Abstract
The class II Histone deacetylase (HDAC), HDAC4, is expressed in a tissue-specific manner, and it represses differentiation of specific cell types. We demonstrate here that HDAC4 is expressed in the proliferative zone in small intestine and colon and that its expression is down-regulated during intestinal differentiation in vivo and in vitro. Subcellular localization studies demonstrated HDAC4 expression was predominantly nuclear in proliferating HCT116 cells and relocalized to the cytoplasm after cell cycle arrest. Down-regulating HDAC4 expression by small interfering RNA (siRNA) in HCT116 cells induced growth inhibition and apoptosis in vitro, reduced xenograft tumor growth, and increased p21 transcription. Conversely, overexpression of HDAC4 repressed p21 promoter activity. p21 was likely a direct target of HDAC4, because HDAC4 down-regulation increased p21 mRNA when protein synthesis was inhibited by cycloheximide. The importance of p21 repression in HDAC4-mediated growth promotion was demonstrated by the failure of HDAC4 down-regulation to induce growth arrest in HCT116 p21-null cells. HDAC4 down-regulation failed to induce p21 when Sp1 was functionally inhibited by mithramycin or siRNA-mediated down-regulation. HDAC4 expression overlapped with that of Sp1, and a physical interaction was demonstrated by coimmunoprecipitation. Chromatin immunoprecipitation (ChIP) and sequential ChIP analyses demonstrated Sp1-dependent binding of HDAC4 to the proximal p21 promoter, likely directed through the HDAC4–HDAC3–N-CoR/SMRT corepressor complex. Consistent with increased transcription, HDAC4 or SMRT down-regulation resulted in increased histone H3 acetylation at the proximal p21 promoter locus. These studies identify HDAC4 as a novel regulator of colon cell proliferation through repression of p21.
INTRODUCTION
The acetylation of lysine residues in histones, and/or of transcription factors, is an important posttranslational mechanism of transcriptional regulation (Peterson and Laniel, 2004). Histone deacetylases (HDACs) catalyze the deacetylation of histone and nonhistone substrates, and in general act to repress transcription as part of larger corepressor complexes (Glozak et al., 2005; Yang and Gregoire, 2005). Eighteen mammalian HDACs have been identified to date, and they are grouped into four classes based on their respective homology to yeast deacetylases (Bolden et al., 2006). The class II HDACs can be further subdivided into class IIa (HDAC4, -5, -7, and -9) and class IIb (HDAC6 and -10), based on the presence in class IIa members of conserved motifs in the N-terminal domain, and extended C terminal tails, that are essential in regulating their function (Yang and Gregoire, 2005).
HDACs have emerged as critical regulators of cell growth, differentiation, and apoptotic programs (Bolden et al., 2006). A large body of literature indicates that HDAC inhibitors induce cell cycle arrest, differentiation, and apoptosis in colon cancer cell lines in vitro (Heerdt et al., 1994; Mariadason et al., 1997; Archer et al., 1998; Litvak et al., 1998; Mariadason et al., 2000, 2001b; Gurvich et al., 2004; Wilson et al., 2006). Based on such studies, several HDAC inhibitors, including depsipeptide, SAHA, MS-275, and valproic acid, are in clinical trials for the treatment of hemopoetic and solid tumors, including colon cancer (Mei et al., 2004; Bolden et al., 2006; Rasheed et al., 2007), with SAHA recently being approved for the treatment of patients with cutaneous T cell lymphoma.
Several studies have demonstrated increased expression of the class I HDACs, HDAC1, HDAC2, and HDAC3, in multiple human cancers, including colon cancer (Zhu et al., 2004; Huang et al., 2005; Wilson et al., 2006; Khabele et al., 2007). Consistent with the ability of HDAC inhibitors to induce maturation in colon cancer cell lines, individual class I HDACs can repress p21 expression, promote cell proliferation and survival, and inhibit differentiation in vitro (Lagger et al., 2002; Glaser et al., 2003; Zhu et al., 2004; Huang et al., 2005; Wilson et al., 2006). Class I HDACs are maximally expressed in the proliferative compartment of the intestine in vivo (Wilson et al., 2006), consistent with a role in promoting cell growth, and they are important mediators of intestinal development and differentiation in vivo (Tou et al., 2004). In contrast, comparatively little is known about the role that class II HDACs play in intestinal biology.
Similar to class I HDACs, class IIa HDACs are transcriptional corepressors (Yang and Gregoire, 2005). However, important functional differences exist between class I and class IIa HDACs. First, the corepressor function of the latter is regulated by phosphorylation-dependent changes in subcellular localization (Grozinger and Schreiber, 2000; Zhou et al., 2000; Vega et al., 2004a). Second, HDAC4 is known to exist in distinct corepressor complexes than the class I HDACs HDAC1 and HDAC2 (Guenther et al., 2000; Li et al., 2000; Fischle et al., 2002; Jepsen and Rosenfeld, 2002; Codina et al., 2005). The difference in function between class I and II HDACs is also exemplified in various knockout mouse models. For example, whereas knockout of HDAC1 and HDAC2 results in embryonic and perinatal lethality, respectively (Montgomery et al., 2007), HDAC4 knockout mice are viable, but develop skeletal abnormalities (Vega et al., 2004b).
Several observations implicate the class IIa HDAC HDAC4 in regulating differentiation in nonintestinal tissue. HDAC4 knockout mice display premature ossification of developing bones as a result of early onset of chondrocyte hypertrophy, an effect mediated by loss of HDAC4-induced repression of Runx2 (Vega et al., 2004b). HDAC4 also regulates skeletal muscle differentiation through its interaction with the myogenic transcription factor, MEF2 (Miska et al., 1999; McKinsey et al., 2000; Miska et al., 2001). Here, we show that HDAC4 is maximally expressed in the proliferative compartment in normal colonic and small intestinal epithelium and that its expression is down-regulated during differentiation. Direct evidence of a proproliferative role for nuclear HDAC4 was demonstrated in colon cancer cells in vitro, a component of which was Sp1-dependent recruitment of HDAC4 to the p21 promoter, likely directed through the HDAC4-HDAC3-N-CoR/SMRT corepressor complex, resulting in transcriptional repression. The role for HDAC4 in the maintenance of colon cancer cell proliferation is consistent with the ability of HDAC inhibitors to induce growth arrest in these cells. Coupled with a similar role for class I HDACs in these processes, our findings suggest that inhibition of both class I and II HDACs is likely to be more effective as a chemotherapeutic strategy.
MATERIALS AND METHODS
Cell Culture
HCT116 and Caco-2 colon cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA). HCT116 cells with and without deletion of both p21 alleles were kind gifts from Dr Bert Vogelstein (The Johns Hopkins Oncology Center, Baltimore, MD) (Bunz et al., 1999). All were maintained as described previously (Wilson et al., 2006). LS174T DNTCF4 clone N2 cells were a kind gift from Dr. Hans Clevers (Hubrecht Laboratory and Utrecht University, Utrecht, The Netherlands) (van de Wetering et al., 2002). To induce expression of DNTCF4, doxycycline (MP Biomedicals, Solon, OH) was added to a final concentration of 1 μg/ml at time of cell plating, and it was replaced every 24 h in fresh medium over a 96-h period. The sodium salt of the short-chain fatty acid butyrate was obtained from Sigma-Aldrich (St. Louis, MO).
Expression Vectors, Luciferase Reporter Constructs, and Transient Transfection Assays
HCT116 cells were transiently transfected using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). The p21 promoter constructs pWP-133 and pWP-101 were kindly provided by Dr. Yosihiro Sowa (Sowa et al., 1997; Wilson et al., 2006). Sp1/Sp3 reporter constructs have been described previously (Sowa et al., 1997; Wilson et al., 2006). Myc-tagged pcDNA3-HDAC4 and pcDNA3 HDAC4-GFP (1-1084) were kind gifts from Dr. Tony Kouzarides (Wellcome/Cancer Research Campaign Institute, Cambridge, United Kingdom) (Miska et al., 1999, 2001; Miska et al., 2001). The green fluorescent protein (GFP)-tagged HDAC4 deletion mutants 1-326, 206-1040, and 621-1040 were generously provided by Dr. Xiang-Jiao Yang (McGill University, Montreal, Canada) (Wang et al., 1999; Wang and Yang, 2001). In experiments involving butyrate, untreated and treated cells were transfected identically and compared by relative -fold induction by the drug after correction for total cellular protein. Luciferase activity was determined in cell lysates using the luciferase or dual-luciferase assay kits (Promega, Madison, WI). Transfection efficiency was controlled by cotransfection with TK-Renilla (Promega, Madison, WI).
Small Interfering RNA (siRNA) Experiments
Expression of HDAC4 was selectively down-regulated using a pool of two predesigned siRNA duplexes, siHDAC4#1 (sense strand, CGACAGGCCUCGUGUAUGAUU) and siHDAC4#2 (sense strand, AAAUUACGGUCCAGGCUAAUU) (Dharmacon RNA Technologies, Lafayette, CO). This pool is described throughout the manuscript as “siHDAC4.” An additional pool of three siRNA duplexes targeting HDAC4 (siHDAC4 sc) was obtained from an independent source (sc-35540; Santa Cruz Biotechnology, Santa Cruz, CA). Expression of Sp1 was selectively down-regulated using a pool of four predesigned siRNAs (sense strands, GCCAAUAGCUACUCAACUAUU, GAAGGGAGGCCCAGGUGUAUU, GGGCAGACCUUUACAACUCUU, and GGAGUGAUGCCUAAUAUUCUU; Dharmacon RNA Technologies). A pool of four predesigned siRNAs targeting SMRT was obtained from Dharmacon RNA Technologies (M-020145-01). Five different nontargeting control siRNAs were used: nontargeting (NT) pool (Dharmacon RNA Technologies), siNEG and siGFP (Invitrogen), and NT “A” and NT “C” (Santa Cruz Biotechnology). HCT116 cells were transiently transfected with 100 nM siRNA for 24–168 h, by using the Profection (calcium phosphate) transient transfection system (Promega).
Immunohistochemistry
Immunohistochemical detection of HDAC4 was performed as described previously (Velcich et al., 2002), with the addition of an antigen retrieval step by using Target Retrieval Solution (Dako North America, Carpinteria, CA). Formalin-fixed paraffin-embedded sections of human duodenum, or mouse distal colon, were incubated with rabbit anti-HDAC4 (1:25 dilution; Santa Cruz Biotechnology) or rabbit anti-Ki67 (1:1000 dilution; Vector Laboratories, Burlingame, CA) for 1 h. Detection of primary signal was with the anti-rabbit EnVision reagent (Dako North America), followed by incubation with 3′,3′-diaminobenzidine substrate (Cell Marque, Hot Springs, AR) and hematoxylin counterstaining.
Immunofluorescence
Immunofluorescence staining of HCT116 cells has been described previously (Wilson et al., 2003). HDAC4-GFP was visualized in the fluorescein isothiocyanate channel. Endogenous Sp1 was detected with a rabbit anti-Sp1 from Millipore (Billerica, MA). Confocal microscopy confirming the colocalization of HDAC4-GFP and Sp1 was performed using a Leica AOBS laser scanning confocal microscope (Leica Microsystems, Bannockburn, IL).
Subcellular Fractionation and Western Blotting
Protein isolation, Western blotting, and signal detection were performed as described previously (Mariadason et al., 2001a; Wilson et al., 2006). Nuclear and cytosolic fractions were harvested from cell extracts by using the Qproteome Cell Compartment kit (QIAGEN, Valencia, CA). For experiments measuring protein expression along the mouse small intestinal crypt-villus axis, enterocytes were sequentially isolated from a 13-wk-old male C57BL/6 mouse, as described previously (Weiser, 1973a,b; Ferraris et al., 1992; Mariadason et al., 2005). Rabbit anti-HDAC4 (1:100 dilution) was from Santa Cruz Biotechnology. Additional antibodies used were rabbit anti-GFP tag (Invitrogen), rabbit anti-HDAC1 (1:500; Imgenex, San Diego, CA), rabbit anti-HDAC2 (1:2000; Santa Cruz Biotechnology), rabbit anti-HDAC3 (1:2000; Novus Biologicals, Littleton, CO), rabbit anti-HDAC6 (1:100; Santa Cruz Biotechnology), mouse anti-proliferating cell nuclear antigen (PCNA) (1:1000; Santa Cruz Biotechnology), mouse anti-β-tubulin (1:1000; Sigma-Aldrich), mouse anti-β-actin (1:4000; Sigma-Aldrich), rabbit anti-p21 (1:200; Santa Cruz Biotechnology), rabbit anti-Sp1 (1:1000; Millipore), and rabbit anti-SMRT (1:500; Millipore).
Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Quantitative real-time RT-PCR (QPCR) was performed as described previously (Wilson et al., 2002). Relative values were determined from a standard curve of HCT116 cDNA and expressed relative to β-actin. Primer sequences used were HDAC1 (forward [F], 5′-AGCTCCACATCAGTCCTTCCA-3′; reverse [R], 5′-GTGCGGCAGCATTCTAAGGT-3′), HDAC2 (F, 5′-AAACTGCATATTAGTCCTTCAA-3′; R, 5′-TGAGGTAACATGCGCAAATTTT-3′), HDAC3 (F, 5′-GGAGCTGGACACCCTATGAA-3′; R, 5′-TATTGGTGGGGCTGACTCTC-3′), HDAC4 (F, 5′-GGTTTGAGAGCAGGCAGAAC-3′; R, 5′-CAGAGAATGAGGCCAAGGAG-3′), HDAC5 (F, 5′-ATGTATGCTGTGCTGCCTTG-3′; R, 5′-GTAGGAGTTTTGCGGTGATG-3′), HDAC6 (F, 5′-CATTAGGCCTCCTGGACATCA-3′; R, 5′-CGGTGTTTCTGTTGAGCATAG-3′), HDAC7 (F, 5′-TGTCTGCTGGATTTGATGC-3′; R, 5′-TGAGGTTGGGTTTCTGTTT-3′), HDAC8 (F, 5′-AGTCCCGAGTATGTCAGTATG-3′; R, 5′-AAGCATCAGTGTGGAAGGTG-3′), HDAC9 (F, 5′-GCTGTGAAGGTCAAGGAGGA-3′; R, 5′-TTGCTGGGTGAGGTAAAACA-3′), HDAC10 (F, 5′-ACCCCAGCGTCCTTTACTTC-3′; R, 5′-GTAGTCAGCGTTTCCCATCC-3′), p21 (F, 5′-ATGTGTCCTGGTTCCCGTTTC-3′; R, 5′-CATTGTGGGAGGAGCTGTGA-3′), and β-actin (F, 5′-CACCTTCACCGTTCCAGTTT-3′; R, 5′-GATGAGATTGGCATGGCTTT-5′).
Coimmunoprecipitation Experiments
For coimmunoprecipitation experiments, 300 μg of nuclear extract was precleared with protein A agarose beads (Santa Cruz Biotechnology) for 2 h at 4°C and incubated with the following antibodies: 5 μg of rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology), 5 μg of rabbit polyclonal GFP antibody (Invitrogen) to detect HDAC4-GFP, and 5 μg of a rabbit polyclonal anti-Sp1 (Millipore). After overnight incubation, protein complexes were pulled down with protein A agarose beads (4 h at 4°C). The beads were washed twice, pelleted by gentle centrifugation, resuspended in 20 μl of 2× Laemmli SDS sample buffer, and submitted to SDS-polyacrylamide gel electrophoresis as described above.
Chromatin Immunoprecipitation (ChIP)
For ChIP experiments, 2 × 107 HCT116 cells were used. For experiments using the pWP101 transient template, cells were transfected overnight with 2 μg/ml medium of the plasmid. Cells were cross-linked with 1% formaldehyde (Sigma-Aldrich), harvested into cell lysis buffer, and sonicated with 2 × 5-s pulses. The sonicated chromatin was immunoprecipitated with 5 μg of rabbit polyclonal HDAC4 antibody (Imgenex), 5 μg of rabbit polyclonal HDAC3 antibody (Novus Biologicals), 5 μg of a rabbit polyclonal Sp1 antibody (Millipore), 5 μg of a rabbit polyclonal acetylated histone H3 antibody (Millipore), or 5 μg of normal rabbit IgG (Santa Cruz Biotechnology). DNA–protein complexes were isolated with protein A/G Plus agarose beads (Santa Cruz Biotechnology) for 4 h at 4°C, washed, eluted in 0.1% SDS, 0.1M NaHCO3 elution buffer, and cross-links were reversed overnight at 65°C in 0.3 M NaCl. Input samples were also incubated in this way. DNA was purified using phenol:chloroform extraction and ethanol precipitation.
The transient template (pWP101, 4.18kB) was analyzed using two sets of primer pairs (see Figure 9A). The first, designated pWP101-p21, was designed such that the forward primer contains vector backbone sequence immediately distal to the 101-base p21 promoter sequence, and the reverse primer contains luciferase sequence immediately proximal to the 101-base p21 promoter sequence. The second primer pair, designated pWP101-up, was designed against vector backbone sequence ∼2 kb away from the 101-base p21 promoter sequence. The primers used were as follows: pWP101-p21: F, 5′-AAGGTACGGGAGGTACTTGGA-3′ and R, 5′-GTTCCATCTTCCAGCGGATA-3′); and pWP101-up: F, 5′-GGCGCTTTCTCATAGCTCAC-3′ and R, 5′-CCTCGCTCTGCTAATCCTGT-3′).
Figure 9.
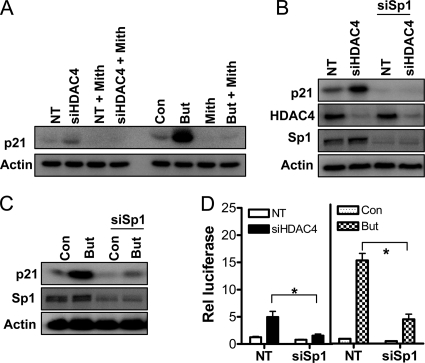
Role of Sp1 in HDAC4-mediated repression of p21. (A) Effect of the Sp1 inhibitor mithramycin on siHDAC4 and butyrate-mediated p21 induction. p21 protein levels in HCT116 cells treated with mithramycin (1 μg/ml) for 24 h in combination with either a 72-h exposure to NT siRNA or siHDAC4 (100 μM), or a 24-h exposure to 2 mM butyrate. The effect of down-regulation of Sp1 on induction of p21 by siHDAC4 or butyrate is shown in B and C, respectively. p21, HDAC4, and Sp1 protein levels in HCT116 cells were determined after 72-h treatment with NT siRNA or siHDAC4 with or without concomitant treatment with siRNA targeting Sp1. All cells were treated with an equal final concentration of siRNA (100 nM). (D) Effect of Sp1 down-regulation on p21 promoter activity stimulated by siHDAC4 or butyrate, as determined by luciferase assay. HCT116 cells were transfected with pWP-133 (0.25 μg) and either NT siRNA siHDAC4 or siSp1 (total siRNA transfected was 100 nM), and cultured for 72h. In separate experiments, cells were transfected with pWP-133 (0.25 μg) and either NT siRNA or siSp1 (both 100 nM), and they were cultured with and without 2 mM butyrate for 24 h. TK-Renilla (0.1 μg) was cotransfected in all treatment groups to control for transfection efficiency. Values shown are mean + SEM of three independent experiments; *p < 0.05, Student's t test.
Endogenous template DNA was analyzed using two independent primer sets designated p21-1 and p21-2. These primers were designed to amplify DNA adjacent to the transcription start site of the p21 promoter containing the six Sp1 binding sites (Wilson et al., 2006). Primer sets were also designed to amplify a region within the p21 promoter 4 kb upstream of the transcription start site, which does not contain Sp1/Sp3 sites, and also the β-actin promoter. Quantitative real-time RT-PCR was performed, with DNA content in Input and immunoprecipitation samples measured relative to a standard curve of HCT116 cell genomic DNA. All experimental values were expressed relative to relevant Input DNA content. Primer sequences were as follows: p21-1: F, 5′-CATTCTGGCCTCAAGATGCT-3′ and R, 5′-CACGAAGACCCTCTCCACTG-3′); p21-2: F, 5′-AGTGCCAACTCATTCTCCAAG-3′ and R, 5′-GACACATTTCCCCACGAAGT-3′); p21 up: F, 5′-AGTCTTGCCTGCCTTCAGAG-3′ and R, 5′-ACGAAGGGCTTGTTTTAGG-3′); and β-actin: F, 5′-AGTGTGGTCCTGCGACTTCT-3′ and R, 5′-ACTGGGTGGGTCGTGTAAAT-3′).
Sequential ChIP
Cross-linked chromatin from HCT116 cells was immunoprecipitated with an antibody against HDAC4 as described in Chromatin Immunoprecipitation, except that chromatin was eluted in 10 mM dithiothreitol for 30 min at 37°C. Eluted chromatin was diluted, subjected to a second immunoprecipitation with antibody against Sp1, and then eluted with standard elution buffer (see above). A “reverse“ sequential ChIP was carried out in which chromatin was immunoprecipitated first with anti-Sp1 and then with anti-HDAC4. The isolated DNA was extracted, purified, and analyzed as described above. Input DNA was calculated from an aliquot of diluted chromatin obtained from the first elution.
Flow Cytometry
For cell cycle analyses, HCT116 cells were stained with propidium iodide and analyzed by flow cytometry as described previously (Wilson et al., 2006).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) Assay
The MTT assay in HCT116 cells was performed as described previously (Wilson et al., 2006).
Xenograft Assay
HCT116 cells were transfected with NT or siHDAC4 (100 nM) overnight, trypsinized, counted, and resuspended in an equal volume of High Concentration Matrigel (BD Biosciences, San Jose, CA). Cells (5 × 106) were then injected into the right flank of SCID mice, and tumor growth was monitored over the course of 7 d. Eight mice per group (NT or siHDAC4) were used. Animals were killed after this time, the tumors were resected, and the volume was calculated from caliper measurements of the smallest and longest tumor diameter (Benimetskaya et al., 2006).
RESULTS
Robust HDAC4 Expression in Normal Intestine and in Colon Cancer Cells
Because class IIa HDACs have been reported to be expressed in a tissue-specific manner (Wang et al., 1999; Liu et al., 2006), we first confirmed the expression of HDAC4 in normal intestinal epithelium. Although considerable variability in HDAC4 protein expression was observed among a panel of different mouse tissues, robust HDAC4 expression was detected in the small intestine and colon (Figure 1A). Furthermore, there was strong expression of HDAC4 in each of a panel of 26 colon cancer cell lines (Supplemental Figure 1A), demonstrating its expression is maintained in transformed colonic epithelial cells. We then determined the subcellular localization of endogenous HDAC4 in colon cancer cells by immunofluorescence analysis. Consistent with reports in other cell types (Wang and Yang, 2001; Liu et al., 2004), HDAC4 expression in the HCT116 colon cancer cell line was observed in both the nucleus and cytoplasm (Supplemental Figure 1B). In contrast, the class I HDAC HDAC1 exhibited an exclusively nuclear staining pattern, whereas localization of the class IIb HDAC HDAC6 was predominantly cytoplasmic, consistent with previous studies (Hubbert et al., 2002; Waltregny et al., 2004).
Figure 1.
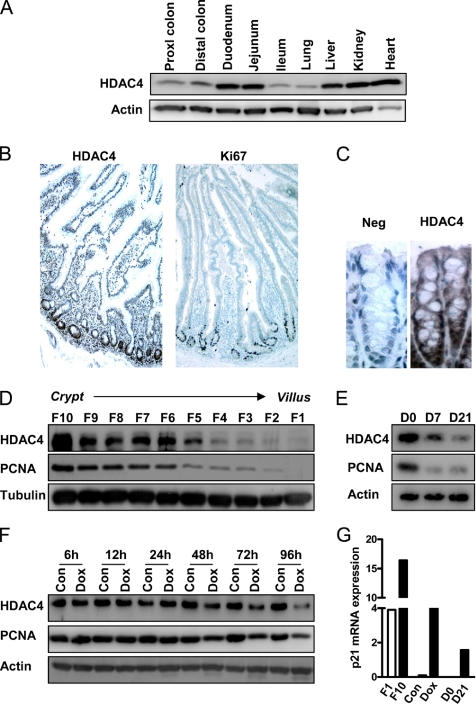
HDAC4 protein expression in normal intestinal epithelium and colon cancer cell lines. (A) Total protein was extracted from various tissues from a wild-type C57BL/6 mouse and levels of HDAC4 quantified by Western blot. Blots were reprobed for actin to ensure equal loading. Immunohistochemical detection of HDAC4 and Ki67 expression in a low-power (10×) longitudinal section of normal human small intestine (B) and HDAC4 in the distal colon of a C57BL/6 mouse (C). (D) Epithelial cells were sequentially isolated along the mouse crypt–villus axis as 10 individual fractions (F), with F1 and F10 representing cells isolated from the villus tip and crypt base, respectively, and HDAC4 and PCNA protein levels determined. (E) Caco-2 cells were allowed to spontaneously differentiate over 21 d, and levels of HDAC4 and PCNA were determined at confluence (day 0) or 7 and 21 d after confluence. (F) LS174T-N2 cells, which are stably transfected with doxycycline-inducible dominant-negative TCF4, were cultured with and without doxycycline for 6–96 h. Doxycycline was replaced every 24 h in fresh medium. Levels of HDAC4 and PCNA protein were quantified by Western blot. (G) Comparison of steady-state levels of p21 mRNA in Weiser F1 and F10 fractions, control and doxycycline-treated LS174T cells (96 h) and day 0 and day 21 Caco-2 cells. Expression of p21 mRNA was calculated relative to an actin mRNA internal standard by QPCR.
HDAC4 Expression Is Down-Regulated during Intestinal Cell Differentiation In Vivo and In Vitro
The epithelium of the small intestine is compartmentalized into the crypt, which contains stem and proliferative progenitor cells, and the villus, containing nonproliferating differentiated epithelial cells. HDAC4 expression was maximal in human small intestinal crypts, an expression pattern similar to that of the established proliferation marker Ki67 (Figure 1B). HDAC4 expression was also detected in mouse distal colon, and it was predominantly nuclear in localization and more highly expressed at the base of the crypts, which contain the proliferative cells (Figure 1C). We confirmed the differential pattern of HDAC4 expression along the mouse small intestinal crypt-villus axis by using a cell fractionation method in which cells are sequentially isolated beginning at the villus tip and culminating in the crypts (Mariadason et al., 2005). As shown in Figure 1D, HDAC4 protein expression was maximal in the crypt cell fraction (fraction 10) and was significantly reduced in cells isolated from the villus tip (fraction 1), a pattern of expression which paralleled that of PCNA. Therefore, expression of HDAC4 was associated with intestinal cell proliferation, and it was down-regulated during cell differentiation, in vivo.
Consistent with these in vivo findings, we confirmed that expression of HDAC4 was down-regulated during growth arrest and differentiation of colon cancer cells in vitro. First, in Caco-2 cells that undergo contact inhibition-driven spontaneous differentiation along the absorptive cell lineage over a 21-d period in culture (Mariadason et al., 2000), HDAC4 expression was markedly reduced during the differentiation program (Figure 1E). Second, we examined HDAC4 expression in LS174T colon cancer cells engineered to undergo growth arrest and differentiation as a result of down-regulation of β-catenin-TCF signaling by inducible expression of dominant negative TCF4 (van de Wetering et al., 2002). As shown in Figure 1F, HDAC4 was down-regulated 48 h after addition of doxycycline. In vivo and in the in vitro models of intestinal differentiation used, HDAC4 down-regulation closely paralleled down-regulation of the proliferation marker PCNA (Figure 1, D–F), whereas there was an inverse correlation between the mRNA levels of the cyclin-dependent kinase inhibitor p21, an established marker of cell differentiation, and HDAC4 (Figure 1G).
HDAC4 Promotes Colon Cell Growth and Survival
The confinement of expression of HDAC4 to the proliferative compartment of the normal small intestinal and colonic epithelium, and its down-regulation during colon cell maturation in vitro suggested it may have a physiological function in maintaining cell proliferation. To directly determine the link between HDAC4 expression and colon cell growth, we examined the effect of HDAC4 down-regulation by RNA interference on cell number and cell cycle indices in HCT116 colon cancer cells.
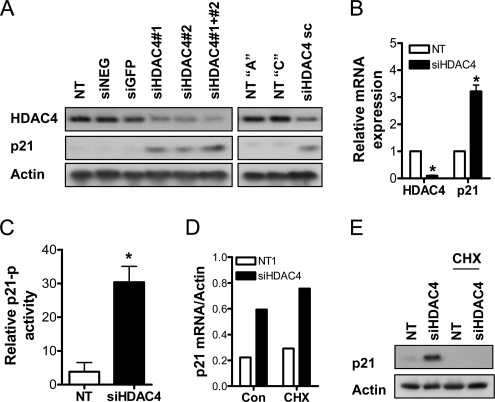
First, we tested the effect of several siRNA duplexes targeting HDAC4 on HDAC4 expression (see Figure 6A for these experiments). To ensure specificity, we used two individual duplexes (siHDAC4#1 and siHDAC4#2), as well as a pool of three independent duplexes (siHDAC4 sc). Transfection with siHDAC4#1 or siHDAC4#2 reduced HDAC4 expression compared with three commercially available negative control siRNAs: NT pool, siNEG, and siGFP. Notably, when siHDAC4#1 and siHDAC4#2 were added as a pool, greater down-regulation of HDAC4 expression was achieved. The down-regulation achieved by transfection with siHDAC4 sc compared with two controls from Santa Cruz Biotechnology, NT A and NT C, was similar to that of siHDAC4#1 or siHDAC4#2 alone. Therefore, we chose to conduct all future HDAC4 down-regulation experiments by using the siHDAC4#1 pool, denoted as siHDAC4 throughout the remainder of the article.
Figure 6.
HDAC4 regulation of p21 expression in colon cancer cells in vitro. (A) The effect of two independent siHDAC4 duplexes #1 and #2 and a pool of #1 and #2 (designated siHDAC4 and used throughout the study) on HDAC4 and p21 protein expression, compared with the negative controls, NT, siNEG, and siGFP (all 100 nM for 72 h). Additionally, the effect of an independent pool of three siRNA duplexes targeting HDAC4 (siHDAC4 sc) on HDAC4 and p21 protein expression was compared with the negative controls, NT A and NT C (all 100 nM for 72 h). See Materials and Methods for a detailed description of these siRNAs. Protein levels of p21 and HDAC4 were determined by Western blot. (B) The steady-state mRNA levels of HDAC4 and p21 in HCT116 cells treated for 36 h with NT or siHDAC4 (both 100 nM), determined by QPCR. Values are mean + SEM of three replicates, and they are expressed relative to β-actin. *p < 0.05 relative to NT1, Student's t test. (C) Effect of HDAC4 down-regulation on p21 promoter activity, as determined by luciferase assay. HCT116 cells were transfected with the p21 luciferase reporter plasmid, pWP-133 (0.25 μg), and either NT or siHDAC4 (100 nM) for 72 h. TK-Renilla (0.1 μg) was cotransfected in all treatment groups to control for transfection efficiency. Values shown are mean + SEM of three independent experiments; *p < 0.05 relative to NT1, Student's t test. (D) The steady-state mRNA levels of p21 in HCT116 cells treated for 24 h with NT or siHDAC4 (both 100 nM), with and without concomitant addition of 5 μg/ml cycloheximide. mRNA expression was determined by QPCR. Values are mean of a representative experiment, and are expressed relative to β-actin. *p < 0.05 relative to NT1, Student's t test. (E) The effect of protein synthesis inhibition on p21 protein induction mediated by HDAC4 down-regulation. HCT116 cells were treated for 48 h with NT or siHDAC4 (both 100 nM), with and without concomitant addition of 5 μg/ml cycloheximide for a subsequent 24 h.
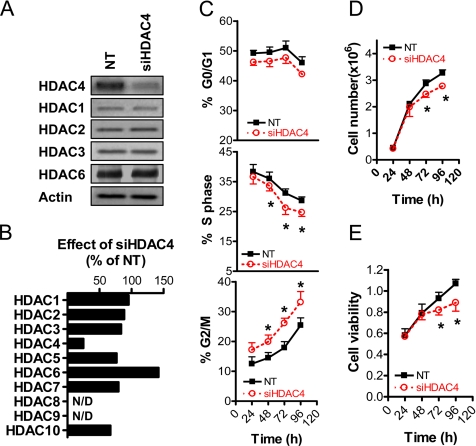
First, we demonstrated that siRNA-mediated targeting of HDAC4 mRNA (siHDAC4) selectively down-regulated HDAC4 expression among both class I and class II HDACs. As shown in Figure 2A, siHDAC4 markedly down-regulated protein expression of HDAC4 but not that of the class I HDACs HDAC1, HDAC2, or HDAC3 or the class IIb HDAC HDAC6. We also demonstrated that siHDAC4 selectively down-regulated HDAC4 expression at the mRNA level, as shown in Figure 2B. The steady-state levels of HDAC4 mRNA were reduced by ∼80% compared with NT siRNA. In contrast, mRNA levels of HDAC1, HDAC2, HDAC3, the class IIa HDACs HDAC5 and HDAC7, and the class IIb HDACs HDAC6 and HDAC10 were not reduced by siHDAC4. The mRNA expression of HDAC8 and HDAC9 was not detected in HCT116 cells.
Figure 2.
Effect of HDAC4 down-regulation on growth of colon cancer cells in vitro. (A) Selective down-regulation of HDAC4 expression in HCT116 cells treated for 72 h with the nontargeting siRNA duplex (NT) or a pool of two siRNAs targeting HDAC4 (siHDAC4). Both NT and siHDAC4 were added to a final concentration of 100 nM. Protein levels of HDAC1, HDAC2, HDAC3, HDAC4, and HDAC6 were determined by Western blot. (B) QPCR analysis of the effect of siHDAC4 on mRNA expression of HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC6, HDAC7, and HDAC10. N/D denotes no mRNA expression for HDAC8 and HDAC9 detected. Results are expressed relative to actin mRNA and are expressed as the percentage of mRNA expression in control (NT) transfected samples. (C) Time course analysis of the effects of NT and siHDAC4 (both 100 nM) on the percentage of cells in G0/G1, S and G2/M phase, determined by flow cytometry. (D) Time course analysis of the effects of NT and siHDAC4 (both 100 nM) on adherent cell number in HCT116 cells. (E) Time course analysis of the effects of NT and siHDAC4 (both 100 nM) on MTT incorporation in HCT116 cells. C–E show the results of three experiments, expressed as mean + SEM; *p < 0.05 relative to relevant control, Student's t test.
We then assessed the effect of siHDAC4 on indices on cell growth compared with that of the control NT siRNA. As shown in Figure 2C, siHDAC4 induced a reduction in the percentage of cells in S phase, with a concomitant increase in the percentage of cells at the G2/M phase of the cell cycle, and a modest decrease in the percentage of cells in G0/G1 phase. Down-regulation of HDAC4 induced an ∼20% reduction in adherent cell number 72–96 h after transfection (Figure 2D). The magnitude of growth inhibition mediated by HDAC4 down-regulation was further increased when the cells were cultured in decreasing amounts of serum, with ∼40% reduction in adherent cell number compared with NT controls observed under serum-free conditions (Supplemental Figure 2). Finally, down-regulation of HDAC4 induced a reduction in cell viability, as assessed by the MTT assay (Figure 2E), collectively demonstrating a proproliferative role for HDAC4 in colon cancer cells.
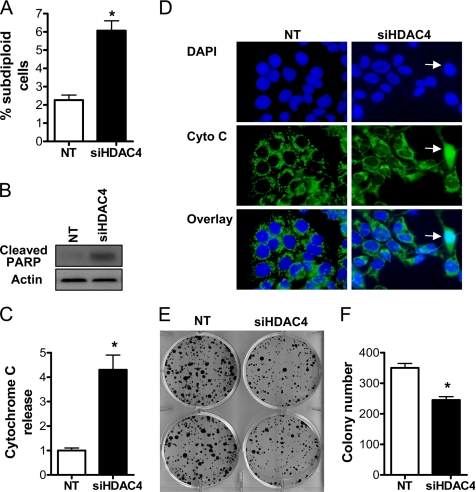
We also demonstrated that HDAC4 promoted an enhancement of colon cell survival (Figure 3). First, treatment of HCT116 cells with siHDAC4 for 72 h resulted in a modest, although statistically significant, increase in the subdiploid cell population (Figure 3A), which was consistent with a parallel increase in cleavage of poly(ADP-ribose) polymerase (PARP) (Figure 3B). We also assayed the release of cytochrome c from mitochondria by immunofluorescence analysis. Nonapoptotic cells are characterized by a punctuate staining pattern of cytochrome c, which we have shown previously to colocalize with the mitochondrial marker Hsp60 (Wilson et al., 2003). In contrast, apoptotic cells display a diffuse staining pattern of cytochrome c, indicative of its release into the cytosol (Wilson et al., 2003). As shown in Figure 3, C and D, siHDAC4 increased the number of cells releasing cytochrome c fourfold relative to cells transfected with NT siRNA. Second, we performed clonogenic assays on cells transfected with the appropriate siRNA for 72 h. As shown in Figure 3, E and F, down-regulation of HDAC4 resulted in the formation of ∼25% fewer colonies compared with HCT116 cells transfected with the NT siRNA.
Figure 3.
Effect of HDAC4 down-regulation on survival of colon cancer cells in vitro. (A) The effect of NT and siHDAC4 (both 100 nM) on subdiploid (apoptotic) cell population assayed by flow cytometry. Values shown are mean + SEM of three independent experiments; p < 0.05 relative to NT, Student's t test. (B) Effect of HDAC4 down-regulation on cleavage of PARP. (C) Cells were treated with NT or siHDAC4 (100 nM) for 72 h, and cytochrome c release was assayed by immunofluorescence. Values were obtained from counts of 200 cells and are expressed as the mean + SEM of three independent experiments; *p < 0.05 relative to NT, Student's t test. (D) Representative fields of NT and siHDAC4-treated cells. 4,6-Diamidino-2-phenylindole (DAPI)-stained nuclei and cytochrome c (cyto C) were detected, and an overlay was generated. The white arrowhead shows a cell releasing cytochrome c into the cytosol, as indicated by a diffuse staining pattern. (E) Clonogenic assay showing the effect of a 72-h incubation with siHDAC4 (100 nM) on formation of cell colonies compared with NT-treated cells. (F) Quantification of colony number from E. Results shown are mean + SEM of three independent experiments; *p < 0.05 relative to NT, Student's t test.
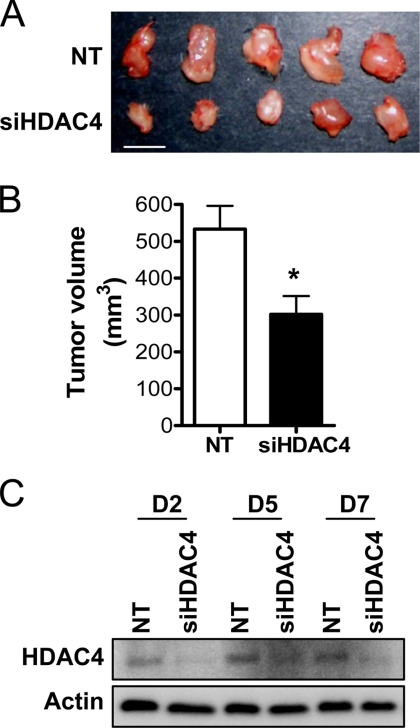
We then examined the effect of down-regulation of HDAC4 on HCT116 cell growth in vivo. Cells transfected with NT or siHDAC4 were injected into SCID mice as xenografts, and growth of the resultant tumor was measured after 7 d. The volume of the siHDAC4-transfected tumors was significantly smaller than tumors deriving from control cells (Figure 4, A and B). HDAC4 protein levels were measured in HCT116 cells cultured in parallel in vitro to confirm that down-regulation was maintained throughout the duration of the experiment. As shown in Figure 4C, HDAC4 down-regulation was maintained over the 7-d experimental period.
Figure 4.
Effect of HDAC4 down-regulation on growth of colon cancer cells in vivo. (A) Representative tumors obtained at sacrifice after 7 d of growth of HCT116 cell xenografts in 4-wk-old male SCID mice (bar, 1 cm). HCT116 (5 × 106) cells were transfected with NT siRNA or siHDAC4 (both 100 nM) and injected into the right flank with an equal volume of Matrigel. (B) Tumor volume was quantified from measurements of the longest and smallest diameter of the tumor. Values shown are mean + SEM, n = 8; *p < 0.05 relative to NT, Student's t test. (C) HDAC4 protein levels in HCT116 cells transfected with NT or siHDAC4 and cultured in parallel in vitro for up to 7 d.
Nuclear Localization of HDAC4 during Cell Proliferation
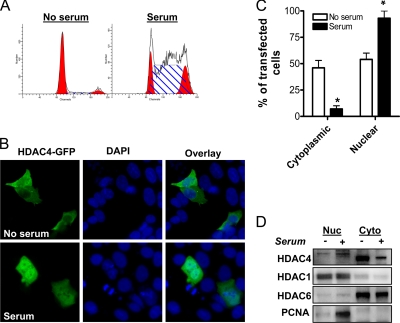
Our immunohistochemical analyses demonstrated maximal HDAC4 expression in the proliferative crypt region in vivo, where it was predominantly nuclear in location. Given the established role of shuttling of HDAC4 between the nucleus and cytoplasm in regulating its effects (Grozinger and Schreiber, 2000), we examined the link between HDAC4 subcellular localization and cell proliferation in HCT116 cells induced to proliferate by a serum pulse after 24-h serum starvation.
For these studies, HCT116 cells were transfected with a full-length HDAC4-GFP (1-1084) expression vector, and then they were serum-starved for 24 h. Subsequently, cells were either pulsed for 16 h with medium containing 10% serum or further incubated under serum-free conditions. As shown in Figure 5A, 64% of the cells were in S phase after the 16-h serum pulse, compared with only 4% in serum-starved cells. Two hundred cells positive for HDAC4-GFP were counted for each condition, and distribution of the construct was analyzed. Representative cell fields are shown in Figure 5B. The proportion of transfected cells displaying exclusively cytoplasmic localization of HDAC4-GFP was markedly higher in the serum-starved, growth-arrested cells (Figure 5, B and C). In addition, there was a concomitant increase in cells displaying nuclear HDAC4-GFP staining in the proliferating cell population (Figure 5, B and C). These results establish a link between the growth-promoting effects of HDAC4 and its nuclear localization.
Figure 5.
Effect of cell proliferation on subcellular localization of HDAC4. (A) HCT116 cells were transfected with HDAC4-GFP, and serum-starved for 24 h. Cells were then either maintained in 0% serum, or they were serum-pulsed (10%) for 16 h. Representative histograms after flow cytometric analysis are shown. (B) Representative fields of HDAC4-GFP 1-1084-transfected cells (green) after serum starvation, or after serum pulse. Nuclei are stained with DAPI (blue). (C) The distribution of HDAC4-GFP after serum starvation, or serum pulse was analyzed by counting 200 GFP-positive cells per treatment. The distribution was categorized as either nuclear or exclusively cytoplasmic. Values shown are mean + SEM of three independent experiments; *p < 0.05 relative to no serum, Student's t test. (D) Endogenous HDAC4, HDAC1, HDAC6, and PCNA protein levels in nuclear and cytosolic fractions were determined by Western blot after subcellular fractionation of serum-starved and serum-pulsed cells.
We validated our findings by measuring endogenous HDAC4 expression in nuclear and cytosolic fractions extracted from serum-starved and serum-pulsed cells. Consistent with our immunofluorescence analyses, cytosolic HDAC4 expression was enriched in growth-arrested, serum-starved cells, whereas nuclear HDAC4 was increased in proliferating, serum-pulsed cells (Figure 5D). Strikingly, the nuclear form of HDAC4 had a slightly higher molecular weight than the cytosolic form. We confirmed that siHDAC4 down-regulated both the high and low molecular forms of HDAC4 (data not shown). The basis for this shift in molecular weight, possibly due to differential posttranslational modification, is worthy of further investigation. The efficiency of subcellular fractionation was confirmed by examining expression of the exclusively nuclear protein HDAC1 and the exclusively cytoplasmic protein HDAC6 (Figure 5D). Validating the increased proliferation of the serum-pulsed treatment group, PCNA expression was markedly increased.
HDAC4 Represses p21 Transcription in Colon Cancer Cells
To determine the mechanism of HDAC4-mediated growth promotion in colon cancer cells, we examined the role of HDAC4 in regulating expression of p21, a cyclin-dependent kinase inhibitor that is a well established target of HDAC inhibitors. Because treatment of human cancer cells with HDAC inhibitors consistently leads to up-regulation of p21 expression (Sowa et al., 1997; Archer et al., 1998; Kim et al., 2001; Wilson et al., 2006), we sought to determine whether down-regulation of HDAC4 had a similar effect.
As shown in Figure 6A, transfection with the HDAC4 siRNAs described above all induced p21 protein expression, with the magnitude of induction correlating well with their respective ability to down-regulate HDAC4 expression. The specificity of HDAC4 down-regulation–mediated p21 induction was confirmed by comparison with the negative control siRNAs described above. Measurement of p21 mRNA expression by QPCR confirmed that HDAC4 down-regulation increased p21 transcription, with a threefold increase in p21 mRNA levels after 36 h (Figure 6B). Consistent with these results, there was a greater than threefold induction of p21 reporter activity in HCT116 cells transfected with siHDAC4 (Figure 6C). We then confirmed that p21 was likely a direct target of HDAC4, because siHDAC4 was able to induce p21 mRNA expression (Figure 6D), but not protein expression (Figure 6E), when protein synthesis was inhibited with 5 μg/ml cycloheximide.
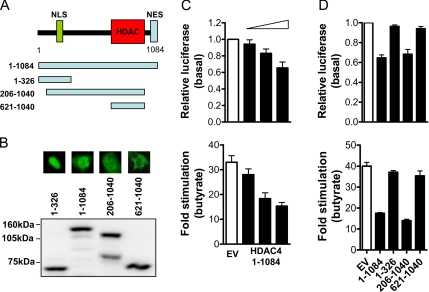
To complement these findings, the effect of overexpression of HDAC4 on p21 promoter activity was examined. The HDAC4 protein comprises 1084 amino acids, with a nuclear localization signal (NLS) located at the amino terminus (amino acids [aa] 244–279), a catalytic HDAC domain (aa 621-1040), and a nuclear export signal (NES) (aa 1044–1069) located at the carboxy terminus (Wang and Yang, 2001; Liu et al., 2004). A schematic diagram is shown in Figure 7A.
Figure 7.
HDAC4 regulation of p21 promoter activity in colon cancer cells. (A) Schematic representation of the HDAC4 protein, showing the NLS, NES, and HDAC catalytic domain. (B) Expression of the full-length and deletion HDAC4 GFP-tagged constructs (all 1 μg) in HCT116 cells was determined by anti-GFP Western blot. Also shown are immunofluorescence photomicrographs of the localization of the various HDAC4-GFP deletion mutants used. (C) Effect of HDAC4 overexpression on basal or 2 mM butyrate-induced p21 promoter activity after 24 h. HCT116 cells were cotransfected with pWP-133 (0.25 μg), TK-Renilla (0.1 μg), and increasing concentrations of HDAC4-GFP (1-1084) or the empty vector control (0–1 μg). Values shown are mean + SEM, and they are expressed as a percentage of pWP-133 activity relative to appropriate empty vector controls (empty bars) for basal p21 activity, and as -fold stimulation of p21 promoter activity for butyrate induction. (D) Effect of the HDAC4-GFP deletion constructs on basal or 2 mM butyrate-induced (24-h) p21 promoter activity. HCT116 cells were cotransfected with pWP-133 (0.25 μg), TK-Renilla (0.1 μg), and 1 μg of the HDAC4-GFP full-length and deletion constructs, or the empty vector control (1 μg). Values shown are mean + SEM, and they are expressed as a percentage of pWP-133 activity relative to corresponding empty vector controls (empty bars) for basal p21 activity, and as -fold stimulation of p21 promoter activity for butyrate induction.
As shown in Figure 7C, transfection of HCT116 cells with full-length HDAC4-GFP (1-1084) inhibited both basal and butyrate-stimulated p21 reporter activity in a concentration-dependent manner. An independent HDAC4 expression plasmid, tagged with myc/his, exerted similar repressive effects on basal and butyrate-stimulated p21 reporter activity (Supplemental Figure 3, A and B).
Likewise, a 206-1040 HDAC4 deletion construct (containing the NLS, and predominantly nuclear in localization) was able to fully repress p21 transcription (Figure 7D). In contrast, the 621-1040 HDAC4 deletion construct (lacking the NLS and predominantly cytoplasmic in localization), and the 1-326 HDAC4 deletion construct (predominantly nuclear in localization, but lacking the deacetylase domain), failed to inhibit p21 reporter activity (Figure 7D). Subcellular localization and equal expression of the respective HDAC4-GFP constructs in HCT116 cells were confirmed by immunofluorescence and Western blot, respectively (Figure 7, A and B). These results demonstrated that nuclear localization and an intact deacetylase domain were required for HDAC4-mediated repression of p21.
Repression of p21 Is a Component of HDAC4-mediated Promotion of Colon Cancer Cell Growth In Vitro
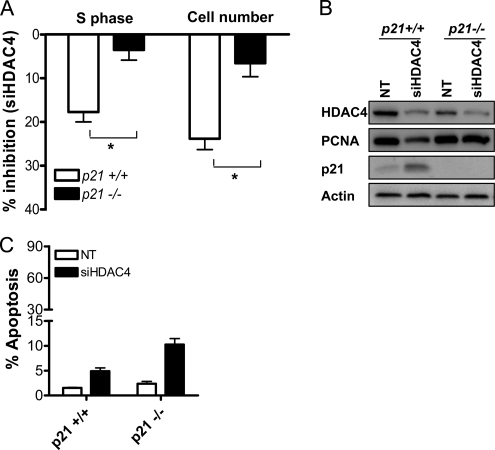
To investigate the functional significance of p21 induction in the growth arrest observed after HDAC4 down-regulation, we examined whether siHDAC4 could induce growth arrest in HCT116 cells in which p21 was deleted. As shown in Figure 8A, siHDAC4-mediated inhibition of the percentage of cells in S phase, and adherent cell number were significantly reduced in HCT116 p21 null cells compared with wild-type cells. Similar efficiency of HDAC4 down-regulation in HCT116 p21 wild-type and null cells was confirmed by Western blot (Figure 8B). Consistent with the effects on cell growth, HDAC4 down-regulation in p21 wild-type cells reduced expression of PCNA, whereas a minimal effect was observed in p21-null cells (Figure 8B). Collectively, these data indicate that repression of p21 is an important component of HDAC4-mediated growth promotion. In contrast, apoptosis induction after HDAC4 down-regulation was not impaired in p21-deficient cells (Figure 8C), demonstrating that p21 was necessary for the growth promoting, but not the prosurvival, effects of HDAC4.
Figure 8.
Role of p21 in HDAC4 siRNA-induced growth arrest and apoptosis induction. (A) Analysis of the effects of NT siRNA and siHDAC4 (both 100 nM) on adherent cell number and the percentage of cells in S phase determined by flow cytometry, in p21-null and wild-type HCT116 cells. Results are expressed as percentage of inhibition mediated by siHDAC4 72 h after transfection, and they are mean + SEM of three experiments. *p < 0.05 relative to effect in p21 wild-type cells, Student's t test. (B) Protein levels of HDAC4, PCNA, and p21 in p21 wild-type and null cells transfected with either NT siRNA or siHDAC4 (100 nM). (C) The role of p21 in apoptosis induction mediated by HDAC4 down-regulation for 72 h. Apoptosis was measured as subdiploid HCT116 cell content by flow cytometry. Values are mean + SEM of three experiments.
Down-Regulation of Sp1 Abrogates HDAC4-mediated Repression of p21
Because we and others have previously demonstrated that HDAC inhibitors induce p21 in a Sp1/Sp3-dependent manner (Sowa et al., 1997; Wilson et al., 2006), the role of the Sp1 transcription factor in HDAC4-mediated repression of p21 was examined. We first determined whether HDAC4 down-regulation could induce p21 in HCT116 cells when Sp1 was functionally inhibited. Mithramycin is an established inhibitor of the binding of transcription factors, such as Sp1, to GC-rich elements (Liu et al., 2006). As shown in Figure 9A, siHDAC4 was unable to stimulate p21 expression in the presence of mithramycin. Similarly, the stimulatory effect of the HDAC inhibitor butyrate on p21 expression was ablated by mithramycin.
To more specifically determine the role of Sp1 in HDAC4-mediated repression of p21, we down-regulated its expression in HCT116 cells with Sp1-targeting siRNA. Approximately 70% reduction of Sp1 expression was achieved. As shown in Figure 9B, Sp1 down-regulation inhibited basal p21 expression, and it inhibited the p21 induction mediated by siHDAC4. Similarly, Sp1 down-regulation inhibited p21 induction by the HDAC inhibitor butyrate (Figure 9C). These effects were also observed for p21 promoter reporter experiments after down-regulation of Sp1 (Figure 9D). Collectively, these findings suggest a role for Sp1 in p21 repression mediated by HDAC4 and in HDAC inhibitor-mediated derepression of p21.
HDAC4 Expression Is Linked to Reduced Histone Acetylation at the Proximal p21 Promoter
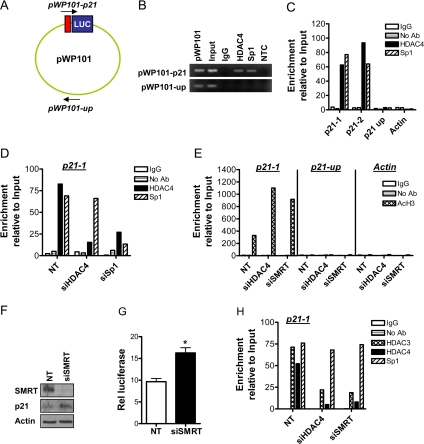
We then performed ChIP experiments to directly demonstrate HDAC4 localization to the p21 promoter. First, we transfected HCT116 cells with the pWP-101 p21 luciferase reporter plasmid (Sowa et al., 1997), which contains the 101 bases downstream of the p21 transcriptional start site. This region of the p21 promoter contains four Sp1 binding sites known to be important for HDAC inhibitor induction of p21 (Sowa et al., 1997). We designed primers to the vector backbone and luciferase sequences flanking the 101-base pair p21 promoter sequence to interrogate whether HDAC4 associates with this locus (designated pWP-101-p21; Figure 10A). As a control, we also designed primers interrogating a region of the vector backbone ∼2000-base pairs downstream (designated pWP-101-up; Figure 10A). As shown in Figure 10B, enrichment of HDAC4 binding to the proximal p21 promoter, but not the vector backbone, was observed after PCR amplification of ChIP DNA.
Figure 10.
HDAC4 binds to the p21 promoter. (A) ChIP analysis of HDAC4 binding to the pWP101 p21 luciferase reporter plasmid template. HCT116 cells were transiently transfected with pWP101 and harvested after 48 h. Primer sets were designed to interrogate the 101 base pairs immediately proximal to the transcription start site of the p21 promoter, which contains four of the six Sp1 binding sites (pWP101-p21) and a separate region of the vector backbone ∼2 kb away (pWP101-up). (B) ChIP DNA was analyzed by PCR amplification and 1% agarose gel electrophoresis. NTC, no template control. (C) ChIP analysis of HDAC4 binding to the endogenous p21 promoter. Primer sets probing the proximal region of the p21 promoter, adjacent to the six Sp1 binding sites, were used (p21-1 and p21-2), as were primers probing a region of the p21 promoter 4Kb upstream from the transcriptional start site (p21-up), or the actin promoter. DNA content after immunoprecipitation with HDAC4 or Sp1 antibodies, or no antibody or nonspecific antibody (IgG) controls, were determined by real-time RT-PCR. All values were expressed relative to Input DNA content. (D) ChIP analysis of HDAC4 and Sp1 binding to the p21-1 promoter locus following transfection of cells with NT siRNA, siHDAC4 or siSp1 (all 100 nM) for 72 h. All values were expressed relative to Input DNA content. (E) ChIP analysis of histone H3 acetylation at the loci described above after transfection of cells with NT siRNA, siHDAC4, or siSMRT (all 100 nM) for 72 h. All values were expressed relative to Input DNA content. (F) Selective down-regulation of SMRT protein and parallel induction of p21 protein expression in HCT116 cells. HCT116 cells were transfected with 100 nM NT siRNA of siSMRT for 72 h. (G) Effect of SMRT down-regulation on p21 promoter activity, as determined by luciferase assay. HCT116 cells were transfected with pWP-133 (0.25 μg) and 100 nM of either NT siRNA or siSMRT and cultured for 72 h. TK-Renilla (0.1 μg) was cotransfected in all treatment groups to control for transfection efficiency. Values shown are mean + SEM of three independent experiments; *p < 0.05, Student's t test. (H) ChIP analysis of the binding of HDAC3 and HDAC4 to the p21-1 promoter locus after transfection of cells with 100 nM NT siRNA, siHDAC4, or siSMRT for 72 h. All values were expressed relative to Input DNA content.
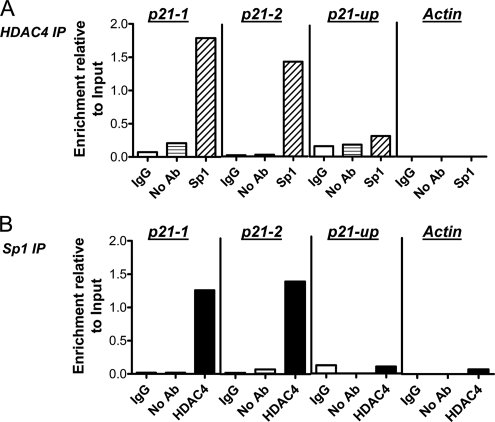
We then used an alternative strategy to interrogate the endogenous proximal p21 promoter. We designed two independent primer sets (p21-1 and p21-2), which were efficient in amplifying ChIP DNA in quantitative real-time PCR (Q-ChIP). As shown in Figure 10C, ∼30-fold and 40-fold enrichment of HDAC4 binding to the p21-1 and p21-2 promoter loci, respectively, was observed in comparison with IgG and no antibody controls. We demonstrated the specificity of HDAC4 binding to the proximal p21 promoter in two ways. First, there was no such enrichment of HDAC4 binding to a 4-kb upstream region of the p21 promoter, or to the actin promoter (Figure 10C). Second, as shown in Figure 10D, there was markedly reduced enrichment of HDAC4 binding to the proximal p21 promoter (p21-1 locus) in cells transfected with siHDAC4 compared with NT-transfected cells (Figure 10D). Similar results were obtained utilizing the p21-2 primer set. Consistent with the transcriptional activation of p21 linked to down-regulation of HDAC4, we observed a marked increase in acetylation of histone H3 at the p21 proximal promoter, but not at the 4-kb upstream region of the p21 promoter, or at the actin promoter, after treatment with siHDAC4 (Figure 10E).
Because HDAC4 is known to associate with the HDAC3–N-CoR/SMRT complex (Fischle et al., 2002), we used siRNA-targeting SMRT to test the role of SMRT in HDAC4 recruitment to the proximal p21 promoter. As shown in Figure 10D, a 72-h treatment with siSMRT significantly down-regulated SMRT expression. Importantly, down-regulation of SMRT induced p21 protein expression and promoter activity to a similar extent as did HDAC4 down-regulation (Figure 10, F and G). As shown in Figure 10H, and consistent with their localization in the N-CoR/SMRT complex, the binding of HDAC4 and HDAC3 to the proximal p21 promoter was reduced after SMRT down-regulation. Down-regulation of HDAC4 also markedly reduced HDAC3 association with this locus, further linking these HDACs in p21 regulation (Figure 10H).
HDAC4 Associates with Sp1 at the Proximal p21 Promoter
We then sought to directly link the binding of HDAC4 with Sp1 at the proximal p21 promoter. We confirmed that Sp1 binding paralleled that of HDAC4 at both the transient pWP101 template (Figure 10B) and at the proximal p21-1 and p21-2 promoter loci, but not at the upstream p21 locus (Figure 10C). The specificity of Sp1 occupancy of the proximal p21 promoter was further demonstrated by the greatly reduced occupancy of Sp1 in cells transfected with siSp1 compared with NT-transfected cells (Figure 10D). The effect of down-regulation of Sp1 on HDAC4 occupancy of the proximal p21 promoter was then examined. As shown in Figure 10D, there was markedly reduced HDAC4 binding to the p21-1 and p21-2 promoter regions after transfection of cells with siSp1.
To demonstrate the simultaneous presence of HDAC4 and Sp1 at the proximal p21 promoter, we performed sequential ChIP experiments. As shown in Figure 11A, when anti-Sp1 immunoprecipitation was performed on eluted chromatin obtained from an initial anti-HDAC4 immunoprecipitation, there was specific enrichment of DNA corresponding to the p21-1 promoter locus compared with IgG and no antibody controls. Similar results were observed at the p21-2 promoter locus (data not shown). Importantly, there was no specific enrichment associated with successive HDAC4 and Sp1 immunoprecipitations at the 4-kb upstream p21 promoter locus or the actin promoter, consistent with the results of the single ChIPs presented above. We also performed the reverse sequence of immunoprecipitations (that is, successive Sp1 and HDAC4 immunoprecipitations). As shown in Figure 11B, the co-occupancy of Sp1 and HDAC4 was again only observed at the proximal p21 promoter locus.
Figure 11.
HDAC4 and Sp1 co-occupancy of the proximal p21 promoter. Sequential ChIP analysis in HCT116 cells (see Materials and Methods) after initial immunoprecipitation with anti-HDAC4, followed by a second immunoprecipitation with IgG, no antibody, or anti-Sp1 (A), or initial immunoprecipitation with anti-Sp1 followed by a second immunoprecipitation with IgG, no antibody, or anti-HDAC4 (B). Whether HDAC4 and Sp1 were simultaneously present at the proximal p21 promoter (p21-1), at a region 4-kb upstream from the transcriptional start site of the p21 promoter (p21-up) or at the actin promoter was tested by QPCR. All values were initially expressed relative to relevant Input DNA content.
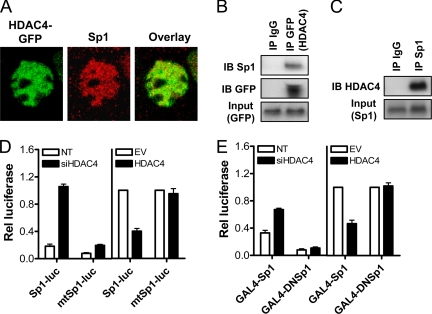
We then used immunofluorescence analysis and confocal microscopy to confirm that HDAC4 and Sp1 colocalize in cell nuclei. As shown in Figure 12A, significant overlap of transiently transfected HDAC4-GFP with endogenous Sp1 was evident in HCT116 cells. To demonstrate this interaction biochemically, coimmunoprecipitation experiments in HCT116 cells were performed. As shown in Figure 12B, an immunoprecipitation with anti-GFP successfully pulled down Sp1. To demonstrate this interaction at the endogenous level, we then showed that endogenous HDAC4 was immunoprecipitated by a Sp1 antibody, but not by a nonspecific antibody (IgG) (Figure 12C).
Figure 12.
HDAC4 associates with and represses Sp1 transactivity. (A) Overlap of HDAC4-GFP and endogenous Sp1 in HCT116 cells as determined by immunofluorescence analysis utilizing confocal microscopy. (B) HCT116 cells were transfected with HDAC4-GFP (1-1084) for 4 8h, and 300 μg of nuclear extract was immunoprecipitated with anti-GFP or IgG control antibodies. Immunoprecipitates were probed for Sp1 and GFP content by Western blot. Equal amounts of input protein were demonstrated by GFP Western blot. (C) HCT116 cells in the growing phase were harvested, and 300 μg of nuclear extract immunoprecipitated with anti-Sp1 or IgG control antibodies. Immunoprecipitates were probed for HDAC4 content by Western blot. Equal amounts of input protein were demonstrated by Sp1 Western blot. (D) Effect of HDAC4 down-regulation or HDAC4 overexpression on activity of Sp1/Sp3 reporter constructs containing either three consensus Sp1/Sp3 binding sites (Sp1-luc) or three mutant binding sites (mtSp1-luc). HCT116 cells were cotransfected with Sp1-luc or mtSp1-luc (0.1 μg) in combination with TK-Renilla (0.1 μg), NT siRNA or siHDAC4 (100 nM) for 72 h. In separate experiments, HCT116 cells were cotransfected with Sp1-luc or mtSp1-luc (0.1 μg) in combination with HDAC4-GFP 1-1084 or empty vector control (1 μg), and TK-Renilla (0.1 μg) for 24 h. Values shown are the mean + SEM of a representative experiment. (E) A one-hybrid system was also used in which active Sp1 (GAL4-Sp1) or inactive (GAL4-DNSp1) was fused to GAL4, and transactivation was measured using a luciferase reporter linked to five consensus GAL4 DNA binding sites. HCT116 cells were cotransfected with GAL4-Sp1 or GAL4-DNSp1 (0.1 μg) in combination with TK-Renilla (0.1 μg), NT or siHDAC4 (100 μM) for 72 h. In separate experiments, HCT116 cells were cotransfected with GAL4-Sp1 or GAL4-DNSp1 (0.1 μg) in combination with HDAC4-GFP 1-1084 or empty vector control (1 μg), and TK-Renilla (0.1 μg) for 24 h. Values shown are mean + SEM of a representative experiment.
HDAC4 Represses Sp1 Transcriptional Activity
Finally, we sought to directly establish a link between HDAC4 and Sp1 transcriptional activity. To do this, we used a Sp1/Sp3 luciferase reporter construct, containing five consensus Sp1 binding sites, or a control reporter containing mutated Sp1/Sp3 binding sites (Wilson et al., 2006). As shown in Figure 12D, siHDAC4 induced an approximately fivefold increase in Sp1/Sp3 reporter activity compared with the NT control, but it had minimal effects on the control mt-Sp1-Luc reporter. Furthermore, and consistent with the ability of HDAC4 to repress Sp1/Sp3-driven transcription, overexpression of HDAC4 markedly reduced basal Sp1/Sp3 reporter activity (Figure 12D).
Because the Sp1/Sp3 reporter cannot discriminate between the binding of Sp1 or Sp3, we then used a one-hybrid system in which Sp1 was fused to GAL4 (GAL4-Sp1), and transactivation was measured using a luciferase reporter driven by a minimal promoter linked to five consensus GAL4 DNA binding sites (Sowa et al., 1999). As shown in Figure 12E, down-regulation of HDAC4 induced a modest transactivation of Sp1 in HCT116 cells. Consistent with this result, overexpression of HDAC4 inhibited basal Sp1 transactivity (Figure 12E). The specificity of these results was demonstrated by minimal transactivation effects after transfection with a fusion construct containing the dominant-negative form of Sp1 (GAL4-DNSp1), which lacks the transactivation domain.
DISCUSSION
We have identified the class II HDAC HDAC4 as a proproliferative and prosurvival factor for colon cancer cells in vitro. Furthermore, HDAC4 is maximally expressed in the proliferative compartment in normal human and mouse small intestinal and colonic epithelium, establishing a link between HDAC4 expression and cell proliferation in vivo. The proproliferative effect of HDAC4 in colon cancer cells and the confinement of its expression to the proliferative crypt compartment is consistent with that reported previously for the class I HDACs, HDAC1, -2, and -3 (Wilson et al., 2006). These findings are also consistent with the growth inhibitory effects of HDAC inhibitors on colon cancer cells (Archer et al., 1998; Wilson et al., 2006). Although it is known that class I HDACs are consistently up-regulated in human colon cancer and in other solid tumors at the mRNA and/or protein levels (Zhu et al., 2004; Huang et al., 2005; Wilson et al., 2006; Khabele et al., 2007), relatively less is known about HDAC4 expression in colon cancer. Although down-regulation of HDAC4 mRNA in human colon tumor tissue compared with normal has been shown (Lleonart et al., 2006), we detected no overall difference in HDAC4 protein expression between in a panel of matched human colon tumor-normal pairs (unpublished observations) in which clear overexpression of HDAC1, HDAC2, and HDAC3 was detected previously (Wilson et al., 2006).
Our demonstration of a link between cell proliferation and nuclear localization of HDAC4, in contrast to its predominantly cytoplasmic localization in cells undergoing cell cycle arrest, is consistent with the importance of nucleocytoplasmic shuttling of class II HDACs as a mechanism of their functional regulation (Grozinger and Schreiber, 2000; McKinsey et al., 2000; Miska et al., 2001). Our findings are also consistent with a study demonstrating phosphorylation and nuclear localization of HDAC4 after activation of the Ras/extracellular signal regulated kinase pathway through the proproliferative stimulus oncogenic Ras (Zhou et al., 2000) and with reduced growth in non–small-cell lung cancer cells after the cytoplasmic containment of HDAC4 by treatment with an HDAC inhibitor in conjunction with irradiation (Geng et al., 2006).
Several lines of evidence collectively demonstrated the importance of Sp1-mediated repression of p21 in the growth-promoting effects of HDAC4 in colon cancer cells. Down-regulation of HDAC4 increased histone H3 acetylation at the Sp1 binding site-rich proximal promoter, consistent with our observations of increased p21 transcription and protein expression. In turn, that this induction of p21 was inhibited by pharmacological and molecular inhibition of Sp1 was consistent with biochemical and imaging data demonstrating an association between HDAC4 and Sp1 at the subcellular level, and ChIP and sequential ChIP experiments localizing this interaction to the proximal p21 promoter. Although it is presently unknown whether HDAC4-mediated repression of p21 plays a similar role in promoting cell growth in vivo, such a role would be consistent with our observation of p21 induction along the crypt–villus axis in parallel to HDAC4 down-regulation (Mariadason et al., 2005; this study). Furthermore, the down-regulation of HDAC4 during spontaneous Caco-2 cell differentiation, or after DNTCF4-induced differentiation of LS174T cells, was paralleled by a concomitant increase in p21 expression, consistent with previous studies (Mariadason et al., 2002; van de Wetering et al., 2002).
Further investigation elaborated a potential mechanism of Sp1-dependent targeting of HDAC4 to the proximal p21 promoter in colon cancer cells. Collectively, induction of p21 expression and promoter activity after SMRT down-regulation, loss of association of HDAC3 and HDAC4 with the proximal p21 promoter after down-regulation of SMRT, and loss of HDAC3 association with this locus after HDAC4 down-regulation, were consistent with a model of p21 repression mediated by HDAC4 through association with the catalytically active HDAC3, within the N-CoR/SMRT corepressor complex. We have previously demonstrated that HDAC3 represses p21 in a Sp1-dependent manner (Wilson et al., 2006). Furthermore, it has been shown that HDAC4 acts as a “scaffold” protein with the HDAC3–NCo-R/SMRT complex without contributing to the overall deacetylase activity of the complex (Fischle et al., 2002), consistent with the weak catalytic activity of HDAC4 and other class IIa HDACs on standard acetyl-lysine substrates as reported recently (Lahm et al., 2007).
Here, we add the HDAC4–HDAC3–N-CoR/SMRT corepressor complex to an emerging model of coordinate p21 regulation in colon cancer cells mediated by Sp1- and/or Sp3-dependent recruitment of HDACs. This likely involves recruitment of multiple corepressor complexes to the proximal p21 promoter, because HDAC1 and HDAC2, which also repress p21 (Wilson et al., 2006), exist in complexes distinct to N-CoR/SMRT, namely, Sin3A and NuRD (Jepsen and Rosenfeld, 2002). The potential relevance of HDAC regulation of p21 expression also needs to be viewed in the context that there are multiple key factors and pathways perturbed in colon cancer that can also potentially modulate expression of p21, such as TGF-β/SMAD, β-catenin-TCF/c-myc, and p53. In particular, because p53 and HDAC4 have been directly linked in several reports (Berns et al., 2004; Imbriano et al., 2005; Basile et al., 2006), the role of p53 in HDAC4-mediated repression is worthy of further investigation.
Our demonstration that siRNA-mediated down-regulation of HDAC4 expression induced growth arrest and apoptosis in HCT116 cells is consistent with the anti-proliferative and proapoptotic effects of HDAC inhibitors in human cancer cell lines (Heerdt et al., 1994; Mariadason et al., 1997; Litvak et al., 1998; Mariadason et al., 2000, 2001b; Gurvich et al., 2004), and with the previously reported observation that HDAC4 down-regulation reduces clonogenic survival and induces apoptosis in HeLa cells (Kao et al., 2003). However, previous observations clearly indicate that the role(s) of HDAC4 in regulating cell growth and survival is complex, and cell type and stimulus dependent. In a high-throughput short hairpin RNA screening study in fibroblasts, HDAC4 was identified as a mediator of p53-dependent growth arrest (Berns et al., 2004). Furthermore, overexpression of HDAC4 induces apoptosis in various human cell lines (Liu et al., 2004; Paroni et al., 2004; Bolger and Yao, 2005), an effect ascribed to caspase-mediated cleavage of HDAC4, and the subsequent nuclear accumulation of its amino-terminal fragment (Liu et al., 2004; Paroni et al., 2004). A role for HDAC4 in mediating DNA repair has also been reported, with its targeting to specific nuclear foci observed in response to radiation-induced DNA damage in several cell types (Kao et al., 2003; Basile et al., 2006; Geng et al., 2006). These DNA repair and apoptosis-promoting functions of HDAC4 may be mutually exclusive, as suggested by a recent study demonstrating that non–small-cell lung cancer cells are radiosensitized to apoptosis after disruption of DNA repair by HDAC inhibitor-induced cytoplasmic retention of HDAC4 (Geng et al., 2006). We speculate that HDAC4 acts as a proproliferative and prosurvival factor under basal conditions, mediated, respectively, by repression of p21, and by potentially promoting DNA damage repair and/or repressing the expression of proapoptotic genes, as shown for HDAC3 (Escaffit et al., 2007). In contrast, under stressful conditions and after caspase cleavage, HDAC4 acts to facilitate apoptosis.
In addition to its role in cell proliferation and survival, a role for HDAC4 in inhibiting differentiation of bone and muscle cells through interaction with the transcription factors Runx2 and MEF2, respectively, has also been described previously (Miska et al., 2001; Vega et al., 2004b). Whether HDAC4 plays a similar role in inhibiting intestinal cell differentiation is presently unknown. However, the confinement of HDAC4 expression to the proliferative, undifferentiated cell compartment in the normal intestinal epithelium would be consistent with such a role, and is worthy of further investigation.
A role for a class II HDAC, HDAC4, in promoting growth of colon cancer cells has important implications for cancer therapy, because HDAC inhibitors currently undergoing clinical trial in stage IV colon cancer patients have established class specificity. For example, depsipeptide (FK228), in contrast to hydroxamic acid-based HDAC inhibitors such as SAHA (vorinostat), only weakly inhibits class II HDACs. (Bolden et al., 2006; Rasheed et al., 2007). Therefore, our demonstration of an important role for HDAC4 in colon cell growth would suggest that inhibitors of class I and II HDACs are likely to be more efficient at inhibiting colon cancer cell growth than class I-specific inhibitors.
In conclusion, these findings identify HDAC4 as an important regulator of proliferation of colon cancer cells in vitro, through Sp1-dependent repression of p21. The localization of HDAC4 to the proliferative zone in the intestinal epithelium is consistent with its physiological role in maintaining cell proliferation and inhibiting maturation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Health grants CA-100823 and CA-123316 (to J.M.M.) and CA-88104 (to L.H.A.) and Cancer Center grant P30-13330.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0139) on July 16, 2008.
REFERENCES
- Archer S. Y., Meng S., Shei A., Hodin R. p21WAF1 is required for butyrate-mediated growth inhibition of colon cancer cells. Proc. Natl. Acad. Sci. USA. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile V., Mantovani R., Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J. Biol. Chem. 2006;281:2347–2357. doi: 10.1074/jbc.M507712200. [DOI] [PubMed] [Google Scholar]
- Benimetskaya L., et al. Bcl-2 protein in 518A2 melanoma cells in vivo and in vitro. Clin. Cancer Res. 2006;12:4940–4948. doi: 10.1158/1078-0432.CCR-06-1002. [DOI] [PubMed] [Google Scholar]
- Berns K., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Bolden J. E., Peart M. J., Johnstone R. W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Bolger T. A., Yao T. P. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Hwang P. M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K. W., Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codina A., Love J. D., Li Y., Lazar M. A., Neuhaus D., Schwabe J. W. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA. 2005;102:6009–6014. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffit F., Vaute O., Chevillard-Briet M., Segui B., Takami Y., Nakayama T., Trouche D. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol. Cell. Biol. 2007;27:554–567. doi: 10.1128/MCB.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris R. P., Villenas S. A., Diamond J. Regulation of brush-border enzyme activities and enterocyte migration rates in mouse small intestine. Am. J. Physiol. 1992;262:G1047–G1059. doi: 10.1152/ajpgi.1992.262.6.G1047. [DOI] [PubMed] [Google Scholar]
- Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Geng L., Cuneo K. C., Fu A., Tu T., Atadja P. W., Hallahan D. E. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of {gamma}-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res. 2006;66:11298–11304. doi: 10.1158/0008-5472.CAN-06-0049. [DOI] [PubMed] [Google Scholar]
- Glaser K. B., Li J., Staver M. J., Wei R. Q., Albert D. H., Davidsen S. K. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem. Biophys. Res. Commun. 2003;310:529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Glozak M. A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Schreiber S. L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Gurvich N., Tsygankova O. M., Meinkoth J. L., Klein P. S. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- Heerdt B. G., Houston M. A., Augenlicht L. H. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res. 1994;54:3288–3293. [PubMed] [Google Scholar]
- Huang B. H., Laban M., Leung C. H., Lee L., Lee C. K., Salto-Tellez M., Raju G. C., Hooi S. C. Inhibition of histone deacetylase 2 increases apoptosis and p21(Cip1/WAF1) expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol. Cell. Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K., Rosenfeld M. G. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Kao G. D., McKenna W. G., Guenther M. G., Muschel R. J., Lazar M. A., Yen T. J. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabele D., Son D. S., Parl A. K., Goldberg G. L., Augenlicht L. H., Mariadason J. M., Rice V. M. Drug-induced inactivation or gene silencing of class i histone deacetylases suppresses ovarian cancer cell growth: implications for therapy. Cancer Biol. Ther. 2007:6. doi: 10.4161/cbt.6.5.4007. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lee S., Lee T., Lee Y. W., Trepel J. B. Transcriptional activation of p21(WAF1/CIP1) by apicidin, a novel histone deacetylase inhibitor. Biochem. Biophys. Res. Commun. 2001;281:866–871. doi: 10.1006/bbrc.2001.4434. [DOI] [PubMed] [Google Scholar]
- Lagger G., et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A., et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak D. A., Evers B. M., Hwang K. O., Hellmich M. R., Ko T. C., Townsend C. M., Jr. Butyrate-induced differentiation of Caco-2 cells is associated with apoptosis and early induction of p21Waf1/Cip1 and p27Kip1. Surgery. 1998;124:161–169. discussion 169–170. [PubMed] [Google Scholar]
- Liu F., Dowling M., Yang X. J., Kao G. D. Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 2004;279:34537–34546. doi: 10.1074/jbc.M402475200. [DOI] [PubMed] [Google Scholar]
- Liu F., Pore N., Kim M., Voong K. R., Dowling M., Maity A., Kao G. D. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol. Biol. Cell. 2006;17:585–597. doi: 10.1091/mbc.E05-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleonart M. E., et al. New p53 regulated genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol. Rep. 2006;16:603–608. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- Mariadason J. M., Arango D., Corner G. A., Aranes M., Hotchkiss K., A., Yang W., Augenlicht L. H. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002;62:4791–4804. [PubMed] [Google Scholar]
- Mariadason J. M., Barkla D. H., Gibson P. R. Effect of short-chain fatty acids on paracellular permeability in Caco-2 intestinal epithelium model. Am. J. Physiol. 1997;272:G705–G712. doi: 10.1152/ajpgi.1997.272.4.G705. [DOI] [PubMed] [Google Scholar]
- Mariadason J. M., Bordonaro M., Aslam F., Shi L., Kuraguchi M., Velcich A., Augenlicht L. H. Down-regulation of beta-catenin TCF signaling is linked to colonic epithelial cell differentiation. Cancer Res. 2001a;61:3465–3471. [PubMed] [Google Scholar]
- Mariadason J. M., et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Mariadason J. M., Rickard K. L., Barkla D. H., Augenlicht L. H., Gibson P. R. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J. Cell. Physiol. 2000;183:347–354. doi: 10.1002/(SICI)1097-4652(200006)183:3<347::AID-JCP7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Mariadason J. M., Velcich A., Wilson A. J., Augenlicht L. H., Gibson P. R. Resistance to butyrate-induced cell differentiation and apoptosis during spontaneous Caco-2 cell differentiation. Gastroenterology. 2001b;120:889–899. doi: 10.1053/gast.2001.22472. [DOI] [PubMed] [Google Scholar]
- McKinsey T. A., Zhang C. L., Lu J., Olson E. N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S., Ho A. D., Mahlknecht U. Role of histone deacetylase inhibitors in the treatment of cancer (review) Int. J. Oncol. 2004;25:1509–1519. [PubMed] [Google Scholar]
- Miska E. A., Karlsson C., Langley E., Nielsen S. J., Pines J., Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E. A., Langley E., Wolf D., Karlsson C., Pines J., Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001;29:3439–3447. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R. L., Davis C. A., Potthoff M. J., Haberland M., Fielitz J., Qi X., Hill J. A., Richardson J. A., Olson E. N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroni G., Mizzau M., Henderson C., Del Sal G., Schneider C., Brancolini C. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell. 2004;15:2804–2818. doi: 10.1091/mbc.E03-08-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Laniel M. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Rasheed W. K., Johnstone R. W., Prince H. M. Histone deacetylase inhibitors in cancer therapy. Expert Opin. Investig. Drugs. 2007;16:659–678. doi: 10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]
- Sowa Y., Orita T., Minamikawa-Hiranabe S., Mizuno T., Nomura H., Sakai T. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 1999;59:4266–4270. [PubMed] [Google Scholar]
- Sowa Y., Orita T., Minamikawa S., Nakano K., Mizuno T., Nomura H., Sakai T. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem. Biophys. Res. Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- Tou L., Liu Q., Shivdasani R. A. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol. Cell. Biol. 2004;24:3132–3139. doi: 10.1128/MCB.24.8.3132-3139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 2004a;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R. B., et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004b;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- Waltregny D., North B., Van Mellaert F., de Leval J., Verdin E., Castronovo V. Screening of histone deacetylases (HDAC) expression in human prostate cancer reveals distinct class I HDAC profiles between epithelial and stromal cells. Eur. J. Histochem. 2004;48:273–290. [PubMed] [Google Scholar]
- Wang A. H., Bertos N. R., Vezmar M., Pelletier N., Crosato M., Heng H. H., Th'ng J., Han J., Yang X. J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Yang X. J. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser M. M. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J. Biol. Chem. 1973a;248:2536–2541. [PubMed] [Google Scholar]
- Weiser M. M. Intestinal epithelial cell surface membrane glycoprotein synthesis. II. Glycosyltransferases and endogenous acceptors of the undifferentiated cell surface membrane. J. Biol. Chem. 1973b;248:2542–2548. [PubMed] [Google Scholar]
- Wilson A. J., Arango D., Mariadason J. M., Heerdt B. G., Augenlicht L. H. TR3/Nur77 in colon cancer cell apoptosis. Cancer Res. 2003;63:5401–5407. [PubMed] [Google Scholar]
- Wilson A. J., et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- Wilson A. J., Velcich A., Arango D., Kurland A. R., Shenoy S. M., Pezo R. C., Levsky J. M., Singer R. H., Augenlicht L. H. Novel detection and differential utilization of a c-myc transcriptional block in colon cancer chemoprevention. Cancer Res. 2002;62:6006–6010. [PubMed] [Google Scholar]
- Yang X. J., Gregoire S. Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Richon V. M., Wang A. H., Yang X. J., Rifkind R. A., Marks P. A. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc. Natl. Acad. Sci. USA. 2000;97:14329–14333. doi: 10.1073/pnas.250494697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K. P., Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.