Abstract
Nuclear export of mRNA requires several key mRNA-binding proteins that recognize and remodel the mRNA and target it for export via interactions with the nuclear pore complex. In Saccharomyces cerevisiae, the shuttling heterogeneous nuclear ribonucleoprotein, Nab2, which is essential for mRNA export, specifically recognizes poly(A) RNA and binds to the nuclear pore-associated protein, myosin-like protein 1 (Mlp1), which functions in mRNA export and quality control. Specifically, the N-terminal domain of Nab2 (Nab2-N; residues 1–97) interacts directly with the C-terminal globular domain of Mlp1 (CT-Mlp1: residues 1490–1875). Recent structural and binding studies focused on Nab2-N have shown that Nab2-N contains a hydrophobic patch centered on Phe73 that is critical for interaction with Mlp1. Engineered amino acid changes within this patch disrupt the Nab2/Mlp1 interaction in vitro. Given the importance of Nab2 and Mlp1 to mRNA export, we have examined the Nab2/Mlp1 interaction in greater detail and analyzed the functional consequences of disrupting the interaction in vivo. We find that the Nab2-binding domain of Mlp1 (Mlp1-NBD) maps to a 183-residue region (residues 1586–1768) within CT-Mlp1, binds directly to Nab2 with micromolar affinity, and confers nuclear accumulation of poly(A) RNA. Furthermore, we show that cells expressing a Nab2 F73D mutant that cannot interact with Mlp1 exhibit nuclear accumulation of poly(A) RNA and that this nab2 F73D mutant genetically interacts with alleles of two essential mRNA export genes, MEX67 and YRA1. These data provide in vivo evidence for a model of mRNA export in which Nab2 is important for targeting mRNAs to the nuclear pore for export.
Production of mature mRNA for translation is a complex multistep process involving an array of RNA-binding proteins (1–3). From its transcriptional inception in the nucleus, pre-mRNA that emerges from RNA polymerase II must be bound by processing factors for 5′-end capping, splicing, and 3′-end cleavage and polyadenylation to reach maturation (4). Alongside these processing events, maturing mRNA must be co-transcriptionally loaded with heterogeneous nuclear ribonucleoproteins (hnRNPs),2 mRNA export adaptors, and an mRNA export receptor to facilitate its active export from the nucleus to the cytoplasm through nuclear pore complexes (NPCs), large proteinaceous channels that perforate the nuclear membrane and mediate nucleocytoplasmic transport (1–3, 5). There are a number of nuclear quality controls that safeguard against improperly processed mRNA being exported and translated into deleterious proteins (6, 7). In particular, this quality control machinery likely monitors the state of processed mRNA by checking for the presence or absence of mRNA processing and export factors.
One of the key players in mRNA export is the conserved heterodimeric mRNA export receptor, Mex67/Mtr2 (NXF1/NXT1 or TAP/p15 in metazoans), which plays an essential role in bulk mRNA export in Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster and stimulates export of transcripts in vertebrate cells (8–12). In S. cerevisiae, a temperature-sensitive mutant of Mex67, mex67-5, shows significant nuclear accumulation of poly(A) RNA at the non-permissive temperature (8). The Mex67/Mtr2 heterodimer interacts directly with phenylalanine-glycine (FG) repeat-containing nuclear pore proteins (nucleoporins or Nups) in the NPC, which facilitates translocation of the receptor with its mRNA cargo through the transport channel of the pore (12–17).
In addition to the Mex67/Mtr2 receptor, efficient mRNA export also requires mRNA adaptors. The most well characterized mRNA adaptor is the evolutionarily conserved protein, Yra1 (Aly/REF in metazoans), which binds to RNA and recruits Mex67/Mtr2 to transcripts (18–20). In S. cerevisiae, Yra1 is essential for mRNA export and thermosensitive alleles, including yra1–8, show nuclear accumulation of poly(A) RNA at the non-permissive temperature (18, 21). In vertebrates, Aly/REF also mediates mRNA export (22, 23). Interestingly, however, Aly/REF is not essential for bulk mRNA export in Drosophila or C. elegans and Yra1 only binds to about 1000 mRNA transcripts, representing one-fifth of the genome, in S. cerevisiae (24–26). These results suggest that different mRNA adaptors can operate in metazoans and yeast. An example of an alternative mRNA adaptor is the S. cerevisiae hnRNP, Npl3, which is essential for mRNA export in yeast, associates with mRNA co-transcriptionally, and can bind to the Mex67 export protein (27–31).
Another potential mRNA export adaptor is the S. cerevisiae hnRNP, Nab2, which is a zinc finger protein essential for mRNA export in yeast (32–34). Nuclear abundant poly(A) RNA-binding protein 2 (Nab2) shuttles between the nucleus and cytoplasm (31, 32, 35) and contains three key functional domains: an N-terminal domain (residues 1–97) involved in mRNA export (36), an RGG domain (residues 201–265) that acts as the nuclear import signal for Nab2 (37), and a C-terminal zinc finger domain (residues 262–477) that specifically binds polyadenosine RNA (31, 34, 36). A putative human orthologue of Nab2, ZC3H14, has a similar domain structure (34). The N-terminal domain of Nab2 (Nab2-N) is critical for Nab2 function in vivo and a Nab2 mutant that lacks the N-terminal domain (residues 4–97), nab2-1/nab2 ΔN, confers a severe growth defect and nuclear accumulation of poly(A) RNA when expressed as the sole cellular copy of Nab2 (36).
For efficient transport of mRNA, export adaptors and hnRNPs may contribute to mRNA export through facilitating interactions with components of the nuclear pore. In S. cerevisiae, the nuclear pore-associated protein, myosin-like protein 1 (Mlp1), acts as a docking platform for hnRNPs and other mRNA factors and also plays a role in mRNA export and quality control (38–40). Mlp1 and its close relative Mlp2 are large proteins attached to the nucleoplasmic face of the nuclear pore that are homologous to the mammalian Tpr (translocated promoter region) protein and are thought to act like control gates at the NPC portal to grant or deny mRNA access to the cytoplasm (38, 40–43). The Mlp1/2/Tpr proteins contain a coiled-coil N-terminal domain that tethers them to the pore via specific nucleoporins and a globular C-terminal domain that binds to mRNA export factors (38, 43–45). Notably, the C-terminal domain of Mlp1 (CT-Mlp1) binds to the hnRNPs, Nab2 and Npl3, and expression of CT-Mlp1 or the C-terminal domain of human Tpr causes nuclear accumulation of poly(A) RNA (38, 46), linking the Mlp1/Tpr proteins with mRNA export. The Mlp1/2 proteins also bind to the mRNA export adaptor, Yra1 (39). Significantly, Mlp2 binds more strongly to the Yra1–8 export mutant protein than to wild-type Yra1, suggesting that Mlp2 recruits Yra1 for docking of mRNA-protein complexes, but retains malformed complexes that contain the Yra1–8 mutant protein (39). The yeast MLP1 and MLP2 genes are not essential for cell viability and mlp1Δ or mlp2Δ single deletion mutants or mlp1Δ mlp2Δ double deletion mutants are viable and show no significant growth defects (41, 47, 48). However, deletion of MLP1 impairs nuclear retention of intron-containing mRNAs and permits the escape of pre-mRNA to the cytoplasm, supporting the notion that Mlp1 plays a pivotal role in mRNA quality control (40).
The N-terminal domain of Nab2 (Nab2-N) is both necessary and sufficient to bind to Mlp1 (49). Structural studies show that this domain has a fold based on a five α-helix bundle analogous to the proline-tryptophan-isoleucine (PWI) fold, which is found in several RNA-binding proteins (49, 50). Although the PWI fold is thought to mediate binding to RNA and DNA in other RNA-binding proteins acids (50), Nab2-N does not appear to bind to nucleic acids (49). The Nab2-N structure revealed a hydrophobic patch centered on residue Phe73 that could potentially serve as an interface for protein-protein interactions (49). Previously, we showed that Nab2 residue Phe73 is important for the interaction between Nab2 and Mlp1, but not between Nab2 and Gfd1, another Nab2-binding partner (49, 51).
Given the way in which Nab2 and Mlp1 function in mRNA export and quality control, we have investigated the interaction between these two proteins in greater detail and show here that the Nab2-binding domain of Mlp1 (Mlp1-NBD) maps to a 183-amino acid region (residues 1586–1768) within CT-Mlp1. The Mlp1-NBD can bind directly to Nab2 and expression of this domain is sufficient to cause nuclear accumulation of poly(A) RNA, confirming the tight link between Nab2, CT-Mlp1, and poly(A) RNA. We then use a structure-based engineered mutant of Nab2 that cannot interact with Mlp1 to explore the functional consequences of disrupting the Nab2/Mlp1 interaction in vivo. Overall, these studies provide in vivo evidence for a model of mRNA export in which Nab2 is important for targeting mRNAs to the nuclear pore for export.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Chemicals—All DNA manipulations were performed according to standard methods (52) and all media were prepared by standard procedures (53). Yeast strains and plasmids used are described in Table 1. All chemicals were obtained from Sigma, U. S. Biological Corp. (Swampscott, MA), or Fisher Scientific (Pittsburgh, PA) unless otherwise noted.
TABLE 1.
Yeast strains and plasmids
| Strain/plasmid | Description | Source or Ref. |
|---|---|---|
| W303 (ACY233) | MATα ura3Δ leu2Δ trp1Δ his3Δ | |
| NAB2Δ (ACY427) | MATa leu2Δ ura3Δ his3Δ NAB2::HIS3 (pAC636) | 72 |
| NAB2ΔMEX67Δ (SWY3601) | MATa trp1Δ leu2Δ his3Δ ura3Δ NAB2::HIS3 MEX67::KAN (pRS314-mex67-5) | 70 |
| NAB2ΔYRA1Δ (FSY2327) | MATa ura3Δ his3Δ leu2Δ trp1Δ YRA1::HIS3 NAB2::HIS3 (pFS1876, pAC636) | F. Stutz |
| pGEX4T-3 | GST, AMPR, bacterial expression vector | GE Healthcare |
| pAC1340 | GST-CT-Mlp1 (residues 1490–1875) in pGEX4T-3, AMPR | 38 |
| pAC1678 | GST-CT5 (residues 1490–1585) in pGEX4T-3, AMPR | This study |
| pAC1679 | GST-CT4 (residues 1490–1682) in pGEX4T-3, AMPR | This study |
| pAC1680 | GST-CT2 (residues 1490–1779) in pGEX4T-3, AMPR | This study |
| pAC1681 | GST-CT6 (residues 1586–1779) in pGEX4T-3, AMPR | This study |
| pAC1723 | GST-CT7 (residues 1677–1779) in pGEX4T-3, AMPR | This study |
| pGEX4T-1 | GST, AMPR, bacterial expression vector | GE Healthcare |
| pAC2058 | GST-Nab2-N-WT (residues 1–97) in pGEX4T-1, AMPR | 49 |
| pAC2349 | GST-Nab2-N-F72D (residues 1–97) in pGEX4T-1, AMPR | 49 |
| pAC2350 | GST-Nab2-N-F73D (residues 1–97) in pGEX4T-1, AMPR | 49 |
| pAC2453 | GST-Nab2-N-F73W (residues 1–97) in pGEX4T-1, AMPR | 49 |
| pET28a | His6, KANR, bacterial expression vector | Novagen |
| pAC785 | His6-Nab2 in pET28a, KANR | 32 |
| pET30a | His6, S-Tag, KANR, bacterial expression vector | Novagen |
| pAC2052 | His6-Nab2-N-WT (residues 1–97) in pET30a, KANR | This study |
| pAC2054 | His6-S-Tag-Mlp1-NBD (residues 1586–1768) in pET30a, KANR | This study |
| pPS295 (pAC18) | pGAL1, 2μ, URA3 | 73 |
| pAC2075 | pGAL1-SV40 bipartite NLS-MLP1-NBD (residues 1586–1768), 2μ, URA3, AMPR | This study |
| pAC1690 | pGAL1-SV40 bipartite NLS-myc-CT-MLP1 (residues 1490–1875), 2μ, URA3, AMPR | This study |
| pAC1692 | pGAL1-SV40 bipartite NLS-myc-CT5 (residues 1490–1585), 2μ, URA3, AMPR | This study |
| pAC1693 | pGAL1-SV40 bipartite NLS-myc-CT4 (residues 1490–1682), 2μ, URA3, AMPR | This study |
| pAC1694 | pGAL1-SV40 bipartite NLS-myc-CT3 (residues 1490–1717), 2μ, URA3, AMPR | This study |
| pAC1695 | pGAL1-SV40 bipartite NLS-myc-CT2 (residues 1490–1779), 2μ, URA3, AMPR | This study |
| pAC1696 | pGAL1-SV40 bipartite NLS-myc-CT1 (residues 1490–1811), 2μ, URA3, AMPR | This study |
| pAC636 | NAB2, CEN, URA3, AMPR | 32 |
| pAC717 | NAB2, CEN, LEU2, AMPR | 36 |
| pAC1152 | nab2 ΔN, CEN, LEU2, AMPR | 36 |
| pAC2351 | nab2 F72D, CEN, LEU2, AMPR | This study |
| pAC2352 | nab2 F73D, CEN, LEU2, AMPR | This study |
| pAC1036 | ΔRGG NAB2-GFP, CEN, LEU2, AMPR | 36 |
| pFS1876 | YRA1, CEN, URA3, AMPR | 19 |
| pFS2152 (pAC2266) | yra1–8, CEN, TRP1, AMPR | 21 |
| pRS314-mex67-5 | mex67-5, CEN, TRP1, AMPR | 8 |
Generation of Nab2 Mutants—Nab2 mutants GST-NT F72D (pAC2349), GST-NT F73D (pAC2350), and GST-NT-F73W (pAC2453) were previously described (49). Nab2 mutants nab2 F72D (pAC2351) and nab2 F73D (pAC2352) were generated by site-directed mutagenesis using oligonucleotides (Integrated DNA Technologies) encoding the F72D or F73D amino acid substitution, NAB2 (pAC717) plasmid template, and QuikChange Site-directed Mutagenesis Kit (Stratagene). All constructs were sequenced to ensure the presence of each desired mutation and the absence of any additional mutations.
Protein Expression and Purification—Recombinant GST fusion and His-tagged proteins were expressed in bacteria and purified for in vitro solution and solid phase binding assays. GST (pGEX4T-3), GST-CT-Mlp1 (pAC1340), GST-CT2 (pAC1680), GST-CT4 (pAC1679), GST-CT5 (pAC1678), GST-CT6 (pAC1681), GST-CT7 (pAC1723), and His-Nab2 (pAC785) were expressed in Escherichia coli DE3 cells and purified by batch purification. GST (pGEX4T-1), GST-Nab2-N-WT (pAC2058), GST-Nab2-N-F72D (pAC2349), GST-Nab2-N-F73D (pAC2350), GST-Nab2-N-F73W (pAC2453), His-Nab2-N-WT (pAC2052), and His-S-Tag-Mlp1-NBD (pAC2054) were expressed in E. coli DE3 cells and purified by column chromatography. Overnight cultures were used to inoculate 100 ml of LB media for batch purification or 1 liter of LB media for purification by column chromatography. Cultures were grown at 37 °C to an A600 of 0.6–0.8, induced with 200 μm isopropyl 1-thio-d-galactopyranoside, and grown at 30 °C for 5 h. For batch purification of GST fusion proteins, cells were collected and lysed in 10 ml of phosphate-buffered saline (PBS) supplemented with protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 3 ng/ml pepstatin A, leupeptin, aproptinin, and chymostatin) by incubation with lysozyme (100 mg/ml) for 30 min on ice and sonication. Lysates were cleared by centrifugation and incubated with glutathione-Sepharose 4B (GE Healthcare) for 2 h at 4°C with mixing. The beads were then washed once with 10 ml of PBS supplemented with 0.5% Triton X-100 and twice with 10 ml of PBS. For purification of GST fusion proteins by column chromatography, cells were collected and lysed in 20 ml of buffer A (PBS, 2 mm 2-mercaptoethanol) supplemented with protease inhibitor mixture by a French press and cleared lysates were loaded onto a GSTrap 4B column (GE Healthcare) pre-equilibrated with buffer A. Bound protein was eluted with a linear gradient of buffer B (PBS, 2 mm 2-mercaptoethanol, 10 mm reduced glutathione) and fractions containing GST protein were dialyzed into PBS supplemented with 2 mm dithiothreitol (DTT). For batch purification of Histagged proteins, cells were collected and lysed in 10 ml of lysis buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 10 mm imidazole) supplemented with protease inhibitor mixture by incubation with lysozyme and sonication. Lysates were cleared by centrifugation and incubated with nickel-nitrilotriacetic acid-agarose (Qiagen) in lysis buffer for 2 h at 4 °C with mixing. The beads were then washed twice with 10 ml of wash buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 20 mm imidazole). His-tagged proteins were eluted from agarose with 1 ml of elution buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 250 mm imidazole) and buffer exchanged into PBS supplemented with 2 mm DTT using a Centricon centrifugal filter device (Millipore). For purification of His-tagged proteins by column chromatography, cells were collected and lysed in 20 ml of buffer A (50 mm NaH2PO4, pH 7.4, 500 mm NaCl, 20 mm imidazole, 2 mm 2-mercaptoethanol) supplemented with protease inhibitor mixture by a French press and cleared lysates were loaded onto a HiTrap IMAC HP column (GE Healthcare) pre-equilibrated with buffer A. Bound protein was eluted with a linear gradient of buffer B (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 250 mm imidazole, 2 mm 2-mercaptoethanol) and fractions containing His-tagged protein were dialyzed into PBS supplemented with 2 mm DTT.
Solution Binding Assays—For Nab2 binding to CT-Mlp1 and CT-Mlp1 truncation mutants, Sepharose-bound GST, GST-CT-Mlp1, or GST-CT2 and -4–7 (6 μg) was incubated with 2 μg of purified His-Nab2 at 4 °C in 1 ml of PBS supplemented with protease inhibitor mixture, 0.1 mg/ml BSA competitor, and 0.5% Triton X-100 for 1 h 30 min at 4°C with mixing. Unbound fractions were collected and the beads were washed three times with 1 ml of PBS for 5 min each. Bound fractions were eluted with loading buffer and samples were analyzed by SDS-PAGE followed immunoblotting with an anti-His antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The intensities of the His-Nab2 bands in the bound fractions were quantitated by integration of the pixel densities of the bands using ImageJ version 1.4 software (National Institute of Health, MD; rsb.info.nih.gov/ij/) and percentage bound relative to CT-Mlp1 was calculated by dividing the His-Nab2 band intensity in each bound fraction by the His-Nab2 band intensity in the bound fraction of GST-CT-Mlp1. For Mlp1-NBD binding to Nab2-N, His-Nab2-N-WT, or ovalbumin coupled to CNBr-Sepharose beads (30 μl), as described previously (49), was incubated with purified His-Mlp1-NBD (20 μg) in 1 ml of PBS supplemented with protease inhibitor mixture, 2 mm DTT, and 0.5 mg/ml BSA for 1 h 30 min at 4 °C with mixing. Unbound fractions were collected and the beads were washed three times with 1 ml of PBS for 5 min each. Bound fractions were eluted with loading buffer and samples were analyzed by SDS-PAGE followed by Coomassie staining. For co-purification of Mlp1-NBD with Nab2-N, 50-ml cultures of DE3 cells expressing GST-Nab2-N or His-Mlp1-NBD were combined, collected, and lysed in 10 ml of PBS supplemented with protease inhibitor mixture by sonication. Lysates were cleared and incubated with glutathione-Sepharose for 2 has described previously for batch purification of GST fusion proteins. The beads were washed three times with 5 ml of PBS for 2 min each. Unbound, bound, and wash samples were analyzed by SDS-PAGE followed by Coomassie staining.
Solid Phase Binding Assay—Solid phase binding assays were performed essentially as described by Bayliss et al. (54). Microtiter Immulux HB plates (Dynex) were coated with 100 μl/well of 125 nm GST, GST-Nab2-N (residues 1–97), GST-Nab2-N-F72D, GST-Nab2-N-F73D, or GST-Nab2-N-F73W in coating buffer (PBS supplemented with 2 mm DTT and 0.2 mm phenylmethylsulfonyl fluoride) for 16 h at 4 °C on a rocker. After adsorption, plates were washed three times by immersion in PBS and incubated in 100 μl of binding buffer (coating buffer supplemented with 3% BSA and 0.1% Tween 20) overnight at 4 °C on a rocker. Binding reactions were carried out for 2 h at 4 °C with 100 μl/well of 1–5 μm S-Tag-Mlp1-NBD (residues 1586–1768) protein in binding buffer. After binding, plates were washed three times by immersion in binding buffer without BSA and proteins were cross-linked for 15 min at room temperature by incubation in 1 mg/ml 1-ethyl-3-(3-dimethyl-aminopropyl)carboiimide (Pierce) in the same buffer. The wells were then washed for 20 min in PBS-T (PBS supplemented with 0.2% Tween 20), 10 min with PBS-T containing 100 mm ethanolamine, and finally incubated for 10 min in PBS-T containing 3% BSA. The bound S-tagged proteins were detected by incubation with S-protein-horseradish peroxidase conjugate (Novagen) in coating buffer containing 1% BSA and 0.1% Tween 20 for 1 h at 4°C on rocker. The plates were washed three times by immersion in PBS. Then, 100 μl/well of horseradish peroxidase substrate (1-Step™ Turbo TMB (3,3′5,5′-tetramethylbenzidine)-ELISA (Pierce)) was added for 30 min at room temperature and the reaction was stopped by addition of 100 μl of 2 m H2SO4. The absorbance of the samples was measured at 450 nm with an ELX808 Ultra Microplate Reader with KCjunior software (Bio-Tek Instruments, Inc.). For each plate, average absorbance values at A450 were determined for GST and GST-Nab2-N proteins at each S-Tag-Mlp1-NBD concentration from triplicate samples. Background GST absorbance values were subtracted from those of the GST-Nab2 proteins. From three plates, absorbance values for GST-Nab2-N proteins at each S-Tag-Mlp1-NBD concentration were used to generate saturation binding curves by non-linear regression using Prism 4 software (GraphPad Software, Inc.) and calculate the mean apparent Kd. Standard error of the mean (S.E.; standard deviation divided by the square root of n, where n = 3) values of A450 readings from triplicate samples of GST-Nab2-N proteins at each S-Tag-Mlp1-NBD concentration for binding curves in Fig. 4B and S.E. values of triplicate mean Kd values for GST-Nab2-N protein binding to S-Tag-Mlp1-NBD in Table 2 were calculated using Prism 4 software.
FIGURE 4.
The Nab2-binding domain of Mlp1 binds to the N-terminal domain of Nab2 with micromolar affinity and Nab2 F73D substitution disrupts the Nab2/Mlp1 interaction. A, surface representation of the N-terminal domain of Nab2 (Nab2-N) showing the putative Mlp1 interaction site (gray) identified by Grant et al. (49). Phe73 (dark gray) is central to this site and its hydrophobic aromatic side chain is completely exposed to the solvent. B, the binding affinity of the Nab2-binding domain of Mlp1 (Mlp1-NBD; residues 1586–1768) for Nab2-N wild-type and mutants F72D, F73D, and F73W (residues 1–97) was measured quantitatively in a solid-phase binding assay (54). Binding of S-Tag-Mlp1-NBD (Mlp1-NBD) to GST-Nab2-N wild-type (Nab2-N-WT), GST-Nab2-N-F72D (Nab2-N-F72D), GST-Nab2-N-F73D (Nab2-N-F73D), or GST-Nab2-N-F73W (Nab2-N-F73W) at increasing concentrations of S-Tag-Mlp1-NBD was measured at optical density of 450 nm (A450) as described under “Experimental Procedures.” Measurements at A450 (amount bound) were used to generate saturation binding curves by non-linear regression. Standard error bars for A450 readings from triplicate samples of GST-Nab2-N-WT and mutants at each S-Tag-Mlp1-NBD concentration are indicated. These binding curves were used to calculate apparent dissociation constants for binding between Mlp1-NBD and Nab2-N wild-type (Kd ∼ 1.3 ± 0.4 μm), Nab2-N-F72D (Kd ∼ 2.3 ± 1.1 μm), or Nab2-N-F73W (Kd ∼ 0.8 ± 0.2 μm) (see Table 2). The weak binding between Mlp1-NBD and Nab2-N-F73D was indistinguishable from nonspecific binding, indicating that the F73D substitution essentially disrupts the Nab2/Mlp1 interaction.
TABLE 2.
Dissociation constants for Nab2-N wild-type/mutants and Mlp1-NBD Data represent the Kd means ± S.E. from triplicate experiments.
| Nab2-N | Mlp1 | Kd apparent |
|---|---|---|
| μm | ||
| GST-Nab2-N-WT | Mlp1-NBD | 1.26 ± 0.38 |
| GST-Nab2-N-F72D | Mlp1-NBD | 2.31 ± 1.06 |
| GST-Nab2-N-F73D | Mlp1-NBD | Not detectable |
| GST-Nab2-N-F73W | Mlp1-NBD | 0.83 ± 0.18 |
| GST | Mlp1-NBD | Not detectable |
ΔRGG-Nab2-GFP Relocalization Assay—To examine the effect of Mlp1 expression on Nab2 localization in yeast cells, we exploited a previously developed assay where relocalization of a largely cytoplasmic Nab2 mutant fused to GFP, ΔRGG-Nab2-GFP, to the nucleus is monitored upon Mlp1 expression (38). For analysis of the effect of expression of CT-Mlp1, CT-Mlp1 truncation mutants CT1–5 or Mlp1-NBD on the localization of ΔRGG-Nab2-GFP, wild-type W303 cells expressing ΔRGG-Nab2-GFP (pAC1036) and containing galactose-inducible CT-MLP1 (pAC1690), CT1 (pAC1696), CT2 (pAC1695), CT3 (pAC1694), CT4 (pAC1693), CT5 (pAC1692), MLP1-NBD (pAC2075), or vector control (pPS295) were grown in minimal media with 2% glucose overnight at 30 °C, transferred to minimal media with 2% galactose, and grown to log phase at 30 °C. For all samples, the localization of ΔRGG-Nab2-GFP was examined by direct fluorescence microscopy.
Fluorescence in Situ Hybridization (FISH)—The intracellular localization of poly(A) RNA was assayed by FISH (32). Briefly, wild-type W303 cells containing galactose-inducible CT-MLP1 (pAC1690), CT1 (pAC1696), CT2 (pAC1695), CT3 (pAC1694), CT4 (pAC1693), CT5 (pAC1692), MLP1-NBD (pAC2075), or vector control (pPS295) were grown in minimal media with 2% glucose overnight at 30 °C, transferred to minimal media with 2% galactose, and grown to log phase at 30 °C. NAB2Δ cells or NAB2Δ mex67-5 cells containing NAB2 (pAC717), nab2 ΔN (pAC1152), nab2 F72D (pAC2351), or nab2 F73D (pAC2352) were grown in minimal media with 2% glucose overnight, transferred to minimal media with 2% glucose, and grown to log phase at 30 °C. Cells were prepared as previously described by Wong et al. (55). A digoxigenin-labeled oligo(dT) probe and fluorescein isothiocyanate-conjugated anti-digoxigenin antibody (1:200 dilution, Roche Molecular Biochemicals) were used to localize poly(A) RNA. For all samples, cells were examined by fluorescence microscopy. Cells were also stained with 4′,6-diamidino-2-phenylindole-dihydrochloride (DAPI) to detect chromatin and hence the position of the nucleus.
To quantitate the percentage of cells processed for FISH that showed nuclear accumulation of poly(A) RNA, NAB2, nab2 F72D, nab2 F73D, mex67-5 NAB2, mex67-5 nab2 F72D, and mex67-5 nab2 F73D cells probed with oligo(dT) to visualize poly(A) RNA and 10 fields of cells for each strain were imaged. For each field of cells, cells were counted and scored using ImageJ version 1.4 software, and the percentage of cells with nuclear poly(A) RNA was calculated. Cells were scored as showing nuclear poly(A) RNA if the fluorescent signal in the nucleus was greater than the signal in the cytoplasm. The mean percentage of cells with nuclear poly(A) RNA was calculated from the percentages in the 10 fields of cells. Variation in the percentage nuclear poly(A) RNA in each field from the mean was assessed by standard deviation (square root of the mean squared). All scoring of nuclear poly(A) RNA was performed blind to avoid biasing results. In each field of cells, at least 195 NAB2 and nab2 mutant cells and at least 43 mex67-5 NAB2 and mex67-5 nab2 mutant cells were counted. The fold increase in nuclear poly(A) RNA accumulation in nab2 mutant cells relative to that in NAB2 or mex67-5 NAB2 cells was calculated by dividing the mean percentage of nuclear poly(A) RNA in nab2 mutant cells by the mean percentage in corresponding NAB2 wild-type cells.
Microscopy—Cells expressing ΔRGG-Nab2-GFP or processed for FISH were visualized by using filters from Chroma Technology (Brattleboro, VT) and an Olympus BX60 direct fluorescence microscope equipped with a photometric Quantix digital camera from Roper Scientific (Tucson, AZ). All images were captured using IP Lab Spectrum software.
Nab2 Functional Assay—In vivo functional analysis of Nab2 was carried out through a standard plasmid shuffle assay combined with serial dilution and spotting or growth curves. To generate single mutants of nab2 or double mutants of nab2 and mex67-5 or yra1–8, NAB2Δ cells (ACY427), NAB2Δ mex67-5 cells (NAB2Δ MEX67Δ cells (SWY3601) containing mex67-5 TRP1 plasmid (pRS314-mex67-5)) or NAB2Δ yra1–8 cells (NAB2Δ YRA1Δ cells (FSY2327) containing yra1–8 TRP1 plasmid (pFS2152)) containing NAB2 URA3 maintenance plasmid (pAC636) were transformed with NAB2 (pAC717), nab2 ΔN (pAC1152), nab2 F72D (pAC2351), or nab2 F73D (pAC2352) LEU2 plasmids and selected on Leu-Ura- minimal media with 2% glucose. Cells were grown overnight at 30 °C to saturation in Leu-Ura- minimal media with 2% glucose. Cell concentrations were normalized by A600 and cultures were serially diluted in sterile H2O to obtain ∼10,000, 1000, 100, 10, or 1 cell per 3-μl volume. These dilutions were spotted onto control Leu-Ura- minimal media, where the NAB2 URA3 maintenance plasmid is maintained, or Ura- minimal media containing 5-fluoroorotic acid (5-FOA), which selects for cells that have lost the NAB2 URA3 maintenance plasmid (56). Growth of NAB2Δ, NAB2Δ mex67-5, or NAB2Δ yra1–8 cells harboring NAB2, nab2 ΔN, nab2 F72D, or nab2 F73D as the sole copy of nab2 was examined at 30 °C. For growth curve analysis, NAB2Δ or NAB2Δ mex67-5 cells harboring NAB2, nab2 F72D, or nab2 F73D as the sole copy of nab2 were grown overnight at 30 °C to saturation in Leu-Ura- minimal media with 2% glucose. Cell concentrations were normalized by A600, diluted 100-fold in 100 μl of Leu-Ura- minimal media with 2% glucose and added to wells of a MicroWell F96 microtiter plate (Nunc). Samples were loaded in duplicate. Cells in plate wells were grown at 30 °C with shaking and absorbance at A600 was measured every 30 min for 40 h in an ELX808 Ultra Microplate Reader with KCjunior software (Bio-Tek Instruments, Inc.). Duplicate sample absorbance for time points were averaged and plotted using Microsoft Excel for Mac 2004 (Microsoft Corp.).
RESULTS
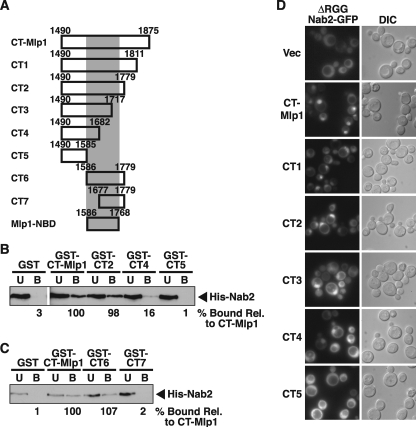
Identification of a Minimal Nab2-binding Region in Mlp1—Previous work demonstrated that the globular, acidic C-terminal domain of Mlp1 (CT-Mlp1, residues 1490–1875) interacts with the hnRNP, Nab2, both in vitro and in vivo and that ectopic expression of CT-Mlp1 causes nuclear accumulation of poly(A) RNA (38). These data indicate that Mlp1 is involved in poly(A) RNA export and suggest that one function of Nab2 is to serve as a molecular link between Mlp1 and poly(A) RNA. To test this model and gain further insight into the molecular basis for the Nab2/Mlp1 interaction, we employed a series of CT-Mlp1 truncation mutants with progressive deletions from the C-terminal end of Mlp1 (truncation mutants CT1–5, see Fig. 1A) to map the region within CT-Mlp1 that interacts directly with Nab2. These truncation mutants were used in both in vitro and in vivo binding assays with Nab2. For in vitro binding, recombinant GST, GST-CT-Mlp1, GST-CT2, CT4, or CT5 bound to glutathione beads was incubated with soluble recombinant His-Nab2 and the bound and unbound fractions were analyzed by immunoblotting with anti-His antibody to detect bound Nab2. The results were quantitated as described under “Experimental Procedures.” As shown in Fig. 1B, we found that CT-Mlp1 CT2 (residues 1490–1779) binds to Nab2 with similar affinity (98% bound) as intact CT-Mlp1 (residues 1490–1875), whereas CT4 (residues 1490–1682) binds more weakly to Nab2 (16% bound) compared with CT-Mlp1, and CT5 (residues 1490–1585) shows no binding to Nab2 (1% bound). As a control, GST alone does not bind to Nab2. This result suggests that the Nab2-binding region within CT-Mlp1 lies within residues 1490–1779.
FIGURE 1.
Mapping of CT-Mlp1 reveals a Nab2-binding region between residues 1586 and 1779. A, schematic depicting sizes of CT-Mlp1 truncation mutants (CT1–7) and the Nab2-binding domain of Mlp1 (Mlp1-NBD) constructed. B, CT-Mlp1 truncation mutant CT2 (residues 1490–1779) binds to Nab2 in vitro, but mutant CT5 (residues 1490–1585) does not bind to Nab2. Recombinant GST, GST-CT-Mlp1 (CT-Mlp1), or the GST-CT-Mlp1 truncation mutant (CT2, CT4, CT5) bound to glutathione-Sepharose beads were incubated with recombinant His-tagged Nab2 (Nab2) and unbound (U) and bound (B) fractions were analyzed by immunoblotting with anti-His antibody. The percentage of His-Nab2 band intensity in each bound fraction relative to the His-Nab2 band intensity in the bound fraction of GST-CT-Mlp1 is indicated below the immunoblot. C, CT-Mlp1 truncation mutant CT6 (residues 1586–1779) binds to Nab2 in vitro. Recombinant GST, GST-CT-Mlp1 (CT-Mlp1), or the GST-CT-Mlp1 truncation mutant (CT6, CT7) bound to glutathione-Sepharose beads was incubated with recombinant His-tagged Nab2 (Nab2) and unbound (U) and bound (B) fractions were analyzed by immunoblotting with anti-His antibody. The percentage of His-Nab2 band intensity in each bound fraction relative to the His-Nab2 band intensity in the bound fraction of GST-CT-Mlp1 is indicated below the immunoblot. D, expression of CT-Mlp1 truncation mutant CT2 (residues 1490–1779) causes relocalization of ΔRGG-Nab2-GFP to the nucleus. CT-Mlp1 and CT-Mlp1 truncation mutants (CT1–5) were expressed in yeast cells expressing a Nab2 mutant fused to GFP (ΔRGG-Nab2-GFP), which displays localization throughout the cell (36), and ΔRGG-Nab2-GFP was visualized by direct fluorescence microscopy. Empty vector (Vec) was used as a control. Corresponding differential interference contrast (DIC) images are shown.
The observation that CT-Mlp1 residues 1490–1779 (CT2) binds to Nab2, but residues 1490–1585 (CT5) does not interact defines the C-terminal end of the minimal Nab2-binding region within CT-Mlp1. To further delineate the N-terminal end of the minimal Nab2-binding region, we generated two additional CT-Mlp1 truncation mutants, CT6 (residues 1586–1779) and CT7 (residues 1677–1779) (Fig. 1A). Recombinant GST, GST-CT-Mlp1, GST-CT6, or GST-CT7 bound to glutathione beads was incubated with soluble recombinant His-Nab2 and bound and unbound fractions were analyzed by immunoblotting with anti-His antibody to detect bound Nab2. As shown in Fig. 1C, we found that CT-Mlp1 mutant CT6 (residues 1586–1779) binds to Nab2 to the same extent (>100% bound) as the intact CT-Mlp1 (residues 1490–1875), whereas CT7 (residues 1677–1779) does not bind to Nab2 (2% bound). These data indicate that the Nab2-binding region within CT-Mlp1 resides within residues 1586–1779.
We used a secondary in vivo Nab2 binding assay to confirm the in vitro data mapping the Nab2-binding region of Mlp1 to residues 1586–1779. Previously, we developed a ΔRGG-Nab2 relocalization assay that reports on the interaction between Nab2 and Mlp1 in vivo (38). In contrast to wild-type Nab2, which shows nuclear localization at steady-state (35), ΔRGG-Nab2 shows diffuse nuclear and cytoplasmic localization at steady-state due to lack of the RGG domain (36), which is required for interaction with the Nab2 import receptor Kap104 (37, 57). Expression of CT-Mlp1, which localizes to the nucleus (38), causes nuclear relocalization of ΔRGG-Nab2 fused to GFP (ΔRGG-Nab2-GFP), which normally locates throughout the yeast cell (36). CT-Mlp1-induced relocalization of ΔRGG-Nab2-GFP to the nucleus is consistent with CT-Mlp1 binding to Nab2 in vivo. Therefore, to confirm the in vitro binding data using this in vivo binding assay, we expressed CT-Mlp1 and CT-Mlp1 truncation mutants CT1–5 in yeast from a galactose-inducible GAL1–10 promoter. To ensure nuclear targeting of each of the CT-Mlp1 deletions, the CT1–5 sequences were fused to an engineered bipartite nuclear localization signal based on the simian virus 40 (SV40) nuclear localization signal (58). Wild-type yeast cells expressing ΔRGG-Nab2-GFP were induced to express CT-Mlp1 and CT1–5 by the addition of galactose and the localization of ΔRGG-Nab2-GFP was assessed by direct fluorescence microscopy. The expression and localization of CT-Mlp1 and CT1–5 were confirmed by immunoblotting and GFP fusion localization (data not shown). As shown in Fig. 1D, CT-Mlp1 truncation mutant CT1 (residues 1490–1811) and CT2 (residues 1490–1779) relocalize ΔRGG-Nab2-GFP to the nucleus to a similar degree as the intact CT-Mlp1 (residues 1490–1875). Truncation mutants CT3 (residues 1490–1717) and CT4 (residues 1490–1682) partially relocalize ΔRGG-Nab2-GFP to the nucleus compared with CT-Mlp1 and truncation mutant CT5 (residues 1490–1585) does not relocalize ΔRGG-Nab2-GFP to the nucleus. This result indicates that CT-Mlp1 residues 1490–1779 binds to Nab2, whereas CT-Mlp1 residues 1490–1585 do not interact with Nab2 in vivo. Taken together, the Nab2 binding assays strongly suggest the Nab2-binding region within CT-Mlp1 maps to residues 1586–1779.
The Nab2-binding Region of Mlp1 Causes Nuclear Accumulation of Poly(A) RNA—Expression of CT-Mlp1 (residues 1490–1875) causes nuclear accumulation of poly(A) RNA (38). To map the region within CT-Mlp1 that causes poly(A) RNA accumulation and assess whether it corresponds to the Nab2-binding region, we expressed CT-Mlp1 and CT-Mlp1 truncation mutants CT1–5 in yeast from a galactose-inducible promoter. Wild-type yeast cells were induced to express CT-Mlp1 and CT1–5 by the addition of galactose and samples were processed for FISH using an oligo(dT) probe to visualize bulk poly(A) RNA. CT-Mlp1 truncation mutants CT1 (residues 1490–1811) and CT2 (residues 1490–1779) cause nuclear accumulation of poly(A) RNA to a similar extent as the intact CT-Mlp1 (residues 1490–1875), CT3 (residues 1490–1717) causes partial nuclear accumulation of poly(A) RNA compared with CT-Mlp1, and CT4 (residues 1490–1682) and CT5 (residues 1490–1585) do not cause poly(A) RNA accumulation in the nucleus (Fig. 2). This result indicates that the region within CT-Mlp1 that is sufficient to cause nuclear accumulation of poly(A) RNA maps to residues 1490–1779. Thus, both the Nab2-binding and poly(A) RNA interacting regions of CT-Mlp1 reside within residues 1490–1779.
FIGURE 2.
Mapping of CT-Mlp1 defines a region between residues 1490 and 1779 sufficient to cause nuclear poly(A) RNA accumulation. CT-Mlp1 and CT-Mlp1 truncation mutants (CT1–5) were expressed in yeast cells and poly(A) RNA (Poly(A)) was visualized by fluorescence in situ hybridization with an oligo(dT) probe as described under “Experimental Procedures.” Cells were stained with DAPI to visualize the position of the nucleus. Corresponding differential interference contrast (DIC) images are shown.
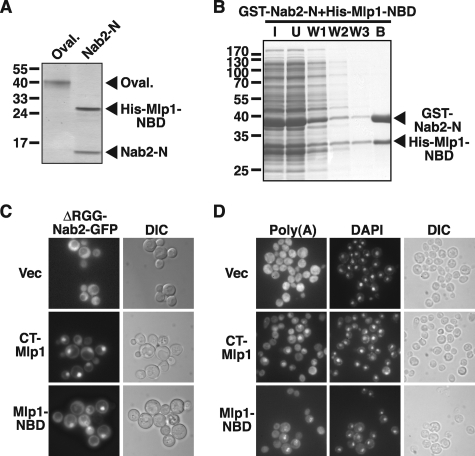
Delineation of the Mlp1-Nab2 Binding Domain (Mlp1-NBD)—As a complement to our mapping using engineered deletions, we also isolated a proteolytic fragment of the intact CT-Mlp1 that bound to Nab2 in vitro and identified the Mlp1 tryptic peptides within this fragment by mass spectroscopy (see supplemental Fig. S1 and supplemental Table 1). We mapped these peptides within CT-Mlp1 and used the information to refine the C-terminal end of the Nab2-binding region of Mlp1 to residue 1768. We termed the final Nab2-binding region of Mlp1: Mlp1-Nab2-binding domain (Mlp1-NBD; residues 1586–1768) (Fig. 1A).
The Mlp1-NBD Interacts with the Nab2 N-terminal Domain—Recent domain analysis of the hnRNP Nab2 has established that the N-terminal domain of Nab2 (Nab2-N, residues 1–97) binds to both CT-Mlp1 and full-length Mlp1 in vitro (49). To determine whether the Nab2-binding domain of Mlp1 (Mlp1-NBD, residues 1586–1768) interacts directly with Nab2-N, we analyzed the interaction by an in vitro binding assay using purified recombinant proteins. A control protein, ovalbumin, or recombinant His-Nab2-N-WT (residues 1–97) coupled to Sepharose beads was incubated with soluble recombinant His-Mlp1-NBD and bound fractions were analyzed by SDS-PAGE and Coomassie staining. As shown in Fig. 3A, Mlp1-NBD binds to Nab2-N-WT, but does not bind to control ovalbumin beads. To further support this direct interaction and test its specificity, we tested whether Mlp1-NBD can be co-purified with Nab2-N from a complex protein mixture. We combined cell lysates from E. coli expressing His-Mlp1-NBD and E. coli expressing GST-Nab2-N, incubated the mixed lysates with glutathione beads and analyzed the bound fraction by SDS-PAGE and Coomassie staining. We find that Mlp1-NBD does indeed co-purify with Nab2-N from a complex mixture of bacterial proteins, highlighting the specificity of this interaction (Fig. 3B). Together, the experiments demonstrate that the Nab2-binding domain of Mlp1 (residues 1586–1768) interacts directly with the N-terminal domain of Nab2 (residues 1–97). Importantly, previous studies show that the interaction between CT-Mlp1 and Nab2 in yeast lysate is insensitive to RNase treatment and therefore RNA-independent and also that Nab2-N does not interact with poly(A) RNA in vitro (38, 49). The direct interaction between Nab2-N and Mlp1-NBD is thus most likely RNA-independent as well.
FIGURE 3.
The Nab2-binding domain of Mlp1 directly interacts with Nab2 and causes nuclear accumulation of poly(A) RNA. A, the Nab2-binding domain of Mlp1 (Mlp1-NBD; residues 1586–1768) binds to the N-terminal domain of Nab2 (Nab2-N; residues 1–97) in vitro. His-Nab2-N (Nab2-N) or, as a control, ovalbumin (Oval.) coupled to Sepharose beads was incubated with His-Mlp1-NBD and bound fractions were analyzed by SDS-PAGE and Coomassie staining. B, the Nab2-binding domain of Mlp1 (Mlp1-NBD) co-purifies with the N-terminal domain of Nab2 (Nab2-N). Cell lysates from E. coli expressing His-Mlp1-NBD and E. coli expressing GST-Nab2-N were combined, incubated with glutathione beads, and beads were washed three times. Input combined lysate (I), unbound (U) fraction, three wash fractions (W1, W2, W3), and bound (B) fraction were analyzed by SDS-PAGE and Coomassie staining. C, expression of the Nab2-binding domain of Mlp1 (Mlp1-NBD) relocalizes ΔRGG-Nab2-GFP to the nucleus. CT-Mlp1 and Mlp1-NBD were expressed in yeast cells expressing ΔRGG-Nab2-GFP, which displays localization throughout the cell (36), and ΔRGG-Nab2-GFP was visualized by direct fluorescence microscopy. Empty vector (Vec) was used as control. Corresponding differential interference contrast (DIC) images are shown. D, expression of the Nab2-binding domain of Mlp1 (Mlp1-NBD) causes nuclear accumulation of poly(A) RNA. CT-Mlp1 and Mlp1-NBD were expressed in yeast cells and poly(A) RNA (Poly(A)) was visualized by fluorescence in situ hybridization with an oligo(dT) probe as described under “Experimental Procedures.” Cells were stained with DAPI to visualize the position of the nucleus. Corresponding differential interference contrast images are shown.
To further compare the Mlp1-NBD (residues 1586–1768) with the intact CT-Mlp1, we asked whether the Mlp1-NBD can both interact with Nab2 and cause nuclear accumulation of poly(A) RNA in vivo. In the ΔRGG-Nab2-GFP relocalization assay, we expressed nuclear-targeted CT-Mlp1 and Mlp1-NBD from a galactose-inducible promoter and found that Mlp1-NBD (residues 1586–1768) relocalizes most ΔRGG-Nab2-GFP to the nucleus, although not quite to the same extent as CT-Mlp1 (residues 1490–1875) (Fig. 3C). To test if the expression of Mlp1-NBD affects poly(A) RNA localization, we expressed Mlp1-NBD and CT-Mlp1 in yeast cells and localized the poly(A) RNA by FISH with an oligo d(T) probe. We find that the Mlp1-NBD does induce accumulation of poly(A) RNA within the nucleus, although not quite to the same degree as CT-Mlp1 (Fig. 3D). These results indicate that the Mlp1-NBD (residues 1586–1768) within CT-Mlp1 is primarily responsible for CT-Mlp1-induced nuclear poly(A) RNA accumulation, but that small contributions are also made by other residues outside this region of CT-Mlp1. Overall, these data confirm that the Mlp1-NBD, like the intact CT-Mlp1, has the capacity both to bind Nab2 in vivo and to impact poly(A) RNA localization.
To measure quantitatively the interaction between the Mlp1-NBD and Nab2-N, we examined the binding of S-Tag-Mlp1-NBD to GST-Nab2-N wild-type (WT) at increasing concentrations of S-Tag-Mlp1-NBD in a solid-phase microtiter plate binding assay (54). This analysis generated a saturation binding curve and an apparent dissociation constant (Kd) value for Mlp1-NBD binding to Nab2-N-WT. We find that Mlp1-NBD binds to wild-type Nab2-N with an apparent Kd of ∼1.3 ± 0.4 μm (Fig. 4B, Table 2), indicating that the relative binding affinity between Mlp1-NBD and Nab2-N is in the micromolar range. The rather weak affinity between Mlp1 and Nab2 likely reflects the need for the Nab2/Mlp1 interaction to be transient during mRNA export.
Specific Amino Acid Substitutions Disrupt the Nab2-N/Mlp1-NBD Interaction—The crystal structure of the N-terminal domain of Nab2 (Nab2-N, residues 1–97) shows that this domain is based on a five α-helix bundle with a PWI-like fold (49). A Nab2 hydrophobic patch centered on a phenylalanine residue, Phe73 (Fig. 4A), was proposed to be an important constituent of the Nab2/Mlp1 interaction interface (49). Consistent with this hypothesis, we showed that recombinant Nab2-N-F73D does not bind to Mlp1, whereas Nab2-N-F72D is still competent to bind Mlp1, in an in vitro bead binding assay. To assess the impact of amino acid substitutions at Nab2 Phe72 or Phe73 on the relative binding affinity of Nab2 for Mlp1, we measured the binding of S-Tag-Mlp1-NBD to GST-Nab2-N mutants F72D, F73D, or F73W at increasing concentrations of S-Tag-Mlp1-NBD in the solid-phase binding assay. We find that the interaction between Nab2-N-F73D and Mlp1-NBD is undetectable in this assay, whereas Mlp1-NBD binds to Nab2-N-F72D with roughly 2-fold lower relative affinity (Kd ∼ 2.3 ± 1.1 μm) compared with wild-type (Fig. 4B, Table 2). Furthermore, we find that Mlp1-NBD binds to Nab2-N-F73W with 1.5-fold higher affinity (Kd ∼ 0.8 ± 0.2 μm) compared with wild-type Nab2-N (Fig. 4B, Table 2). The data reveal that substituting Nab2-N residue Phe73 with Asp significantly reduces the relative binding affinity between Nab2-N and the Mlp1-NBD, supporting the idea that Nab2 Phe73 is critical for interaction with Mlp1. The impaired binding of Nab2-N to Mlp1-NBD seen with the hydrophilic substitution, F73D, together with the enhanced binding seen with the hydrophobic substitution, F73W, is consistent with the proposal (49) that hydrophobic interactions contribute significantly to the Nab2/Mlp1 interface.
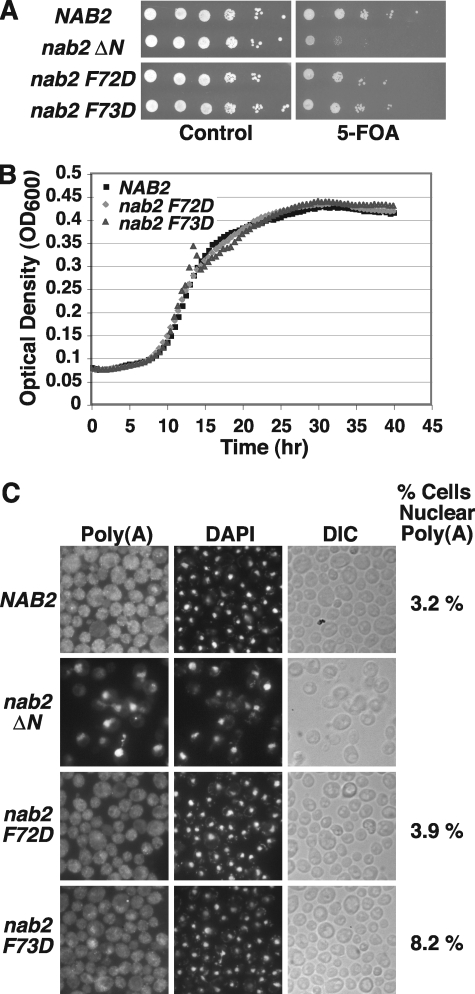
Functional Significance of the Nab2/Mlp1 Interaction—With the knowledge gained from the Nab2-N structure and engineered Nab2 mutants, we proceeded to assess the functional consequence of disrupting the Nab2/Mlp1 interaction in vivo and define the way in which this interaction contributes to mRNA export. As the NAB2 gene is essential for viability (35), we examined whether Nab2-F73D or Nab2-F72D (as a control) could replace wild-type Nab2 in yeast cells. As a further control in these assays, we used a previously characterized Nab2 mutant, ΔN, lacking the Nab2 N-terminal domain altogether (residues 4–97), which confers a severe slow growth phenotype when expressed as the only cellular copy of Nab2 (36). Using a plasmid shuffle assay, NAB2Δ yeast cells maintained by a NAB2 URA3 plasmid and also containing NAB2, nab2 ΔN, F72D or F73D LEU2 test plasmids were grown to saturation, serially diluted in 10-fold dilutions, and spotted on control plates (where the NAB2 URA3 maintenance plasmid is retained) and plates containing 5-FOA to remove the NAB2 URA3 maintenance plasmid and leave the test plasmids as the sole cellular copies of Nab2. Both nab2 F72D and nab2 F73D cells grow similarly to wild-type NAB2 cells on 5-FOA plates (Fig. 5A). However, as anticipated, nab2 ΔN cells show a slow growth phenotype. We confirmed that nab2 F72D and nab2 F73D cells grow similarly to wild-type NAB2 cells by measuring the optical density of the cells in liquid culture over time and generating quantitative growth curves for the cells (Fig. 5B). This result indicates that substituting Nab2 residue Phe73 with Asp and disrupting the Nab2/Mlp1 interaction does not significantly affect cell growth in a wild-type background. Additionally, given the knowledge that the N-terminal domain of Nab2 is required for proper Nab2 function (including binding to other proteins such as Gfd1 (49, 51)), the data suggest that the F73D substitution does not severely impact the overall structure of the Nab2 protein.
FIGURE 5.
nab2 F73D mutant cells grow similarly to wild-type cells but display nuclear accumulation of poly(A) RNA. A, nab2 F73D cells are viable and grow similarly to wild-type NAB2 cells. NAB2Δ cells maintained by a NAB2 URA3 plasmid and containing NAB2, nab2 ΔN, nab2 F72D, or nab2 F73D LEU2 test plasmids were grown to saturation, serially diluted in 10-fold dilutions, and spotted on control and 5-FOA plates. Cells were grown at 30 °C. B, growth curve analysis of nab2 F73D cells confirms that they grow similarly to wild-type NAB2 cells. NAB2Δ cells carrying only NAB2, nab2 F72D, or nab2 F73D plasmids were grown to saturation, diluted, and their optical density was measured at A600 for 40 h as described under “Experimental Procedures.” C, nab2 F73D cells display nuclear accumulation of poly(A) RNA. NAB2Δ cells carrying only NAB2, nab2 ΔN, nab2 F72D, or nab2 F73D plasmids were grown in liquid culture at 30 °C and poly(A) RNA (Poly(A)) was visualized by FISH using an oligo(dT) probe as described under “Experimental Procedures.” Cells were stained with DAPI to visualize the position of the nucleus. Corresponding differential interference contrast (DIC) images are shown. FISH results were quantitated as described under “Experimental Procedures” to assess the mean percentage of cells with nuclear accumulation of poly(A) RNA within 10 fields of at least 195 cells each (indicated to the right of DIC images). Variation in the percentage of cells with nuclear poly(A) RNA in each field of cells from the mean was assessed by calculating the standard deviation: NAB2, 3.2 ± 1.0%; nab2 F72D, 3.9 ± 1.5%; and nab2 F73D, 8.2 ± 2.0%. Thus, nab2 F73D cells exhibit a 2.5-fold increase in nuclear poly(A) RNA accumulation relative to NAB2 cells, whereas nab2 F72D cells show only a modest 1.2-fold increase. The percentages of nuclear poly(A) RNA accumulation observed in nab2 F73D cells were typical of multiple independent experiments.
To determine whether disruption of the Nab2/Mlp1 interaction impacts mRNA export, we next assessed whether nab2 F72D or nab2 F73D cells show nuclear accumulation of poly(A) RNA in vivo. We examined poly(A) RNA localization in wild-type NAB2, nab2 F72D, and nab2 F73D cells by FISH and then quantitated the percentage of cells with nuclear poly(A) RNA accumulation as described under “Experimental Procedures.” As a control, we also examined poly(A) RNA localization in nab2 ΔN cells, which show nuclear accumulation of poly(A) RNA (36). NAB2Δ cells expressing NAB2, nab2 ΔN, F72D, or F73D were processed for FISH using an oligo(dT) probe to visualize bulk poly(A) RNA. The nab2 F73D cells show a 2.5-fold increase in nuclear accumulation of poly(A) RNA compared with wild-type cells, whereas nab2 F72D cells show accumulation of poly(A) RNA in the nucleus comparable with wild-type cells (Fig. 5C). Control nab2 ΔN cells show significant nuclear accumulation of poly(A) RNA as expected (36). This result suggests that the Nab2/Mlp1 interaction facilitates but is not absolutely required for mRNA export.
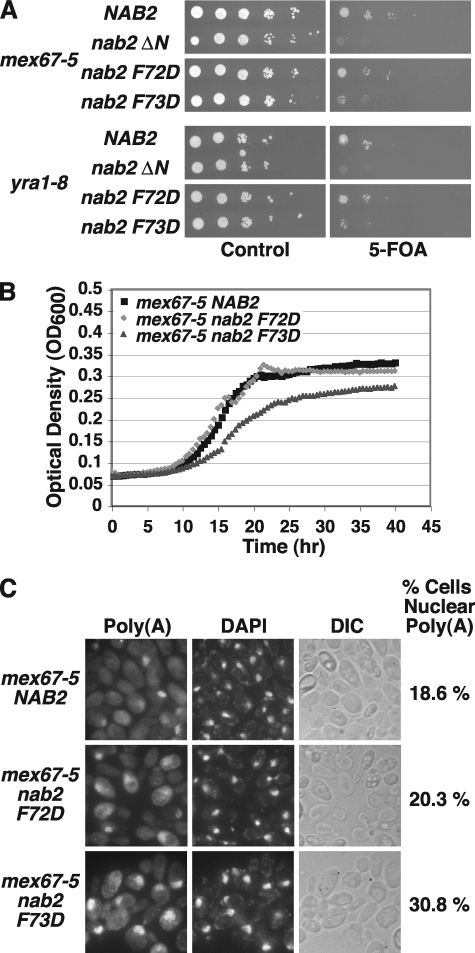
Because multiple mRNA export factors contribute to efficient mRNA export, we considered the possibility that the interaction between Nab2 and Mlp1 might contribute to a step in mRNA export that is not normally rate-limiting in a wild-type background. Thus, an effect on overall cellular function as assayed by cell growth might only be evident in a genetic background where additional steps in mRNA export are compromised or suboptimal. We therefore tested for genetic interactions between nab2 F72D or nab2 F73D and mRNA export mutants, mex67-5 (8) or yra1–8 (21), at the permissive growth temperature for these conditional mutants. Using the plasmid shuffle assay, NAB2Δ mex67-5 yeast cells harboring the NAB2 URA3 maintenance plasmid and containing NAB2, nab2 ΔN, F72D or F73D LEU2 test plasmids were grown to saturation, serially diluted, and spotted on control and 5-FOA plates. The mex67-5 nab2 F73D double mutant cells show a slow growth phenotype compared with mex67-5 NAB2 cells, whereas mex67-5 nab2 F72D cells show growth similar to mex67-5 NAB2 cells (Fig. 6A). Further supporting a functional link between Mex67 and Nab2, mex67-5 nab2 ΔN cells are not viable. We confirmed the slow growth phenotype of mex67-5 nab2 F73D cells relative to mex67-5 NAB2 cells by measuring the optical density of the cells in liquid cultures and generating quantitative growth curves (Fig. 6B). In a similar manner, NAB2Δ yra1–8 yeast cells harboring the NAB2 URA3 maintenance plasmid and containing NAB2, nab2 ΔN, F72D or F73D LEU2 test plasmids were grown to saturation and spotted on control and 5-FOA plates. The yra1–8 nab2 F73D double mutant cells show a slow growth phenotype compared with yra1–8 NAB2 cells (Fig. 6A), whereas yra1–8 nab2 F72D cells show growth similar to yra1–8 NAB2 cells. These results demonstrate that nab2 F73D genetically interacts with mRNA export mutants mex67-5 and yra1–8 at their permissive temperatures. Together, the data suggest that although disrupting the Nab2 interaction with Mlp1 does not detectably impact yeast cell growth when the mRNA export pathway is fully functional, it does impact cell growth when mRNA export is suboptimal.
FIGURE 6.
The nab2 F73D mutant displays genetic interactions with mRNA export mutants mex67-5 and yra1–8 and mex67-5 nab2 F73D cells show nuclear poly(A) RNA accumulation. A, mex67-5 nab2 F73D cells or yra1–8 nab2 F73D cells have a slow growth phenotype. NAB2Δ mex67-5 or NAB2Δ yra1–8 cells maintained by a NAB2 URA3 plasmid and containing NAB2, nab2 ΔN, nab2 F72D, or nab2 F73D LEU2 test plasmids were grown to saturation, serially diluted in 10-fold dilutions and spotted on control and 5-FOA plates. Cells were grown at 30 °C. B, growth curve analysis of mex67-5 nab2 F73D cells confirms that they grow more slowly than mex67-5 NAB2 cells. mex67-5 NAB2Δ cells carrying only NAB2, nab2 F72D, or nab2 F73D plasmids were grown to saturation, diluted, and their optical density was measured at A600 for 40 h as described under “Experimental Procedures.” C, mex67-5 nab2 F73D cells show nuclear accumulation of poly(A) RNA. NAB2Δ mex67-5 cells carrying only NAB2, nab2 F72D, or nab2 F73D plasmids were grown in liquid culture at 30 °C and poly(A) RNA (Poly(A)) was visualized by FISH using an oligo(dT) probe as described under “Experimental Procedures.” Cells were stained with DAPI to visualize the position of the nucleus. Corresponding differential interference contrast (DIC) images are shown. FISH results were quantitated as described under “Experimental Procedures” to assess the mean percentage of cells with nuclear accumulation of poly(A) RNA within 10 fields of at least 43 cells each (indicated to the right of the DIC images). Variation in the percentage of cells with nuclear poly(A) RNA in each field of cells from the mean was assessed by calculating the standard deviation: mex67-5 NAB2, 18.6 ± 4.8%; mex67-5 nab2 F72D, 20.3 ± 6.0%; and mex67-5 nab2 F73D, 30.8 ± 6.9. Thus, mex67-5 nab2 F73D cells exhibit a 1.7-fold increase in nuclear poly(A) RNA accumulation relative to mex67-5 NAB2 cells, whereas mex67-5 nab2 F72D cells show only a slight 1.1-fold increase. The percentages of nuclear poly(A) RNA accumulation observed in mex67-5 nab2 F73D cells were typical of multiple independent experiments.
Given that the nab2 F73D cells showed a mild mRNA export defect in a wild-type background, we wished to assess whether cells expressing Nab2 F73D in a mex67-5 mRNA export mutant background, which showed a severe cell growth defect, exhibit a similar or more pronounced mRNA export defect. To address this possibility, we examined poly(A) RNA localization in the mex67-5 nab2 F73D cells and quantitated the percentage of cells with nuclear poly(A) RNA accumulation. Importantly, we examined the cells at the permissive temperature of 30 °C where mex67-5 cells show minimal nuclear accumulation of poly(A) RNA (8). NAB2Δ mex67-5 cells expressing NAB2, nab2 F72D, or nab2 F73D were processed for FISH with an oligo(dT) probe to visualize bulk poly(A) RNA. The mex67-5 nab2 F73D cells show a 1.7-fold increase in nuclear accumulation of poly(A) RNA compared with mex67-5 NAB2 cells (Fig. 6C). The combined results suggest that Nab2-F73D, which disrupts the Nab2/Mlp1 interaction, impacts mRNA export in vivo.
DISCUSSION
In this study, we have examined the functional significance of the interaction between the poly(A) RNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, to test a model where Nab2 facilitates targeting of mRNA for export via interaction with Mlp1. We find that disruption of the Nab2/Mlp1 interaction in vivo causes nuclear accumulation of poly(A) RNA. Furthermore, the nab2 F73D mutant shows genetic interactions with two genes essential for mRNA export, MEX67 and YRA1, supporting the hypothesis that Nab2 binding to Mlp1 contributes to efficient mRNA export. In addition, we have characterized the Nab2/Mlp1 interaction in detail and find that the Nab2-binding domain of Mlp1 (Mlp1-NBD) is sufficient to bind to the N-terminal domain of Nab2 (Nab2-N) and cause nuclear accumulation of poly(A) RNA, suggesting that Nab2 is an important molecular link between Mlp1 and poly(A) RNA. We also find that Mlp1-NBD binds to Nab2-N with micromolar affinity, indicating that the Nab2/Mlp1 interaction, like other mRNA export factor-nucleoporin interactions (59, 60), is comparatively weak. Together, the results are consistent with the notion that Nab2 facilitates targeting of mRNA for export via transient interaction with Mlp1 and acts in concert with other mRNA export factors to coordinate proper mRNA export.
We have mapped the Nab2-binding domain of Mlp1 (Mlp1-NBD) to a 183-amino acid region (residues 1586–1768) within CT-Mlp1. Analysis of the Mlp1-NBD protein sequence using Phyre (www.sbg.bio.ic.ac.uk/phyre/html/index.html) suggests that it forms a predominantly α-helical secondary structure, with the initial 90 residues (1586–1676) likely to form two helices (61). Sequence alignment of the Mlp1-NBD peptide with Mlp1 peptides from other Saccharomyces species highlights key hydrophobic residues within the first 90 residues of Mlp1-NBD that have been evolutionarily conserved and perhaps retained because they form α-helical secondary structure or serve as functional contact sites. It will be necessary to determine the structure of Mlp1-NBD and its complex with Nab2-N to define the Mlp1-NBD fold more precisely and identify the residues within Mlp1-NBD that make contact with the hydrophobic patch and residue Phe73 in Nab2-N.
Analysis of the binding between Nab2-N and Mlp1-NBD indicates that Mlp1-NBD binds to Nab2-N with a relatively weak affinity that is in the micromolar range (Kd ∼ 1.3 μm). Apparent binding affinities between the nuclear transport factor, NTF2, or the mRNA receptor heterodimer, TAP-p15, and FG repeat-containing nucleoporins have been measured and are also in the micromolar range (59, 60). For example, NTF2 binds to FG repeat nucleoporins with an apparent Kd of 1–4 μm and TAP-p15 binds to FG repeat nucleoporins with an apparent Kd of 4–11 μm (59, 60). Therefore, the binding constant of 1.3 μm observed between Nab2 and Mlp1 is comparable with the binding constants measured between other nuclear transport factors and FG-nucleoporins. Like the low-affinity transport factor-nucleoporin interactions, the rather weak Nab2/Mlp1 interaction is therefore likely to be transient in vivo. In this way, Nab2 may bind transiently to Mlp1 with a relatively high off-rate consistent with rapid transport of the properly assembled mRNA export complex.
Quantitative analysis of the interaction between Nab2-N-F73D and Mlp1-NBD shows that Mlp1-NBD binding to Nab2-N-F73D is too weak to measure. This result supports previous data showing that CT-Mlp1 cannot interact with Nab2-N-F73D in vitro and confirms that Nab2 residue Phe73 is critical for the Nab2/Mlp1 interaction. In addition, we find that Mlp1-NBD binds to a second Nab2 mutant, Nab2-N-F73W, with 1.5-fold higher affinity (Kd ∼ 0.8 μm) compared with wild-type protein (Kd ∼ 1.3 μm). This result strengthens the idea that large hydrophobic residues within Nab2-N are important for binding to Mlp1 and underscores the hydrophobic nature of the Nab2/Mlp1 interaction.
A key finding of this study is that expression of Nab2 F73D and the consequent disruption of the Nab2/Mlp1 interaction in vivo causes nuclear accumulation of poly(A) RNA and that this nab2 F73D mutant displays genetic interactions with alleles of two genes essential for mRNA export, MEX67 and YRA1. This result indicates that the Nab2/Mlp1 interaction is functionally important and contributes to mRNA export. nab2 F73D cells show a 2.5-fold increase in nuclear accumulation of poly(A) RNA relative to wild-type NAB2 cells, but do not exhibit a detectable growth defect. A potential explanation for this result is that the Nab2/Mlp1 interaction increases the rate and/or efficiency of one step of the mRNA export pathway that is not normally rate-limiting and therefore the Nab2/Mlp1 interaction is not absolutely required for cell growth in a wild-type background. mex67-5 nab2 F73D cells, however, show a 1.7-fold increase in nuclear accumulation of poly(A) RNA relative to mex67-5 NAB2 cells and clearly exhibit a decrease in cell growth. One possible explanation for the growth defect in mex67-5 nab2 F73D cells is that the Nab2/Mlp1 interaction becomes critical for a cellular function, such as efficient mRNA export, when additional steps in mRNA export and/or mRNA metabolism are compromised due to impairment of the Mex67 mRNA export receptor. A second potential explanation for the growth defect in mex67-5 nab2 F73D cells is that a threshold number of cells that exhibit mRNA export defects within the population must be reached before any effects on the overall growth properties of the culture are observed. Thus, the ∼30% of nab2 F73D mex67-5 cells that show nuclear accumulation of poly(A) RNA may be sufficient to impact the overall growth properties of the cell population, whereas the ∼8% of nab2 F73D cells that show nuclear poly(A) RNA accumulation may not be sufficient. Further study will be required to ascertain precisely how defects in cell growth are linked to impaired mRNA export.
The analysis of the Nab2 F73D mutant presented here and previous work on components of the mRNA export pathway suggest a model for Nab2 function in mRNA export in which Nab2 helps to target mRNA to the nuclear pore for export (Fig. 7A). In this simplified model, mRNA is co-transcriptionally loaded with the mRNA export adaptor, Yra1 (21, 62), and the poly(A) tails of the mRNA are specifically bound by Nab2 during or after polyadenylation (34–36). Yra1 binds to the mRNA export protein, Mex67 (18, 19), and recruits the Mex67/Mtr2 heterodimeric receptor to the mRNA (16). Mex67/Mtr2 then interacts with FG-nucleoporins at the pore to translocate the mRNA to the cytoplasm (17). Alongside Yra1 and Mex67, Nab2 binds to Mlp1 at the pore (38), via interaction between the Nab2-N hydrophobic patch and the Mlp1-NBD, to help concentrate the mRNA at the pore for interaction with other mRNA export factors and transport to the cytoplasm. The assembled mRNA complex containing Nab2 is then translocated through the NPC channel by Mex67/Mtr2, which interacts with FG-nucleoporins in the channel (17, 63). Upon reaching the cytoplasmic face of the nuclear pore, the mRNA export complex encounters the DEAD-box helicase, Dbp5 (64, 65), which is bound to and activated by the nucleoporin, Gle1 (66–69), and Dbp5 remodels the mRNP to trigger dissociation of Nab2 from the RNA (70), release of Mex67, and disassembly of the complex (71). Importantly, examination of Dbp5 function suggests that disassembly of the mRNA export complex is probably a rate-limiting step in mRNA export (70, 71). In a more detailed mRNA export model, several other mRNA export adaptors, hnRNPs, and mRNA processing factors can be added that aid in the concentration of mRNA at the nuclear face of the pore, the translocation of that mRNA through the pore and its remodeling and disassembly at the cytoplasmic face of the pore.
FIGURE 7.
Models for Nab2 function in mRNA export and the observed growth and mRNA export defects in mex67-5 nab2 F73D cells at the permissive temperature. A, model for Nab2 function in mRNA export in which Nab2 helps to target mRNA to the nuclear pore for export. Initially, mRNA is co-transcriptionally loaded with the mRNA export adaptor, Yra1 (21, 62), and bound by Nab2 at the poly(A) tail (34–36). Yra1 then binds to the mRNA export protein, Mex67 (18, 19), recruiting the Mex67/Mtr2 heterodimeric receptor to the mRNA (16) and Mex67/Mtr2 interacts with FG-nucleoporins at the pore to translocate the mRNA to the cytoplasm (17). Alongside Yra1 and Mex67, Nab2 binds to Mlp1 at the pore (38), via the Nab2-N/Mlp1-NBD interaction. The assembled mRNA complex containing Nab2 is then translocated through the NPC channel by Mex67/Mtr2 (17, 63). Upon reaching the cytoplasmic face of the nuclear pore, the mRNA export complex encounters the DEAD-box helicase, Dbp5 (64, 65), tethered and activated by the nucleoporin, Gle1 (66–69), which remodels the mRNP to trigger dissociation of Nab2 from the RNA (70), release of Mex67, and disassembly of the complex (71). B, model to explain the observed growth and mRNA export defects in mex67-5 nab2 F73D cells at the permissive temperature in which combined impairment of Mex67 and Nab2 reduces mRNA export. Here, the Mex67-5 mutant, which cannot interact with Mtr2 and FG-nucleoporins at the non-permissive temperature (8, 16), may also be impaired in binding to FG-nucleoporins at the permissive temperature, potentially resulting in a moderate reduction in the rate of translocation. Similarly, the Nab2 F73D mutant, which cannot interact with Mlp1, may reduce the rate of mRNA concentration at the pore. Combination of the reduction in mRNA translocation rate produced by Mex67-5 and reduction in the mRNA concentration rate caused by Nab2 F73D could become rate-limiting and result in a decreased rate of mRNA export coupled with nuclear accumulation of poly(A) RNA.
Fig. 7B presents a plausible model to account for the observed growth and mRNA export defects in mex67-5 nab2 F73D cells. We envisage that the Mex67-5 mutant, which cannot interact with Mtr2 and FG-nucleoporins at the non-permissive temperature (8, 16), may also be impaired in binding to FG-nucleoporins even at the permissive temperature. As the interaction between Mex67 and the FG-nucleoporins is necessary for the translocation step of mRNA export (17), the Mex67-5 mutant may reduce the rate of translocation through the nuclear pore. At the permissive temperature, this modest reduction in the rate of translocation produced by Mex67-5 may not impact the overall rate of mRNA export as the translocation step is probably not normally rate-limiting. Similarly, the Nab2 F73D mutant, which cannot interact with Mlp1, may reduce the rate of mRNA concentration at the pore. At the permissive temperature, this moderate reduction in the rate of mRNA concentration produced by Nab2 F73D may not affect the total rate of mRNA export as the concentration step is also probably not normally rate-limiting. However, the cumulative effect of impairing both mRNA translocation and concentration steps could be more marked and reduce the overall rate of mRNA export. At the permissive temperature, in mex67-5 nab2 F73D cells, the combined reduction in the mRNA translocation rate produced by Mex67-5 and the reduction in mRNA concentration rate caused by Nab2 F73D could become rate-limiting and result in a decreased rate of mRNA export coupled with nuclear accumulation of poly(A) RNA. Alternatively, in mex67-5 nab2 F73D cells, malformed mRNP complexes containing Nab2 F73D and Mex67-5 may be generated that are not recognized and removed by the surveillance machinery because Nab2 F73D does not bind to Mlp1 efficiently. Further work will be required to distinguish between these alternative models.
The combined data presented here highlight the importance of the Nab2/Mlp1 interaction in vivo and underscore the extent of cooperation between mRNA export factors, Nab2, Yra1, and Mex67, that is necessary for efficient mRNA export. In addition, the data suggest that although the mRNA export system in yeast can tolerate single component mutations, such as nab2 F73D, double component mutations, like mex67-5 nab2 F73D, produce a growth defect and significant nuclear accumulation of poly(A) RNA, consistent with the step in the export pathway in which they participate becoming rate-limiting, even under permissive conditions. The results of this study are therefore consistent with a model in which Nab2 helps to target the mRNP complex to the nuclear pore via interaction with Mlp1 to facilitate mRNA export. In this model, it is clear that Nab2 acts in concert with other mRNA export factors, particularly Yra1 and Mex67, to form an mRNP that is competent of export. Future studies on the genetic and physical interactions between Nab2 and other mRNA export components will help to elucidate the underlying mechanism of mRNA export in eukaryotic organisms.
Supplementary Material
Acknowledgments
We are most grateful to members of the Corbett laboratory for helpful discussions and comments. We also thank Richard P. Grant and Neil J. Marshall of the Stewart laboratory for structural analysis of the N-terminal domain of Nab2. In addition, we thank Susan R. Wente and Françoise Stutz for generously providing reagents.
This work was supported, in whole or in part, by a National Institutes of Health grant (to A. H. C.). This work was also supported by a Wellcome Trust Programme Grant (to M. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: hnRNP, heterogeneous nuclear ribonucleoprotein; NPC, nuclear pore complex; FG-nucleoporin, phenylalanine-glycine repeat-containing nuclear pore protein; Mlp1, myosin-like protein 1; CT-Mlp1, C-terminal domain of Mlp1; Nab2-N, N-terminal domain of Nab2; Mlp1-NBD, Nab2-binding domain of Mlp1; Tpr, translocated promoter region; GST, glutathione S-transferase; PBS, phosphate-buffered saline; BSA, bovine serum albumin; GFP, green fluorescent protein; DAPI, 4′,6-diamidino-2-phenylindole-dihydrochloride; 5-FOA, 5-fluoroorotic acid; FISH, fluorescence in situ hybridization; DTT, dithiothreitol.
References
- 1.Vinciguerra, P., and Stutz, F. (2004) Curr. Opin. Cell Biol. 16 285-292 [DOI] [PubMed] [Google Scholar]
- 2.Köhler, A., and Hurt, E. (2007) Nat. Rev. Mol. Cell. Biol. 8 761-773 [DOI] [PubMed] [Google Scholar]
- 3.Stutz, F., and Izaurralde, E. (2003) Trends Cell Biol. 13 319-327 [DOI] [PubMed] [Google Scholar]
- 4.Proudfoot, N. J., Furger, A., and Dye, M. J. (2002) Cell 108 501-512 [DOI] [PubMed] [Google Scholar]
- 5.Suntharalingam, M., and Wente, S. R. (2003) Dev. Cell 4 775-789 [DOI] [PubMed] [Google Scholar]
- 6.Fasken, M. B., and Corbett, A. H. (2005) Nat. Struct. Mol. Biol. 12 482-488 [DOI] [PubMed] [Google Scholar]
- 7.Sommer, P., and Nehrbass, U. (2005) Curr. Opin. Cell Biol. 17 1-8 [DOI] [PubMed] [Google Scholar]
- 8.Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R., and Hurt, E. (1997) EMBO J. 16 3256-3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan, W., Zolotukhin, A. S., Bear, J., Patenaude, D. J., and Felber, B. K. (2000) RNA (Cold Spring Harbor) 6 1762-1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herold, A., Klymenko, T., and Izaurralde, E. (2001) RNA (Cold Spring Harbor) 7 1768-1780 [PMC free article] [PubMed] [Google Scholar]
- 11.Grüter, P., Tabernero, C., vonKobbe, C., Schmitt, S., Saavedra, C., Bachi, A., Wilm, M., Felber, B. K., and Izaurralde, E. (1998) Mol. Cell 1 649-659 [DOI] [PubMed] [Google Scholar]
- 12.Braun, I. C., Herold, A., Rode, M., Conti, E., and Izaurralde, E. (2001) J. Biol. Chem. 276 20536-20543 [DOI] [PubMed] [Google Scholar]
- 13.Guzik, B. W., Levesque, L., Prasad, S., Bor, Y. C., Black, B. E., Paschal, B. M., Rekosh, D., and Hammarskjold, M. L. (2001) Mol. Cell. Biol. 21 2545-2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fribourg, S., Braun, I. C., Izaurralde, E., and Conti, E. (2001) Mol. Cell 8 645-656 [DOI] [PubMed] [Google Scholar]
- 15.Wiegand, H. L., Coburn, G. A., Zeng, Y., Kang, Y., Bogerd, H. P., and Cullen, B. R. (2002) Mol. Cell. Biol. 22 245-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Rosa, H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Panté, N., and Hurt, E. (1998) Mol. Cell. Biol. 18 6826-6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strässer, K., Bassler, J., and Hurt, E. (2000) J. Cell Biol. 150 695-706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strässer, K., and Hurt, E. (2000) EMBO J. 19 410-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stutz, F., Bachi, A., Doerks, T., Braun, I. C., Seraphin, B., Wilm, M., Bork, P., and Izaurralde, E. (2000) RNA (Cold Spring Harbor) 6 638-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zenklusen, D., Vinciguerra, P., Strahm, Y., and Stutz, F. (2001) Mol. Cell. Biol. 21 4219-4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zenklusen, D., Vinciguerra, P., Wyss, J. C., and Stutz, F. (2002) Mol. Cell. Biol. 22 8241-8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues, J. P., Rode, M., Gatfield, D., Blencowe, B. J., Carmo-Fonseca, M., and Izaurralde, E. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1030-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou, Z., Luo, M. J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. (2000) Nature 407 401-405 [DOI] [PubMed] [Google Scholar]
- 24.Gatfield, D., and Izaurralde, E. (2002) J. Cell Biol. 159 579-588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longman, D., Johnstone, I. L., and Cáceres, J. F. (2003) RNA (Cold Spring Harbor) 9 881-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hieronymus, H., and Silver, P. A. (2003) Nat. Genet. 33 155-161 [DOI] [PubMed] [Google Scholar]
- 27.Gilbert, W., and Guthrie, C. (2004) Mol. Cell 13 201-212 [DOI] [PubMed] [Google Scholar]
- 28.Henry, M., Borland, C. Z., Bossie, M., and Silver, P. A. (1996) Genetics 142 103-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singleton, D., Chen, S., Hitomi, M., Kumagai, C., and Tartakoff, A. (1995) J. Cell Sci. 108 265-272 [DOI] [PubMed] [Google Scholar]
- 30.Russell, I., and Tollervey, D. (1995) Eur. J. Cell Biol. 66 293-301 [PubMed] [Google Scholar]
- 31.Wilson, S. M., Datar, K. V., Paddy, M. R., Swedlow, J. R., and Swanson, M. (1994) J. Cell Biol. 127 1173-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green, D. M., Marfatia, K. A., Crafton, E. B., Zhang, X., Cheng, X., and Corbett, A. H. (2002) J. Biol. Chem. 277 7752-7760 [DOI] [PubMed] [Google Scholar]
- 33.Hector, R. E., Nykamp, K. R., Dheur, S., Anderson, J. T., Non, P. J., Urbinati, C. R., Wilson, S. M., Minvielle-Sebastia, L., and Swanson, M. S. (2002) EMBO J. 21 1800-1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly, S. M., Pabit, S. A., Kitchen, C. M., Guo, P., Marfatia, K. A., Murphy, T. J., Corbett, A. H., and Berland, K. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12306-12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson, J. T., Wilson, S. M., Datar, K. V., and Swanson, M. S. (1993) Mol. Cell. Biol. 13 2730-2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marfatia, K. A., Crafton, E. B., Green, D. M., and Corbett, A. H. (2003) J. Biol. Chem. 278 6731-6740 [DOI] [PubMed] [Google Scholar]
- 37.Lee, D. C., and Aitchison, J. D. (1999) J. Biol. Chem. 274 29031-29037 [DOI] [PubMed] [Google Scholar]
- 38.Green, D. M., Johnson, C. P., Hagan, H., and Corbett, A. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1010-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinciguerra, P., Iglesias, N., Camblong, J., Zenklusen, D., and Stutz, F. (2005) EMBO J. 24 813-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004) Cell 116 63-73 [DOI] [PubMed] [Google Scholar]
- 41.Strambio-de-Castillia, C., Blobel, G., and Rout, M. P. (1999) J. Cell Biol. 144 839-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cordes, V. C., Reidenbach, S., Rackwitz, H. R., and Franke, W. W. (1997) J. Cell Biol. 136 515-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrd, D. A., Sweet, D. J., Panté, N., Konstantinov, K. N., Guan, T., Saphire, A. C., Mitchell, P. J., Cooper, C. S., Aebi, U., and Gerace, L. (1994) J. Cell Biol. 127 1515-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hase, M. E., and Cordes, V. C. (2003) Mol. Biol. Cell 14 1923-1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feuerbach, F., Galy, V., Trelles-Sticken, E., Fromont-Racine, M., Jacquier, A., Gilson, E., Olivo-Marin, J. C., Scherthan, H., and Nehrbass, U. (2002) Nat. Cell Biol. 4 214-221 [DOI] [PubMed] [Google Scholar]
- 46.Bangs, P., Burke, B., Powers, C., Craig, R., Purohit, A., and Doxsey, S. (1998) J. Cell Biol. 143 1801-1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kölling, R., Nguyen, T., Chen, E. Y., and Botstein, D. (1993) Mol. Gen. Genet. 237 359-369 [DOI] [PubMed] [Google Scholar]
- 48.Kosova, B., Panté, N., Rollenhagen, C., Podtelejnikov, A., Mann, M., Aebi, U., and Hurt, E. (2000) J. Biol. Chem. 275 343-350 [DOI] [PubMed] [Google Scholar]
- 49.Grant, R. P., Marshall, N. J., Yang, J. C., Fasken, M. B., Kelly, S. M., Harreman, M. T., Neuhaus, D., Corbett, A. H., and Stewart, M. (2008) J. Mol. Biol. 376 1048-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szymczyna, B. R., Bowman, J., McCracken, S., Pineda-Lucena, A., Lu, Y., Cox, B., Lambermon, M., Graveley, B. R., Arrowsmith, C. H., and Blencowe, B. J. (2003) Genes Dev. 17 461-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suntharalingam, M., Alcazar-Roman, A. R., and Wente, S. R. (2004) J. Biol. Chem. 279 35384-35391 [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Second Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 53.Adams, A., Gottschling, D. E., Kaiser, C. A., and Stearns, T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 54.Bayliss, R., Littlewood, T., Strawn, L. A., Wente, S. R., and Stewart, M. (2002) J. Biol. Chem. 277 50597-50606 [DOI] [PubMed] [Google Scholar]
- 55.Wong, D. H., Corbett, A. H., Kent, H. M., Stewart, M., and Silver, P. A. (1997) Mol. Cell. Biol. 17 3755-3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boeke, J. D., Truehart, J., Natsoulis, G., and Fink, G. (1987) Methods Enzymol. 154 164-175 [DOI] [PubMed] [Google Scholar]
- 57.Aitchison, J. D., Blobel, G., and Rout, M. P. (1996) Science 274 624-627 [DOI] [PubMed] [Google Scholar]
- 58.Hodel, M. R., Corbett, A. H., and Hodel, A. E. (2001) J. Biol. Chem. 276 1317-1325 [DOI] [PubMed] [Google Scholar]
- 59.Bayliss, R., Ribbeck, K., Akin, D., Kent, H. M., Feldherr, C. M., Görlich, D., and Stewart, M. (1999) J. Mol. Biol. 293 579-593 [DOI] [PubMed] [Google Scholar]
- 60.Katahira, J., Straesser, K., Saiwaki, T., Yoneda, Y., and Hurt, E. (2002) J. Biol. Chem. 277 9242-9246 [DOI] [PubMed] [Google Scholar]
- 61.Bennet-Lovsey, R. M., Herbert, A. D., Sternberg, M. J., and Kelley, L. A. (2008) Proteins 70 611-625 [DOI] [PubMed] [Google Scholar]
- 62.Lei, E. P., Krebber, H., and Silver, P. A. (2001) Genes Dev. 15 1771-1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terry, L. J., and Wente, S. R. (2007) J. Cell Biol. 178 1121-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tseng, S. S., Weaver, P. L., Liu, Y., Hitomi, M., Tartakoff, A. M., and Chang, T. H. (1998) EMBO J. 17 2651-2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snay-Hodge, C. A., Colot, H. V., Goldstein, A. L., and Cole, C. N. (1998) EMBO J. 17 2663-2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strahm, Y., Fahrenkrog, B., Zenklusen, D., Rychner, E., Kantor, J., Rosbach, M., and Stutz, F. (1999) EMBO J. 18 5761-5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodge, C. A., Colot, H. V., Stafford, P., and Cole, C. N. (1999) EMBO J. 18 5778-5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alcazar-Roman, A. R., Tran, E. J., Guo, S., and Wente, S. R. (2006) Nat. Cell Biol. 8 711-716 [DOI] [PubMed] [Google Scholar]
- 69.Weirich, C. S., Erzberger, J. P., Flick, J. S., Berger, J. M., Thorner, J., and Weis, K. (2006) Nat. Cell Biol. 8 668-676 [DOI] [PubMed] [Google Scholar]
- 70.Tran, E. J., Zhou, Y., Corbett, A. H., and Wente, S. R. (2007) Mol. Cell 28 850-859 [DOI] [PubMed] [Google Scholar]
- 71.Lund, M. K., and Guthrie, C. (2005) Mol. Cell 20 645-651 [DOI] [PubMed] [Google Scholar]
- 72.Lange, A., Mills, R. E., Devine, S. E., and Corbett, A. H. (2008) J. Biol. Chem. 283 12926-12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schlenstedt, G., Saavedra, C., Loeb, J. D., Cole, C. N., and Silver, P. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 225-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.