Abstract
The S phase-specific activation of NEIL1 and not of the other DNA glycosylases responsible for repairing oxidatively damaged bases in mammalian genomes and the activation of NEIL1 by proliferating cell nuclear antigen (PCNA) suggested preferential action by NEIL1 in oxidized base repair during DNA replication. Here we show that NEIL1 interacts with flap endonuclease 1 (FEN-1), an essential component of the DNA replication. FEN-1 is present in the NEIL1 immunocomplex isolated from human cell extracts, and the two proteins colocalize in the nucleus. FEN-1 stimulates the activity of NEIL1 in vitro in excising 5-hydroxyuracil from duplex, bubble, forked, and single-stranded DNA substrates by up to 5-fold. The disordered region near the C terminus of NEIL1, which is dispensable for activity, is necessary and sufficient for high affinity binding to FEN-1 (KD ≅ 0.2 μm). The interacting interface of FEN-1 is localized in its disordered C-terminal region uniquely present in mammalian orthologs. Fine structure mapping identified several Lys and Arg residues in this region that form salt bridges with Asp and Glu residues in NEIL1. NEIL1 was previously shown to initiate single nucleotide excision repair, which does not require FEN-1 or PCNA. The present study shows that NEIL1 could also participate in strand displacement repair synthesis (long patch repair (LP-BER)) mediated by FEN-1 and stimulated by PCNA. Interaction between NEIL1 and FEN-1 is essential for efficient NEIL1-initiated LP-BER. These studies strongly implicate NEIL1 in a distinct subpathway of LP-BER in replicating genomes.
Reactive oxygen species are the most pervasive genotoxic agents in mammals (and other aerobic organisms), because they are continuously generated in vivo as by-products of respiration. Reactive oxygen species are also produced during various pathophysiologic states and oxidative metabolism of xenobiotics (1–3). Reactive oxygen species have been implicated in the etiology of cancer and various diseases and also in aging (4); these induce a plethora of oxidatively modified bases in DNA, together with strand breaks via oxidation of the deoxyribose (5). Many oxidized bases and abasic (AP)2 sites generated by enzymatic removal of these damaged bases are mutagenic and likely to be responsible for sporadic mutations leading to cancer and other diseases (4, 6). For example, 5-hydroxyuracil (5-OHU; an abundant DNA lesion generated by oxidative deamination of C) behaves like T, and its replication would lead to a G:C → A:T mutation. 5-OHU is excised from DNA by all mammalian glycosylases to varying degrees. Oxidized bases (and single-stranded breaks) in DNA are primarily repaired via the base excision repair (BER) pathway, a highly conserved process that is initiated with excision of the damaged base by a DNA glycosylase. Four oxidized base-specific DNA glycosylases, namely OGG1 (8-oxoguanine-DNA glycosylase 1), NTH1 (endonuclease III homolog), and NEIL1 and -2 (endonuclease VIII-like 1 and 2), have so far been characterized in mammalian cells and are responsible for repair of more than a few dozen oxidatively damaged bases. Based on structure and reaction mechanism, OGG1 and NTH1 belong to one family, whose prototype is Escherichia coli Nth (7, 8), whereas E. coli Nei is the prototype of NEIL1 and NEIL2, which we and others have recently identified and characterized (9–13).
Oxidized base-specific DNA glycosylases are bifunctional, with intrinsic AP lyase activity that cleaves the sugar phosphate bond of the AP site after base excision, leading to a DNA strand break at the damage site. The Nei-type enzymes NEIL1 and NEIL2 carry out successive βδ lyase reaction to cleave the deoxyribose from the AP site and produce 3′-phosphate and 5′-phosphate termini (14, 15). In contrast, OGG1 and NTH1 generate 5′-phosphate and 3′-phosphodeoxyribose (α,β-unsaturated aldehyde) via a β-lyase reaction at the AP site. We have recently established that NEIL-dependent BER in mammalian cells utilizes polynucleotide kinase (PNK) rather than the mammalian AP endonuclease, APE1 (14, 16).
We have recently shown that NEIL1 is activated by WRN (Werner syndrome protein), a member of the Rec Q family of helicases whose deficiency is associated with the aging phenotype and cancer predisposition (17), and by PCNA, a sliding clamp involved in DNA replication (18). Additionally, in collaboration with us, Lu's group recently showed that the Rad9-Rad1-Hus1 (9-1-1) complex, a DNA damage-activated alternative sliding clamp, also stably interacts with and stimulates NEIL1 (19). Our earlier data showing binary interaction between NEIL1 and DNA polymerase β (Polβ), DNA ligase IIIα, or XRCC1 suggested the presence of a BER complex in vivo that is likely to be formed after infliction of genomic damage (14). Subsequently, we showed that such a complex can be isolated from cell extracts that is competent to carry out complete repair of an oxidized base and whose level is enhanced after oxidative stress (16). Several other non-BER proteins have also been shown to be involved in NEIL-initiated BER; however, their precise in vivo role in repair is yet to be understood. For example, NEIL2 interacts with YB-1, a Y-box-binding protein, and we suggested that YB-1 may be required for the fine tuning of repair (20).
Mammalian cells have two distinct BER subpathways defined by the repair patch size (21, 22). Our repair studies with NEIL1 and NEIL2 so far suggested that NEIL-initiated repair involves Polβ that results in short patch or single nucleotide excision repair (SN-BER). However, based on our initial observation that among the four oxidized base-specific DNA glycosylases only NEIL1 expression is S-phase-dependent, we suspected that NEIL1 may be preferentially involved in repair of base damage in the replicative genome and thus utilizes replication DNA polymerases δ/ε (Polδ/ε) (9). The unique ability of NEILs to excise base lesions from single-stranded regions of DNA further supported this hypothesis (23). Our more recent studies showing stable interaction between NEIL1 and PCNA provided additional support for NEIL1 involvement in long patch (LP)-BER (18). LP-BER was discovered during repair studies of reduced AP sites whose cleavage by APE1 generates a 5′-blocking reduced AP group that could not be removed by the 5′-lyase activity of Polβ and requires the 5′-endo/exonuclease activity of flap endonuclease 1 (FEN-1) (22, 24, 25). In that case, 2–6 nucleotides are removed in addition to the 5′ reduced deoxyribose phosphate by the 5′-endo/exonuclease activity of FEN-1 (26, 27). The resulting gap is subsequently filled in by Polδ/ε or Polβ (22, 28). We show in this report that NEIL1 stably interacts with and is stimulated by FEN-1 and provide the first evidence for NEIL1-initiated LP-BER.
EXPERIMENTAL PROCEDURES
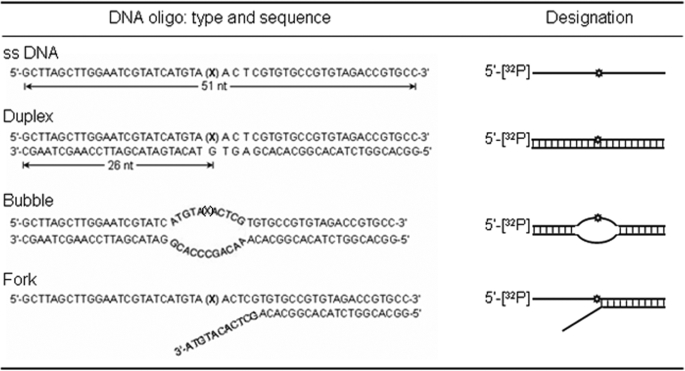
Oligonucleotide Substrates and Labeling—A 51-mer oligonucleotide containing 5-OHU at position 26 from the 5′-end and the complementary oligonucleotide containing G opposite the lesion and oligonucleotides for producing bubble and fork oligonucleotides (Table 1) were purchased from Invitrogen. Annealing was carried out by heating complementary oligonucleotides at 95 °C for 3–5 min, followed by slow cooling to room temperature. To produce 5′-32P-labeled substrates, the single-stranded oligonucleotides were labeled at their 5′ termini with [γ-32P]ATP (GE Healthcare) using T4 PNK before annealing. To generate bubble or fork substrates, appropriate oligonucleotide pairs were annealed (Table 1). The labeled substrates were purified to remove unincorporated label by size exclusion on a Sephadex G25 column.
TABLE 1.
Oligonucleotide substrates used in this study and their designations X, the oxidized base, 5-OHU.
Expression and Purification of Recombinant Proteins—Recombinant, untagged wild-type (WT) NEIL1 and its C-terminal deletion mutants were purified to homogeneity from E. coli as described previously (9, 17). The GST-fused NEIL1 domains, 312–349 and 312–389, and GST-fused FEN-1 C-terminal fragment (residues 328–380) were expressed in E. coli and purified from the cell extracts via affinity chromatography and eluting the bound proteins with 10 mm reduced glutathione. The proteins were digested with thrombin to cleave the GST tag, followed by removal of GST by chromatography on SP-Sepharose. Human WT FEN-1, its C-terminal deletion polypeptides with residues 1–336, 1–344, 1–355, and 1–367, as well as point mutants were purified as described previously (29, 30).
Site-directed Mutagenesis of NEIL1 and FEN-1—The point mutant constructs for NEIL1 were generated using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). Mutagenesis reactions were performed using pET22(b) vector harboring NEIL1 cDNA using appropriate primers. Mutations were confirmed by direct sequencing. After expressing in E. coli transformed with the plasmid, the recombinant His tag proteins were purified according to the protocol used for WT NEIL1.
Cell Culture and Co-immunoprecipitation—The human colororectal carcinoma cell line HCT116 was grown in McCoy's 5A medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml; Invitrogen). The cell cultures were individually transfected with C-terminally FLAG-tagged mammalian expression constructs for NEIL1 and empty FLAG vector (1 μg each) (31). Cells were lysed at 48 h after transfection by sonication (15% output, 10 s) in a lysis buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100) containing phosphatase inhibitors (Sigma) and protease inhibitor mixture (Roche Applied Science). The cell extracts were immunoprecipitated with anti-FLAG M2 antibody cross-linked to agarose beads (Sigma) and analyzed for the presence of FEN-1 in the FLAG-NEIL1 immunocomplex by immunoblotting with FEN-1 antibody (Bethyl Laboratories).
Far Western Analysis—For far Western analysis, proteins were separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane, treated with 6 m guanidine HCl, and then renatured with successive dilutions of guanidine HCl in PBS, 1 mm dithiothreitol (32). After blocking with 5% nonfat dry milk in PBS, 0.5% Tween 20, the membrane was incubated with interacting partner proteins in the blocking buffer for 3 h at 4°C before immunoblot analysis with appropriate antibodies.
In Vitro Affinity Pull-down (Co-elution) Assay—His tag WT or truncated FEN-1 mutants (10 pmol) were mixed with His-select nickel-agarose beads (20 μl; Sigma) for 1 h at 4°C and then incubated with untagged WT or deletion mutants of NEIL1 (20 pmol) for 1 h at 4 °C. The beads were washed extensively with 0.5% Tween 20 in PBS, and the bound proteins were eluted with SDS sample buffer for immunoblotting analysis.
GST pull-down assays were performed as described previously (17, 18, 33). Briefly, proteins were mixed with glutathione-Sepharose beads (20 μl) alone or bound to 10 pmol of GST-tagged NEIL1 domains (312–349 and 312–389), incubated with purified WT/truncated FEN-1 (5 pmol) in 0.5 ml, and the bound proteins were separated by SDS-PAGE and probed with anti-FEN-1 antibody. Affinity pull-down assays using CNBr-Sepharose beads were performed as previously published (30).
Surface Plasmon Resonance (SPR) Analysis—Interaction between NEIL1 and FEN-1 was analyzed by SPR using Biacore 3000 (GE Healthcare). Full-length NEIL1 (35 RU) was directly immobilized to a CM5 sensor chip via amine coupling. FEN-1 (62.5 nm to 2 μm) was injected over the sensor chip at 50 μl/min for 2 min in 10 mm Tris (pH 7.5), 0.1 m NaCl, 1 mm MgCl2, and 1 mm dithiothreitol. The response units were corrected for the blank from the reference flow cell and analyzed using a 1:1 (Langmuir) binding model (BIAevaluation software; GE Healthcare).
Fluorescence Analysis—Interaction of FEN-1 with the NEIL1 C-terminal peptide (residues 312–349, which lacks aromatic residues) was monitored by the increase in the intrinsic tryptophan fluorescence of FEN-1 (λex = 295 nm, λem = 300–450 nm) upon titration in an LS50 spectrofluorimeter (PerkinElmer Life Sciences). For all binding experiments, the proteins in 10 mm PBS, pH 7.5, 5% glycerol were incubated at 25 °C for 5 min. The binding constant KD was calculated by plotting ΔF (change in FEN-1 fluorescence at 345 nm) versus ligand concentration according to the equation ΔF =ΔFmax·[ligand]/KD + [ligand].
Analysis of NEIL1 Activity—The strand cleavage of substrate DNA after lesion excision by NEIL1 was carried out using 5-OHU-containing oligonucleotide substrates at 37 °C for 15 min in a 10-μl reaction mixture containing 40 mm Hepes, pH 7.5, 50 mm KCl, 100 μg/ml bovine serum albumin, and 5% glycerol. The reaction was stopped with formamide dye mix (80% formamide, 20 mm NaOH, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol), and the products were separated by electrophoresis in 20% polyacrylamide gel containing 8 m urea in 1× Tris borate-EDTA buffer, pH 8.4. The radioactivity was quantitated using a PhosphorImager and ImageQuant software (Amersham Biosciences).
For kinetic analysis, the substrate (2.5–120 nm) was incubated with 20 nm NEIL1 or NEIL1 plus FEN-1 (10 nm) for 4 min at 37 °C. Reactions were initiated by adding NEIL1 to the reaction mixture. Km, Vmax, and kcat were calculated from the linear range of the reaction by regression analysis using Sigma Plot.
In Vitro Reconstitution of BER— Ten pmol of 5-OHU-containing duplex oligonucleotide was incubated with various repair/replication proteins whose concentrations optimized for maximum product formation were as follows: 100 fmol each of NEIL1, PNK, Polβ, FEN-1, PCNA, and 200 fmol of DNA ligase IIIα. The reaction mixture (20 μl) also containing 1 mmol of ATP, 10 μmol of [α-32P]dNTP, and unlabeled dNTPs (25 mmol) was incubated for 30 min at 37 °C. Appropriate controls lacked various components.
Immunostaining and Confocal Microscopy—HCT116 cells (5 × 104/dish) transiently expressing FLAG-NEIL1 were grown to 50% confluence on glass coverslips (25 mm) in 5% CO2 at 37 °C. After washing with PBS, drying, and fixing in acetone-methanol 1:1 (v/v), the cells were incubated with rabbit FEN-1 antibody and counterstained with fluorescein-conjugated anti-rabbit IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The nuclear staining was carried out for 15 min with 10 ng/ml 4,6-diamidino-2-phenylindole dihydrochloride. Confocal microscopy was performed using a Zeiss LSM510 META system according to published protocol (17).
RESULTS
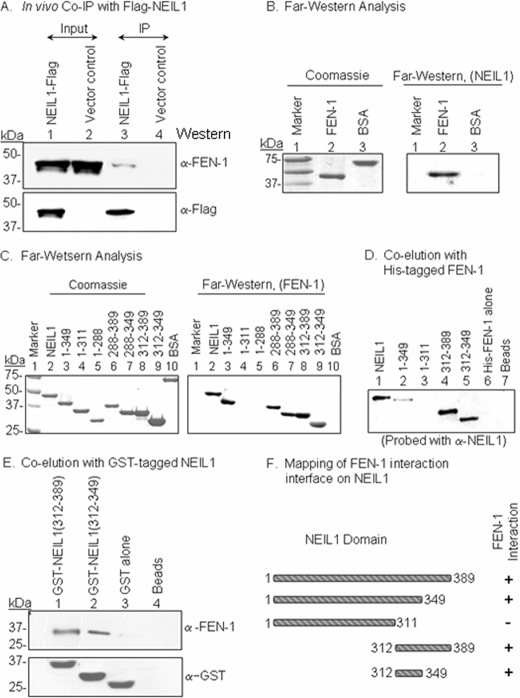
Association of NEIL1 with FEN-1 in Vivo—We had previously shown that NEIL1 forms a stable complex with PNK, Polβ, DNA ligase IIIα, and XRCC1 that is able to carry out efficient SN-BER of oxidized bases (14). We have now identified FEN-1 in the FLAG-NEIL1 immunocomplex from HCT 116 cells but not in the vector control (Fig. 1A).
FIGURE 1.
Identification of FEN-1 in the NEIL1 immunocomplex and mapping of the FEN-1-interacting interface on NEIL1. A, extracts of HCT116 cells expressing either FLAG-NEIL1 (lanes 1 and 3) or FLAG vector (lanes 2 and 4) or were immunoprecipitated with FLAG antibody (lanes 3 and 4). Top, Western analysis with FEN-1 antibody; bottom, anti-FLAG antibody. B, far Western analysis of FEN-1 interaction with NEIL1. FEN-1 (25 pmol) in the membrane was incubated with NEIL1 and subsequently immunoblotted with NEIL1 antibody. Bovine serum albumin was used as a control. C, far Western analysis of FEN-1 interaction with WT NEIL1 (lane 2), deletion mutants of NEIL1 (lanes 3–5), or GST-fused C-terminal domains of NEIL1 (lanes 6–9). Left, Coomassie staining after SDS-PAGE; Right, far Western immunoblot with FEN-1, probed with FEN-1 antibody. D, co-elution of WT (lane 1) and deletion mutants (lanes 2–5) of NEIL1 with His-tagged FEN-1 coupled to His-select nickel-beads. His-FEN-1 alone (lane 6) and beads alone (lane 7) served as controls. E, co-elution (pull-down) analysis of FEN-1 with GST-tagged C-terminal segments of NEIL1 coupled to glutathione-Sepharose beads. Top, immunoblot analysis of eluted FEN-1; bottom, immunoblot analysis for GST to confirm comparable levels of GST-tagged proteins. F, schematic representation of the NEIL1 interaction region for FEN-1.
In vivo association of NEIL1 and FEN-1 was further confirmed by immunocytochemical analysis. NEIL1 and FEN-1 were both localized to the nucleus of HCT116 cells transiently transfected with FLAG-NEIL1, and superimposition of confocal images fluorescence-stained for NEIL1 and FEN-1 showed substantial co-localization (supplemental Fig. 1).
Direct Interaction between FEN-1 and NEIL1; Mapping the Interacting Interface of NEIL1—The binary interaction between recombinant NEIL1 and FEN-1 was confirmed using far Western analysis and an affinity co-elution assay. NEIL1 interacted with FEN-1, whereas no interaction was observed with bovine serum albumin used as a control (Fig. 1B). We next mapped the NEIL1-interacting domain for FEN-1. All of the interacting partners for NEIL1 identified so far bind to its disordered C-terminal region (residues 289–389), which is dispensable for activity (17, 18). Using nested deletion mutants of NEIL1, we observed stable interaction of FEN-1 with residues 1–349 and 289–389, 289–349, 312–389, and 312–349 but not with 1–311 and 1–288 in NEIL1 (Fig. 1C). We concluded that the minimum interacting interface for FEN-1 lies within NEIL1 residues 312–349. Co-elution of NEIL1 and its deletion polypeptides with His-FEN-1 showed that FEN-1 stably binds with full-length and truncation mutants with residues 1–349, 312–389, and 312–349 but not with residues 1–311 (Fig. 1D). The beads or FEN-1 alone (lanes 5 and 6, respectively) were used as controls. Reciprocal co-elution of FEN-1 with GST-fused C-terminal peptides of NEIL1 showed that NEIL1 residues 312–349 by itself were necessary and independently sufficient for interaction (Fig. 1E). Control GST did not interact with FEN-1, as expected (Fig. 1E, lane 3).
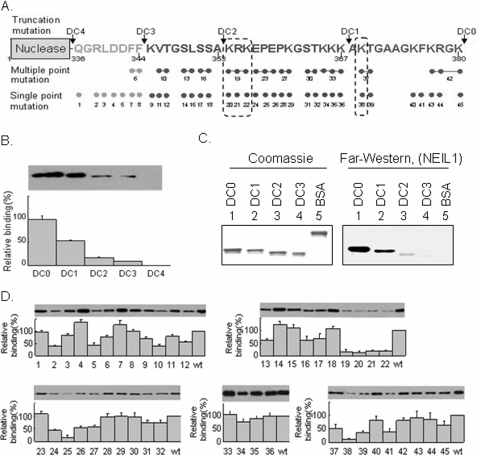
Mapping the NEIL1 Interaction Site to the FEN-1 C-terminal Region—Like NEIL1, FEN-1 has an unconserved region at its C terminus that is disordered (34) and is involved in interaction with several partners (30). We tested whether the same segment is also involved in interaction with NEIL1 by using WT FEN-1 (DC0) and its C-terminal deletion mutants named DC1 (residues 1–367), DC2 (residues 1–353), DC3 (residues 1–344), and DC4 (residues 1–336) (Fig. 2A). Deletion of the C terminus does not affect folding or stability of the FEN-1 core domain (30), but CNBr pull-down analysis showed that NEIL1 stably bound with DC0 (arbitrarily set at 100% relative binding) and DC1 (∼50%) and weakly with both DC2 (∼10%) and DC3 (∼5%) and did not interact with DC4 at all (Fig. 2B). This result was confirmed by far Western analysis (Fig. 2C). Taken together, these results suggest that NEIL1 stably interacts with residues 354–380 of FEN-1 and makes weak contacts with residues 336–353.
FIGURE 2.
Mapping of the interacting domain of FEN-1 and residues for NEIL1. A, design of the scanning mutations in the C-terminal region of FEN-1. WT (DC0), nested deletions, DC1, DC2, DC3, DC4, and single- and multiple-point mutations used in this study are represented. The arrows indicate the truncation sites. B, mapping the interaction domain of FEN-1 with NEIL1 by pull-down assay. NEIL1 was immobilized on Sepharose beads and incubated with WT and C-terminal deletion mutants of FEN-1. The bound proteins were separated on a 4–15% gradient SDS-PAGE and detected using FEN-1 antibody. C, far Western analysis of membrane-bound WT and deletion mutants of FEN-1 with NEIL1 in solution, as in Fig. 1C. D, identification of interacting residues in FEN-1 with NEIL1 by pull-down assay. Upper panels, immunoblots with FEN-1 antibody; lower panels, binding of single- and multiple-point mutants of FEN-1 with NEIL1, relative to WT FEN-1.
Identification of the FEN-1 NEIL1-binding Residues—We used site-specific mutants of FEN-1 to identify the residues in the interacting region that make contact with NEIL1. The residues in the FEN-1 C-terminal region critical for interaction with PCNA, APE1, WRN, Rad9-Rad1-Hus1 (9-1-1), and endonuclease G were recently identified using alanine-scanning mutagenesis (30). These mutant proteins showed nuclease activity comparable with that of the WT protein (30). Similar studies were carried out to identify the FEN-1 residues involved in NEIL1 binding (Fig. 2A). Several residues in the 356–380 region of FEN-1 were important for NEIL1 interaction. Specifically, single Ala mutants 20 (Lys354), 21 (Arg355), 22 (Lys356), 25 (Pro358), and 38 (Lys369) and multiple mutant 19 (Lys356/Arg357/Lys357) showed less than 25% affinity for NEIL1 relative to the WT protein (Fig. 2D). The low affinity of the Pro358 mutant could be due to the lack of proper folding rather than specific involvement of Pro in binding. In general, the Lys and Arg residues in the FEN-1 C-terminal region were found to be involved in binding, suggesting electrostatic interactions with acidic residues in NEIL1.
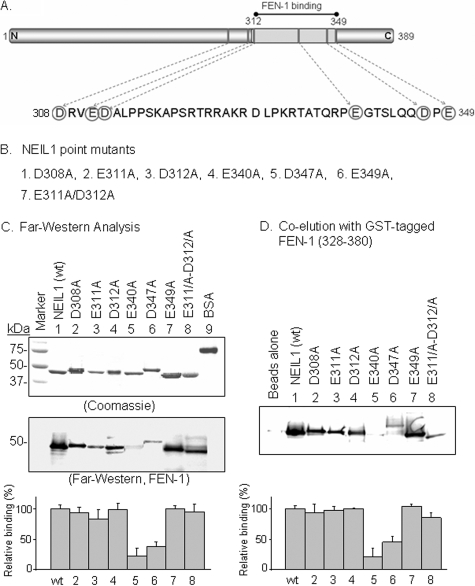
Identification of FEN-1 Interaction Residues in NEIL1—We then generated point mutants of Asp and Glu in residues 311–349 of NEIL1 by substitution with Ala (Fig. 3, A and B). The mutant proteins were purified to near homogeneity, as indicated by SDS-PAGE analysis (Fig. 3C, top). All of these mutants had activity comparable with that of WT NEIL1 (data not shown). Far Western and GST pull-down analysis showed that the NEIL1 mutants E340A and D347A had less than 50% binding relative to WT NEIL1 (Fig. 3C). However, the other mutants showed similar binding as WT NEIL1. Thus, basic residues in FEN-1 (Lys354, Arg355, Lys356, and Lys369) and acidic residues in NEIL1 (Glu340 and Asp347) contribute to the binding between these two proteins via salt bridges.
FIGURE 3.
Identification of critical NEIL1 residues for FEN-1 interaction. A, circled Asp and Glu residues were mutated singly or multiply to Ala, as indicated in B. C, far Western analysis, membrane-bound WT (lane 1), and point mutants (lanes 2–8) of NEIL1 with FEN-1 in solution (middle), probed with anti-FEN-1. SDS-PAGE of purified NEIL1 proteins (top). Relative binding of FEN-1 is given at the bottom. D, co-elution of NEIL1 with GST-tagged FEN-1 C-terminal fragment (residues 328–380) bound to glutathione-Sepharose beads; the eluted NEIL1 was immunoblotted using NEIL1 antibody. The bottom panel shows the relative binding.
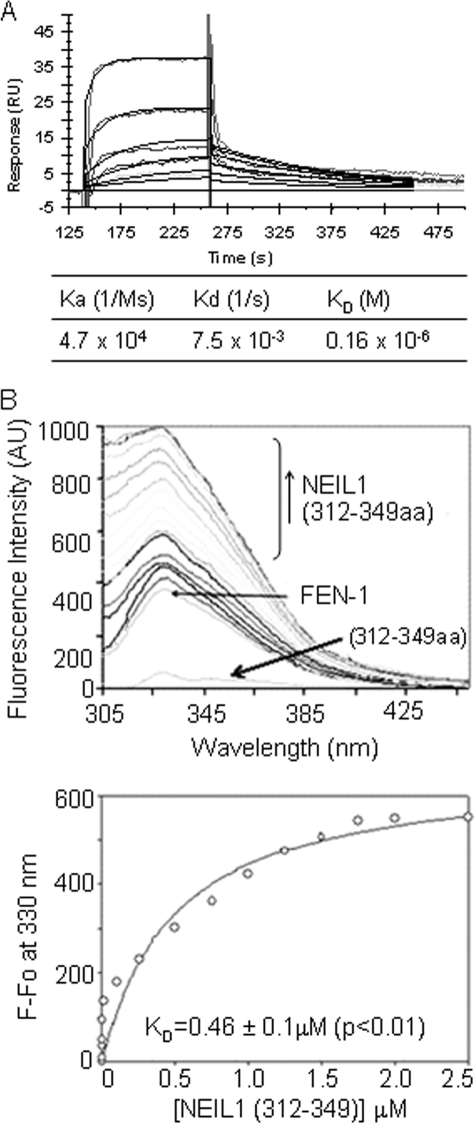
Binding Affinity Analysis—We utilized Biacore SPR analysis to further characterize the interaction between NEIL1 and FEN-1. By directly immobilizing WT NEIL1 onto the sensor chip and injecting increasing concentrations of FEN-1, we could observe an increase in binding in the sensorgrams (Fig. 4A). We calculated the kinetic parameters based on a 1:1 (Langmuir) binding model using BIAevaluation software. FEN-1 showed high affinity for NEIL1 (KD ≅ 0.16 μm).
FIGURE 4.
Quantitation of NEIL1 affinity for FEN-1. A, SPR analysis as described under “Experimental Procedures.” The kinetic parameters were calculated using BIAevaluation software. B, intrinsic fluorescence analysis. Plot of changes in the intrinsic tryptophan fluorescence of FEN-1 alone or when titrated with NEIL1 C-terminal peptide (residues 312–349) versus wavelength. The binding parameters are shown (bottom).
We also measured changes in the intrinsic fluorescence of FEN-1 in the presence of the NEIL1 C-terminal segment. The fluorescence emission maximum of FEN-1 increased upon titration with the NEIL1 peptide (residues 312–349) before reaching a plateau. The NEIL1 peptide lacking aromatic residues contributed negligibly to the fluorescence signal (Fig. 4B). An apparent dissociation constant (KD) of 0.46 μm was calculated for the binding of FEN-1 with the NEIL1 peptide (residues 312–349), which provides a minimum interaction interface for FEN-1 (Fig. 4B). The binding constants calculated from SPR using full-length NEIL1 and intrinsic fluorescence studies using truncated NEIL1 polypeptides are not identical, suggesting additional contacts with full-length NEIL1 beyond residues 312–349. On the other hand, the small discrepancy could also be due to differences in the buffer conditions used in these experiments. In any case, both methods indicate strong binding between NEIL1 and FEN-1.
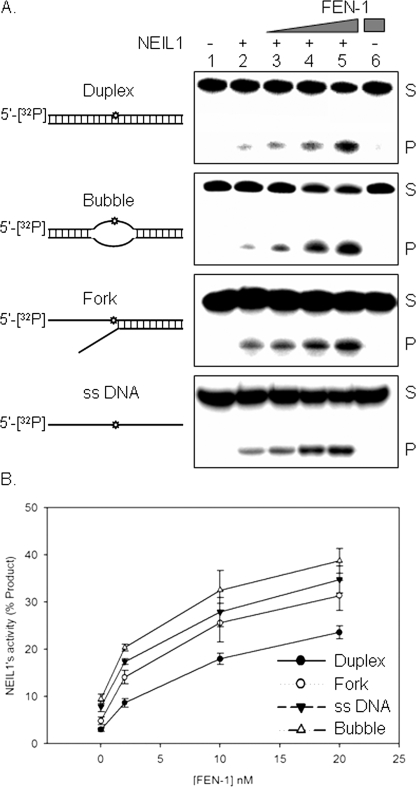
FEN-1 Increases NEIL1 Turnover—To assess the physiological significance of FEN-1 interaction with NEIL1, we examined its effect on NEIL1 activity. The strand incision activity of NEIL1 on a 5-OHU-containing duplex oligonucleotide substrate was stimulated ∼2-fold by FEN-1 at a 1:1 molar ratio (Fig. 5A). Bubble and fork oligonucleotides showed similar levels of stimulation (∼3-fold at a 1:1 molar ratio), whereas single-stranded DNA showed the highest stimulation (∼5-fold). As expected, FEN-1 had no activity on the 5-OHU-containing oligonucleotide (Fig. 5A, lane 6), and the interference from 5′-endo/exo activities of FEN-1 was minimal under the experimental conditions.
FIGURE 5.
Stimulation of NEIL1 DNA glycosylase activity by FEN-1. A, effect of FEN-1 (2–20 nm) on NEIL1 (10 nm) with duplex, bubble, fork and single-stranded DNA substrates containing 5-OHU (Table 1). S and P, the 5′ 32P-labeled uncleaved substrate (51 nt) and NEIL1 incision product (25 nt), respectively. Lane 6, FEN-1 alone as control for possible substrate cleavage by FEN-1. B, dose-dependent stimulation of NEIL1 activity for various substrates by FEN-1.
We calculated the kinetic parameters of NEIL1 using a fork substrate in the presence or absence of FEN-1 (Fig. 5B and Table 2). The lack of change in the Km in the presence of FEN-1 suggested that its binding did not affect the loading of NEIL1 onto the substrate. However, an ∼6-fold increase in the kcat indicated that FEN-1 enhanced release of the product from NEIL1 after lesion excision and strand cleavage.
TABLE 2.
Kinetic parameters for NEIL1 and its stimulation by FEN-1
| Reaction | Km | kcat | kcat/Km |
|---|---|---|---|
| nm | ×10–2/min | ×10–2/nm/min | |
| NEIL1 only | 15.9 ± 1.1 | 3.7 ± 0.9 | 0.23 |
| NEIL1 plus FEN-1 | 11.8 ± 0.7 | 22.7 ± 1.8 | 1.9 |
We reciprocally tested the effect of NEIL1 on the activity of FEN-1. FEN-1 has several nuclease activities, including 5′-flap endonuclease, 5′-exonuclease, and gap cleavage, all of which may have important biological roles (35). The presence of NEIL1 stimulated the 5′-flap endonuclease activity of FEN-1 slightly (∼1.5-fold) over FEN-1 alone (data not shown).
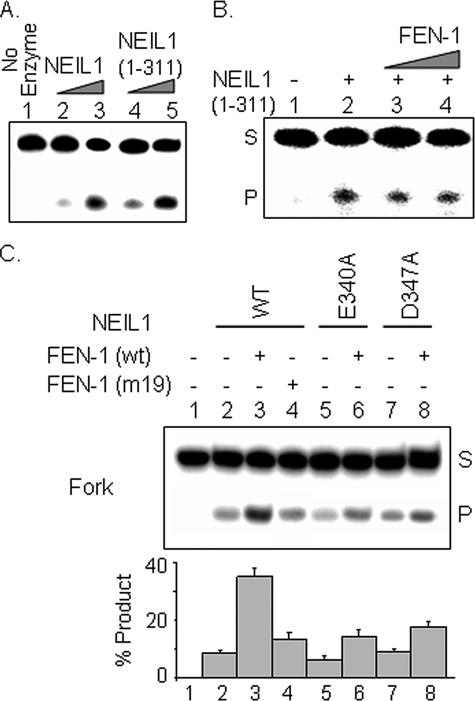
Interaction with FEN-1 Is Required for NEIL1 Stimulation— The truncated NEIL1-(1–311) lacking the primary FEN-1 binding interface exhibited similar DNA glycosylase activity as WT NEIL1 (Fig. 6A), indicating the dispensability of the C-terminal 78 residues for activity and confirming our previous observations that the deletion mutants 1–349 and 1–288 had activities comparable with that of WT NEIL1 (14). Here we show that FEN-1 failed to stimulate the 1–311 mutant (Fig. 6B). Furthermore, the NEIL1 point mutants E340A and D347A were stimulated to a much lesser extent than the WT NEIL1 (Fig. 6C). Similarly, the FEN-1 mutant 19 (K354/R355/K356) lacking affinity for NEIL1 stimulated its activity only slightly (Fig. 6C, lane 4). Together, these results indicate that stable binding of FEN-1 is essential for enhanced turnover of NEIL1.
FIGURE 6.
Physical association of NEIL1 and FEN-1 is required for stimulation of NEIL1 activity. A, activities of 2 and 20 nm each of WT NEIL1 and 1–311 mutant with fork substrate. B, effect of FEN-1 on DNA glycosylase activity of NEIL1 C-terminal deletion peptide (residues 1–311). C, effect of noninteracting point mutants of FEN-1 on NEIL1 activity with fork oligonucleotide. The E340A and D347A point mutants of NEIL1 were stimulated to a lesser extent by FEN-1 in comparison with WT NEIL1.
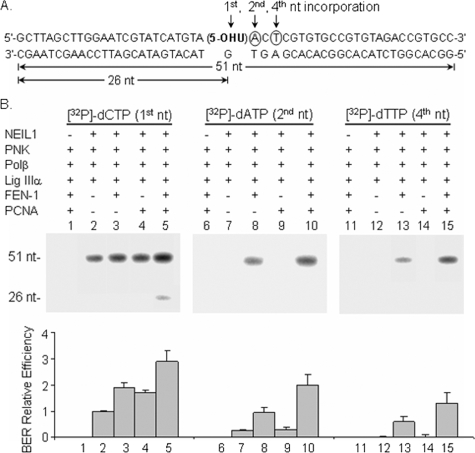
FEN-1 Mediates NEIL1-initiated LP-BER—We investigated next if NEIL1 initiates strand displacement and long patch repair of the 5-OHU lesion site in the presence of FEN-1. We measured incorporation of [32P]dAMP, the second nt, or [32P]-dTMP, fourth nt, 3′ to the 5-OHU lesion site (Fig. 7, lanes 6–15). In the absence of FEN-1, no strand displacement synthesis and LP-BER was observed (Fig. 7B, lanes 7 and 12), even in the presence of PCNA (lanes 9 and 14). However, in the presence of FEN-1, incorporation at the second and fourth nt was observed for NEIL1-initiated repair (lanes 8 and 13). Moreover, PCNA stimulated NEIL1-initiated LP-BER about 2-fold (Fig. 7B, lanes 10 and 15). We recently showed that PCNA stimulates NEIL1 by enhancing the loading of NEIL1 onto its substrate (18). In addition, PCNA was also shown to interact with Polβ and FEN-1 and to stimulate their activities (36, 37). Together, these results show that NEIL1 participates in both SN- and LP-BER subpathways, depending on the presence of FEN-1. This is the first experimental evidence for NEIL1-initiated LP-BER.
FIGURE 7.
FEN-1-mediated, NEIL1-initiated LP-BER and its stimulation by PCNA. A, incorporation of radiolabeled [32P]dCMP indicates SN-BER and LP-BER, whereas incorporation of [32P]dAMP and [32P]dTMP indicate LP-BER with strand displacement from the lesion site with patch size of 2 and 4 nucleotides, respectively. B, effects of FEN-1 and PCNA on NEIL1-initiated BER. Left, incorporation of [32P]dCMP; middle, incorporation of [32P]dAMP; right, incorporation of [32P]dTMP. In the absence of FEN-1, no strand displacement synthesis was observed with NEIL1 (lanes 7 and 12), even in the presence of PCNA (lanes 9 and 14). FEN-1-mediated, NEIL1-initiated LP-BER was stimulated by PCNA (lanes 10 and 15). The bottom panel shows a graphic representation of the relative BER efficiencies.
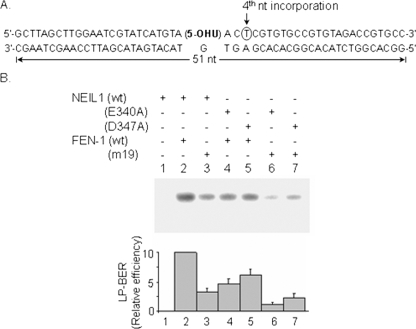
We then assessed the requirement of NEIL1-FEN-1 interaction in LP-BER by using the point mutants of both proteins that abrogate physical contacts. As shown in Fig. 8, the ability of FEN-1 mutant-19 (K354/R355/K356) to carry out NEIL1-initiated LP-BER was reduced by about 60% relative to WT FEN-1. Similarly, the relative LP-BER efficiency was significantly decreased for NEIL1 mutants E340A and D347A versus WT NEIL1. Thoroughly disrupting the interaction between NEIL1 and FEN-1 profoundly reduced the efficiency of LP-BER (Fig. 8, lanes 6 and 7).
FIGURE 8.
NEIL1 interaction with FEN-1 is essential for efficient NEIL1-initiated strand displacement repair synthesis. A, incorporation of labeled [32P]dTMP was used as a measure of strand displacement repair. B, inhibition of NEIL1-initiated LP-BER in the presence of point mutants of NEIL1 and/or FEN-1 with decreased interaction (lanes 3–7). PNK, Polβ, DNA ligase IIIα, and PCNA were present in all reactions.
DISCUSSION
The emergence of the “cellular interactome” concept has led to a new paradigm in which collaboration of multiple proteins involving binary interactions in a coordinated fashion leads to enhanced efficiency of metabolic pathways. It is now evident that the cellular processes for repair of both endogenous and induced genomic damage are essential for maintaining genomic integrity and homeostasis, which involve dynamic and complex interactions among a multitude of proteins and with DNA in the repair interactome.
It was previously shown that NEILs carry out SN-BER mediated by PNK, Polβ, DNA ligase IIIα, and XRCC1 (14, 16). Both NEIL1 and NEIL2 interact pairwise with downstream repair proteins in the SN-BER: Polβ, DNA ligase IIIα, and XRCC1. Both NEIL immunocomplexes from cell extracts also contain PNK, which does not directly interact with the NEILs (14, 16). We have recently documented that NEIL1 interacts with and is stimulated by PCNA and WRN, proteins involved in DNA replication (17, 18). These observations led us to postulate that NEIL1 is preferentially involved in replication-associated BER. Cross-talk between DNA replication and DNA repair proteins may be critical, because the repair of oxidatively damaged bases in replicating DNA is more urgent than in quiescent genomes, in order to prevent mutation fixation. The oxidized base lesions do not invariably block replication. Thus, the repair co-opts the replication machinery for repair synthesis. This model suggests that the early enzymes in repair (a DNA glycosylase or APE1) recruit the replication proteins via physical interaction. Here we show direct interaction of NEIL1 with FEN-1 in carrying out LP-BER.
Stimulation of NEIL1 by FEN-1 can be explained by the following two possible scenarios. FEN-1, an indispensable participant in DNA replication that is required for Okazaki primer removal, collaborates with NEIL1 only when damage is encountered in the template. However, since both NEIL1 and FEN-1 are stimulated by PCNA (18, 36), a more likely scenario is that NEIL1 and FEN-1 are components of a larger complex with other replication-associated proteins. Our previous observation of the presence of PCNA and RFC in the NEIL1 immunocomplex supports this possibility (18). However, the model of a larger preformed complex raises the question of stoichiometry and steric interference. This makes mapping the interaction interfaces among these proteins vital to understanding the nature of these complexes. We have shown here that NEIL1 binds to residues 344–380, at the C terminus of FEN-1 (Fig. 2), whereas PCNA binds to residues 325–334 (30, 34, 38). Thus, distinct regions of FEN-1 are involved in its interaction with PCNA and NEIL1. We propose that PCNA-bound NEIL1 initiates repair of an oxidized base by recognizing the lesion in the leading or lagging strand template ahead of the replicative DNA polymerase. Initiation of repair interrupts the fork movement, and the collapsed replication fork forms a “chicken foot” structure (39, 40). This repair is likely to occur via LP-BER, where NEIL1 collaborates with FEN-1, PCNA, and possibly other replication proteins. We are currently exploring the interactions of NEIL1 with Polδ, RFC, and DNA ligase I, all of which are involved in both cellular DNA replication and LP-BER. Earlier studies showed that LP-BER could also be mediated by Polβ and FEN-1 (37). The stable interaction of Polβ with FEN-1 and PCNA further supports involvement of Polβ in LP-BER (36, 37, 41).
Interestingly, the co-localization studies showed substantial foci of red (FEN-1 alone) and yellow (FEN-1 co-localized with NEIL1; supplemental Fig. 1). This observation is consistent with multiple roles for FEN-1, only some of which involve NEIL1.
The present study, together with previous studies in our laboratory (14, 17, 18), shows that the disordered C-terminal extension of NEIL1 acts as the common interaction interface for diverse partners. It is important to note that E. coli Nei, the prokaryotic ortholog of NEIL1, does not contain this extension, underscoring the physiological significance of protein-protein interactions in mammalian repair (42). The interaction of NEIL1 with multiple partners via a common interaction interface (C terminus) could be explained based on the conformational flexibility of its C-terminal region. The flexible interface in a protein may provide high specificity/low affinity binding, rapid protein turnover, and the opportunity for multiple targets (43). We mapped the NEIL1 interaction segment to residues 354–380 at the C terminus of FEN-1, with weak contact to residues 344–354 (Fig. 2). As with NEIL1, the C-terminal region of human FEN-1 appears to be an unfolded extension with disordered structure, which is absent in the prokaryotic members of the FEN-1 family (30) and was proposed to regulate activity through modulation of binding to DNA and interactions with other proteins (44, 45). As in the case of NEIL1, distinct sequences in the FEN-1 C-terminal region are involved in interaction with WRN, Bloom protein, and PCNA. PCNA binds to residues 335–344, whereas WRN binds to residues 363–380 (44, 46).
Finally, we have observed that NEIL2 also binds to FEN-1, but with no apparent functional consequences.3 Interestingly, despite many common interacting partners and enzymatic properties, NEIL1 and NEIL2 have distinct binding specificities, supporting our hypothesis that NEIL1 is preferentially involved in replication-associated BER, whereas NEIL2 may be involved in transcription-coupled BER. The differential functional association of NEIL1 and NEIL2 with FEN-1 is consistent with the above scenario.
Acknowledgments
We thank Alex Kurosky and Steven Smith of the Biomedical Resources Facility at the University of Texas, Medical Branch (UTMB), for protein and DNA sequencing services. Thanks are due to Rodrigo Maillard and James Lee (UTMB) for fluorescence spectroscopy and data analysis. We also thank Eugene Knutson (UTMB Optical Imaging Core Facility) for confocal microscopy. We express gratitude to Priscilla Cooper and Miaw-Sheue Tsai (Lawrence Berkeley National Laboratory, CA) for the Biacore facility. We thank members of the Mitra Laboratory for various scientific discussions, Wanda Smith for expert secretarial assistance, and David Konkel for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health United States Public Health Service Research Grants P01 CA092584, R01 CA81063, R01 CA53791, and P30 ES06676 (to S. M.); R01 CA102271 (to T. K. H.); and R01 CA073764 (to B. H. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: AP, abasic; 5-OHU, 5-hydroxyuracil; FEN-1, flap endonuclease 1; PNK, polynucleotide kinase; Polδ, DNA polymerase δ; Polβ, DNA polymerase β; RFC, replication factor C; BER, base excision repair; SN- and LP-BER, single nucleotide and long patch base excision repair, respectively; SPR, surface plasmon resonance; WT, wild-type; PCNA, proliferating cell nuclear antigen; GST, glutathione S-transferase; PBS, phosphate-buffered saline; nt, nucleotide.
Hegde et al., unpublished experiment.
References
- 1.Cadet, J. L., and Brannock, C. (1998) Neurochem. Int. 32 117-131 [DOI] [PubMed] [Google Scholar]
- 2.Breen, A. P., and Murphy, J. A. (1995) Free Radic. Biol. Med. 18 1033-1077 [DOI] [PubMed] [Google Scholar]
- 3.Grisham, M. B., Hernandez, L. A., and Granger, D. N. (1986) Am. J. Physiol. 251 G567-G574 [DOI] [PubMed] [Google Scholar]
- 4.Ames, B. N., Shigenaga, M. K., and Hagen, T. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7915-7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dizdaroglu, M., Jaruga, P., Birincioglu, M., and Rodriguez, H. (2002) Free Radic. Biol. Med. 32 1102-1115 [DOI] [PubMed] [Google Scholar]
- 6.Krokan, H. E., Nilsen, H., Skorpen, F., Otterlei, M., and Slupphaug, G. (2000) FEBS Lett. 476 73-77 [DOI] [PubMed] [Google Scholar]
- 7.Radicella, J. P., Dherin, C., Desmaze, C., Fox, M. S., and Boiteux, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8010-8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda, S., Biswas, T., Roy, R., Izumi, T., Boldogh, I., Kurosky, A., Sarker, A. H., Seki, S., and Mitra, S. (1998) J. Biol. Chem. 273 21585-21593 [DOI] [PubMed] [Google Scholar]
- 9.Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., Dizdaroglu, M., and Mitra, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3523-3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazra, T. K., Kow, Y. W., Hatahet, Z., Imhoff, B., Boldogh, I., Mokkapati, S. K., Mitra, S., and Izumi, T. (2002) J. Biol. Chem. 277 30417-30420 [DOI] [PubMed] [Google Scholar]
- 11.Bandaru, V., Sunkara, S., Wallace, S. S., and Bond, J. P. (2002) DNA Repair (Amst.) 1 517-529 [DOI] [PubMed] [Google Scholar]
- 12.Morland, I., Rolseth, V., Luna, L., Rognes, T., Bjoras, M., and Seeberg, E. (2002) Nucleic Acids Res. 30 4926-4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takao, M., Kanno, S., Shiromoto, T., Hasegawa, R., Ide, H., Ikeda, S., Sarker, A. H., Seki, S., Xing, J. Z., Le, X. C., Weinfeld, M., Kobayashi, K., Miyazaki, J., Muijtjens, M., Hoeijmakers, J. H., van der Horst, G., and Yasui, A. (2002) EMBO J. 21 3486-3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiederhold, L., Leppard, J. B., Kedar, P., Karimi-Busheri, F., Rasouli-Nia, A., Weinfeld, M., Tomkinson, A. E., Izumi, T., Prasad, R., Wilson, S. H., Mitra, S., and Hazra, T. K. (2004) Mol. Cell 15 209-220 [DOI] [PubMed] [Google Scholar]
- 15.Zharkov, D. O., Shoham, G., and Grollman, A. P. (2003) DNA Repair (Amst.) 2 839-862 [DOI] [PubMed] [Google Scholar]
- 16.Das, A., Wiederhold, L., Leppard, J. B., Kedar, P., Prasad, R., Wang, H., Boldogh, I., Karimi-Busheri, F., Weinfeld, M., Tomkinson, A. E., Wilson, S. H., Mitra, S., and Hazra, T. K. (2006) DNA Repair (Amst.) 5 1439-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, A., Boldogh, I., Lee, J. W., Harrigan, J. A., Hegde, M. L., Piotrowski, J., de Souza Pinto, N., Ramos, W., Greenberg, M. M., Hazra, T. K., Mitra, S., and Bohr, V. A. (2007) J. Biol. Chem. 282 26591-26602 [DOI] [PubMed] [Google Scholar]
- 18.Dou, H., Theriot, C. A., Das, A., Hegde, M. L., Matsumoto, Y., Boldogh, I., Hazra, T. K., Bhakat, K. K., and Mitra, S. (2008) J. Biol. Chem. 283 3130-3140 [DOI] [PubMed] [Google Scholar]
- 19.Guan, X., Bai, H., Shi, G., Theriot, C. A., Hazra, T. K., Mitra, S., and Lu, A. L. (2007) Nucleic Acids Res. 35 2463-2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das, S., Chattopadhyay, R., Bhakat, K. K., Boldogh, I., Kohno, K., Prasad, R., Wilson, S. H., and Hazra, T. K. (2007) J. Biol. Chem. 282 28474-28484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortini, P., Pascucci, B., Parlanti, E., Sobol, R. W., Wilson, S. H., and Dogliotti, E. (1998) Biochemistry 37 3575-3580 [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, Y., Kim, K., Hurwitz, J., Gary, R., Levin, D. S., Tomkinson, A. E., and Park, M. S. (1999) J. Biol. Chem. 274 33703-33708 [DOI] [PubMed] [Google Scholar]
- 23.Dou, H., Mitra, S., and Hazra, T. K. (2003) J. Biol. Chem. 278 49679-49684 [DOI] [PubMed] [Google Scholar]
- 24.Klungland, A., and Lindahl, T. (1997) EMBO J. 16 3341-3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frosina, G., Fortini, P., Rossi, O., Carrozzino, F., Raspaglio, G., Cox, L. S., Lane, D. P., Abbondandolo, A., and Dogliotti, E. (1996) J. Biol. Chem. 271 9573-9578 [DOI] [PubMed] [Google Scholar]
- 26.Lieber, M. R. (1997) BioEssays 19 233-240 [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589-615 [DOI] [PubMed] [Google Scholar]
- 28.Pascucci, B., Stucki, M., Jonsson, Z. O., Dogliotti, E., and Hubscher, U. (1999) J. Biol. Chem. 274 33696-33702 [DOI] [PubMed] [Google Scholar]
- 29.Liu, R., Qiu, J., Finger, L. D., Zheng, L., and Shen, B. (2006) Nucleic Acids Res. 34 1772-1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, Z., Chavez, V., Singh, P., Finger, L. D., Hang, H., Hegde, M. L., and Shen, B. (2008) J. Mol. Biol. 377 679-690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhakat, K. K., Mokkapati, S. K., Boldogh, I., Hazra, T. K., and Mitra, S. (2006) Mol. Cell. Biol. 26 1654-1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman, L., Moorthy, N. C., Murthy, K. G., Manley, J. L., Bustin, M., and Prives, C. (1998) Genes Dev. 12 462-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leppard, J. B., Dong, Z., Mackey, Z. B., and Tomkinson, A. E. (2003) Mol. Cell. Biol. 23 5919-5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakurai, S., Kitano, K., Yamaguchi, H., Hamada, K., Okada, K., Fukuda, K., Uchida, M., Ohtsuka, E., Morioka, H., and Hakoshima, T. (2005) EMBO J. 24 683-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen, B., Singh, P., Liu, R., Qiu, J., Zheng, L., Finger, L. D., and Alas, S. (2005) BioEssays 27 717-729 [DOI] [PubMed] [Google Scholar]
- 36.Dianova, I. I., Bohr, V. A., and Dianov, G. L. (2001) Biochemistry 40 12639-12644 [DOI] [PubMed] [Google Scholar]
- 37.Podlutsky, A. J., Dianova, I. I., Podust, V. N., Bohr, V. A., and Dianov, G. L. (2001) EMBO J. 20 1477-1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gary, R., Ludwig, D. L., Cornelius, H. L., MacInnes, M. A., and Park, M. S. (1997) J. Biol. Chem. 272 24522-24529 [DOI] [PubMed] [Google Scholar]
- 39.Heller, R. C., and Marians, K. J. (2006) Nature 439 557-562 [DOI] [PubMed] [Google Scholar]
- 40.Heller, R. C., and Marians, K. J. (2006) Nat. Rev. Mol. Cell. Biol. 7 932-943 [DOI] [PubMed] [Google Scholar]
- 41.Kedar, P. S., Kim, S. J., Robertson, A., Hou, E., Prasad, R., Horton, J. K., and Wilson, S. H. (2002) J. Biol. Chem. 277 31115-31123 [DOI] [PubMed] [Google Scholar]
- 42.Hegde, M. L., Hazra, T. K., and Mitra, S. (2008) Cell Res. 18 27-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilser, V. J., and Thompson, E. B. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8311-8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma, S., Sommers, J. A., Gary, R. K., Friedrich-Heineken, E., Hubscher, U., and Brosh, R. M., Jr. (2005) Nucleic Acids Res. 33 6769-6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapados, B. R., Hosfield, D. J., Han, S., Qiu, J., Yelent, B., Shen, B., and Tainer, J. A. (2004) Cell 116 39-50 [DOI] [PubMed] [Google Scholar]
- 46.Sharma, S., Otterlei, M., Sommers, J. A., Driscoll, H. C., Dianov, G. L., Kao, H. I., Bambara, R. A., and Brosh, R. M., Jr. (2004) Mol. Biol. Cell 15 734-750 [DOI] [PMC free article] [PubMed] [Google Scholar]