Abstract
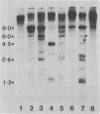
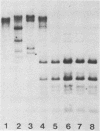
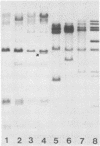
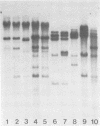
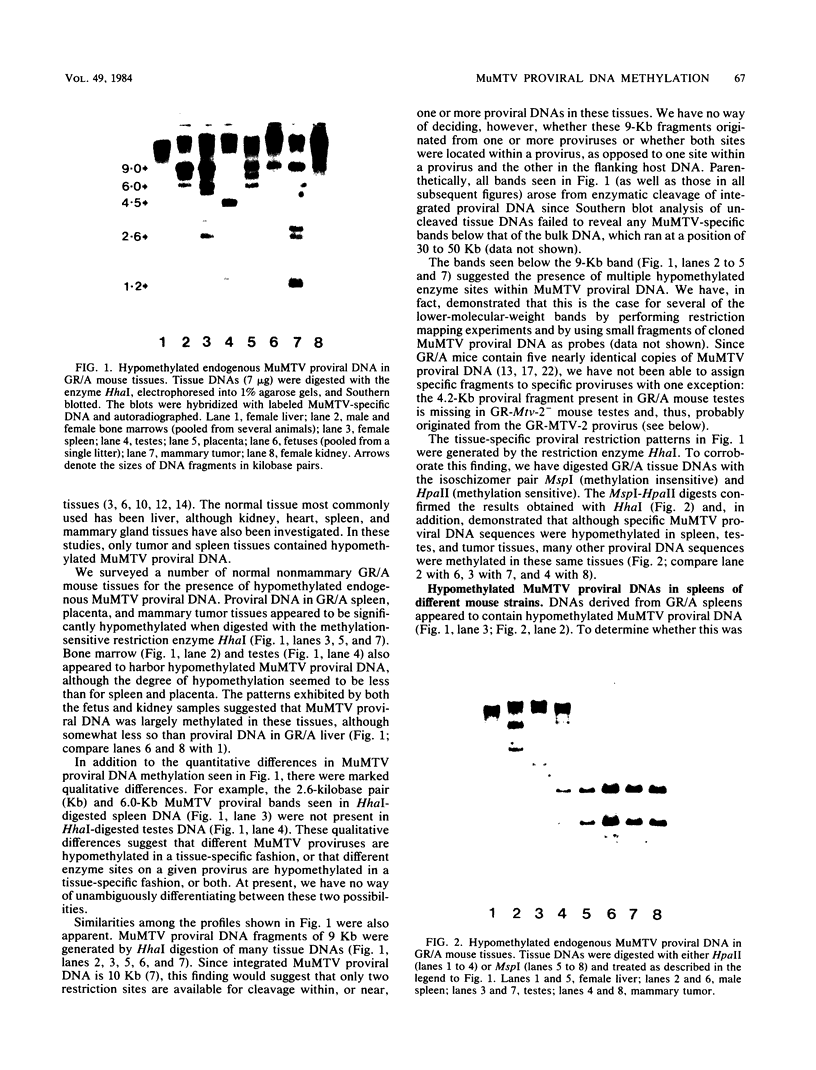
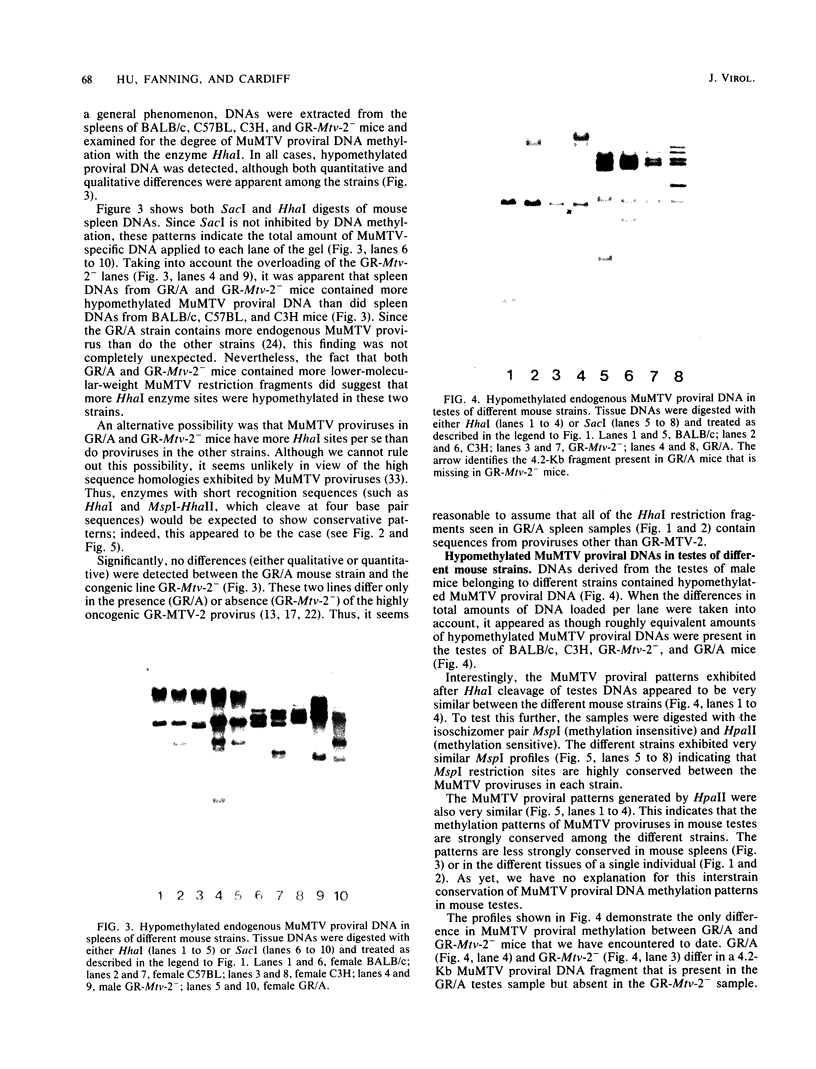
The methylation state of endogenous mouse mammary tumor virus (MuMTV) proviral DNA was examined in normal mouse tissues. DNAs from various tissues were cleaved with the methylation-sensitive enzymes HhaI and HpaII and analyzed by Southern blotting. Tissue-specific MuMTV proviral DNA methylation patterns were found in the BALB/c, C3H, C57BL, GR/A, and GR-Mtv-2- mouse strains. MuMTV proviral DNA was hypomethylated in DNAs from the spleens and testes of all strains examined. The GR/A mouse strain, which was most thoroughly studied, also showed hypomethylation of MuMTV proviral DNA in bone marrow and placental tissues. Analysis of RNAs extracted from GR/A liver, mammary tumor, testes, placenta, and spleen tissues demonstrated that MuMTV proviral hypomethylation need not reflect significant proviral transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznik T., Cohen J. C. Altered methylation of endogenous viral promoter sequences during mammary carcinogenesis. Nature. 1982 Jan 21;295(5846):255–257. doi: 10.1038/295255a0. [DOI] [PubMed] [Google Scholar]
- Brown D. D. Gene expression in eukaryotes. Science. 1981 Feb 13;211(4483):667–674. doi: 10.1126/science.6256857. [DOI] [PubMed] [Google Scholar]
- Callahan R., Drohan W., Gallahan D., D'Hoostelaere L., Potter M. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4113–4117. doi: 10.1073/pnas.79.13.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C. Methylation of milk-borne and genetically transmitted mouse mammary tumor virus proviral DNA. Cell. 1980 Mar;19(3):653–662. doi: 10.1016/s0092-8674(80)80042-0. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Traina V. L., Breznik T., Gardner M. Development of a mouse mammary tumor virus-negative mouse strain: a new system for the study of mammary carcinogenesis. J Virol. 1982 Dec;44(3):882–885. doi: 10.1128/jvi.44.3.882-885.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Varmus H. E. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979 Mar 29;278(5703):418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- Drohan W. N., Benade L. E., Graham D. E., Smith G. H. Mouse mammary tumor virus proviral sequences congenital to C3H/Sm mice are differentially hypomethylated in chemically induced, virus-induced, and spontaneous mammary tumors. J Virol. 1982 Sep;43(3):876–884. doi: 10.1128/jvi.43.3.876-884.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Etkind P. R., Sarkar N. H. Integration of new endogenous mouse mammary tumor virus proviral DNA at common sites in the DNA of mammary tumors of C3Hf mice and hypomethylation of the endogenous mouse mammary tumor virus proviral DNA in C3Hf mammary tumors and spleens. J Virol. 1983 Jan;45(1):114–123. doi: 10.1128/jvi.45.1.114-123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G., Puma J. P., Cardiff R. D. Selective amplification of mouse mammary tumor virus in mammary tumors of GR mice. J Virol. 1980 Oct;36(1):109–114. doi: 10.1128/jvi.36.1.109-114.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T. G., Vassos A. B., Cardiff R. D. Methylation and amplification of mouse mammary tumor virus DNA in normal, premalignant, and malignant cells of GR/A mice. J Virol. 1982 Mar;41(3):1007–1013. doi: 10.1128/jvi.41.3.1007-1013.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein S. C., Ross S. R., Yamamoto K. R. Chromosomal position effects determine transcriptional potential of integrated mammary tumor virus DNA. J Mol Biol. 1982 Apr 15;156(3):549–565. doi: 10.1016/0022-2836(82)90266-2. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N. E., Groner B., Diggelmann H., Van Nie R., Michalides R. Genomic location of mouse mammary tumor virus proviral DNA in normal mouse tissue and in mammary tumors. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1161–1168. doi: 10.1101/sqb.1980.044.01.125. [DOI] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- Mermod J. J., Bourgeois S., Defer N., Crépin M. Demethylation and expression of murine mammary tumor proviruses in mouse thymoma cell lines. Proc Natl Acad Sci U S A. 1983 Jan;80(1):110–114. doi: 10.1073/pnas.80.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., van Nie R., Nusse R., Hynes N. E., Groner B. Mammary tumor induction loci in GR and DBAf mice contain one provirus of the mouse mammary tumor virus. Cell. 1981 Jan;23(1):165–173. doi: 10.1016/0092-8674(81)90281-6. [DOI] [PubMed] [Google Scholar]
- Morris V. L., Medeiros E., Ringold G. M., Bishop J. M., Varmus H. E. Comparison of mouse mammary tumor virus-specific DNA in inbred, wild and Asian mice, and in tumors and normal organs from inbred mice. J Mol Biol. 1977 Jul;114(1):73–91. doi: 10.1016/0022-2836(77)90284-4. [DOI] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Cassio D., Levilliers J., Sala-Trepat J., Weiss M. C. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 1982 Oct;30(3):825–833. doi: 10.1016/0092-8674(82)90287-2. [DOI] [PubMed] [Google Scholar]
- Razin A., Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Yamamoto K. R., Bishop J. M., Varmus H. E. Glucocorticoid-stimulated accumulation of mouse mammary tumor virus RNA: increased rate of synthesis of viral RNA. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2879–2883. doi: 10.1073/pnas.74.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traina V. L., Taylor B. A., Cohen J. C. Genetic mapping of endogenous mouse mammary tumor viruses: locus characterization, segregation, and chromosomal distribution. J Virol. 1981 Dec;40(3):735–744. doi: 10.1128/jvi.40.3.735-744.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardimon L., Kressmann A., Cedar H., Maechler M., Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Medeiros E., Bishop J. M., Nowinski R. C., Sarkar N. H. Transcription of mouse mammary tumor virus genes in tissues from high and low tumor incidence mouse strains. J Mol Biol. 1973 Oct 5;79(4):663–679. doi: 10.1016/0022-2836(73)90070-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]