Abstract
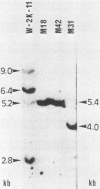
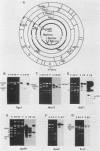
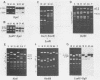
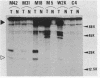
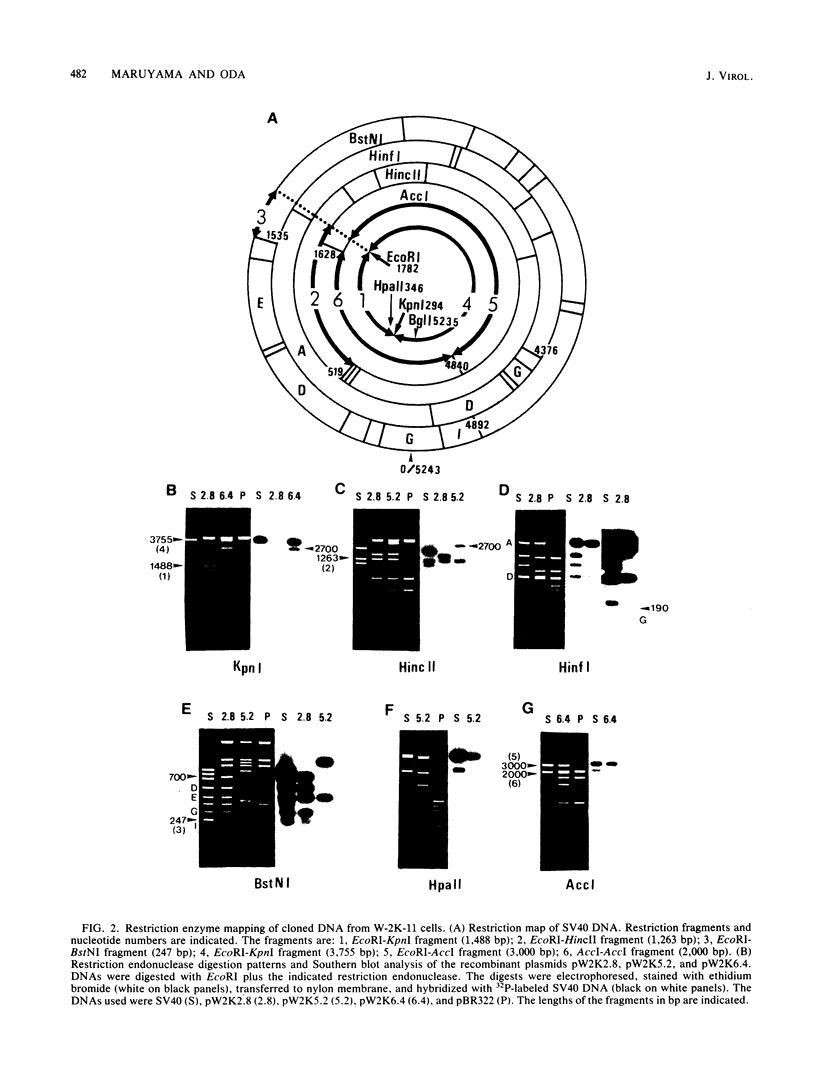
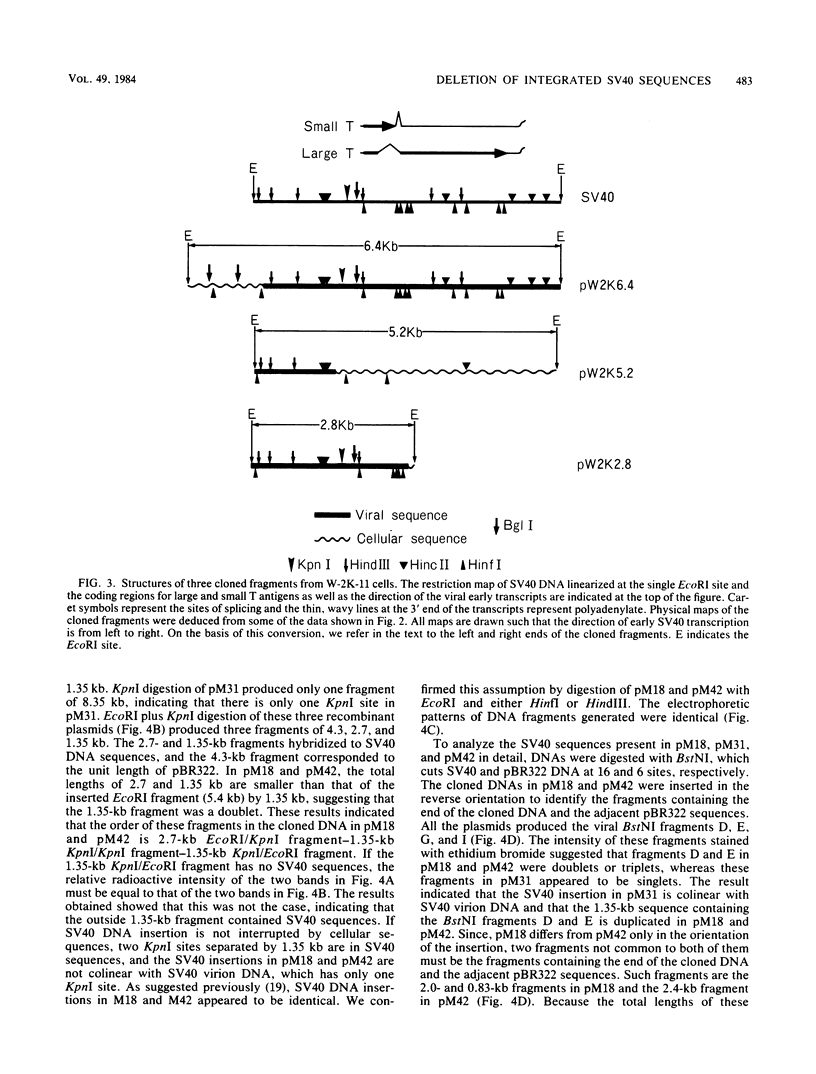
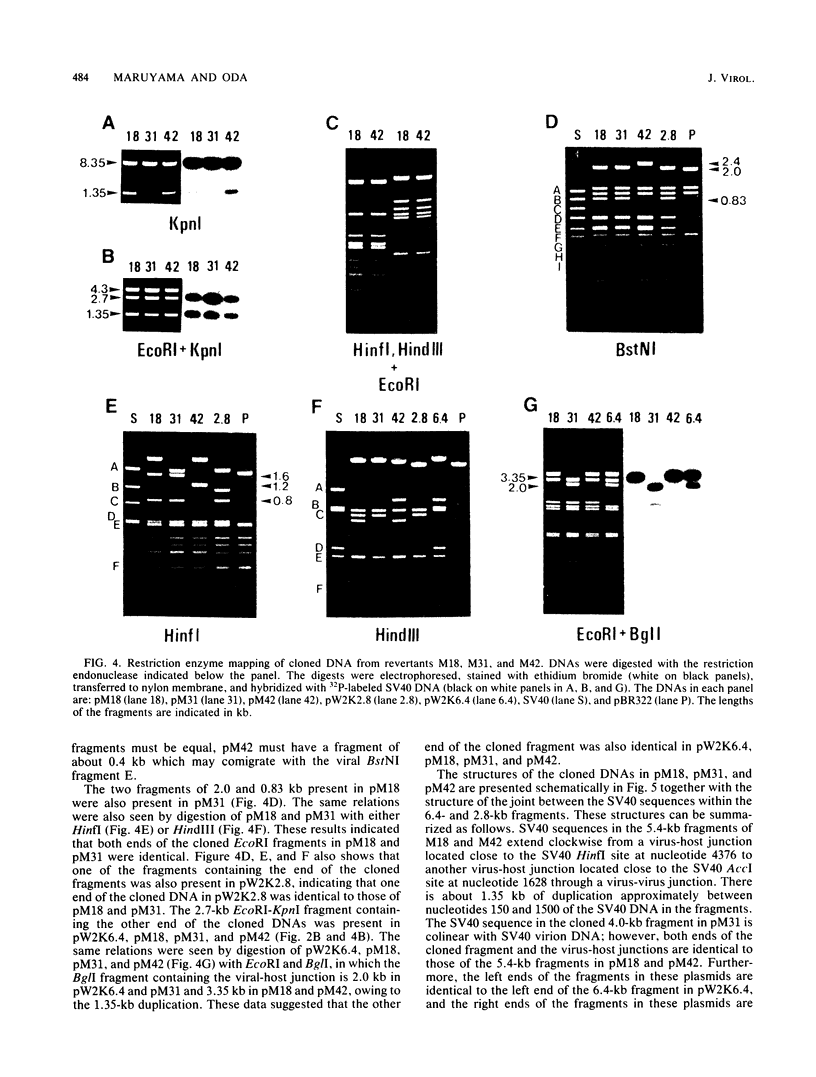
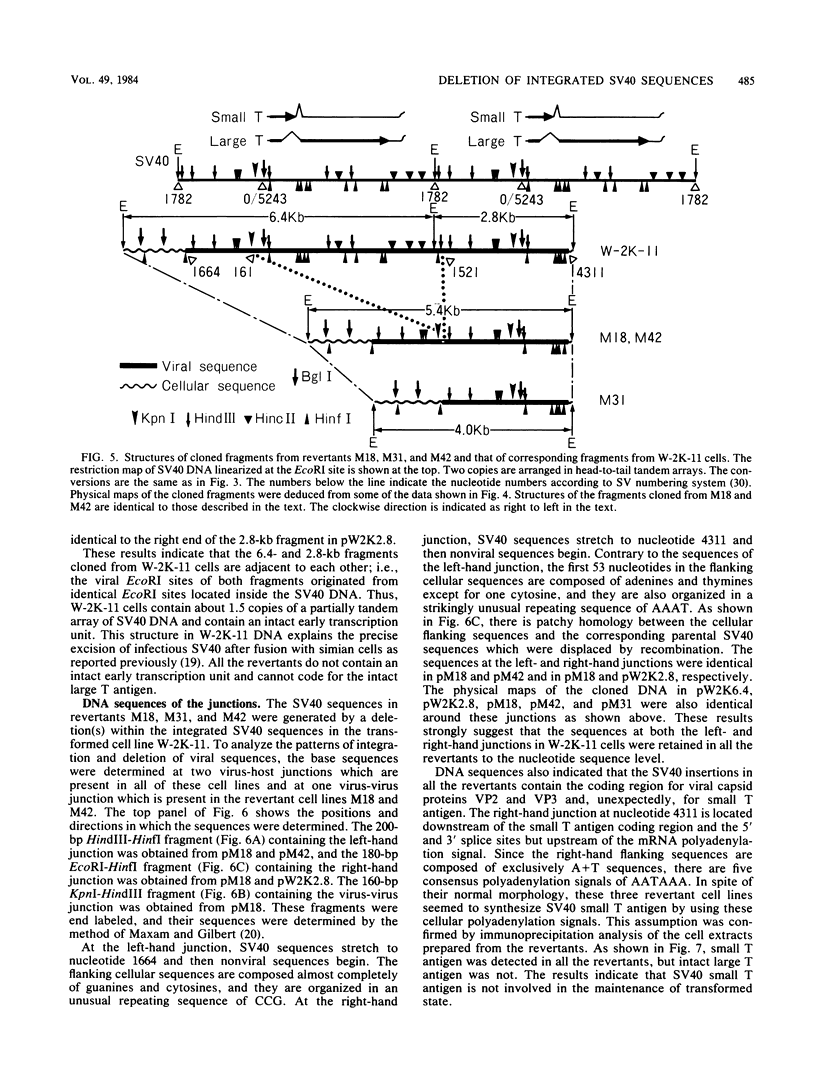
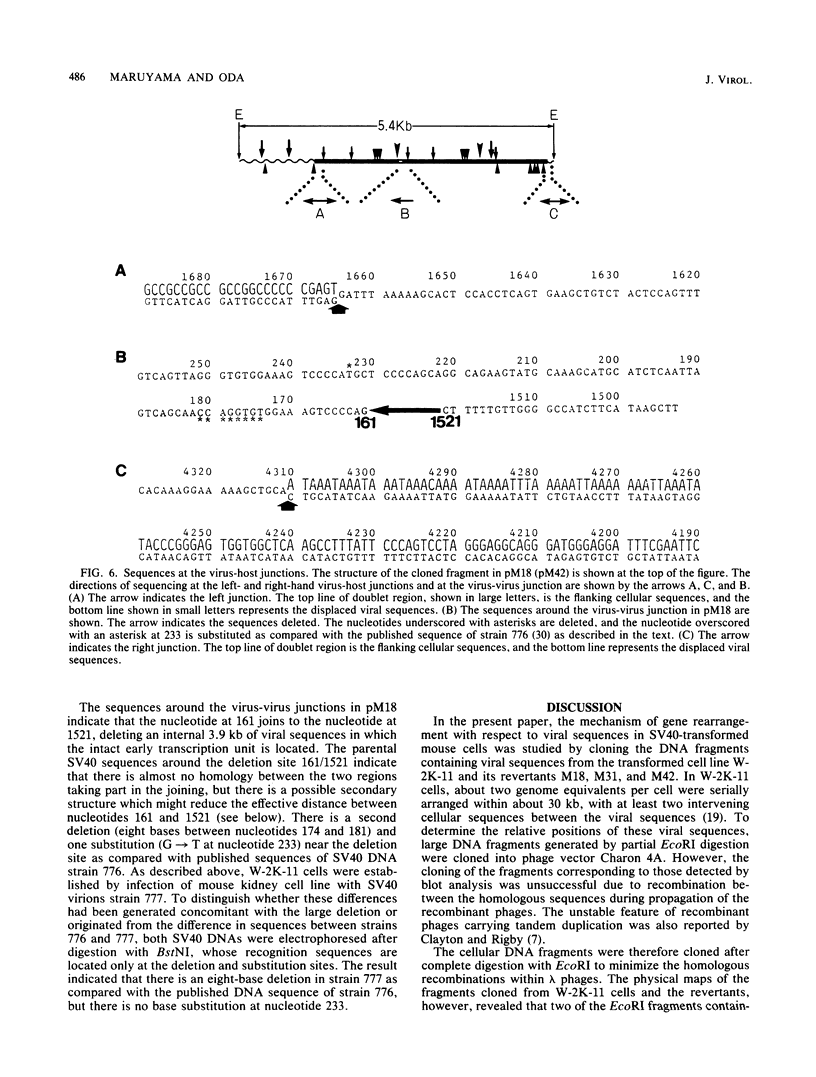
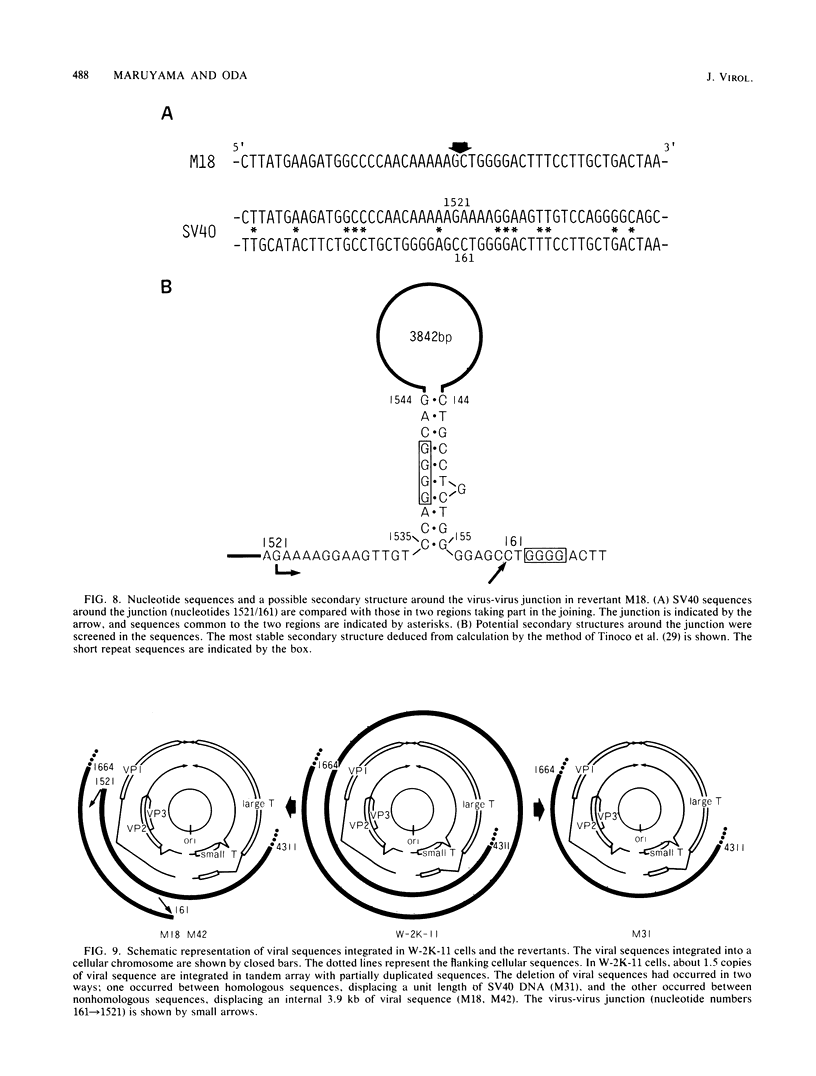
Simian virus 40 (SV40) DNA insertions from SV40-transformed mouse cell line W-2K-11 and its revertants M18, M31, and M42 were cloned. W-2K-11 cells contain 1.5 copies of the SV40 sequences in a partially tandem duplicated form. The endpoints of the viral sequences at the virus-host junctions are located very close to those reported by others, indicating that there are some preferred sites for integration and rearrangement in SV40 sequences. One flanking cellular sequence is a long stretch of adenine and thymine with repeated AAAT, and the other is a stretch of guanine and cytosine with repeated CCG. There are patchy homologies between the flanking cellular sequences and the corresponding parental SV40 sequences. The sequences around both junctions were retained in all the revertants, whereas most of the internal SV40 sequences coding for large T antigen were deleted. The coding sequences for small T antigen are intact, and small T antigen was expressed in all the revertants. The fragments cloned from M18 and M42 were identical and 3.9 kilobases of SV40 sequences were deleted. The parental SV40 sequences around the deletion site have sequences capable of forming a secondary structure which might reduce the effective distance between the two regions. The SV40 DNA retained in M31 is colinear with SV40 virion DNA, and a unit length of SV40 DNA was deleted within the SV40 sequences present in W-2K-11 cells. These results indicated that two types of deletion occurred during the reversion, one between homologous sequences and the other between nonhomologous sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Blechl A. E., Denniston-Thompson K., Faber H. E., Richards J. E., Slightom J. L., Tucker P. W., Smithies O. Cloning human fetal gamma globin and mouse alpha-type globin DNA: preparation and screening of shotgun collections. Science. 1978 Dec 22;202(4374):1279–1284. doi: 10.1126/science.725603. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Clayton C. E., Rigby P. W. Cloning and characterization of the integrated viral DNA from three lines of SV40-transformed mouse cells. Cell. 1981 Aug;25(2):547–559. doi: 10.1016/0092-8674(81)90073-8. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Nucleotide sequence of a conserved SV40 DNA segment containing the origin of viral DNA replication as an inverted repetition. J Mol Biol. 1978 Dec 5;126(2):259–274. doi: 10.1016/0022-2836(78)90362-5. [DOI] [PubMed] [Google Scholar]
- Hiscott J. B., Murphy D., Defendi V. Instability of integrated viral DNA in mouse cells transformed by simian virus 40. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1736–1740. doi: 10.1073/pnas.78.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L., Boast S., Fried M. Excision of polyoma virus genomes from chromosomal DNA by homologous recombination. Nature. 1982 Jan 28;295(5847):349–350. doi: 10.1038/295349a0. [DOI] [PubMed] [Google Scholar]
- Lavi S., Winocour E. Acquisition of sequences homologous to host deoxyribonucleic acid by closed circular simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):309–316. doi: 10.1128/jvi.9.2.309-316.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. N., Brockman W. W., Nathans D. Evolutionary variants of simian virus 40: cloned substituted variants containing multiple initiation sites for DNA replication. Virology. 1975 Jul;66(1):53–69. doi: 10.1016/0042-6822(75)90178-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Furuta T., Ueda K., Oda K. Properties of T antigen-positive phenotypic revertants isolated from SV40-transformed rat and mouse cells. Microbiol Immunol. 1982;26(1):31–40. doi: 10.1111/j.1348-0421.1982.tb00151.x. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Hiwasa T., Oda K. I. Characterization of flat revertant cells isolated from simian virus 40-transformed mouse and rat cells which contain multiple copies of viral genomes. J Virol. 1981 Mar;37(3):1028–1043. doi: 10.1128/jvi.37.3.1028-1043.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T., Singer M., Rosenberg M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. II. The structure of variant 1103 and its comparison to variant CVPS/1P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3592–3597. [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Greene R., Stringer J., Mitchison T., Hu S. L., Botchan M. Analysis of the sites of integration of viral DNA sequences in rat cells transformed by adenovirus 2 or SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):569–584. doi: 10.1101/sqb.1980.044.01.059. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Westphal H., Srinivasan P. R., Dulbecco R. The integrated state of viral DNA in SV40-transformed cells. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1288–1295. doi: 10.1073/pnas.60.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemer D., Enquist L., Leder P. Improved derivative of a phage lambda EK2 vector for cloning recombinant DNA. Nature. 1976 Oct 7;263(5577):526–527. doi: 10.1038/263526a0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: nucleotide sequences at viral-viral recombinant joints in naturally arising variants. Virology. 1981 Mar;109(2):344–352. doi: 10.1016/0042-6822(81)90505-5. [DOI] [PubMed] [Google Scholar]