Abstract
MicroRNAs (miRNAs) regulate the expression of numerous genes and are implicated in the pathogenesis of many human diseases. miRNAs act as specificity determinants to guide deposition of microRNPs (miRNPs) onto miRNA recognition elements (MREs) that are found in mRNA targets. We have adapted a site-specific crosslinking approach, previously used in the analysis of splicing, to interrogate protein factors that physically associate with MREs in human cells. We find that Ago proteins are the only proteins that bind to the MRE in a miRNA-dependent manner. Our method may be used in various experimental systems to analyze protein factors that influence MRE accessibility by miRNAs and the composition of MRE-bound miRNPs.
Keywords: microRNA, miRNA, microRNP, miRNP, RISC, RNAi, Argonaute, Ago
INTRODUCTION
MicroRNAs (miRNAs) and short interfering RNAs (siRNAs) are ∼22-nucleotide (nt), noncoding, regulatory RNAs. miRNAs assemble with Argonaute (Ago) proteins into miRNPs (functionally equivalent to RNA induced silencing complexes [RISCs]), which are the effector complexes that recognize miRNA recognition elements (MREs) in mRNAs targeted by miRNAs and mediate miRNA functions (for reviews, see Bartel 2004; Liu et al. 2008). Argonaute proteins have molecular masses of ∼95 kDa and are divided into Ago and Piwi subclades (Carmell et al. 2002). There are four Ago proteins (Ago1–4) in humans and mice (Liu et al. 2008). An important principle underlying miRNP–MRE recognition is the need for perfect complementarity between the proximal region (positions 2–8) of the miRNA with the MRE; this area of base pairing is known as the “seed” or “proximal area” or “nucleus” (Lewis et al. 2003; Kiriakidou et al. 2004; Rajewsky and Socci 2004). Such partial binding between a miRNP and its mRNA target promotes translational repression or accelerated degradation of the target mRNA (for reviews, see Zamore and Haley, 2005; Nilsen 2007; Liu et al. 2008). When the complementarity between the miRNA and an MRE extends continuously beyond the seed and if the miRNA is bound to Ago2, the targeted mRNA is cleaved by Ago2 (for reviews, see Zamore and Haley 2005; Nilsen 2007; Liu et al. 2008).
Uncovering the comprehensive picture of miRNA-mediated functions will require the development of methods that lead to the isolation and identification of factors that participate in direct interactions between miRNAs and their targets. Such methods may also be useful for the biochemical isolation of mRNA targets that physically associate with cognate miRNAs. Although mRNA targets that coimmunoprecipitate with endogenous Ago proteins (Beitzinger et al. 2007) or epitope-tagged Ago proteins (Easow et al. 2007; Karginov et al. 2007) have been isolated without prior crosslinking, it is unknown whether the interpretation of such approaches might be complicated from reassociation artifacts between miRNPs and mRNAs, induced after cell lysis (Mili and Steitz 2004). The use of epitope-tagged Ago proteins may also limit studies of miRNPs and their targets since a large fraction of epitope-tagged Ago protein is devoid of miRNAs (Maniataki and Mourelatos 2005a,b).
Previous studies have used photocrosslinking to study RISC assembly (Tomari et al. 2004) and formaldehyde crosslinking to identify RISC/miRNP proteins that bind to mRNAs (Vasudevan et al. 2007). We have adapted a site-specific crosslinking approach, previously used in the analysis of splicing (Moore and Sharp 1992; Wyatt et al. 1992; Newman et al. 1995; Wu et al. 1999), to interrogate protein factors that are in direct contact with an MRE that is specifically recognized by the let-7 miRNA. We find that Ago proteins are the only proteins that bind to the MRE in a let-7-dependent manner. This method may be used in various experimental systems to analyze protein factors that influence MRE accessibility by miRNAs and the composition of MRE-bound miRNPs.
RESULTS AND DISCUSSION
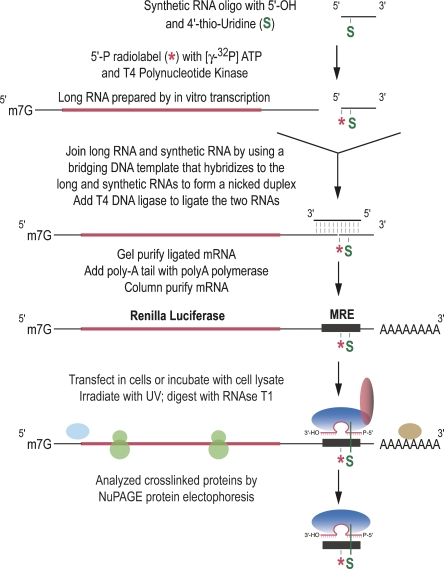
To isolate endogenous proteins that bind directly to an MRE, we have adapted a site-specific crosslinking approach previously used in the analysis of splicing (Moore and Sharp 1992; Wyatt et al. 1992; Newman et al. 1995; Wu et al. 1999). With this method we can induce ultraviolet (UV) crosslinks between an MRE present in the 3′-UTR of a reporter mRNA and the proteins that are in direct contact (Fig. 1) based on the strategy of site-specific incorporation of modified, radiolabeled nucleotides (Moore and Query 1998). Because a single MRE for let-7b, naturally found in the 3′-UTR of human and mouse lin-28 mRNAs, is necessary and sufficient to confer miRNA-dependent translational repression in HeLa cells (Kiriakidou et al. 2004), we prepared Renilla Luciferase reporter mRNAs containing a single MRE for let-7b, and used total HeLa cell lysates to identify proteins that physically associate with the MRE (Figs. 1, 2A). Two site-specific modifications were incorporated in the reporter mRNAs: a single, photoreactive modified base, 4-thio-Uridine (s4U), and a single radiolabel 32P adjacent to the s4U. When protein factors are bound to the reporter mRNA and irradiated by UV light, proteins that are in direct contact with s4U form covalent bonds and become radiolabeled. After RNase treatment, only the s4U-containing fragment of the mRNA that is bound to proteins remains and is detected along with the bound proteins by protein electrophoresis.
FIGURE 1.
Strategy for identification of factors that associate directly with an MRE.
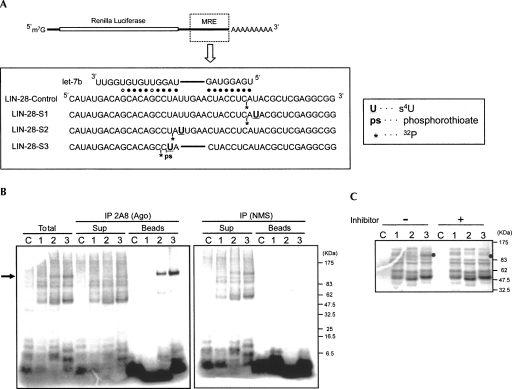
FIGURE 2.
miRNA-guided and direct contacts between Ago proteins and an MRE. (A) mRNAs used to detect proteins that bind to an MRE for let-7b. Base pairing between LIN-28 MRE with let-7b is shown. Single s4U (U) and an adjacent, single 32P radiolabel (*) are incorporated in defined positions within the MRE, as shown. After UV irradiation, a covalent bond is formed between the s4U and cellular factors, and the 32P radiolabel is transferred to the bound factors. LIN-28 S3 mRNA, whose MRE sequence is perfectly complementary to let-7b, contains a phosphorothioate (ps) between position 10 and 11 to reduce cleavage by Ago 2. (B) Ago proteins bind directly to MRE. Indicated mRNAs (LIN-28-Control [C], LIN-28-S1 [1], LIN-28-S2 [2], and LIN-28-S3 [3]) were incubated in total HeLa cell extract (Total) or with supernatants or beads after immunoprecipitation (IP) with 2A8 anti-Ago monoclonal antibody or nonimmune mouse serum (NMS, negative control). After UV irradiation and RNase treatment, samples were analyzed by NuPAGE protein electrophoresis, and radiolabeled proteins were visualized by storage phosphor autoradiography. (Arrow) Ago proteins. (C) Crosslinking of Ago proteins to a target mRNA requires the cognate miRNA. The crosslinking experiments using total HeLa cell extract were performed with (+) or without (−) let-7b inhibitor. (Filled circles) Position of Ago proteins.
As shown in Figure 2A, we constructed four reporter mRNAs: “LIN-28-Control” with a radiolabel but without s4U, “LIN-28-S1” with s4U at position −1, “LIN-28-S2” with s4U at the loop region between positions 8 and 9, and “LIN-28-S3” with s4U at position 10. Since the MRE of LIN-28-S3 is perfectly complementary to let-7b in positions 1–18, we incorporated a phosphorothioate bond between positions 10 and 11 to prevent cleavage by Ago2 and thus retain the association of Ago2/let-7 miRNPs. The mRNAs were incubated with total HeLa cell lysate, irradiated by UV light, treated with RNase T1 (G-specific), and analyzed by NuPAGE protein electrophoresis (Fig. 2B, “Total”) (see Materials and Methods). Numerous bands between 50 and 150 kDa were observed with LIN-28-S1, LIN-28-S2, and LIN-28-S3. Because no crosslinking was observed with LIN-28-Control, which lacked a photoreactive s4U, these bands represented proteins specifically crosslinked to the s4U in the MRE region. After RNase T1 treatment, 14 nt, 7 nt, and 16 nt of RNA were excised from LIN-28-S1, LIN-28-S2, and LIN-28-S3, respectively, as a s4U- and 32P-containing region. Thus, the proteins that crosslinked to LIN-28-S2 migrated a little faster than those that crosslinked to LIN-28-S1 and LIN-28-S3. The patterns of the crosslinked proteins were identical between LIN-28-S1, LIN-28-S2, and LIN-28-S3, with the exception of a ∼100-kDa band (Fig. 2B, indicated by an arrow), corresponding in size to Ago proteins, which was observed in LIN-28-S2 or LIN-28-S3, but not in LIN-28-S1.
Next, we performed immunoprecipitation with 2A8, a monoclonal antibody that recognizes all four human Ago proteins (Nelson et al. 2007), or nonimmune mouse serum (NMS) as a negative control. The supernatant after immunoprecipitation or the beads were incubated with the labeled mRNAs, UV-irradiated, treated with RNase T1, and analyzed by NuPAGE. As shown in Figure 2B, Ago proteins became radiolabeled only when the s4U was within the MRE (Fig. 2B, “beads,” lanes 2,3), and there was corresponding reduction of the intensity of labeled Agos in the supernatant after immunoprecipitation (Fig. 2B, “sup”). There were no radiolabeled proteins in the NMS immunoprecipitates, and radiolabeled Ago proteins remained in the NMS supernatant (Fig. 2B, right panel). These findings indicate that although many proteins can bind to the MRE region, Ago proteins bind specifically to the MRE. To assess whether the Ago binding to the MRE is dependent on the cognate miRNA (let-7), we pre-incubated total HeLa cell lysate with a miRIDIAN let-7b inhibitor and then performed the crosslinking experiment. Previous studies have shown that 2′-O-methylated RNA molecules that are antisense to miRNAs are potent and specific inhibitors of miRNAs (Hutvagner et al. 2004; Meister et al. 2004). miRIDIAN inhibitors incorporate additional, structured, double-stranded regions that enhance their potency (Vermeulen et al. 2007). As shown in Figure 2C, the let-7b inhibitor caused a marked reduction of the intensity of the ∼100-kDa Ago protein band, while the intensity of other protein bands was not affected, indicating that Ago binding to the MREs was let-7-dependent.
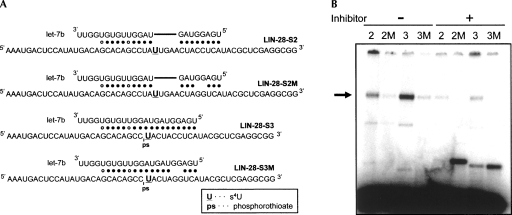
We next tested whether immunopurified miRNPs from HeLa cells could crosslink to MREs in a miRNA-dependent and seed-dependent manner. Four synthetic, 5′-end-labeled RNAs carrying either the MRE region of LIN-28-S2 or LIN-28-S3 that bind to let-7 with seed or perfect complementarity, respectively, or corresponding mutants that disrupt the seed (LIN-28-S2M and LIN-28-S3M) were used as target RNAs (Fig. 3A). The RNAs were incubated with 2A8 immunoprecipitates (containing Ago/let-7 miRNPs) either in the absence or presence of let-7b-inhibitor, followed by UV-irradiation. Because the target RNAs were small and 5′-end-labeled, we did not treat the samples with RNase T1 after crosslinking but proceeded with protein electrophoresis. As shown in Figure 3B, binding of Ago proteins to the MREs was let-7-dependent, while mismatches in the seed region dramatically reduced binding. The faster migrating bands toward the bottom of the gel are radiolabeled RNA targets, not bound to any proteins. These findings indicate that Ago/miRNA miRNPs are the predominant determinants of target RNA recognition by miRNAs. Since 2A8 recognizes all four Ago proteins, we do not know whether all Ago proteins are bound to the MRE or whether they all recognize it with the same affinity. As Ago-specific antibodies are being developed (Rudel et al. 2008) it will be interesting to examine whether there is preferential binding of specific MREs by specific Ago proteins.
FIGURE 3.
Mismatches in the miRNA proximal (seed) region disrupt the binding between Ago proteins and their target RNA. (A) Target RNA sequences. (U) s4U, (ps) phosphorothioate. (B) 5′-end-labeled target RNAs were incubated with 2A8 (anti-Ago) immunoprecipitates, in the presence (+) or absence (−) of let-7-inhibitor, irradiated with UV, and analyzed by NuPAGE electrophoresis. (Arrow) Crosslinked Ago proteins. The faster migrating bands toward the bottom of the gel are radiolabeled RNA targets, not bound to any proteins.
An important conclusion of our study is that the only proteins that bind to the MRE in a miRNA-dependent manner are Ago proteins, and such binding is very specific, as demonstrated, for example, by the lack of Ago crosslinking to the MRE in the presence of a miRNA inhibitor or when 4-thio-Uridine is placed just 1 nt upstream of the seed. Our finding that Ago/miRNA miRNPs, either immunopurified or in a whole cell lysate, are able to specifically associate with an MRE in a miRNA-dependent manner, indicates that mRNAs isolated from Ago immunoprecipitates (Beitzinger et al. 2007; Easow et al. 2007; Karginov et al. 2007) very likely represent authentic targets.
Another important observation is that many proteins bind to the MRE in a miRNA-independent manner, and our approach can be used to interrogate whether any of these proteins influences binding of Ago/miRNAs to the MRE. In that regard it is interesting to note that the Dnd1 RNA binding protein inhibits the activity of several miRNAs by binding to their cognate mRNA targets and precluding miRNA binding (Kedde et al. 2007).
Further uses of this method would be to interrogate whether other miRNP-associated proteins, such as GW182 or proteins of the decapping complex (Eulalio et al. 2007) or FXR1 (Vasudevan et al. 2007), are deposited in a miRNA-dependent manner onto cognate MREs. This method may also be used to interrogate whether the protein composition of miRNPs differs when miRNAs are bound to their targets in polysomes (Maroney et al. 2006; Nottrott et al. 2006), and can be adapted to analyze the protein composition of miRNPs bound to MREs in systems that recapitulate miRNA translational repression in vitro (Mathonnet et al. 2007; Thermann and Hentze 2007; Wakiyama et al. 2007).
MATERIALS AND METHODS
Construction of reporter mRNAs containing s4U and 32P radiolabel
Site-specifically modified target mRNAs were constructed by splinted ligation of two RNAs according to methods described by Moore and Query (1998): (RNA-1) was a 5′-32P-labeled synthetic RNA oligo containing s4U near its 5′-end, and (RNA-2) was a capped transcript containing Renilla luciferase mRNA sequences. For (RNA-1), synthetic RNAs containing s4U were obtained from Dharmacon Co.: 5′-AUACGCUCGAGGCGG-3′ for LIN-28-Control, 5′-A(s4U)ACGCUCGAGGCGG-3′ for LIN-28-S1, 5′-A(s4U)UGAACUACCUCAUACGCUCGAGGCGG-3′ for LIN-28-S2, and 5′-C(phosphorothioate)(s4U)ACUACCUCAUACGCUCGAGGCGG-3′ for LIN-28-S3. These synthetic RNAs were 5′-end labeled using [γ-32P] ATP and T4 polynucleotide kinase (T4 PNK, New England Biolabs), followed by incubation for 10 min at 70°C to deactivate PNK. (RNA-2) was prepared by in vitro transcription. First, we synthesized DNA templates from the pRL-TK plasmid bearing a let-7b MRE in the 3′-UTR (Kiriakidou et al. 2004) by PCR using following primers:
Forward primer: 5′-TAATACGACTCACTATAGGCTAGCC-3′;
Reverse primer: 5′-TGAGGTAGTTCAATAGGCTGTGCTG-3′ for LIN-28-Control and LIN-28-S1;
Reverse primer: 5′-TAGGCTGTGCTGTCATATGGAGTC-3′ for LIN-28-S2; and
Reverse primer: 5′-GCTGTGCTGTCATATGGAGTCATTTAG-3′ for LIN-28-S3.
A capped (RNA-2) was then synthesized by using the mMESSAGE mMACHINE kit (Ambion) and then purified by MEGAclear (Ambion). (RNA-1) and (RNA-2) were joined by T4 DNA ligase. T4 DNA ligase ligates nicks in duplex regions, and we used the following bridging DNA templates that hybridize to the (RNA-1) and (RNA-2) to form nicked duplexes:
5′-CTCGAGCGTATGAGGTAGTTCAATAGGCTGTGC-3′ for LIN-28-Control, LIN-28-S1, and LIN-28-S2; and
5′-GAGGTAGTAGGCTGTGCTGT-3′ for LIN-28-S3.
The (RNA-1) (100 pmol), the (RNA-2) (20 pmol), and the DNA template (100 pmol) were first incubated for 5 min at 70°C in a reaction mixture (50 μL) consisting of 1× buffer for T4 DNA Ligase (USB Co.), and then annealed at room temperature. Ligation was performed simultaneously for 4 h at 37°C by adding 25 units of T4 DNA Ligase (USB Co.) and 0.8 mM of ATP. The ligated mRNAs were PAGE purified (using 5% UREA PAGE). Poly(A) tail was then added to the purified mRNAs using the Poly(A) Tailing Kit (Ambion).
For the binding analysis using target RNAs with mismatches in the seed region, the following synthetic RNAs containing s4U were obtained from Dharmacon:
5′-AAAUGACUCCAUAUGACAGCACAGCCUA(s4U)UGAACUACCUCAUACGCUCGAGGCGG-3′ for LIN-28-S2;
5′-AAAUGACUCCAUAUGACAGCACAGCCUA(s4U)UGAACUAGGUCAUACGCUCGAGGCGG-3′ for LIN-28-S2M;
5′-AAAUGACUCCAUAUGACAGCACAGCC(s4U)ACUACCUCAUACGCUCGAGGCGG-3′ for LIN-28-S3; and
5′-AAAUGACUCCAUAUGACAGCACAGCC(s4U)ACUAGGUCAUACGCUCGAGGCGG-3′ for LIN-28-S3M.
These RNAs were 5′-end labeled using [γ-32P] ATP and T4 PNK (New England Biolabs).
Immunoprecipitation
Total HeLa cell lysate was prepared by brief sonication in lysis buffer containing 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 2.5 mM MgCl2, 0.05% NP-40, protease inhibitor (Complete, Roche) and RNase inhibitor (0.1 units/μL, Promega); lysates were clarified by centrifugation at 20,000g for 10 min. Immunoprecipitation was performed as previously described (Mourelatos et al. 2002) using the 2A8 anti-Ago monoclonal antibody (Nelson et al. 2007), or nonimmune mouse serum as negative control.
Crosslinking
32P-labeled RNA (∼10,000 cpm) were incubated for 70 min at 28°C in 10 μL of total HeLa lysate, or beads or supernatant in the lysis buffer obtained from 2A8 IP. Where indicated, the lysate or the beads were pre-incubated for 30 min at 28°C with 25 pmol of let-7b inhibitor (miRIDIAN MicroRNA Inhibitor, Dharmacon) before the incubation with the labeled RNAs. Crosslinking was performed for 30 min on ice by irradiation with a 365-nm hand-held lamp (EL series UV lamp, UVP). When the reporter mRNAs were used, reactions were digested with 30 units of RNase T1 (Roche) for 20 min at 37°C. Cross-linked proteins were separated by NuPAGE (NuPAGE 4%–12% Bis-Tris, Invitrogen) and detected by storage-phosphor autoradiography.
ACKNOWLEDGMENTS
We thank members of our laboratory for stimulating discussions. This research was supported by a Human Frontier Science Program Long Term Fellowship to Y.K. and by NIH grants GM0720777, NS056070, and a URF grant from the University of Pennsylvania to Z.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1133808.
REFERENCES
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beitzinger, M., Peters, L., Zhu, J.Y., Kremmer, E., Meister, G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Easow, G., Teleman, A.A., Cohen, S.M. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A., Rehwinkel, J., Stricker, M., Huntzinger, E., Yang, S.F., Doerks, T., Dorner, S., Bork, P., Boutros, M., Izaurralde, E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., Simard, M.J., Mello, C.C., Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov, F.V., Conaco, C., Xuan, Z., Schmidt, B.H., Parker, J.S., Mandel, G., Hannon, G.J. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde, M., Strasser, M.J., Boldajipour, B., Vrielink, J.A., Slanchev, K., le Sage, C., Nagel, R., Voorhoeve, P.M., van Duijse, J., Orom, U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., Hatzigeorgiou, A. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu, X., Fortin, K., Mourelatos, Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki, E., Mourelatos, Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005a;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki, E., Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005b;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Yu, Y., Fisher, J., Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet, G., Fabian, M.R., Svitkin, Y.V., Parsyan, A., Huck, L., Murata, T., Biffo, S., Merrick, W.C., Darzynkiewicz, E., Pillai, R.S., et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Dorsett, Y., Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili, S., Steitz, J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C. Uses of site-specifically modified RNAs constructed by RNA ligation. In: Smith C., editor. RNA–protein interactions: A practical approach. Oxford University Press; Oxford, UK: 1998. pp. 75–108. [Google Scholar]
- Moore, M.J., Sharp, P.A. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., De Planell-Saguer, M., Lamprinaki, S., Kiriakidou, M., Zhang, P., O'Doherty, U., Mourelatos, Z. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A.J., Teigelkamp, S., Beggs, J.D. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nottrott, S., Simard, M.J., Richter, J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N., Socci, N.D. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Rudel, S., Flatley, A., Weinmann, L., Kremmer, E., Meister, G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann, R., Hentze, M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., Zamore, P.D. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Vasudevan, S., Tong, Y., Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vermeulen, A., Robertson, B., Dalby, A.B., Marshall, W.S., Karpilow, J., Leake, D., Khvorova, A., Baskerville, S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama, M., Takimoto, K., Ohara, O., Yokoyama, S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., Green, M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., Steitz, J.A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Haley, B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]