Abstract
Rho and Rab family GTPases play a key role in cytoskeletal organization and vesicular trafficking, but the exact mechanisms by which these GTPases regulate polarized cell growth are incompletely understood. A previous screen for genes that interact with CDC42, which encodes a Rho GTPase, found SWF1/PSL10. Here, we show Swf1p, a member of the DHHC-CRD family of palmitoyltransferases, localizes to actin cables and cortical actin patches in Saccharomyces cerevisiae. Deletion of SWF1 results in misorganization of the actin cytoskeleton and decreased stability of actin filaments in vivo. Cdc42p localization depends upon Swf1p primarily after bud emergence. Importantly, we revealed that the actin regulating activity of Swf1p is independent of its DHHC motif. A swf1 mutant, in which alanine substituted for the cysteine required for the palmitoylation activity of DHHC-CRD proteins, displayed wild-type actin organization and Cdc42p localization. Bgl2p-marked exocytosis was found wild type in this mutant, although invertase secretion was impaired. These data indicate Swf1p has at least two distinct functions, one of which regulates actin organization and Bgl2p-marked secretion. This report is the first to link the function of a DHHC-CRD protein to Cdc42p and the regulation of the actin cytoskeleton.
INTRODUCTION
Polarized secretion supported by an asymmetrically organized actin cytoskeleton promotes polarized morphogenesis in many cell types. Recent studies showed that Cdc42p, a small GTPase of the Rho family with multiple distinct functions, is essential for the proper polarization of the actin cytoskeleton and secretory apparatus (reviewed in Nelson, 2003; Etienne-Manneville, 2004; Van Aelst and Cline, 2004; Park and Bi, 2007). For example, in mouse hippocampal neurons, knockout of Cdc42 leads to organizational defects in the actin cytoskeleton (Garvalov et al., 2007), whereas, expression of dominant Cdc42 alleles lead to a decrease in the polarized distribution of actin and a specific population of exocytotic vesicles marked with tetanus neurotoxin insensitive-vesicle–associated membrane protein (Alberts et al., 2006). Cdc42-dependent defects in exocytosis are not always correlated with defects in the actin cytoskeleton. Expression of dominant-negative Cdc42 in Madin-Darby canine kidney cells, for example, inhibits vesicle trafficking to the basolateral membrane without apparent effect on the actin cytoskeleton (Kroschewski et al., 1999). In Saccharomyces cerevisiae, alleles of cdc42 revealed that Cdc42p is necessary for both the polarization of the actin cytoskeleton and the exocyst complex at sites of polarized growth, i.e., the bud tip (Adamo et al., 2001; Zhang et al., 2001; Roumanie et al., 2005). Even with a steady advance toward understanding the mechanics of Cdc42p-dependent cell polarization in recent years, especially the functional links that coordinate the organization and dynamics of the actin cytoskeleton with vesicle trafficking, many of the genes that promote CDC42-dependent cell polarization remain unidentified. For this reason, several genetic screens were undertaken to identify genes that promote CDC42-dependent cell polarization in S. cerevisiae (Kozminski et al., 2003).
SWF1/PSL10/YDR126W (hereafter SWF1) was one of 30 CDC42-interacting genes identified in S. cerevisiae in a synthetic genetic interaction screen for genes involved in cell polarization (Kozminski et al., 2003). This gene was also identified previously in three independent genetic screens, each of which implicated Swf1p in a specific cellular process. A screen for genes required for meiotic division and recombination identified SWF1 as important for spore wall formation (Enyenihi and Saunders, 2003). In mitotic cells, in a screen based on the missorting of carboxypeptidase Y, SWF1 was found involved in vesicle trafficking and sorting to the vacuole (Bonangelino et al., 2002). Although seeming dissimilar, all of these cellular processes depend in common upon a functioning actin cytoskeleton (Hill et al., 1996; Bonangelino et al., 2002; Taxis et al., 2006; Isgandarova et al., 2007). The idea of a physiological link between SWF1 function and the actin cytoskeleton is further supported by the identification of SWF1 as PSL10 (profilin synthetic lethal). Haarer found that deletion of SWF1 is synthetic lethal with a hypomorphic allele of PFY1 (Bartels et al., 1999), which encodes a potent regulator of the actin cytoskeletal dynamics and organization (Jockusch et al., 2007). However, whether SWF1 regulates the dynamics and organization of the actin cytoskeleton has not been examined.
SWF1 encodes a polypeptide (Swf1p) with five predicted transmembrane domains; it is one of seven proteins in S. cerevisiae that contain a DHHC-CRD motif (Putilina et al., 1999; Linder and Deschenes, 2003; Politis et al., 2005; Mitchell et al., 2006). As with many of the >20 DHHC-CRD–containing proteins in mammals (Fukata et al., 2004), including Huntingtin interacting protein 14 (Ducker et al., 2004), three of the DHHC-CRD proteins in yeast (Erf2p, Pfa3p, and Akr1p) have palmitoyltransferase (PT) activity. Swf1p seems to have PT activity as well (Valdez-Taubas and Pelham, 2005), although in vitro reconstitution of this activity has yet to be demonstrated. Palmitoylation, the result of PT activity, is a posttranslational lipid modification that is essential for the trafficking of signaling molecules such as Ras, and, in the case of mutant huntingtin protein, for the prevention of protein aggregation (see references in Linder and Deschenes, 2007). In budding yeast, Swf1p was shown to palmitoylate the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins Snc1p, Syn8, and Tlg1p (Valdez-Taubas and Pelham, 2005). Palmitoylation protects the Qc-SNARE Tlg1p from ubiquitin-mediated degradation, suggesting that palmitoylation is one mechanism by which a cell spatially and temporally regulates exocytosis (Valdez-Taubas and Pelham, 2005). One prediction of this model that has not been tested is whether loss of the DHHC motif (i.e., the predicted PT activity) results in a secretory defect, and, if so, whether this secretory defect occurs independently of effects on the actin cytoskeleton. This prediction is tested in this study.
MATERIALS AND METHODS
Strains and Microbial Techniques
Yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. S. cerevisiae and Escherichia coli strains were cultured and transformed as described in Kozminski et al. (2003, 2006), unless otherwise noted. Multiple independent transformants or independently derived strains were analyzed in each experiment.
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| DDY1102 (alias KKY49) | MATa/MATα SWF1/SWF1 ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ADE2/ade2-1 lys2-801/LYS2 | Kozminski et al. (2000) |
| KKY281 | MATα CDC42:natMX can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Kozminski et al. (2003) |
| KKY282 | MATα cdc42-101:natMX can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | This studya |
| KKY283 | MATα cdc42-118:natMX can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Kozminski et al. (2003) |
| KKY293 | MATα cdc42-129:natMX can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | This studya |
| KKY1060 | MATa/MATα swf1Δ::kanMX4/swf1Δ::kanMX4 ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ADE2/ade2-1 lys2-801/lys2-801 | KKY1063 × KKY1065 |
| KKY1062 | MATa/MATα SWF1/swf1Δ::kanMX4 ura3-52/ura3-52 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ADE2/ade2-1 lys2-801/lys2-801 | KKY1064 × KKY1065 |
| KKY1063 | MATα swf1Δ::kanMX4 ura3-52 his3Δ200 leu2-3,112 lys2-801 | Meiotic product of KKY1074 |
| KKY1064 | MATα ura3-52 his3Δ200 leu2-3,112 lys2-801 | Meiotic product of KKY1074 |
| KKY1065 | MATaswf1Δ::kanMX4 ura3-52 his3Δ200 leu2-3,112 ade2-1 lys2-801 | Meiotic product of KKY1074 |
| KKY1067 | MATaswf1Δ::kanMX4 CDC42:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 | Meiotic product of KKY1145 |
| KKY1068 | MATaswf1Δ::kanMX4 cdc42-118:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 | Meiotic product of KKY1144 |
| KKY1069 | MATa/MATα swf1Δ::kanMX4/SWF1 CDC42/cdc42-129:natMX his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0/LYS2 CAN1/can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 met15Δ0/MET15 | KKY293 × 1-7-11 |
| KKY1070 | MATaswf1Δ::kanMX4 cdc42-101:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 | Meiotic product of KKY1143 |
| KKY1074 | MATa/MATα swf1Δ::kanMX4/SWF1 his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 ADE2/ade2-1 lys2-801/LYS2 | This studya |
| KKY1084 | KKY1064 [pKK1586, YCplac33(SWF1)] | This studyb |
| KKY1085 | KKY1064 [pKK1873, YCplac33(swf1-DHHA)] | This studyb |
| KKY1086 | KKY1063 [pKK1586,YCplac33(SWF1)] | This studyb |
| KKY1087 | KKY1063 [pKK1873, YCplac33(swf1-DHHA)] | This studyb |
| KKY1088 | MATapfy1-4:LEU2 ura3-52 his3Δ200 leu2-3,112 lys2-801 | This studyc |
| KKY1090 | MATα swf1Δ::kanMX4 ura3-52 his3Δ200 leu2-3,112 lys2-801 | This studyd |
| KKY1104 | MATa/MATα SWF1-GFP:kanMX4/SWF1 his3Δ200/his3Δ200 ura3-52/ura3-52 leu2-3,112/leu2-3,112 ADE2/ade2-1 lys2-801/LYS2 | This studya |
| KKY1120 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 | BY4741 × BY4742 |
| KKY1134 | MATaswf1Δ::kanMX4 cdc42-101:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 [YCplac33] | This studye |
| KKY1135 | MATaswf1Δ::kanMX4 cdc42-101:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 [pKK1586,YCplac33(SWF1)] | This studye |
| KKY1136 | MATaswf1Δ::kanMX4 cdc42-101:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 [pKK1873, YCplac33(swf1-DHHA)] | This studye |
| KKY1137 | KKY1068 [YCplac33] | This studyb |
| KKY1138 | KKY1068 [pKK1586,YCplac33(SWF1)] | This studyb |
| KKY1139 | KKY1068 [pKK1873, YCplac33(swf1-DHHA)] | This studyb |
| KKY1140 | MATaswf1Δ::kanMX4 cdc42-129:natMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 can1Δ::MFA1pr-HIS3-MFα1pr-LEU [pKK1586,YCplac33(SWF1)] | This studyf |
| KKY1141 | KKY1069 [YCplac33] | This studyb |
| KKY1142 | KKY1069 [pKK1873, YCplac33(swf1-DHHA)] | This studyb |
| KKY1143 | MATa/MATα swf1Δ::kanMX4/SWF1 CDC42/cdc42-101:natMX his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0/lys2Δ0 CAN1/can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 met15Δ0/MET15 | KKY282 × 1-7-11 |
| KKY1144 | MATa/MATα swf1Δ::kanMX4/SWF1 CDC42/cdc42-118:natMX his3 Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0/lys2Δ0 CAN1/can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 met15Δ0/MET15 | KKY283 × 1-7-11 |
| KKY1145 | MATa/MATα swf1Δ::kanMX4/SWF1 CDC42/CDC42:natMX his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0/lys2Δ0 CAN1/can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 met15Δ0/MET15 | KKY281 × 1-7-11 |
| KKY1146 | MATaswf1Δ::kanMX4 ura3-52 his3Δ200 leu2-3,112 lys2-801 | Meiotic product of KKY1074 |
| KKY1147 | MATaura3-52 his3Δ200 leu2-3,112 lys2-801 | Meiotic product of KKY1074 |
| DDY663 | MATα ura3-52 leu2-3,112 lys2-801 | D. Drubing |
| DDY664 | MATaura3-52 leu2-3,112 lys2-801 | D. Drubing |
| DDY1300 | MATaCDC42:LEU2 ura3-52 leu2-3,112 his3Δ200 lys2-801 | Kozminski et al. (2000) |
| DDY1304 | MATacdc42-101:LEU2 ura3-52 leu2-3,112 his3Δ200 lys2-801 | Kozminski et al. (2000) |
| DDY1326 | MATacdc42-118:LEU2 ura3-52 leu2-3,112 his3Δ200 lys2-801 | Kozminski et al. (2000) |
| DDY1336 | MATacdc42-123:LEU2 ura3-52 leu2-3,112 his3Δ200 lys2-801 | Kozminski et al., (2000) |
| DDY1344 | MATacdc42-129:LEU2 ura3-52 his3Δ200 leu2-3,112 lys2-801 | Kozminski et al. (2000) |
| DDY2008 | MATapfy1-4:LEU2 ura3-52 his3Δ200 leu2-3,112 lys2-801 | Wolven et al. (2000) |
| 1-7-11 | MATaswf1Δ::kanMX4 ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 | Winzeler et al. (1999) |
| BY4741 | MATahis3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Brachmann et al. (1998) |
| BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Brachmann et al. (1998) |
| NY17 | MATasec6-4 ura3-52 | Novick et al. (1980) |
| Y3611 | MATa/MATα can1Δ::MFA1pr-HIS3-MFα1pr-LEU2/CAN1 his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2 | Kozminski et al. (2003) |
a See Materials and Methods.
b By transformation with indicated plasmid.
c Progeny of backcross of DDY2008 with DDY663.
d Progeny of backcross of KKY1063 with DDY663.
e Meiotic product of KKY1143 transformed with indicated plasmid.
f Meiotic product of KKY1069 transformed with indicated plasmid.
g University of California at Berkeley.
Table 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Source |
|---|---|---|
| pAC129 | pALTER-1(cdc42–118) | Kozminski et al. (2000) |
| pAC183 | pRS305(cdc42–101:LEU2)a | Kozminski et al. (2000) |
| pAC329 | pRS305(cdc42–129:LEU2)a | Kozminski et al. (2000) |
| pGAT2 | pBAT(GST) | Peränen et al. (1996) |
| pML1 | pFA6a-kanMX6 | Longtine et al. (1998) |
| pML4 | pFA6a-GFP(S65T)-kanMX6 | Longtine et al. (1998) |
| YCplac33 | CEN URA3 | Gietz and Sugino (1988) |
| YEplac195 | 2 μ URA3 | Gietz and Sugino (1988) |
| pKK661 | pGAT2(CDC42) | Kozminski et al. (2003) |
| pKK1469 | pRS305(cdc42–101:natMX)a | This study |
| pKK1473 | pRS305(cdc42–129:natMX)a | This study |
| pKK1516 | pGAT2(GST-cdc42–118) | This study |
| pKK1585 | YEplac195(SWF1) | This study |
| pKK1586 | YCplac33(SWF1) | This study |
| pKK1871 | pGAT2(GST-SWF1) | This study |
| pKK1872 | pGAT2(GST-SWF1-C) | This study |
| pKK1873 | YCplac33(swf1-DHHA) | This study |
a Modified pRS305 vector lacking LEU2. Alias of this vector is pKK294 (Kozminski et al., 2000).
KKY282 and KKY293 were derived from Y3611, by using pKK1469 and pKK1473 as sources of integrating DNA, respectively, following the method that was used to construct KKY281 and KKY 283 (Kozminski et al., 2003).
The method of Longtine et al. (1998) was used to delete SWF1 in DDY1102, forming KKY1074. Polymerase chain reaction (PCR) was used to amplify a kanMX4 disruption cassette with Accuzyme (Bioline, Randolph, MA) from template pML1 with the primers oSD2 (CTCTTTAGGAAATTCGTAGCTATCAACAAAGGTAGTGTCATTGTATATATACTTCCGCCCGGATCCCCGGGTTAATTAA) and oKK144 (AATAAACCGCTTGTATAATGAATTTGTTCGACATTTGCGGTTAGAATCTATGGCGGTGAGGAATTCGAGCTCGTTTAAAC; SWF1 flanking sequence underlined). The same method was used to introduce a green fluorescent protein (GFP) tag at the C terminus of Swf1p in DDY1102, forming KKY1104. PCR was used to amplify GFP-kanMX from template pML4, by using Accuzyme (Bioline) and primers oKK143 (CGAAATTCCCAATATATATGACAAAGGTACCTTCCTGGCCAATCTCACAGATTTAATACGGATCCCCGGGTTAATTAA; SWF1 flanking sequence underlined) and oKK144.
Plasmids
To construct a 2μ plasmid containing SWF1 (pKK1585), the region of chromosome IV between base pairs 702754 and 704475 was amplified from KKY283 genomic DNA with BioXact DNA polymerase (Bioline) by using the primers oKK172 (GACTAAGCTTCATTCGCTCATCCTTAAAC; HindIII site underlined) and oKK173 (CTGATCTAGAGTAATATACAATTATCTTACGTAG; XbaI site underlined), and subcloned into the HindIII and XbaI sites of YEplac195. To verify the fidelity of amplification, the subcloned PCR fragment was sequenced. To construct a CEN plasmid containing SWF1 (pKK1586), a HindIII-XbaI fragment from pKK1585 was subcloned into the HindIII and XbaI sites of YCplac33.
To construct pKK1872 (GST-SWF1-C), the coding sequence for the nonconserved C terminus (amino acids 235–336) of Swf1p was amplified by PCR from pKK1586 by using the primers oKK170 (GACCAAGCTTCTATATTAAATCTGTGAGATTGGC; HindIII site underlined) and oKK182 (CTGAGGATCCGCCATTGTAAAGGAGGGAATG; BamHI site underlined) and subcloned into the BamHI and HindIII sites of pKK1871. To verify the fidelity of amplification, the subcloned PCR fragment was sequenced. To construct pKK1871 (GST-SWF1), the coding sequence of SWF1 was amplified by PCR from DDY1102 genomic DNA using primers oKK169 (ACGCGGATCCATGTCATGGAATCTACTATTTGTG; BamHI site underlined) and oKK170 and subcloned into the BamH1 and HindIII sites of pKK1516. To construct pKK1516, the coding sequence of cdc42-118 was amplified by PCR from pAC129 by using primers oKK18 (GCGCGGGATCCATGCAAACGCTAAAGTGTGTTGTTG; BamHI site underlined) and oKK24 (GCGCGCAAGCTTCTACAAAATTGTACATTTTTTACTTTTC; HindIII site underlined) and subcloned into the BamHI and HindIII sites of pKK661.
The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to construct pKK1873 (swf1-DHHA), in which the codon for cysteine 164 was mutagenized to that of alanine. This PCR-based approach used pKK1586 as a template and primers oKK179 (GTCGCCGACCATCATGCCATCTGGATAAATAAC) and oKK180 (GTTATTTATCCAGATGGCATGATGGTCGGCGAC).
Removing a NotI-NotI LEU2-containing fragment from pAC183 and pAC329 and replacing it with a NotI-Not1 natMX-containing fragment constructed pKK1469 and pKK1473, respectively. The natMX fragment was generated by PCR as described in Kozminski et al. (2003).
α Factor Release Assay
α Factor (synthesized at the University of Virginia, Charlottesville, VA) was added to log-phase cells (OD600 = 0.25), grown in rich medium at 25°C, to a final concentration of 5 μg/ml. After culturing for an additional 90 min, the cells were harvested by centrifugation for 5 min at 1.5 krpm in an IEC clinical centrifuge at room temperature. Each cell pellet was then washed twice with 12.5 ml of rich medium and cultured as described above. After the second wash (t = 0), 0.5 ml of each culture was collected every 10 min and fixed with 4% formaldehyde. Before scoring bud size by phase microscopy, each sample was sonicated briefly.
Exocytosis Assays
Bgl2p secretion was assayed as described in Kozminski et al. (2006), following the method of Harsay and Schekman (2007).
To assay for the secretion of invertase (Suc2p), the assay of Bankaitis et al. (1989) was used with modification. For each strain assayed, 5 ml of mid-log phase culture (∼0.25–0.3 OD600) was pelleted at 600 × g for 5 min. The pellet was resuspended in 5 ml of rich medium (5% glucose) and then grown at 25°C for 60 min. To assay NY17 cells at permissive and restrictive temperatures, 10 ml of mid-log phase culture was split equally and processed in parallel, except one culture was preshifted to 37°C for 15 min after 60-min growth at 25°C. Both NY17 cultures were then pelleted as described above and washed with 10 ml of sterile water prewarmed to either 25 or 37°C. After the wash, cells were resuspended in 3 ml of rich medium (0.1% glucose) and incubated at either 25°C or 37°C for 90 min. After this incubation, all cultures, including the NY17 cultures, were harvested as described above. Cells were gently resuspended in 5 ml of ice-cold 10 mM NaN3/10 mM KF, incubated on ice for 10 min, and then diluted with 5 ml of ice-cold water. The cell suspension was further diluted with ice-cold water if the OD600 was >0.7. The cell suspension was centrifuged again as before. Pellets were washed twice with 10 ml of water and then resuspended gently in 1 ml of 0.2 M sodium acetate, pH 4.9. Then, 0.5 ml of each cell suspension was transferred to each of two new microfuge tubes. One tube was maintained on ice, and to the other tube 12 μl of 20% Triton X-100 was added, with gentle mixing followed by one freeze-thaw cycle in a dry ice-ethanol bath. The equivalent of 0.02 OD600 units from each sample was assayed for invertase activity following the method of Goldstein and Lampen (1975).
Electron Microscopy
For the analysis of vesicles within yeast, thin sections for transmission electron microscopy were prepared according to the methods of Salminen and Novick (1987) and Wright (2000), with modifications. In brief, 12.5 ml of mid-log phase yeast culture (∼6 OD600 units), grown in rich medium at 25 or 37°C were rapidly pipetted into an equal volume of 2× fixative (0.2 M sodium cacodylate buffer, pH 6.8, 0.4 M d-sorbitol, 4 mM MgCl2, 4 mM CaCl2, 8% paraformaldehyde, and 6% glutaraldehyde; fixatives from Ted Pella, Redding, CA) and incubated 10 min at room temperature. Cells were then pelleted at 1500g for 5 min, resuspended in 25 ml of fixative, and incubated overnight at 4°C. Fixed cells were pelleted at 1500 × g for 5 min at room temperature, washed twice with 20 ml of 0.1 M cacodylate buffer, pH 6.8, and then washed twice with 25 ml of 50 mM potassium phosphate buffer, pH 7.5. Cells were then pelleted as described above, resuspended in 1 ml of 50 mM potassium phosphate buffer containing 0.25 mg/ml Zymolyase 100T (Seikagaku, Tokyo, Japan), and incubated at 30°C for 45 min. Fixed spheroplasts were then pelleted at 1000 × g for 1 min at room temperature and washed thrice with ice-cold 0.1 M cacodylate buffer. After the wash, pelleted spheroplasts were resuspended in 1 ml of ice-cold 1% (vol/vol) osmium tetroxide and 0.8% (wt/vol) potassium ferricyanide in 0.1 M cacodylate buffer, pH 6.8, for 1 h on ice. The spheroplasts were then washed thrice with deionized water, resuspended in aqueous 0.5% uranyl acetate for 30 min at room temperature in the dark, and then washed twice with deionized water. Graded dehydration with ethanol and then acetone preceded embedment with Spurr's resin. Silver/gold thin sections were cut with a diamond knife and stained with lead citrate (Venable and Coggeshall, 1965) for 5 min, 3% uranyl acetate in 50% acetone for 15 min, and lead citrate again for 5 min. Stained sections were examined on a JEOL 100CXII electron microscope (JEOL, Tokyo, Japan) equipped with a SIA L-3C digital camera (Scientific Instruments and Applications, Atlanta, GA).
Glutathione Transferase (GST)-Swf1-C Expression and Purification
GST-Swf1-C was purified from BL21(DE3) E. coli transformed with pKK1872 after an overnight induction at 25°C with 0.1 mM isopropyl β-d-thiogalactoside. All subsequent procedures were completed at 4°C. Bacteria were harvested from a 1-l culture at 4000 rpm (20 min; Beckman J6 rotor). The pellet was resuspended in 40 ml of ice-cold lysis buffer (50 mM Tris-Cl, pH 8.0, 1 mM EDTA, 1 mM dithiothreitol [DTT], 250 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 0.5 μg/ml each of leupeptin, aprotinin, pepstatin A, chymostatin, and antipain). Bacteria were then lysed on ice with sonication. Lysate was centrifuged at 35,000 rpm for 45 min in a Beckman Ti45 rotor. The supernatant was recovered and nutated with 2 ml of glutathione-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), prewashed in lysis buffer, for 20 min. After this incubation, the bead suspension was poured into a column and successively washed with 25 ml of wash buffer [50 mM Tris-Cl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 200 mM NaCl, 5% (vol/vol) glycerol, and 1 mM PMSF], 250 ml of wash buffer with 1% Triton X-100, 250 ml of wash buffer with 400 mM NaCl, 250 ml of wash buffer with 1% betaine and 0.1% Tween 20, and 250 ml of wash buffer. GST-Swf1-C was eluted as 1-ml fractions with elution buffer [50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 0.1 mM EGTA, 5% (vol/vol) glycerol, and 25 mM glutathione] until the absorbance of the eluate at 280 nm reached 0.02. Peak GST-Swf1-C fractions, as determined by absorbance spectroscopy at 280 nm and SDS-polyacrylamide gel electrophoresis (PAGE), were pooled (∼4 ml total volume) and twice dialyzed against 1.5 l of phosphate-buffered saline (PBS) containing 0.1 mM DTT and 10% (vol/vol) glycerol. Final protein concentration was determined by UV absorbance at 280 nm or by Bradford assay (Bio-Rad, Hercules, CA), by using bovine serum albumin (BSA) as a standard.
Swf1p Antibody Production and Purification
Because thrombin cleavage of the GST-Swf1-C fusion protein resulted in complete degradation of the Swf1 fragment, uncleaved GST-Swf1-C was used as an immunogen. Covance Research Products (Denver, PA) performed all inoculations and bleeds. We brought 250 μg of GST-Swf1-C to 0.5 ml with PBS and mixed with 0.5 ml of Freund's complete adjuvant before subcutaneous injection into New Zealand White rabbits. Booster injections containing 125 μg of GST-Swf1-C and Freund's incomplete adjuvant were administered 3 wk after the initial injection. Bleeds were collected 10 d after each injection and screened for immunoreactivity against Swf1p in whole cell lysates of wild-type yeast (DDY1102). A strain lacking Swf1p (KKY1060) was used as a control. Immunoreactivity was first detected in lysates at week 10. Exsanguination occurred at week 16.
The purification of antibodies against Swf1p proceeded in two steps. First, antibodies against GST were depleted from serum by affinity chromatography, by using a GST column prepared as described in Kozminski et al. (2000). Depletion was confirmed by probing dot blots of recombinant GST with crude serum or with the eluate of the GST column. Second, serum depleted of antibodies against GST and diluted 1:50 in Tris-buffered saline containing 1 mg/ml BSA was preabsorbed three times, at room temperature for 2 h each, against nitrocellulose filters containing whole cell lysates of a swf1Δ/swf1Δ strain KKY1060.
Fluorescence Microscopy
Cells prepared for direct or indirect fluorescence microscopy were observed with epifluorescence on a Nikon E800 microscope equipped with a 100×/1.3 Plan Neofluar objective. Images were captured with an Orca 100ER digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab software (Improvision, Lexington, MA). Unless otherwise noted, exposure and contrast enhancement were constant and linear for each image series.
Cells were prepared for indirect immunofluorescence microscopy as described in Kozminski et al. (2000, 2006), except when cells were probed with antibody against Tpm1p or Cdc11p. In those cases, methanol and acetone replaced SDS at the permeabilization step per the method of Pringle et al. (1989). Polyclonal rabbit antibodies against Swf1p (this study), Cdc42p (Kozminski et al., 2000), GFP (Abcam, Cambridge, MA), Tpm1p (Pruyne et al., 1998), and Cdc11p (Santa Cruz Biotechnology, Santa Cruz, CA) were used at 1:100, 1:625, 1:1000, 1:50, and 1:300, respectively. Guinea pig polyclonal antibody against yeast actin (Palmgren et al., 2001) was used at 1:250. Secondary antibodies goat anti-rabbit Alexa 568-conjugated (Invitrogen, Carlsbad, CA), goat anti-rabbit fluorescein isothiocyanate (FITC)-conjugated (Jackson ImmunoResearch Laboratories, West Grove, PA), and donkey anti-guinea pig rhodamine-conjugated (Jackson ImmunoResearch Laboratories) were used at 1:50 per the manufacturers' instructions.
Actin localization in fixed cells by using rhodamine-conjugated phalloidin (Invitrogen) followed a modified method of Adams and Pringle (1991). One milliliter of mid-log phase yeast culture was fixed for 20 min at room temperature with 5% (vol/vol) formaldehyde. Cells were pelleted in a microfuge for 1 min at 8000 × g, washed twice with 1 ml of PBS, and then resuspended in PBS containing 0.1% (vol/vol) Triton X-100 for 4 min at room temperature. Detergent was removed with two additional PBS washes. Pelleted cells were resuspended in 2.2 μM rhodamine-conjugated phalloidin in PBS and incubated 2 h in the dark at room temperature. To remove excess rhodamine-conjugated phalloidin, stained cells were washed three times with 1 ml of PBS containing 0.1% (vol/vol) Triton X-100. After the last wash, cells were resuspended in 10–20 μl of mounting medium. Then, 3–4 μl of stained cell suspension was applied to a slide, after which a coverslip was affixed with nail polish.
The staining of bud scars with Calcofluor White (Fluorescent Brightner 28; Sigma-Aldrich, St. Louis, MO) followed the method of Pringle (1991).
Immunoblotting
Immunoblotting was performed as described in Kozminski et al. (2000, 2006). Swf1p was detected with purified anti-Swf1p antibodies diluted 1:1000. Tubulin served as a loading control and was detected with AA2, a mouse monoclonal antibody raised against amino acids 412–430 of bovine brain β-tubulin (gift of A. Frankfurter, University of Virginia), diluted to 130 ng/ml.
Assays of Actin Dynamics
To compare the sensitivity of yeast to latrunculin A (LatA), halo assays were performed. For each strain tested, an overnight culture grown in rich medium was diluted to 0.1 OD600 and plated on rich medium. After aspiration of excess culture, plates were dried for 1 h. Each of three different concentrations of LatA (BIOMOL Research Laboratories, Plymouth Meeting, PA), serially diluted with dimethyl sulfoxide, was spotted (10 μl) onto 6-mm concentration disks (B231039; Fisher Scientific, Sparks, MD), which were then placed on the lawn of cells. Plates were incubated at 25° for 2 d. The diameter of the zone of no growth around each disk was measured and used to calculate the relative apparent sensitivity of each strain to LatA following the method of Reneke et al. (1988).
To compare the turnover of actin filaments in vivo among different yeast strains, the method of Lappalainen and Drubin (1997) was used. Briefly, log-phase cultures grown in rich medium at 25°C were diluted to 0.4 OD600. LatA was then added to each 0.4-ml culture to 0.4 mM final. At 0, 2, and 6 min post-LatA addition, a 125-μl aliquot of each culture was fixed with 4.4% (vol/vol) paraformaldehyde for 20 min at room temperature. Fixed cells were then stained with 6.6 μM rhodamine-phalloidin (Invitrogen) in methanol and imaged as described above.
Actin Binding Assays
To determine whether the C-terminal domain of Swf1p binds filamentous actin (F-actin), a supernatant depletion assay (Mullins et al., 1997) was performed. For 200-μl reactions, rabbit muscle F-actin (kind gift of Dr. D. Schafer, University of Virginia) in MKEI-50 (20 mM imidazole-KOH, pH 7.0, 50 mM KCl, 2 mM MgCl2, and 1 mM EGTA) was diluted to a final concentration between 15 and 0.15 μM in MKEI-50 containing 0.5× G buffer (2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.1 mM DTT, and 0.005% NaN3). Then, 2 μl of GST-Swf1-Cp in PBS containing 10% glycerol and 0.1 mM DTT was mixed into each reaction to give a final fusion protein concentration of 1.5 μM. Reactions were incubated 1 h at room temperature and then ultracentrifuged at 70,000 rpm in a TLA120.1 rotor for 60 min at 4°C. The top 120 μl of supernatant was collected from each reaction and mixed with 40 μl of 4× protein sample buffer (4% SDS, 20% glycerol, 0.2M Tris-Cl, pH 7.0, 4 mM EDTA, 0.16 M DTT, and 0.2 mg/ml bromphenol blue). Equivalent volumes were then analyzed on a 13% SDS-PAGE gel stained with Coomassie Blue.
To determine whether the C-terminal domain of Swf1p binds G-actin, a pyrene–actin assembly assay was performed. For a 200-μl reaction, 16 μM rabbit muscle G-actin (5% pyrene-labeled; kind gift of Dr. D. Schafer) in G buffer was diluted to 4 μM in MKEI-50 containing 0.6× G buffer. Then, 10 μl of GST-Swf1-Cp in PBS containing 10% glycerol and 0.1 mM DTT (or buffer alone) was mixed into the reaction. Fluorescence intensity over time at 25°C was measured immediately using a QuantaMaster fluorometer (Photon Technology International, Birmingham, NJ).
RESULTS
Genetic Analysis Reveals a Functional Relationship between SWF1 and CDC42
We identified SWF1 in a synthetic genetic array screen for genes that promote polarized cell growth (Kozminski et al., 2003). SWF1 conferred a growth defect when deleted in combination with cdc42-118ts, an allele conditionally defective in polarity establishment (Kozminski et al., 2000). Of the 30 genes identified in this screen, 20, including SWF1, were not previously associated with polarized cell growth. Therefore, this screen associated SWF1 for the first time with the process of polarized cell growth.
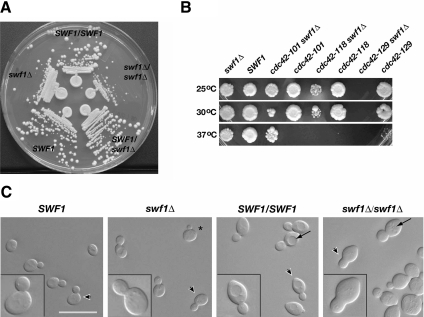
To begin to understand how SWF1 functions in the process of polarized cell growth, we first examined haploid and diploid strains lacking SWF1. We found that haploid cells with a SWF1 deletion doubled in log phase culture at 25°C at one third to one half the rate measured for congenic wild-type cells. A difference in doubling rate was also observed on solid media, in which haploid swf1Δ and homozygous diploid swf1Δ/swf1Δ cells formed smaller colonies than wild-type cells (Figure 1A). This slower rate of growth was not due a partial block at a specific cell cycle phase, but rather it seemed to be due to a slower rate of bud growth in the mutant cells relative to wild-type cells. Asynchronous cultures of the haploid wild-type and swf1Δ strains contained a very similar morphological distribution of cells throughout the cell cycle, as assayed by a visual scoring of bud size (Supplemental Table 1). The only notable difference between these cultures was that the swf1Δ culture contained fewer unbudded cells than the wild-type culture, 30 versus 43%, respectively. A similar, but smaller difference, was also observed in asynchronous cultures of the wild-type and homozygous swf1Δ/swf1Δ diploid cells (Supplemental Table 1). Consistent with a bud growth defect rather than a bud emergence defect, swf1Δ and SWF1 haploid cells initiated bud emergence at approximately the same time after release from α-factor arrest (Supplemental Figure 1). The swf1Δ growth phenotype was recessive; a heterozygous (swf1Δ/SWF1) diploid strain grew at the same rate as a congenic wild-type (SWF1/SWF1) strain (Figure 1A). Calcofluor staining did not reveal any defects in bud site selection in the mutant cells (data not shown).
Figure 1.
Analyses of the growth, genetic interactions, and morphology of cells lacking SWF1. (A) Cells lacking SWF1 formed smaller colonies than wild-type cells. Wild-type and mutant cells were streaked on rich medium and grown for 2 d at 25°C. Shown clockwise, wild-type diploid (DDY1102), homozygous mutant (KKY 1060), heterozygous mutant (KKY1062), wild-type haploid (KKY1064), and mutant haploid (KKY1063). (B) swf1Δ displayed synthetic genetic interaction with cdc42 alleles defective in distinct processes. Equivalent dilutions of SWF1, swf1Δ, cdc42, and swf1Δ cdc42 strains were plated and grown at 25, 30, and 37°C for 2 d on rich medium. The strains shown (from left to right) are KKY1067, 281, 1070, 282, 1068, 283, and 293. No swf1Δ cdc42-129 cells were plated because these two alleles are lethal in combination. (C) Cells that lacked SWF1 had an aberrant morphology, most notably a thickening of the mother-bud neck (compare insets). Shown from left to right are differential interference-contrast images of log-phase congenic haploid wild-type (KKY1064) and swf1Δ cells (KKY1063) as well as diploid wild-type (DDY1102) and swf1Δ/swf1Δ cells (KKY1060) grown in rich medium at 25°C. Arrowheads indicate the cells presented in the insets at an enlarged view. Arrows indicate vacuoles. Bar, 10 μm.
Consistent with the results of the synthetic genetic array screen described above, synthetic sickness was found at 25°C when swf1Δ was absent, in haploid cells, in combination with cdc42-118 (Figure 1B). Mutations that have a synthetic interaction with cdc42-118 often show the same interaction with cdc42-101, an allele that also confers a temperature-conditional polarized growth defect in G1 of the cell cycle. Surprisingly, deletion of SWF1 suppressed the growth defect of cdc42-101 at restrictive temperature (Figure 1B). In addition, swf1Δ was found synthetic lethal with cdc42-129, an allele that inhibits the G2/M switch from apical-to-isotropic bud growth. These synthetic genetic interactions did not seem to be an additive effect. That is, synthetic sickness between swf1Δ and temperature-conditional alleles of CDC42 did not correlate with the restrictive temperature range of the cdc42 alleles (Figure 1B). Although the molecular basis for the specificity of these genetic interactions is unknown, these results indicated that SWF1 functions in or in parallel with one or more CDC42-dependent pathways that regulate polarized cell growth.
SWF1 Is Required for Proper Polarized Cell Growth and Regulates Actin
The genetic interaction found between SWF1 and CDC42 and the dependency of SNARE palmitoylation upon Swf1p in vivo (Valdez-Taubas and Pelham, 2005) suggested that SWF1 is necessary for proper polarized cell growth. Consistent with this prediction, we found that both haploid and diploid cells lacking SWF1 displayed an aberrant morphology. In comparison with wild-type cells, swf1Δ haploid cells seemed rounder (see cell with asterisk in Figure 1C). Diploids homozygous for swf1Δ (Figure 1C) but not heterozygous (data not shown) displayed a similar mutant morphology. The aberrant thickening of the neck was especially noticeable in mutant diploid cells, along with fragmentation of the vacuole, which was less apparent in swf1Δ haploid cells (Figure 1C).
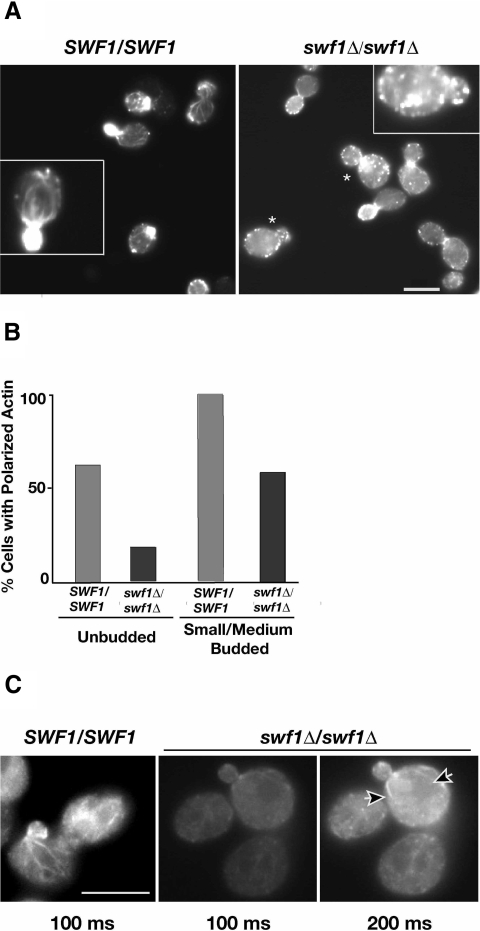
To determine whether cells lacking SWF1 are defective in cell polarization, as implied by the genetic interactions between swf1Δ and cdc42-118ts, we used the cell cycle-dependent organization of the cortical actin cytoskeleton as a readout for cell polarization. In unbudded wild-type cells in late G1, cortical actin patches are concentrated at the incipient bud site. In S and G2 phases of the cell cycle, cortical actin patches are found at the apical tip of small- and medium-sized buds, with actin cables running parallel to the mother-bud axis (Figure 2A, left). This pattern of cytoskeletal organization was significantly different in swf1Δ haploid cells (Figure 8A, top right) and swf1Δ/swf1Δ diploid cells (Figure 2A, right). Actin patches were no longer found polarized at the apical bud tip in mutant cells to the extent found in wild-type cells (Figure 2A). Rather, in swf1Δ/swf1Δ cells, actin patches were found distributed throughout mother cells and buds, often in greatest concentration at the bud neck. Only 58% of small- and medium-budded mutant cells had properly polarized actin patches, in comparison with 100% of the budded wild-type cells (Figure 2B). In mutant cells, loss of cell polarization was not restricted to one phase of the cell cycle. Only 18% of unbudded swf1Δ/swf1Δ cells, grown in rich medium, displayed polarization of the cortical actin cytoskeleton, in comparison with 62% of wild-type cells grown under the same conditions (Figure 2B). Consistent with a previous report (Bonangelino et al., 2002), actin cables were difficult to observe, if observed at all, in both swf1Δ haploid and homozygous diploid mutants (Figures 8A and 2, A and C, respectively). When actin cables were observed in mutant cells, they seemed short and very thin compared with those observed in wild-type cells (Figure 2, A and C). Cables rarely ran the complete length of the mother cell in cells lacking SWF1, although they were frequently found to extend from bud site to opposite pole in wild-type cells. These differences were accentuated when cables were visualized independently of actin patches (Figure 2C), by using an antibody against tropomyosin (Tpm1p), an F-actin binding protein that associates exclusively with actin cables (Pruyne et al., 1998). These results indicate that SWF1 is necessary for proper cell polarization and actin organization.
Figure 2.
Cell polarization was disrupted in the absence of SWF1. (A) Log phase wild-type (DDY1102; left) and homozygous mutant diploid (KKY1060; right) cells, grown in rich medium at 25°C, stained with rhodamine-phalloidin. The inset in the left panel shows a cell with a polarized cortical actin cytoskeleton, and the inset in the right panel shows a cell with a nonpolarized cortical actin cytoskeleton. Polarization of the cortical actin cytoskeleton (actin patches) was absent in many swf1Δ/swf1Δ cells; examples are marked with asterisk. Bar, 5 μm. (B) Comparison of the number of wild-type and mutant cells with a polarized cortical actin cytoskeleton at different cell cycle phases. The cortical actin cytoskeleton was scored as polarized when cortical actin patches were distributed to one pole of unbudded cells or were distributed to the bud in small- and medium-budded cells. All scored cells had single nucleus as visualized with 4,6-diamidino-2-phenylindole (DAPI). n > 325 scored for each morphological class. The cells quantified were from the same experiment shown in A. (C) Indirect immunofluorescence micrographs of log phase wild-type (DDY1102; left) and homozygous mutant diploid (KKY1060; middle and right) cells, grown in rich medium at 25°C, and probed with anti-tropomyosin (Tpm1p) and goat anti-rabbit-Alexa 568 antibodies. Tpm1-marked actin cables were shorter in swf1Δ/swf1Δ cells (arrowheads), compared with wild-type, and required twice the image acquisition time to visualize. The middle and right panels are identical, except for the image acquisition time, which is indicated below each panel. Bar, 5 μm.
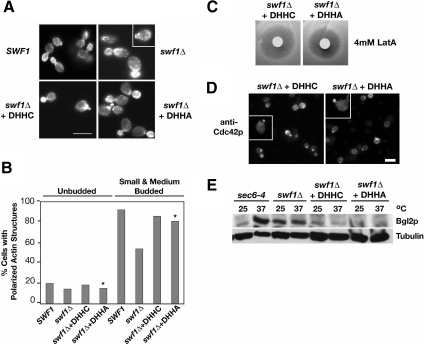
Figure 8.
Actin organization, Cdc42p localization, and Bgl2p-marked secretion were independent of the Swf1p DHHC motif. (A) Micrographs of rhodamine-phalloidin-stained, log phase, haploid, wild-type SWF1 cells (KKY1064), mutant swf1Δcells (KKY 1063), and swf1Δ cells with CEN plasmid that contained wild-type (+DHHC; KKY1086) or mutant SWF1 (+DHHA; KKY1087). All strains were grown at 25°C. SWF1 and swf1Δ stains were grown on rich medium and swf1Δ strains containing a plasmid were grown in SC-URA. Bar, 5 μm. (B) Comparison of the number of cells with polarized cortical actin cytoskeleton, at different cell cycle phases. The cortical actin cytoskeleton was scored as polarized when cortical actin patches were distributed to one pole of unbudded cells or were distributed to the bud in small- and medium-budded cells. All scored cells had a single nucleus as visualized with DAPI. n > 200 cells scored for each morphological class. The cells quantified were from the same experiment shown in A. Asterisks highlight the strain that lacked the DHHC motif. (C) Halo assay with the same strains used in A plated on SC-Ura and incubated at 25°C for 2 d. Filter disks were impregnated with 4 mM LatA. (D) Indirect immunofluorescence micrographs of log-phase swf1Δ cells expressing Swf1-DHHCp (KKY1086; left) or Swf1-DHHAp (KKY1087; right), grown at 25°C in SC-Ura, labeled with anti-Cdc42p and goat anti-rabbbit-Alexa 568 antibodies. Bar, 5 μm. (E) Immunoblot shows from left to right the internal level of secretory marker Bgl2p in log phase sec6-4ts (NY17), swf1Δ (KKY1063), swf1Δ + DHHC (KKY1086), and swf1Δ + DHHA (KKY 1087) cells, cultured for 90 min, at 25 or 37°C in rich and SC-Ura medium. To demonstrate equivalent loading, the same blot was probed with an antibody against β-tubulin.
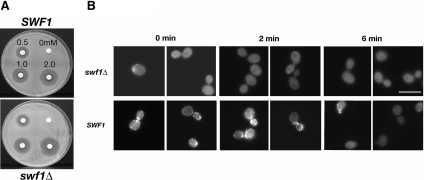
Changes in actin dynamics can cause a misorganization of the actin cytoskeleton (Belmont and Drubin 1998; Belmont et al., 1999). To determine whether actin dynamics was altered in vivo in the absence of Swf1p, we assayed a swf1Δ strain for sensitivity to the drug LatA. Cells that rapidly turnover F-actin structures are known to be hypersensitive to LatA (Ayscough et al., 1997; Lappalainen and Drubin, 1997). Using a halo assay, we found that swf1Δ cells are hypersensitive to LatA. The zone of no colony growth around LatA-impregnated filter disks was of greater diameter on a lawn of swf1Δ cells than on a lawn of wild-type cells (Figure 3A). We calculated the relative apparent LatA sensitivity of the swf1Δ to be two- to threefold greater than that of wild-type cells. To determine whether actin is indeed rapidly turning over in vivo in the absence of Swf1p, we performed a microscopic LatA sensitivity assay. swf1Δ and wild-type cultures were incubated briefly with 400 μM LatA. Aliquots of these cultures were fixed post-LatA addition at 0 min (the time to draw an aliquot and add fixative), 2 min, and 6 min, and then they were processed with rhodamine-phalloidin to visualize F-actin patches by fluorescence microscopy (Figure 3B). Figure 8A shows that in the absence of LatA wild-type and swf1Δ cells have a similar number of cortical F-actin patches. By 2 min post-LatA addition, actin patches were no longer observed in LatA-treated swf1Δ cells, although actin patches were observed readily in LatA-treated wild-type cells at this time point (Figure 3B). Thus, it seems that Swf1p slows actin turnover in vivo and that the organizational defects of the actin cytoskeleton in cells lacking SWF1 may be due to altered actin dynamics.
Figure 3.
Actin turnover was more rapid in the absence of SWF1. (A) Halo assay that showed the sensitivity of a haploid wild type strain (KKY1064) and swf1Δ mutant strain (KKY1063), grown at 25°C on rich medium, LatA. Counterclockwise, filter disks were impregnated with 0, 0.5, 1, and 2 mM LatA. (B) Micrographs of an in vivo actin turnover assay that showed a higher rate of actin turnover in swf1Δ mutant strain (KKY1090) compared with a wild-type strain (DDY663). We added 0.4 mM LatA at time zero, at which time, and at 2 and 6 min postdrug addition, aliquots of each culture were collected and stained with rhodamine-phalloidin. Rhodamine-phalloidin staining of nontreated cells looked like that shown in Figure 8A. Bar, 5 μm.
Swf1p Colocalizes with Cortical Actin Patches
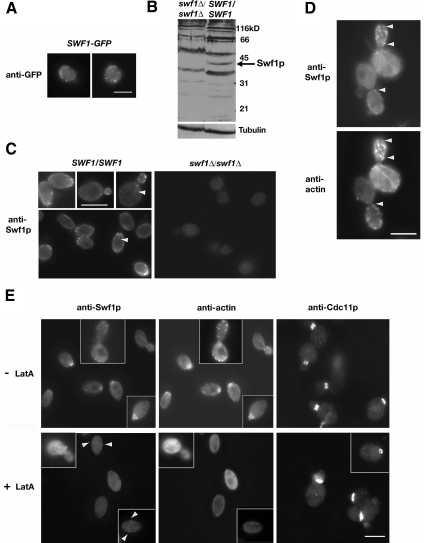
The regulation of the actin cytoskeleton by SWF1 suggested that Swf1p associates in vivo with one or more actin structures. We found that Swf1p colocalizes with cortical actin patches and actin cables (Figure 4). Our initial attempts to localize Swf1p with real-time imaging, by using a GFP fusion tag, proved unsuccessful. Only when wild-type cells were fixed and probed with a polyclonal antibody against GFP was GFP-Swf1p localization possible. We found GFP-Swf1p in cortical puncta of faint intensity (Figure 4A). During, but independent of our study, Valdez-Taubas and Pelham (2005) reported the localization of GFP-Swf1p at the endoplasmic reticulum. This difference in Swf1p localization is considered further in Discussion. To take a different route toward determining where Swf1p localizes, we raised a polyclonal antibody against the C-terminal region of Swf1p. This antibody displayed affinity for the immunogen (GST-Swf1-C) but not GST (Supplemental Figure 2). On immunoblots of whole cell lysates, this antibody recognized a polypeptide of ∼40 kDa, which is the predicted molecular weight of Swf1p and consistent with the migration of myc-tagged Swf1p in SDS-PAGE gels (Ohno et al., 2006). This polypeptide of ∼40 kDa was detectable in whole cell lysates of wild-type cells but not of mutant cells lacking SWF1 (Figure 4B). Even with purification, the antibody we raised against Swf1p exhibited cross-reactivity with other polypeptides on immunoblots. Such cross-reactivity, however, was not apparent in cells prepared for indirect immunofluorescence microscopy. On probing wild-type haploid (data not shown) and diploid (data shown) cells with purified Swf1-C antibody, we found that Swf1p localizes primarily to small cortical puncta (Figure 4C, left). A faint filamentous fluorescent signal was also observed in some cells (arrowheads in Figure 4C). Neither localization pattern was observed in cells lacking Swf1p (Figure 4C, right). The Swf1p localization pattern was consistent among our laboratory strains as well as in a BY4742-derived diploid (KKY1120; data not shown). As with our localization of GFP-Swf1p, antibody against Swf1p did not detect an accumulation of Swf1p in the cell body suggestive of an accumulation at the endoplasmic reticulum.
Figure 4.
Swf1p colocalized with cortical actin patches. (A) Indirect immunofluorescence micrographs of log phase wild-type cells expressing SWF1-GFP (KKY1104) labeled with anti-GFP and goat anti-rabbit-FITC antibodies. (B) Immunoblot of whole cell extract of log phase mutant swf1Δ/swf1Δ (KKY1060; right) and wild-type SWF1/SWF1 (DDY1102) diploid cells, grown at 25°C in rich medium, and probed with a polyclonal antibody against the C terminus of Swf1p. To demonstrate equivalent loading, the same blot was probed with an antibody against β-tubulin. (C) Indirect immunofluorescence micrographs of log phase wild-type SWF1/SWF1 (DDY1102; left) and mutant swf1Δ/swf1Δ (KKY1060; right) diploid cells, grown at 25°C in rich medium, and labeled with the same anti-Swf1p antibody used in B and goat anti-rabbit-Alexa 568. Arrowheads mark filamentous structures detected by the Swf1p antibody. (D) Indirect immunofluorescence micrographs of log phase wild-type cells (DDY1102) double labeled with polyclonal antibodies against Swf1p and actin. Goat anti-rabbit FITC and donkey anti-guinea pig-rhodamine served as secondary antibodies. Arrowheads mark examples of Swf1p and actin colocalization. (E) Indirect immunofluorescence micrographs of log phase wild-type SWF1/SWF1 (DDY1102; left) diploid cells cultured at 25°C in rich medium for 10 min with (bottom row) and without (top row) 400 μM LatA. The left and middle panels in each row show the same cells double labeled as in D. Arrowheads in bottom left panel mark examples of Swf1p localization. The panels on the right show cells from the same culture, with and without LatA, probed with anti-septin (Cdc11p) and goat anti-rabbit-Alexa 568 antibodies. Bars, 5 μm.
After establishing the localization of Swf1p, we double labeled diploid wild-type cells for Swf1p and actin. We found that Swf1p colocalizes with most but not all cortical actin patches (Figure 4D). In control staining reactions that used both secondary antibodies and either of the primary antibodies (data not shown), localization of Swf1p or actin was dependent upon the presence of the cognate primary antibody. No cross-reactivity between mismatched secondary and primary antibodies was observed either. In double label experiments, some cells also exhibited a uniform faint signal for Swf1p along the length of actin cables (Figure 4D). The localization of Swf1p was actin dependent (Figure 4E). When wild-type cells were treated for 10 min with 400 μM LatA, a drug that depolymerizes F-actin structures, Swf1p was no longer observed on cytoplasmic filaments and only a faint Swf1p signal remained at a few cortical puncta (arrowheads in Figure 4E, bottom left). LatA treatment did not globally abolish the localization of proteins at sites of polarized growth. The septin Cdc11p, for example, remained at the incipient bud site and mother-bud neck in the LatA-treated cells (Figure 4E, right). Although these data do not show unequivocally a direct interaction of Swf1p with actin, they do indicate that Swf1p associates with F-actin structures, especially on the cell cortex at sites of endocytosis.
To determine whether Swf1p binds actin directly, we tested whether the C-terminal region (residues 235–336) of Swf1p, which is predicted to be cytoplasmic (Politis et al., 2005), bound F- or G-actin. Due to protein instability, we were unable to assay recombinant full-length Swf1p. In a supernatant depletion assay, we found that the Swf1p C-terminal domain when fused to GST (GST-Swf1-Cp) did not cosediment with F-actin in a concentration-dependent manner (Supplemental Figure 3A). In a pyrene–actin assembly assay, we found that GST-Swf1-Cp in molar excess to G-actin did not affect actin assembly (Supplemental Figure 3B). Both of these results indicate that the C-terminal domain of Swf1p, when bound to GST, is unable to bind either F- or G-actin, suggesting that Swf1p does not bind actin directly.
Cdc42p Mislocalizes in the Absence of Swf1p
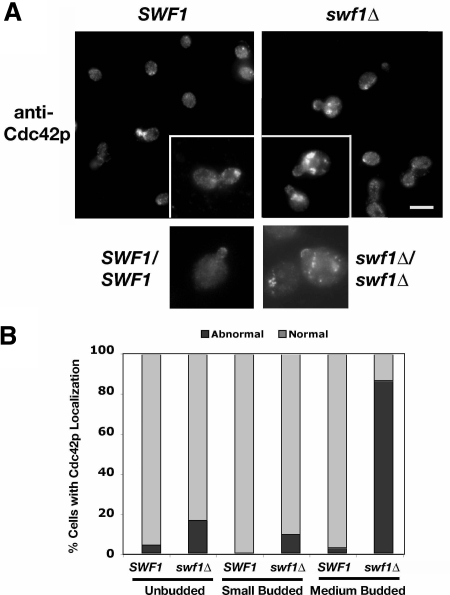
Next, we examined whether the localization of Cdc42p depends upon Swf1p. Cdc42p sits atop a hierarchy of signal transduction events that lead to polarized cell growth (Park and Bi, 2007). Localization of Cdc42p at the incipient bud site in unbudded cells or at the apical bud cortex in small- and medium-budded cells is necessary for proper cell polarization (Ziman et al., 1991). Both the genetic interactions observed between SWF1 and CDC42 and the loss of cytoskeletal polarity in swf1Δ and swf1Δ/swf1Δ mutants suggested that Cdc42p depends upon Swf1p for proper localization. We found little difference in Cdc42p localization in unbudded and small-budded wild-type and swf1Δ cells (Figure 5). Among small-budded cells, 100 and 92% of wild-type and swf1Δ cells, respectively, showed proper Cdc42p localization on the bud cortex. However, a significant difference was observed with medium-budded cells (Figure 5B). We found that 97% of medium-budded wild-type cells had proper Cdc42p localization on the bud cortex, whereas only 13% of the medium-budded swf1Δ cells had properly localized Cdc42p (Figure 5B). In mutant cells, Cdc42p localized normally to the apical bud tip but also abnormally to large patches within the mother cell, proximal to the mother-bud neck (Figure 5A). Similar localization patterns were found in homozygous swf1Δ diploid cells as well (Figure 5A). These data indicate that the polarized localization of Cdc42p is dependent upon Swf1p during a specific phase (S/G2) of the cell cycle.
Figure 5.
Cdc42p mislocalized in the absence of Swf1p. (A) Indirect immunofluorescence micrographs of log-phase haploid SWF1 (KKY1064; top left and inset) and swf1Δ (KKY1063; top right and inset) cells as well as diploid SWF1/SWF1 (DDY 1102; bottom left) and swf1Δ/swf1Δ (KKY1060; bottom right) cells, grown at 25°C in rich medium, labeled with anti-Cdc42p and goat anti-rabbit-Alexa 568 antibodies. Bar, 5 μm. (B) Comparison of the number of haploid SWF1 and swf1Δ cells with a proper localization of Cdc42p at different cell cycle stages. Cdc42p localization was scored as properly localized or normal when it was found as a patch or crescent on the cortex of unbudded cells or at the apical bud tip of small- and medium-budded cells. Bright patches of Cdc42p staining at noncortical locations were scored as abnormal. All scored cells had single nucleus as visualized with DAPI. n >174 cells scored for each strain. The cells quantified were from the same experiment shown in A.
swf1Δ Cells Are Defective in Secretion and Accumulate Vesicles
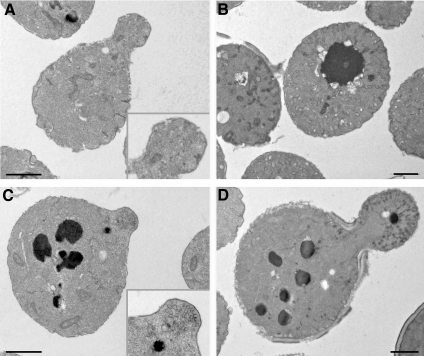
Two observations led to a prediction that cells lacking SWF1 are defective in polarized secretion. First, actin cables, along which secretory vesicles move, were fewer in cells lacking SWF1, compared with wild-type cells (Bonangelino et al., 2002; this study). Second, the SNARE Tlg1p, which is part of the exocytotic apparatus (Holthuis et al., 1998), is degraded in cells lacking SWF1 (Valdez-Taubas and Pelham, 2005). We found two lines of evidence that show swf1Δ cells are defective in polarized secretion. First, thin section electron microscopic analysis revealed an accumulation of 80- to 100-nm vesicles in the buds of two independently derived swf1Δ strains, KKY1063 (Figure 6, bottom) and 1-7-11 (data not shown; BY4742 background). Accumulation of 80- to 100-nm vesicles is a signature phenotype of impaired exocytosis. For example, sec6-4ts cells, which served as our positive control for detecting secretory vesicles in thin section, have a tight exocytotic block at 37°C (Novick et al., 1980; Harsay and Bretscher, 1995). After being shifted from 25°C to 37°C for 120 min, sec6-4ts cells failed to bud and accumulated 80- to 100-nm vesicles (Figure 6, top right). In contrast to the swf1Δ strain, in which vesicles accumulated in the buds, few if any vesicles were observable in the buds of wild-type cells (Figure 6, top left). As with the wild-type strain, the mother cells of the swf1Δ strain did not contain an accumulation of vesicles. These results indicate that SWF1 is not required for vesicle transport to the bud and strongly suggest that SWF1 is necessary for efficient exocytosis from the bud.
Figure 6.
Vesicles (80–100 nm) accumulated in the buds of cells lacking SWF1. Electron micrographs show thin sections of wild-type SWF1 (KKY1064) (A), sec6-4ts (NY17) (B), and swf1Δ (KKY1063) (C and D) log phase cells cultured in rich medium. Strains shown in A, C, and D were cultured at 25°C; the strain shown in B was cultured at 37°C. Insets show an enlarged image of a bud. Bar, 5 μm.
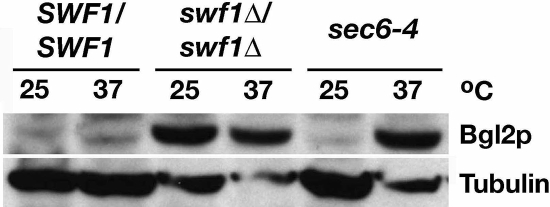
Consistent with this observation and the prediction swf1Δ cells are defective in polarized secretion, we found an internal accumulation of β-1,3-glucanase (Bgl2p) in swf1Δ cells (Figure 7). Bgl2p is a cell wall remodeling enzyme that is secreted to the plasma membrane (Harsay and Bretscher, 1995). On assaying the internal Bgl2p pool in wild-type cells, cells lacking SWF1, and sec6-4ts cells grown at 25°C or shifted from 25 to 37°C, we found no appreciable accumulation of Bgl2p in wild-type cells cultured at either temperature, and we found a significant accumulation of Bgl2p in sec6-4ts control cells only after shift to restrictive temperature. In swf1Δ cells, Bgl2p accumulated to the level observed in sec6-4ts grown at 37°C. This accumulation was not temperature dependent. The results of this Bgl2p assay and our electron microscopy data indicate that swf1Δ cells have a defect in polarized secretion, occurring late in the secretory pathway.
Figure 7.
The secretory marker Bgl2p accumulated internally in the absence of SWF1. Immunoblot shows from left to right the internal level of secretory pathway marker Bgl2p, in log phase wild-type SWF1/SWF1 (DDY1102) and swf1Δ/swf1Δ (KKY 1060), and sec6-4ts (NY17) cell, cultured for 90 min at 25 or 37°C in rich medium. To demonstrate equivalent loading, the same blot was probed with an antibody against β-tubulin.
Actin Organization and Cdc42 Localization Are Independent of the Swf1p DHHC Motif
Valdez-Taubas and Pelham (2005) observed that deletion of the ubiquitin ligase Tul1p did not suppress all swf1Δ phenotypes such as the inability of swf1Δ cells to grow on lactate. This observation suggested that Swf1p has multiple distinct functions. Because of this observation, we asked whether the ability of Swf1p to promote the polarization of the actin cytoskeleton was dependent on the DHHC motif. Earlier studies demonstrated that substitution of cysteine in the DHHC motif with alanine abolished the palmitoyltransferase (PT) activity of DHHC-CRD family palmitoyltransferases (Lobo et al., 2002; Roth et al., 2002), or, in Swf1p, the DHHC motif was necessary for SNARE palmitoylation (Valdez-Taubas and Pelham, 2005). To address this question, we examined by fluorescence microscopy the organization of the actin cytoskeleton in cells solely expressing either wild-type Swf1p (DHHC) or Swf1p with a mutated DHHC motif (DHHA). Wild-type and mutant Swf1p were expressed from a low copy CEN plasmid in swf1Δ cells. We found that cells expressing Swf1-DHHAp had a wild-type morphology and that the actin cytoskeleton remained polarized, similar to cells expressing Swf1-DHHCp (Figure 8, A and B). Likewise little variation in actin polarization was observed among populations of unbudded cells. In the same experiment, we tried to compare actin polarization in cells expressing Swf1-DHHAp to that found in a swf1Δ strain containing only vector. We found, however, that liquid minimal medium did not support the growth of swf1Δ cells containing an empty vector. Therefore, cells expressing Swf1-DHHAp were compared with a swf1Δ strain and a SWF1 strain grown in rich medium. The number of Swf1-DHHCp and Swf1-DHHAp transformants with a polarized actin cytoskeleton closely resembled that of SWF1 cells, indicating that expression of SWF1 from a plasmid did not affect its function. Together, these results showed that the DHHC motif of Swf1p is not necessary for promoting the polarized organization of the actin cytoskeleton.
The polarized organization of the actin cytoskeleton in cells solely expressing Swf1-DHHAp suggested that mutation of the DHHC motif does not affect actin dynamics. To test this idea, we compared the sensitivity of cells expressing either Swf1-DHHCp or Swf1-DHHAp to LatA in a halo assay. As shown in Figure 8C, we found no apparent increase in sensitivity to LatA between a strain expressing Swf1-DHHAp to a strain expressing Swf1-DHHCp. This result suggested that substitution of the cysteine in the DHHC motif does not affect actin dynamics.
Wild-type actin organization in cells expressing only Swf1-DHHAp implied that the DHHC motif is not necessary for the proper localization of Cdc42p at sites of polarized growth. To test whether Cdc42p, a key regulator of actin organization at polarized growth sites, localized properly in cells with a DHHA motif, we probed cells expressing either Swf1-DHHCp or Swf1-DHHAp with an antibody against Cdc42p. We found a wild-type pattern of Cdc42p localization in cells expressing Swf1-DHHAp (Figure 8D). Therefore, the Swf1p DHHC motif is not necessary for the proper localization of Cdc42p.
Loss of the Swf1p DHHC Motif Differentially Affects Secretion
Because abnormalities were not observed in the organization or dynamics of the actin cytoskeleton in cells expressing Swf1p with a mutated DHHC motif (DHHA), we asked whether the secretion of Bgl2p was blocked in these mutant cells. That is, we sought to determine whether the secretion defect in swf1Δ cells was due to a change in the actin cytoskeleton or due to a change in the palmitoylation state of SNAREs. To address this question, we first assayed Bgl2p secretion in cells expressing no Swf1p (swf1Δ), wild-type Swf1p (DHHC), or Swf1p with a mutated DHHC motif (DHHA). As a control for Bgl2p secretion, we included a sec6-4ts strain in our analysis. This strain is temperature-conditional for secretion and accumulates Bgl2p internally after shift from 25 or 37°C (Harsay and Bretscher, 1995). We found that at both 25 or 37°C, the amount of Bgl2p that accumulated internally in swf1-DHHA cells, which display no actin defects, was very similar to the amount of Bgl2p that accumulated in wild-type cells (Figure 8E). Furthermore, the amount of Bgl2p that accumulated internally in cells expressing either wild-type (DHHC) or mutant (DHHA) Swf1p was much lower than the amount of Bgl2p in swf1Δ cells, which lack Swf1p entirely. These results indicate that Swf1p DHHC motif is not necessary for Bgl2p secretion and suggest that the defect in Bgl2p secretion in swf1Δ cells may result from a defect in actin organization.
Wild-type Bgl2p secretion in cells with a mutated Swf1p DHHC motif contradicted the idea that the DHHC motif, and by inference Swf1p PT activity, is physiologically relevant to secretion. To determine whether the Swf1p DHHC motif is relevant to any type of secretion as predicted from the ability of Swf1p to palmitoylate specific SNAREs, we performed an additional secretion assay. With the same strains used for the Bgl2p assay described above, we assayed the secretion of invertase (Table 3). We found that deletion of SWF1 impaired secretion at 25°C, in comparison with wild-type (Student's t test, p = 0.06). Transformation of swf1Δ cells with a plasmid containing wild-type SWF1 rescued the mutant phenotype. Therefore, no significant difference in invertase secretion was observed whether Swf1p was expressed from a low copy plasmid (+ DHHC) in a swf1Δ strain or from a chromosomal copy of the gene (SWF1). In contrast, the swf1Δ phenotype with respect to invertase secretion was not rescued by plasmid-borne swf1-DHHA. The average ratio of secreted invertase to total invertase in cells expressing Swf1p without a DHHC motif (swf1Δ + DHHA) was significantly less (Student's t test, p = 0.01) than that measured for cells expressing wild-type Swf1p (swf1Δ + DHHC), 0.64 versus 0.77, respectively. None of the swf1 mutants showed a secretory block as severe as that of sec6-4 cells shifted from 25to 37°C, in which the average ratio of secreted to total invertase fell from 0.82 to 0.13. Together, these data demonstrate that cells expressing Swf1p without a DHHC motif are defective in secretion in the invertase-marked secretory pathway, but not the Bgl2p-marked pathway.
Table 3.
Invertase secretion in wild-type and swf1 cells
| Genotype | Mean secreted/total invertase | n samples | n independent assays |
|---|---|---|---|
| At 25°C | |||
| SWF1 | 0.78 ± 0.03 | 10 | 6 |
| swf1Δ | 0.69 ± 0.03 | 7 | 5 |
| sec6-4 | 0.82 ± 0.04 | 8 | 5 |
| swf1Δ + DHHC | 0.77 ± 0.04 | 9 | 5 |
| swf1Δ + DHHA | 0.64 ± 0.02 | 10 | 5 |
| At 37°C | |||
| sec6-4 | 0.13 ± 0.02 | 14 | 7 |
Invertase activity (in A540 units) was measured as described in Materials and Methods. A SE is given for each mean. The strains used for this assay were SWF1 (KKY1064), swf1Δ (KKY1063), sec6-4 (NY17), swf1Δ + DHHC (KKY1086), and swf1Δ + DHHA (KKY1087).
The Swf1p DHHC Motif Is Functionally Relevant to a Growth-related Cdc42p Function
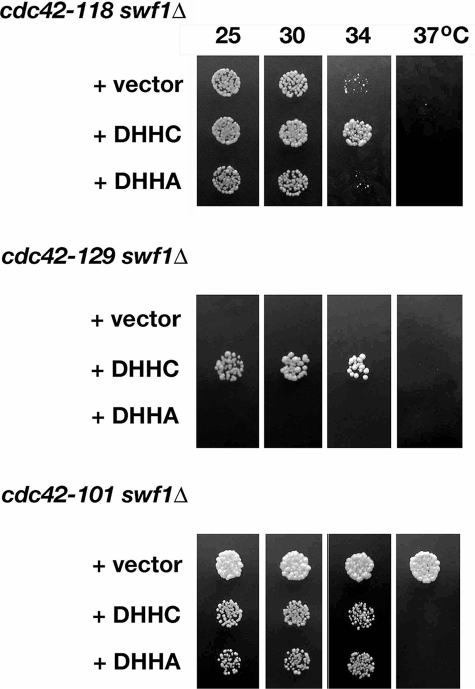
The demonstrated independence of actin organization, Bgl2p-marked secretion, and Cdc42p localization from the Swf1p DHHC motif predicted that the DHHC motif is not functionally relevant to the function of Cdc42p in polarized growth. That is, mutation of the DHHC motif would not enhance cdc42 growth defects. To test this prediction, we compared at a range of temperatures the growth of cdc42ts swf1Δ double mutants that contained an empty plasmid vector with those that contained plasmid-borne swf1-DHHA (Figure 9). Double mutant strains that contained plasmid-borne wild-type SWF1 defined the temperature range of individual cdc42ts alleles under the growth conditions used.
Figure 9.
Synthetic genetic interactions between certain cdc42ts alleles and swf1Δ required the Swf1p DHHC motif. Equivalent dilutions of swf1Δ cdc42-118, swf1Δ cdc42-129, and swf1Δ cdc42-101 strains were plated and grown at 25, 30, and 37°C for 3 d on SC-Ura. These strains contained a CEN plasmid vector that was either empty or filled with wild-type SWF1 (DHHC) or swf1 in which the DHHC motif was mutated (DHHA). The strains shown (from top to bottom) are KKY1137, 1138, 1139, 1140, 1134, 1135, and 1136. Note that no swf1Δ cdc42-129 cells containing vector or plasmid-borne swf1-DHHA were plated because neither the vector nor swf1-DHHA suppressed the synthetic lethal interaction between swf1Δ and cdc42-129.
Contrary to our prediction, we found that the DHHC motif of Swf1p is functionally relevant to Cdc42p function during polarized cell growth. We observed that the Swf1p DHHC motif is necessary for the rescue of two of three different cdc42ts swf1Δ synthetic genetic interactions (Figure 9). A cdc42-118 swf1Δ strain that contained plasmid-borne swf1-DHHA exhibited the same permissive temperature range for growth as one with an empty vector. A similar result was observed with cdc42-129. Because cdc42-129 and swf1Δ are lethal in combination (Figure 1B), we asked whether we could recover viable double mutants with plasmid-borne swf1-DHHA, upon sporulation of a double heterozygote transformed with a plasmid containing swf1-DHHA. We found that we were unable to recover, after meiosis at 25°C, cdc42-129 swf1Δ progeny with either empty vector or plasmid-borne swf1-DHHA. In contrast, cdc42-129 swf1Δ progeny with plasmid-borne SWF1 were easily recoverable. These data indicate that the DHHC motif is functionally relevant to at least one growth-related function of Cdc42p.
In the converse case, where swf1Δ suppresses rather than exacerbates a cdc42 temperature-sensitive growth defect, we observed that a cdc42-101 swf1Δ strain that contained plasmid-borne swf1-DHHA exhibited the same permissive temperature range for growth as a strain with plasmid-borne wild-type SWF1. Although equivalent growth of a cdc42 swf1Δ mutant containing either swf1-DHHA or wild-type SWF1 would normally indicate that a functional DHHC motif is not required for the rescue of the double mutant growth phenotype, it is important to note that the double mutant in this case has an expanded growth range. This result suggests that a part of Swf1p, other than the DHHC motif, counterbalances a Cdc42p function related to growth.
DISCUSSION
Temperature-conditional cdc42 mutants in S. cerevisiae have proven incredibly valuable for the identification of conserved genes and gene families that regulate CDC42-dependent polarized cell growth (Kozminski et al., 2003; Kozminski et al., 2006). Herein, we demonstrated that one such gene, SWF1, is necessary for proper cell polarization. Although the exact biochemical mechanism by which Swf1p regulates cell polarization remains unknown, we found that several cellular processes depend upon Swf1p. Swf1p is necessary for the polarized localization of the Rho-family GTPase Cdc42p, the polarization of the actin cytoskeleton, and for trafficking along a specific secretory pathway. To our surprise, none of these processes depend upon the DHHC motif in Swf1p and, by extension, the PT activity that Swf1p is very likely to possess.
GTPase localization, cytoskeletal polarization, and vesicle trafficking are intimately related processes that are necessary for proper polarized cell growth in yeast (Park and Bi, 2007). In unbudded G1 cells, the localization and activation of Cdc42p at the incipient bud site is essential for the polarized organization of the actin cytoskeleton (Sloat et al., 1981; Adams et al., 1990; Ziman et al., 1991, 1993; Gulli et al., 2000; Richman and Johnson, 2000) and the establishment of polarized secretion (Adamo et al., 2001; Zhang et al., 2001; Roumanie et al., 2005; Zajac et al., 2005). In small- and medium-budded cells, in S and G2/M, respectively, the maintenance of Cdc42p localization at the bud tip depends upon a properly polarized actin cytoskeleton and polarized secretion. Mutations or drugs that disrupt either process disrupt the polarized distribution of Cdc42p (Wedlich-Soldner et al., 2003; Irazoqui et al., 2005; Zajac et al., 2005). With few unbudded and small-budded swf1Δ and swf1Δ/swf1Δ cells displaying a loss of cytoskeletal organization or Cdc42p polarization, relative to medium-budded cells, we conclude that Swf1p does not affect the process of establishing an axis of polarized growth early in the cell cycle. Rather our data suggest that Swf1p functions, with respect to polarized cell growth, after bud emergence as part of the cellular mechanism that maintains a polarized distribution of Cdc42p during bud growth.
Thus far, our data are unable to reveal unequivocally in which process Swf1p acts primarily to promote polarized growth. For example, a loss of polarized secretion in swf1Δ mutants may result from an aberrant organization of the actin cytoskeleton, as is known to occur in tpm1-2 tpm2 mutants (Pruyne et al., 1998). It is also possible that Swf1p functions in more than one process required for polarized cell growth, as suggested by the inability of cdc42-DHHA to rescue all cdc42ts swf1Δ synthetic growth phenotypes. The identification of SWF1 in multiple independent screens (Bartels et al., 1999; Bonangelino et al., 2002; Enyenihi and Saunders, 2003) shows that Swf1p can function in multiple processes (e.g., trafficking to the vacuole, spore wall formation). Taking these data and our own into consideration, the actin cytoskeleton stands out as a common denominator. Therefore, what may seem to be independent processes during polarized cell growth may be due to a single Swf1p activity affecting the actin cytoskeleton. In addition to our data that show Swf1p regulates the actin cytoskeleton, it should not be forgotten that SWF1 was first reported as a gene that is synthetic lethal with a hypomorphic, although undesignated, allele of PFY1 (Bartels et al., 1999). PFY1 encodes profilin, a protein that regulates actin dynamics (Jockusch et al., 2007). We infer from the literature that the allele used in this screen was pfy1-111 (Haarer et al., 1996). We have independently confirmed (our unpublished observations) this genetic interaction by using pfy1-4, an allele that encodes a mutant profilin defective in actin monomer binding (Wolven et al., 2000), whereas finding, within additional although small data sets, no synthetic lethality between swf1Δ and PFY1 alleles specifically defective in functions other than actin monomer binding (e.g., binding to formin homology domain-containing proteins). Because synthetic genetic interactions often reveal a common function between proteins, we must consider whether Swf1p like Pfy1p binds actin monomers. Such biochemical activity is unlikely because Swf1p does not contain any known actin binding motifs. The region of Swf1p most likely to bind actin is the long C-terminal tail, which based on a membrane-spanning topology model of the DHHC-CRD protein Akr1p (Politis et al., 2005) is predicted to be cytoplasmic. We did not find that this C-terminal region binds actin directly. Therefore, it is more probable that the ability of Swf1p to regulate the actin cytoskeleton is by way of protein intermediates. Analogy may be drawn from membrane-anchored cytoskeletons in metazoans, in which Band 4.1 protein and Ezrin/radixin/moesin proteins link ion channels and CD44, respectively, to cortical actin filaments (Denker and Barber, 2002) or in yeast, where during receptor mediated-endocytosis receptors are linked to the actin cytoskeleton by way of Sla1p or the epsin-related proteins Ent1/2p (Engqvist-Goldstein and Drubin, 2003).
Because profilin is a potent regulator of actin dynamics, it is tempting to speculate that the actin organization defects in swf1Δ cells is due to a perturbation of actin dynamics as has been observed in cells expressing mutant profilin (Wolven et al., 2000). Consistent with this idea, we noted, as in a previous study (Bonangelino et al., 2002), the paucity of actin cables in swf1Δ cells. We also observed a more rapid turnover of F-actin structures in swf1Δ cells in vivo in the presence of LatA. However, caution needs to be applied to the interpretation that Swf1p regulates actin dynamics. It is possible that the secretory defect observed in swf1Δ cells has altered the permeability of the cell wall to LatA, relative to wild-type cells. We have, in fact, observed (our unpublished observation) a slight sensitivity of swf1Δ cells to Calcofluor White, indicative of a defect in cell wall formation (Roncero et al., 1988). Unknown is whether this cell wall defect increases cell permeability to LatA to a physiologically significant level. Conversely, it is possible that a defect in the actin cytoskeleton leads to defective wall formation and in turn increased sensitivity to LatA (Gabriel and Kopecka, 1995). What is certain is that the organization, if not the dynamics, of the actin cytoskeleton depends upon Swf1p.
The localization of Swf1p to actin cables and patches is consistent with a role in the organization of the actin cytoskeleton. Why a predicted transmembrane protein is found on actin cables is not known. Cable-associated Swf1p may be derived from a pool of Swf1p associated with transport vesicles that move along actin cables or from a proteolytically processed domain of Swf1p (i.e., the C-terminal tail). Localization, however, is not proof of function at a particular locale. Localization at sites of endocytosis, for example, may solely reflect that Swf1p is concentrated at sites of endocytosis and is above the threshold of detection at these sites. It may be that Swf1p functions elsewhere. For example, Swf1p may function in endosomes or the late Golgi, where its substrate Tlg1p is known to reside (Lewis et al., 2000; Prescianotto-Baschong and Riezman, 2002). The appearance of Swf1p on the plasma membrane and subsequent endocytosis may be due to trafficking between endosomal compartments and the plasma membrane. In contrast, Valdez-Taubas and Pelham (2005) reported a different localization for Swf1p, showing that a GFP-Swf1p fusion protein localizes to the endoplasmic reticulum (ER). We did not find a similar pattern of localization with our indirect immunofluorescence technique, even using the same strain background as Valdez-Taubas and Pelham (2005). We speculate that the observed differences in Swf1p localization may be reflective of protein expression levels. Valdez-Taubas and Pelham (2005) drove expression of their tagged Swf1p with the strong constitutive TPI1 promoter, whereas expression of Swf1p and Swf1-GFP in our study relied upon the endogenous SWF1 promoter.
Our experiments directly showed that Swf1p has a role in secretion. During the course of our study, two independent investigations implicated Swf1p in secretion. In the first investigation, almost 40% (n = 26) of the genes identified in a synthetic lethal screen with swf1Δ by Tong et al. (2004) encode proteins known to function in the Golgi or in post-Golgi trafficking pathways. In the second investigation, Valdez-Taubas and Pelham (2005) showed that the palmitoylation of specific SNARE proteins depends upon Swf1p. In our study, we show that Swf1p is necessary for secretion along two different secretory pathways, one marked by invertase and the other marked by Bgl2p. In the absence of Swf1p, vesicles with a diameter signature of secretory vesicles accumulate in the bud, indicative of a block in late secretory pathway(s). These results are consistent with the study of Valdez-Taubas and Pelham (2005), which showed that Swf1p regulates the stability of the SNARE Tlg1p. When we expressed Swf1p with a mutated DHHC motif, invertase secretion was inhibited. Secretion along the Bgl2p-marked pathway, which is a secretory pathway that supports polarized cell growth, was not inhibited by mutation of the DHHC motif. These results provided several insights into Swf1p and polarized cell growth. First, secretion along the Bgl2p transport pathway, but not the invertase pathway, is independent of the Swf1p DHHC motif, indicating Swf1p has two distinct functions in regard to secretion. The data further suggest that the SNARE Tlg1p functions specifically within the invertase-marked secretory pathway. If, however, Tlg1p also functions within the Bgl2p-marked secretory pathway, it does so in a manner independent of the Swf1p DHHC motif. Second, in swf1 mutant cells, defects in the actin cytoskeleton correlate more closely with defects in Bgl2p-marked secretion than invertase secretion. As stated above, it has not yet been possible to distinguish in swf1 mutants whether a defect in Bgl2p secretion leads to an aberrant actin cytoskeleton or vice versa. What is clear is that Bgl2p-marked secretion and regulation of the actin cytoskeleton functions independently of the DHHC motif and, by inference, PT activity.
Palmitoylation is a posttranslational modification that contributes to the stability and trafficking of specific proteins (Linder and Deschenes, 2007). When in the course of our study it became known that Swf1p contributes to PT activity (Valdez-Taubas and Pelham, 2005), it became apparent to consider whether palmitoylation affected the intracellular distribution of Cdc42p. It is known the distribution of Ras between ER and plasma membrane depends upon the compartment specific palmitoylation of the G protein (Rocks et al., 2005). Furthermore, sequence analysis predicts the palmitoylation of two S. cerevisiae proteins necessary for proper polarized cell growth, Rho1p and Rho3p (Roth et al., 2006). These GTPases are in the same family as Cdc42p. Unlike Rho1 and Rho3, Cdc42p does not have an identifiable palmitoylation site. More importantly, we found that the distribution of Cdc42p in cells with a mutated DHHC motif was indistinguishable from that of cells expressing wild-type Swf1p. Therefore, it does not seem that the polarized localization of Cdc42p at the plasma membrane is dependent on palmitoylation.
Swf1p is among the family of seven transmembrane proteins in S. cerevisiae that contain a DHHC-CRD domain (Linder and Deschenes, 2007). It remains to be seen whether other DHHC-CRD proteins have a role in polarized cell growth, especially the regulation of the actin cytoskeleton. It is intriguing that no other genes that encode DHHC-CRD proteins were found in the synthetic genetic array screen that identified SWF1 as CDC42-interacting, suggesting that the promotion of polarized cell growth may be a specific function of SWF1. In this regard, we have not tested directly the other DHHC-family members. Identifying the complete range of Swf1p functions and that of other DHHC-CRD family members, although outside the scope of the current study, remains an important goal, considering the conservation of the DHHC-CRD protein family among plants, animals, and fungi (Putilina et al., 1999; Linder and Deschenes, 2003; Mitchell et al., 2006).
Supplementary Material
ACKNOWLEDGMENTS
We especially thank Edina Harsay (University of Kansas) for advice on secretion assays; Dorothy Schafer for help with actin binding assays; Anthony Bretscher (Cornell University), Anthony Frankfurter, Pekka Lappalainen (University of Helsinki), Mark Longtine (Washington University in St. Louis), David Pruyne (Cornell University), and Randy Schekman (University of California, Berkeley) for antibodies; David Drubin and P. Lappalainen for strains; and Jan Rednick (University of Virginia Central EM Facility) for assistance with electron microscopy. We also thank P. Lappalainen for a critical reading of the manuscript. National Science Foundation grant 0723342 and the University of Virginia provided support for this study.
Abbreviations used:
- LatA
latrunculin A
- PT
palmitoyltransferase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0252) on August 13, 2008.
REFERENCES
- Adamo J. E., Moskow J. J., Gladfelter A. S., Viterbo D., Lew D. J., Brennwald P. J. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. E., Pringle J. R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- Adams A. E. M., Johnson D. I., Longnecker R. M., Sloat B. F., Pringle J. R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts P., Rudge R., Irinopoulou T., Danglot L., Gauthier-Rouviere C., Galli T. Cdc42 and actin control polarized expression of TI-VAMP vesicles to neuronal growth cones and their fusion with the plasma membrane. Mol. Biol. Cell. 2006;17:1194–1203. doi: 10.1091/mbc.E05-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D. G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis V. A., Malehorn D. E., Emr S. D., Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D. J., Mitchell D. A., Dong X., Deschenes R. J. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L. D., Drubin D. G. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J. Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L. D., Patterson G. M., Drubin D. G. New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J. Cell Sci. 1999;112:1325–1336. doi: 10.1242/jcs.112.9.1325. [DOI] [PubMed] [Google Scholar]
- Bonangelino C. J., Chavez E. M., Bonifacino J. S. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Denker S. P., Barber D. L. Ion transport proteins anchor and regulate the cytoskeleton. Curr. Opin. Cell Biol. 2002;14:214–220. doi: 10.1016/s0955-0674(02)00304-6. [DOI] [PubMed] [Google Scholar]
- Ducker C. E., Stettler E. M., French K. J., Upson J. J., Smith C. D. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–9237. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein A. E., Drubin D. G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Enyenihi A. H., Saunders W. S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42—the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Gabriel M., Kopecka M. Disruption of the actin cytoskeleton in budding yeast results in formation of an aberrant cell wall. Microbiology. 1995;141:891–899. doi: 10.1099/13500872-141-4-891. [DOI] [PubMed] [Google Scholar]
- Garvalov B. K., Flynn K. C., Neukirchen D., Meyn L., Teusch N., Wu X., Brakebusch C., Bamburg J. R., Bradke F. Cdc42 regulates cofilin during the establishment of neuronal polarity. J. Neurosci. 2007;27:13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Gulli M. P., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Haarer B. K., Corbett A., Kweon Y., Petzold A. S., Silver P., Brown S. S. SEC3 mutations are synthetically lethal with profilin mutations and cause defects in diploid-specific bud-site selection. Genetics. 1996;144:495–510. doi: 10.1093/genetics/144.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Bretscher A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Schekman R. Avl9p, a member of a novel protein superfamily, functions in the late secretory pathway. Mol. Biol. Cell. 2007;18:1203–1219. doi: 10.1091/mbc.E06-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. L., Catlett N. L., Weisman L. S. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J. C., Nichols B. J., Dhruvakumar S., Pelham H. R. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Howell A. S., Theesfeld C. L., Lew D. J. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell. 2005;16:1296–1304. doi: 10.1091/mbc.E04-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgandarova S., Jones L., Forsberg D., Loncar A., Dawson J., Tedrick K., Eitzen G. Stimulation of actin polymerization by vacuoles via Cdc42p-dependent signaling. J. Biol. Chem. 2007;282:30466–30475. doi: 10.1074/jbc.M704117200. [DOI] [PubMed] [Google Scholar]
- Jockusch B. M., Murk K., Rothkegel M. The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 2007;159:131–149. doi: 10.1007/112_2007_704. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Alfaro G., Dighe S., Beh C. T. Homologues of oxysterol-binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in S. cerevisiae. Traffic. 2006;7:1224–1242. doi: 10.1111/j.1600-0854.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Beven L., Angerman E., Tong A. H., Boone C., Park H. O. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell. 2003;14:4958–4970. doi: 10.1091/mbc.E03-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Chen A. J., Rodal A. A., Drubin D. G. Functions and functional domains of the GTPase Cdc42p. Mol. Biol. Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschewski R., Hall A., Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D. G. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M. E., Deschenes R. J. New insights into the mechanisms of protein palmitoylation. Biochemistry. 2003;42:4311–4320. doi: 10.1021/bi034159a. [DOI] [PubMed] [Google Scholar]
- Linder M. E., Deschenes R. J. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mitchell D. A., Vasudevan A., Linder M. E., Deschenes R. J. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- Mullins R. D., Stafford W. F., Pollard T. D. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J. Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. J. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ohno Y., Kihara A., Sano T., Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Palmgren S., Ojala P. J., Wear M. A., Cooper J. A., Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peränen J., Rikkonen M., Hyvonen M., Kaariainen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Politis E. G., Roth A. F., Davis N. G. Transmembrane topology of the protein palmitoyltransferase Akr1. J. Biol. Chem. 2005;280:10156–10163. doi: 10.1074/jbc.M411946200. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong C., Riezman H. Ordering of compartments in the yeast endocytic pathway. Traffic. 2002;3:37–49. doi: 10.1034/j.1600-0854.2002.30106.x. [DOI] [PubMed] [Google Scholar]
- Pringle J. R. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Pringle J., Preston R., Adams A., Stearns T., Drubin D., Haarer B., Jones E. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pruyne D. W., Schott D. H., Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Putilina T., Wong P., Gentleman S. The DHHC domain: a new highly conserved cysteine-rich motif. Mol. Cell Biochem. 1999;195:219–226. doi: 10.1023/a:1006932522197. [DOI] [PubMed] [Google Scholar]
- Reneke J. E., Blumer K. J., Courchesne W. E., Thorner J. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Richman T. J., Johnson D. I. Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell Biol. 2000;20:8548–8559. doi: 10.1128/mcb.20.22.8548-8559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Roncero C., Valdivieso M. H., Ribas J. C., Duran A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J. Bacteriol. 1988;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. F., Feng Y., Chen L., Davis N. G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumanie O., Wu H., Molk J. N., Rossi G., Bloom K., Brennwald P. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J. Cell Biol. 2005;170:583–594. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sloat B. F., Adams A., Pringle J. R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Maeder C., Reber S., Rathfelder N., Miura K., Greger K., Stelzer E. H., Knop M. Dynamic organization of the actin cytoskeleton during meiosis and spore formation in budding yeast. Traffic. 2006;7:1628–1642. doi: 10.1111/j.1600-0854.2006.00496.x. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L., Cline H. T. Rho GTPases and activity-dependent dendrite development. Curr. Opin. Neurobiol. 2004;14:297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Venable J. H., Coggeshall R. A simplified lead citrate stain for use in electron microscopy. J. Cell Biol. 1965;25:407–408. doi: 10.1083/jcb.25.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wolven A. K., Belmont L. D., Mahoney N. M., Almo S. C., Drubin D. G. In vivo importance of actin nucleotide exchange catalyzed by profilin. J. Cell Biol. 2000;150:895–904. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. Transmission electron microscopy of yeast. Microsc. Res. Tech. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zajac A., Sun X., Zhang J., Guo W. Cyclical regulation of the exocyst and cell polarity determinants for polarized cell growth. Mol. Biol. Cell. 2005;16:1500–1512. doi: 10.1091/mbc.E04-10-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bi E., Novick P., Du L., Kozminski K. G., Lipschutz J. H., Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Preuss D., Mulholland J., O'Brien J. M., Botstein D., Johnson D. I. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.