-type and homozygous pdi5Δ mutant plants grown on soil. Roots were collected from 4- to 5-week-old seedlings grown in Murashige-Skoog solid medium. Immature seeds were collected from the developing siliques of 6- to 8-week-old plants. Mature seeds were collected from mature siliques. Frozen tissue was ground with a mortar and pestle in liquid nitrogen and homogenized with extraction buffer containing 50 mM Tris-HCl, pH 8.0, 0.25 M sucrose, 2 mM DTT, 2 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail (Sigma-Aldrich). Protein concentrations were determined with a Bio-Rad kit. Proteins were resolved by SDS-PAGE on 10% polyacrylamide gels and electroblotted onto nitrocellulose Hybond-ECL (Amersham Biosciences) using a semidry transfer technique. Protein blots were incubated with the polyclonal anti-PDI5 antiserum at 1:1000 dilution, and conjugates were detected using the ECL anti-rabbit IgG linked to horseradish peroxidase and the ECL chemiluminescence's kit (Amersham Biosciences).
Yeast Two-Hybrid Protein–Protein Interaction Screen
The coding cDNA for residues 21 through 497 of the 501 residues in the PDI5 protein (AAD41430) was inserted into the pBUTE vector (a kanamycin-resistant version of GAL4 bait vector pGBDUC1; James et al., 1996) as a translational fusion to the GAL4 DNA binding domain. The resulting PDI5 bait vector was sequenced to confirm an in-frame fusion, transformed into mating type A of strain PJ694, tested for autoactivation of the β-galactosidase reporter gene, and then used in a yeast two-hybrid screen (Fields and Song, 1989; Durfee et al., 1993) of two Arabidopsis cDNA libraries cloned as fusions to the GAL4 activation domain in pGADT7-rec (Clontech). One library was prepared from 3-d-old etiolated seedlings, and the second was a composite prepared from entire 3-week-old seedlings stressed by various hormones and osmotic and environmental treatments as described by Gingerich et al. (2005) and at the University of Wisconsin Molecular Interaction Facility (www.biotech.wisc.edu/mif/). Approximately 18 million clones were screened via mating at the University of Wisconsin–Madison Molecular Interaction Facility (www.biotech.wisc.edu/mif/). Putative interactors were identified by growth on histidine dropout medium plus 1 mM 3-amino-1,2,4-triazole and by β-galactosidase assays (described below). Negative controls included media only, empty prey (activation domain) vector (pGADC1; James et al., 1996), prey construct expressing mouse epsin, and prey construct expressing human Fbox5, whereas positive controls for the yeast two-hybrid system were two premated interacting pairs: EH:Epsin (Rosenthal et al., 1999). Following selection, 81 yeast wells tested positive (via selection on histidine dropout plus 1 mM 3AT and β-galactosidase assay) for interaction. From these, 62 representative prey plasmids were retransformed into the alpha mating type of PJ694 and validated in a parallel mating and selection assay with the PDI5 bait and the empty bait vector. Fifty-nine clones were positive (grew in interaction selection media and β-gal positive) in the validation test, 51 of which were identified via sequencing. β-Gal activity was assayed in yeast cultures lysed by the addition of YPER (Pierce Biotechnology) combined with chlorophenylred-β-d-galactopyranoside (Roche Diagnostics) as a substrate. Yeast was mated on yeast extract-peptone-dextrose medium plus adenine for 24 h at 30°C and then grown on complete supplemental medium (Q-BIOgene) minus Leu and Trp.
Coimmunoprecipitation of Cys Proteases (RD21 or CP43) with PDI5 Antisera
Equal quantities (200 μg) of recombinant PDI5 plus either recombinant RD21 or CP43 or total E. coli proteins [empty pET-25(+) vector] were mixed with 5 μg anti-PDI5 antiserum in 20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mM DTT and incubated for 2 h at 4°C. Thirty microliters of Protein-A Agarose beads (Invitrogen) was added, and the mixture was rotated (40 rpm) for 1 h at 4°C, centrifuged at 3000g for 10 min at 4°C, and washed three times with the above buffer. The pellet was resuspended in 25 μL of protein loading buffer, heated to 95°C for 5 min, and centrifuged for 30 s at 2000g. Supernatants were loaded onto a 10% polyacrylamide gel for SDS-PAGE. As controls, 40 μg recombinant PDI5, RD21, or CP43 were directly loaded on the gel. The proteins were transferred to nitrocellulose membranes and probed with a 1:750 dilution of anti-HA tag polyclonal primary antibody (BD Biosciences Clontech), and conjugates were detected using the ECL kit (GE Healthcare) as described above for immunoblotting.
Expression and Purification of Recombinant PDI5 and RD21
To construct the expression plasmids, the coding regions of RD21 and PDI5 cDNAs were amplified by RT-PCR from Arabidopsis (Col) wild-type mRNA. The forward and reverse primers of RD21 were 5′-GCCAAGCTTATGGGGTTCCTTAAGCCAACCA-3′ and 5′-GCACTCGAGGGCAATGTTCTTTCTGCCTTGTG-3′ (added HindIII and XhoI sites are underlined, respectively). The forward and reverse primers of PDI5 were 5′-AACGGATCCATGGCGATGAGGGGCTTCACG-3′ and 5′-GCGCTCGAGGAGCTCATCCTTGACTTCCTCA-3′ (added BamHI and XhoI sites are underlined). Each PCR product was digested, ligated into the expression vector pET25b+ (Novagen), and then transformed into DH5α cells for plasmid preparation and confirmation by sequencing. The recombinant proteins were expressed in BL21 (DE3) cells by growing cultures at 22°C for RD21 and 37°C for PDI5 with shaking at 250 rpm to OD600 = 0.6 and then induced with 0.5 mM isopropyl-d-thiogalactoside. Proteins were extracted, purified on Ni-NTA His-Bind columns as described by the manufacturer (Novagen), separated via 10% SDS-PAGE, analyzed by immunoblotting with anti-His-Tag antibodies, and detected by chemiluminescence using the ECL kit (Amersham Biosciences).
In Vitro Protease Assay
Protease activity was assayed using a fluorescence-based (BODiPY) EnzChek protease assay kit (Molecular Probes). Four micrograms of affinity-purified RD21 and/or PDI5 were preincubated at 4°C with and without 5 mM DTT or 5 mM DTNB before processing in the assay. Fluorescence was read in a fluorometer (Turner Bio Systems) with a fluorescein filter (excitation at 485 ± 12.5 nm and emission at 530 ± 15 nm). The reading for PDI5 alone was subtracted from the sample values as background. The assays were conducted in three individual experiments and the data analyzed via analysis of variance.
Light Microscopy
For light and immunofluorescence analysis, Arabidopsis developing flowers, siliques, and seeds were chemically fixed as previously described by Andème-Onzighi et al. (2002). Samples of developing seeds, siliques, and young 12-stage florets prior to bud opening (Smyth et al., 1990; Bowman et al., 1991) of the wild type and pdi5Δ mutant were vacuum-fixed overnight at 4°C with 1% (v/v) glutaraldehyde (EMS) and 2% (v/v) paraformaldehyde (EMS) in 0.1 M sodium phosphate buffered medium, pH 7.2. Samples were washed, dehydrated in ethanol, and embedded in LR White. The infiltrated samples were polymerized overnight at 60°C. LR White resin-embedded thin sections (0.5 to 1.5 μm) were stained with toluidine blue and observed on a Zeiss Axioplan microscope with DIC optics. Immunofluorescence labeling of flowers and developing seeds was performed as previously described (Andème-Onzighi et al., 2002), except that the secondary antibody anti-rabbit IgG was conjugated to Alexa Fluor 555 (Invitrogen, Molecular Probes). The stained flowers, siliques, or seeds were observed with a Zeiss Axioplan2 equipped with epifluorescence irradiation optics and standard filter sets for Alexa 555 (Lambda Technologies Laboratory Service). Immunofluorescence images were acquired with the Image Intelligent digital camera at the same magnification and time exposure. For further processing, the images were imported as TIFF files into Adobe Photoshop 7.0.1.
Electron Microscopy
For immunogold labeling analysis, flowers and developing seeds were preserved by high-pressure freezing/freeze substitution techniques (Otegui et al., 2006). Whole individual 6-week-old Arabidopsis 12-stage florets and developing seeds were placed into an aluminum hat filled with 150 mM sucrose. The samples were frozen in a BALTEC HPM-010 high-pressure freezer (Technotrade) and then transferred to liquid nitrogen for storage. Freeze substitution was performed either in 2% OsO4 for Epon embedding or in 0.1% uranyl acetate plus 0.25% glutaraldehyde in anhydrous acetone in cryo-vials (Nunc) for Lowicryl HM20 embedding for 7 d at −90°C, followed by slow warming to room temperature or −50°C, respectively, over a period of 1 d. After three rinses in acetone, samples were embedded in Epon (Ted Pella), resin/acetone 5, 10, 25, 50, 75, and 2 × 100% (24 h for each concentration), or in Lowicryl HM20 resin (EMS), 33% (24 h), 66% (24 h), and 100% resin (3 d). Polymerization was performed at 60°C for the Epon resin samples and at −50°C under UV light for 2 to 3 d in flat bottom embedding capsules for the Lowicryl HM20 resin samples.
For ultrastructural analysis of seeds, Epon resin-embedded sections were stained with uranyl actetate followed by lead citrate (Otegui et al., 2002). For immunolabeling, Lowicryl HM20 resin-embedded sections were placed on formvar-coated gold or nickel slot grids and blocked for 30 min with 3% (w/v) nonfat dried milk solution in 0.01 M phosphate-buffered saline, pH 7.2, containing 0.1% Tween 20 (PBST-milk). The sections were washed and then incubated with a 10-fold dilution of the following primary polyclonal antibodies (pAbs) for 2 h at room temperature: anti-PDI5, a vacuolar marker; anti-δ-TIP (Jauh et al., 1999), a protein storage marker; anti-2S albumin large chain (Scarafoni et al., 2001); and a Cys protease, anti-RD21 antibodies (Yamada et al., 2001). Sections were washed and transferred to a 25-fold dilution of secondary antibody goat anti-rabbit IgG-conjugated to 10- or 15-nm gold particles (Ted Pella) for 2 h at room temperature. After washing, samples were counterstained by uranyl acetate and lead citrate.
Coimmunogold labeling of anti-PDI5, anti-δ-TIP, anti-2S, and anti-RD21 antibodies in nucellus cells involved the following modifications of the techniques described above. Sections were blocked with PBST-milk for 30 min, washed, blotted dry, and incubated for 1.5 h with the pAbs (anti-δ-TIP or anti-2S or anti-RD21). Grids were washed with 0.5% PBST and then transferred to a droplet of secondary goat anti-rabbit IgG conjugated to 10-nm colloidal gold particles and diluted 25-fold in PBST-milk for 1.5 h. Sections were washed again, fixed with 0.5% (v/v) glutaraldehyde in PBS, pH 7.2, for 30 min, and washed again. Samples were then blocked in PBST-milk again and blotted dry, incubated with either no antisera or specific antisera for 1.5 h with a pAb anti-PDI5, washed, and transferred to a droplet of goat IgG anti-rabbit IgG conjugated directly to either 10- or 15-nm colloidal gold particles for 1.5 h. Sections were washed and then poststained with uranyl acetate and lead citrate. All observations were performed using a Philips CM10 microscope.
Quantitative Analysis of Anti-PDI5 and Anti-RD21 Gold Particle Labeling
The mean surface area (μm2) of the lytic and protein storage vacuoles was determined by stereological means (William, 1977; Seguí-Simarro and Staehelin, 2006). Five independent grids were analyzed, and from each grid 10 randomly chosen lytic and protein storage vacuoles were imaged. Labeling density of the anti-PDI5 or anti-RD21 antibodies was estimated on LVs and PSVs. The density of gold particle labeling was calculated as the average number of particles per μm2 of vacuoles. All controls involved omission of the primary antibody and/or incubation with the preimmune serum.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis Cys proteinase/thiol protease, putative 13 (CP13), At5g43060; Arabidopsis CP19, At3g19390; Arabidopsis CP28, At5g43060; Arabidopsis CP42, At3g19390; Arabidopsis CP43, At5g43060; Arabidopsis PDI5, At1g21750; Arabidopsis Cys protease responsive to dehydration/thiol protease 17 (RD21A17), At1g47128; Arabidopsis Cys protease responsive to dehydration/thiol protease 21 (RD21), At1g47128; Arabidopsis RD21A47, At1g47128; Arabidopsis RD21A52, At1g47128; Arabidopsis RD21A63, At1g47128; Arabidopsis NADH-ubiquinone reductase, At3g62790.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genetic Map of Arabidopsis PDI5 Gene and T-DNA Insertion Mutant Lines.
Supplemental Figure 2. Expression and Immunodetection of Recombinant PDI5 Protein in E. coli Cells.
Supplemental Figure 3. Immunological Controls of PDI5 Antibody Specificity in the Endothelial Cell Layer and in Endothelial Cells.
Supplemental Figure 4. Single- and Double-Immunogold Labeling of PSVs in Endothelial Cells with Anti-2S Albumin with/without Anti-PDI5 or Anti-RD21 Antibodies.
Supplemental Figure 5. Quantitative, Comparative Analysis of the Number and Cross-Sectional Area of LV and PSV, and of Anti-PDI5 Labeling in Senescing Heart-Stage Endothelial and Embryonic Cells.
Supplemental Figure 6. Interaction of PDI5 with Selected Prey in Yeast Two-Hybrid Assay.
Supplemental Figure 7. Immunogold Labeling of BiP in Endothelial Cells.
Supplementary Material
Acknowledgments
We thank J.C. Rogers and Thomas Okita (Washington State University, Pullman, WA), Ikuko Hara-Nishimura (Kyoto University, Japan), Marisa S. Otegui (University of Wisconsin, Madison, WI), and Alessio Scarafoni (University of Milan, Italy) for the generous gifts of antibodies and Thomas Giddings for help in the Molecular, Cellular, and Developmental Biology EM Service Facility. This work was supported by National Science Foundation Grant NSF MCB03-48028 to D.A.C. and L.A.S.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.com) are: Christine Andème Ondzighi (ondzighi@colorado.edu) and David A. Christopher (dchr@hawaii.edu).
Online version contains Web-only data.
References
- Andème-Onzighi, C., Sivaguru, M., Judy-March, J., Baskin, T.I., and Driouich, A. (2002). The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215 949–958. [DOI] [PubMed] [Google Scholar]
- Balmer, Y., Koller, A., del Val, G., Manieri, W., Schurmann, P., and Buchanan, B.B. (2003). Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 100 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., Rycke, R.D., Viane, R., and Inzé, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113 139–148. [Google Scholar]
- Beers, E.P., Woffenden, B.J., and Zhao, C. (2000). Plant proteolytic enzymes: Possible roles during programmed cell death. Plant Mol. Biol. 44 399–415. [DOI] [PubMed] [Google Scholar]
- Beisson, F., Li, Y., Bonaventure, G., Pollard, M., and Ohlrogge, J.B. (2007). The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368. [DOI] [PMC free article] [PubMed]
- Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glocosidase I mutant reveals a critical role of N-glycan trimming in seed development. EMBO J. 20 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman, F. (1975). Integument initiation and testa development in some Cruciferae. Bot. J. Linn. Soc. 70 213–229. [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interaction among floral homeotic genes of Arabidopsis. Development 112 1–20. [DOI] [PubMed] [Google Scholar]
- Bozhkov, P.V., Suarez, M.F., Filonova, L.H., Daniel, G., Zamyatnin, A.A., Rodriguez-Nieto, S., Zhivotovsky, B., and Smertenko, A. (2005). Cysteine protease mcll-Pa executes programmed cell death during plant embryogenesis. Proc. Natl. Acad. Sci. USA 102 14463–14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercos, M., Santamaria, S., and Carbonell, J. (1999). Cloning and characterization of TP4A, a thiol-protease gene induced during ovary senescence and seed germination in pea. Plant Physiol. 119 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez, F., and Cejudo, F.J. (1998). Germinating-related genes encoding proteolytic enzymes are repressed in the nucellus of developing wheat grain. Plant J. 15 569–574. [Google Scholar]
- Domínguez, F., Moreno, J., and Gejudo, F.J. (2001). The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213 352–360. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7 555–569. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977). Anatomy of Seed Plants, 2nd ed. (New York: John Wiley & Sons).
- Fahn, A. (1990). Plant Anatomy. 4th ed. (Oxford, UK: Buttemorth-Heinemann).
- Fath, A., Bethke, P.C., Lonsdale, J., Meza Romero, R., and Jones, R.L. (2000). Programmed cell death in cereal aleurone. Plant Mol. Biol. 44 255–266. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 20 245–246. [DOI] [PubMed] [Google Scholar]
- Freedman, R.B., Hirst, T.R., and Tuite, M.F. (1994). Proteine disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 19 331–336. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2000). Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 44 245–253. [DOI] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1997). Making sense of senescence. Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 113 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich, D.J., Gagne, J.M., Salter, D.W., Hellmann, H., Estelle, M., Ma, L., and Vierstra, R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280 18810–18821. [DOI] [PubMed] [Google Scholar]
- Green, D.R. (1998). Apoptosis pathways: The road to ruin. Cell 94 695–698. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1996). Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 93 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, J.S., Helm, M., and Gietl, C. (2005). Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc. Natl. Acad. Sci. USA 93 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groover, A., DeWitt, N., Heidel, A., and Jones, A. (1997). Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma 196 197–211. [Google Scholar]
- Gunawardena, A.H.L.A.N., Greenwood, J.S., and Dengler, N.G. (2004). Programmed cell death remodels lace plant leaf shape during development. Plant Cell 16 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y.J., Wang, D.H., Peng, Y.B., Bai, S.L., Xu, L.Y., Li, Y.Q., Xu, Z.H., and Bai, S.N. (2003). DNA damage in the early primordial anther is closely correlated with stamen arrest in the female flower of cucumber (Cucumis sativus L.). Planta 217 888–895. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Hatsugai, N., Nakaune, S., Kuroyanagi, M., and Nishimura, M. (2005). Vacuolar processing enzyme: an executor of plant cell death. Curr. Opin. Plant Biol. 8 404–408. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44 321–334. [DOI] [PubMed] [Google Scholar]
- Helm, M., Schmid, M., Hierl, G., Terneus, K., Tan, L., Lottspeich, F., Kieliszewski, M.J., and Gielt, C. (2008). KDEL-tailed cysteine endopedtidases involved in programmed cell death, intercalation of new cells and dismantling of extensin scaffolds 1. Am. J. Bot. (In press). [DOI] [PubMed]
- Hiratsuka, R., Yamada, Y., and Terasaka, O. (2002). Programmed cell death of pinus nucellus in response to pollen tube penetration. J. Plant Res. 115 141–148. [DOI] [PubMed] [Google Scholar]
- Houston, N.L., Chuanszu, F., Xiang, Q.-Y., Schulze, I.-M., Jung, R., and Boston, R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including 18 single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh, G.-Y., Phillips, T.E., and Rogers, J.C. (1999). Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11 1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M. (2001). Programmed cell death in development and defense. Plant Physiol. 125 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M., and Dangl, J.L. (1996). Logjam at Styx: Programmed cell death in plants. Trends Plant Sci. 1 114–119. [Google Scholar]
- Kaiser, C., Susan Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Kapil, R.N., and Tiwari, S.C. (1978). The integumentary tapetum. Bot. Rev. 44 457–490. [Google Scholar]
- Katsuhara, M. (1997). Apoptosis-like cell death in barley root under salt stress. Plant Cell Physiol. 38 1091–1093. [Google Scholar]
- Kim, J., and Mayfield, S.P. (1997). Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278 1954–1957. [DOI] [PubMed] [Google Scholar]
- Kuang, A., Musgrave, M.E., Matthews, S.W., Cummins, D.B., and Tucker, S.C. (1995). Pollen and ovule development in Arabidopsis thaliana under spaceflight conditions. Am. J. Bot. 82 585–595. [PubMed] [Google Scholar]
- Kuriyama, H., and Fukuda, H. (2002). Developmental programmed cell death in plants. Curr. Opin. Plant Biol. 5 568–573. [DOI] [PubMed] [Google Scholar]
- Kutsuna, N., Kumagai, F., Sato, M.H., and Hasezawa, S. (2003). Three-dimensional reconstruction of tubular structure of vacuolar membrane throughout mitosis in living tobacco cells. Plant Cell Physiol. 44 1045–1054. [DOI] [PubMed] [Google Scholar]
- Lam, E. (2005). Vacuolar proteases livening up programmed cell death. Trends Cell Biol. 15 124–127. [DOI] [PubMed] [Google Scholar]
- Levitan, A., Trebitsh, T., Kiss, V., Pereg, Y., Dangoor, I., and Danon, A. (2005). Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102 6225–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.P., and Larkins, B.A. (1996). Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol. Biol. 30 873–882. [DOI] [PubMed] [Google Scholar]
- Li, L., Shimada, T., Takahashi, H., Ueda, H., Fukao, Y., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18 3535–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnestad, C., Doan, D.N.P., Brown, R.C., Lemmon, B.E., Meyer, D.J., Jung, R., and Olsen, O.-A. (1998). Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 118 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D.-P., and Christopher, D.A. (2006). Immunolocalization of a protein disulfide isomerase to Arabidopsis thaliana chloroplasts and its association with starch biogenesis. Int. J. Plant Sci. 167 1–9. [Google Scholar]
- Lu, D.-P., and Christopher, D.A. (2008). Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics., in press. [DOI] [PubMed]
- Martin, S.J., and Green, D.R. (1995). Protease activation during apoptosis: Death by a thousand cuts? Cell 82 349–352. [DOI] [PubMed] [Google Scholar]
- Martínez, M., Abraham, Z., Carbonero, P., and Díaz, I. (2005). Comparative phylogenetic analysis of cystatin gene families from Arabidopsis, rice and barley. Mol Genet Genomics. 273 423–432. [DOI] [PubMed] [Google Scholar]
- Martins, L.M., and Earnshaw, W.C. (1997). Apoptosis: Alive and kicking in 1997. Trends Cell Biol. 7 111–114. [DOI] [PubMed] [Google Scholar]
- Marty, F. (1999). Plant vacuoles. Plant Cell 11 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri, E., Levitan, A., Guo, F., Christopher, D.A., Schaefer, D., Zryd, J.P., and Danon, A. (2002). Characterization of three PDI-like genes in Physcomitrella patens and construction of knockout mutants. Mol. Genet. Genomics 267 231–240. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., Zhang, X.S., Li, J., and O'Neill, S.D. (1996). Ovule development: Identification of stage-specific and tissue-specific cDNAs. Plant Cell 8 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara, K., Kuriyama, H., and Fukuda, H. (2001). Direct evidence of active and rapid nuclear degradation triggerred by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 125 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Capp, R., and Staehelin, L.A. (2002). Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Herdera, R., Schulzec, J., Jungc, R., and Staehelin, L.A. (2006). The proteolytic precessing of seed storage in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18 2567–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Noh, Y.-S., Martínez, D.E., Vila Petroff, M.G., Staehelin, L.A., Amasino, R.M., and Guiamet, J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41 831–844. [DOI] [PubMed] [Google Scholar]
- Otegui, M.S., and Staehelin, L.A. (2000). Syncytial-type cell plates: A novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaéz, D., and Granell, A. (1997). DNA fragmentation is regulated by ethylene during carpel senescence in Pisum sativum. Plant J. 11 137–144. [Google Scholar]
- Overholtzer, M., Mailleux, A.A., Mouneimne, G., Normand, G., Schnitt, S.J., King, R.W., Cibas, E.S., and Brugge, J.S. (2007). A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131 966–979. [DOI] [PubMed] [Google Scholar]
- Raff, M. (1998). Cell suicide for beginners. Nature 396 119–122. [DOI] [PubMed] [Google Scholar]
- Rosenthal, J.A., Chen, H., Slepnev, V.I., Pellegrini, L., Salcini, A.E., Di Fiore, P.P., and De Camilli, P. (1999). The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J. Biol. Chem. 274 33959–33965. [DOI] [PubMed] [Google Scholar]
- Ryerson, D.E., and Heath, M.C. (1996). Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell 8 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarafoni, A., Carzaniga, R., Harris, N., and Croy, R. (2001). Manipulation of the napin primary structure alters its packaging and deposition in transgenic tobacco (Nicotiana tabacum L.) seeds. Plant Mol. Biol. 46 727–739. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Simpson, D.J., Sarioglu, H., Lottspeich, F., and Gietl, C. (2001). The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98 5353–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7 731–749. [Google Scholar]
- Seguí-Simarro, J.M., and Staehelin, L.A. (2006). Cell cycle-dependant changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: A quantitative and spatial analysis. Planta 223 223–236. [DOI] [PubMed] [Google Scholar]
- Serebriiskii, I.G., and Golemis, E.A. (2000). Uses of lacZ to study gene function: Evaluation of β-galactosidase assays employed in the yeast two-hybrid system. Anal. Biochem. 285 1–15. [DOI] [PubMed] [Google Scholar]
- Shimada, H., Mochizuki, M., Ogura, K., Froehlich, J.E., Osteryoung, K.W., Shirano, Y., Shibata, D., Masuda, S., Mori, K., and Takamiyaa, K.I. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19 3157–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni, Y., Zhu, X.-Z., Levanony, H., Segal, G., and Galili, G. (1995). Purification, characterization, and intracellular localization of glycosylated protein disulfide isomerase from Wheat grains. Plant Physiol. 108 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko, A.P., Bozhkov, P.V., Filonova, L.H., von Arnold, S., and Hussey, P.J. (2003). Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J. 33 813–824. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, M., Belenghi, B., Delledonne, M., Menachem, E., and Levine, A. (1999). The involvement of cystein proteinases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., and Alonso, J.M. (2003). T-DNA mutagenesis in Arabidopsis. In Plant Functional Genomics, E. Grotewold, ed (Totowa, NJ: Humana Press), pp. 177–188.
- Suarez, M.F., Filonova, L.H., Smertenko, A., Savenkov, E.I., Clapham, D.H., von Arnold, S., Zhivotovsky, B., and Bozhkov, P.V. (2004). Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 14 R339–R340. [DOI] [PubMed] [Google Scholar]
- Thomas, H., Ougham, H.J., Wagstaff, C., and Stead, A.D. (2003). Defining senescence and death. J. Exp. Bot. 54 1127–1132. [DOI] [PubMed] [Google Scholar]
- Tu, B., Ho-Schleyer, S.C., Travers, K.J., and Weissman, J.S. (2000). Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290 1571–1574. [DOI] [PubMed] [Google Scholar]
- Turano, C., Coppari, S., Altieri, F., and Ferraro, A. (2002). Proteins of the PDI family: Unpredicted non-ER locations and functions. J. Cell. Physiol. 193 154–163. [DOI] [PubMed] [Google Scholar]
- Vercammen, D., Belenghi, B., van de Cotte, B., Beunens, T., Gavigan, J.-A., De Rycke, R., Brackenier, A., Inzé, D., Harris, J.L., and Van Breusegem, F. (2006). Serpin1 of Arabidopsis thaliana is a suicide inhibitor for Metacaspase-9. J. Mol. Biol. 364 625–636. [DOI] [PubMed] [Google Scholar]
- Wan, L., Xia, Q., Qiu, X., and Selvaraj, G. (2002). Early stages of seed development in Brassica napus, a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 30 1–10. [DOI] [PubMed] [Google Scholar]
- Wang, M., Hoekstra, S., Bergen, S., Lamers, G.E.M., Oppedijk, B.A., Heijden, M.W., Priester, W., and Schilperoort, R.B. (1999). Apoptosis in developing anthers and the role of ABA in this process during androgenesis in Hordeum vulgare L. Plant Mol. Biol. 39 489–501. [DOI] [PubMed] [Google Scholar]
- Wedemeyer, W.J., Welker, E., Narayan, M., and Scheraga, H.A. (2000). Disulfide bonds and protein folding. Biochemistry. 39 4207–4216. [DOI] [PubMed] [Google Scholar]
- Wilkinson, B., and Gilbert, H.F. (2004). Protein disulfide isomerase. Biochim. Biophys. Acta 1699 35–44. [DOI] [PubMed] [Google Scholar]
- William, M. (1977). Stereological techniques. In Practical Methods in Electron Microscopy, A.M. Glauert, ed (Amsterdam: North Holland/American Elsevier), pp. 5–84.
- Xu, F.-X., and Chye, M.-L. (1999). Expression of cysteine proteinase during developmental events associated with programmed cell death in brinjal. Plant J. 17 321–327. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Matsushima, R., Nishimura, M., and Hara-Nishimura, I. (2001). A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127 1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Yao, N., and Greenberg, J.T. (2006). Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori, T., Semba, T., Takemoto, H., Akagi, S., Yamamoto, A., and Tashiro, Y. (1990). Protein disulfide-isomerase in rat exocrine pancreatic cells is exported from the endoplasmic reticulum despite possessing the retention signal. J. Biol. Chem. 265 15984–15990. [PubMed] [Google Scholar]
- Young, T.E., Gallie, D.R., and DeMason, D.A. (1997). Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 115 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.