Abstract
Because of their antagonistic catalytic functions, protein-tyrosine phosphatases (PTPs) and protein-tyrosine kinases act together to control phosphotyrosine-mediated signaling processes in mammalian cells. However, unlike for protein-tyrosine kinases, little is known about the cellular substrate specificity of many PTPs because of the lack of appropriate methods for the systematic and detailed analysis of cellular PTP function. Even for the most intensely studied, prototypic family member PTP1B many of its physiological functions cannot be explained by its known substrates. To gain better insights into cellular PTP1B function, we used quantitative MS to monitor alterations in the global tyrosine phosphorylation of PTP1B-deficient mouse embryonic fibroblasts in comparison with their wild-type counterparts. In total, we quantified 124 proteins containing 301 phosphotyrosine sites under basal, epidermal growth factor-, or platelet-derived growth factor-stimulated conditions. A subset of 18 proteins was found to harbor hyperphosphorylated phosphotyrosine sites in knock-out cells and was functionally linked to PTP1B. Among these proteins, regulators of cell motility and adhesion are overrepresented, such as cortactin, lipoma-preferred partner, ZO-1, or p120ctn. In addition, regulators of proliferation like p62DOK or p120RasGAP also showed increased cellular tyrosine phosphorylation. Physical interactions of these proteins with PTP1B were further demonstrated by using phosphatase-inactive substrate-trapping mutants in a parallel MS-based analysis. Our results correlate well with the described phenotype of PTP1B-deficient fibroblasts that is characterized by an increase in motility and reduced cell proliferation. The presented study provides a broad overview about phosphotyrosine signaling processes in mouse fibroblasts and, supported by the identification of various new potential substrate proteins, indicates a central role of PTP1B within cellular signaling networks. Importantly the MS-based strategies described here are entirely generic and can be used to address the poorly understood aspects of cellular PTP function.
Members of the protein-tyrosine phosphatase (PTP)1 family of enzymes function in a coordinated way with protein-tyrosine kinases (PTKs) to control tyrosine phosphorylation signaling pathways that regulate a number of fundamental physiological processes. Alterations in the equilibrium of cellular tyrosine phosphorylation are the cause of multiple human diseases, such as cancer, diabetes, immune deficiencies, and many others. Our knowledge of the physiological and molecular functions of PTKs greatly exceeds that of PTPs, and for most PTPs the identities of their cellular substrates have remained elusive (1). In previous studies, our group undertook various efforts to address this problem and identified cellular substrates for a number of PTPs. We found that receptor-like PTPs (RPTPs), such as RPTPα, dephosphorylate the mature form of the insulin receptor (2) and that RPTPκ regulates β-catenin signaling (3, 4). In addition, we discovered substrates for non-receptor PTPs, like the receptor tyrosine kinase (RTK) negative regulator SIRPα for SHP2 (5), HER2 for BDP1 (6), and the mitogen-activated protein kinases (MAPKs) ERK1, -2, and -5 for the cytosolic form of PTP-SL (7, 8). Moreover in an early study on the prototypic PTP family member PTP1B, we demonstrated a dephosphorylation activity of this phosphatase on precursor forms of several RTKs due to the co-localization of these molecules in the endoplasmic reticulum (9).
Although PTP1B is one of the best understood phosphatases, many of its physiological functions cannot be explained with the already known substrates. PTP1B has received much attention because of its proposed role in type 2 diabetes, obesity, immunity, and cancer (10, 11). Fundamental insights into its physiological function were achieved with loss-of-function mouse genetics models. PTP1B-deficient mice exhibit resistance to diabetes and obesity (12, 13) because of increased activity of its direct substrates, the insulin receptor in liver and skeletal muscle and leptin-induced JAK2 in the hypothalamus (14), respectively. In tumorigenesis, different functions were assigned to PTP1B and depended on its site of action. For example, a tumor-suppressive function was identified in B-cell lymphomas in p53/PTP1B double null mice (15), whereas an oncogenic function was described in mammary tumorigenesis by two laboratories crossing transgenic mice expressing activated forms of ErbB2 with PTP1B null mice (16, 17).
Most of the known PTP1B substrates have been characterized using the substrate-trapping approach (18, 19). This technique takes advantage of catalytically inactive PTP1B mutants, which can form stable interactions with their substrates and then allow subsequent purification of these proteins. However, this approach usually relied on immunoblotting with anti-phosphotyrosine (Tyr(P)) antibodies for protein detection and was therefore rather biased toward the identification of known tyrosine phosphorylated proteins as PTP substrates. A more systematic way to identify proteins interacting with substrate-trapping mutants involves the use of MS-based protein identification (20). Studies were published that used MS analysis in combination with substrate-trapping mutants to identify substrates for PTPs like RPTPσ (21) or PTPN21 (22). Interacting proteins were resolved with SDS-PAGE, and after protein staining, individual bands were excised and analyzed by MS.
Recently modern mass spectrometers have become available that are extremely sensitive and permit measurements with high mass accuracy, such as the hybrid linear ion trap/Fourier transform mass spectrometer (LTQ-FT) (23) or the linear ion trap/orbitrap mass spectrometer (LTQ-Orbitrap) (24). Using nanoscale capillary (nano) ESI LC-MS/MS, these instruments allow an extensive analysis of highly complex cellular proteomes containing a broad range of proteins of different abundances. In addition, these instruments are ideally suited for the analysis of post-translational modifications such as protein phosphorylation on serine, threonine, or tyrosine residues (25). Combination of phosphopeptide MS analysis with quantitative proteomics methodologies, like stable isotope labeling by amino acids in cell culture (SILAC), provides a robust and powerful approach to monitor phosphorylation-dependent signaling in different biological states at the same time (26, 27). With this method, relative protein expression levels and changes in phosphorylation on specific phosphosites can be determined on a global proteome-wide scale (28).
Here we report a new proteomics strategy that uses quantitative mass spectrometry to identify potential cellular substrates for a member of the protein-tyrosine phosphatase family. To this end, we used SILAC labeling to analyze alterations in Tyr(P) signaling of PTP1B-deficient mouse fibroblasts in comparison with wild-type cells under basal, EGF-, or PDGF-stimulated conditions. Additional information about the physical interaction of proteins that are hyperphosphorylated in knock-out (KO) cells to the active site of PTP1B was obtained by using substrate-trapping mutants in a parallel MS analysis. This combined analysis allowed us to propose novel substrates of PTP1B that provide a molecular rationale to understand the described phenotype of its KO mouse embryonic fibroblasts. The MS-based methods and strategies described here also can be used to study other members of the classical PTP family and might have considerable potential to improve the knowledge about their cellular functions.
EXPERIMENTAL PROCEDURES
Antibodies—
Anti-Tyr(P) specific 4G10 monoclonal antibody was purified from hybridoma supernatants using HiTrap protein A columns (GE Healthcare) on an ÁKTA Explorer 100 system (GE Healthcare). The antibody directed against PLCγ1 was from Transduction Laboratories, polyclonal anti-cortactin antibody was raised against the keyhole limpet hemocyanin-coupled peptide KGRYGLFPANYVELRQ, and anti-Fer antibody was described elsewhere (29).
Cell Culture and Ligand Stimulation—
PTP1B wild-type (PTP-1B+/+) and KO (PTP-1B−/−) cells immortalized by infection with the simian virus 40 (SV40) large T antigen have been described previously (30). Both cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics (5 mg/ml penicillin/streptomycin; Invitrogen). For SILAC experiments, cells were grown for 7 days in media containing either normal l-arginine (Arg0) and l-lysine (Lys0) (Sigma), l-[U-13C6,14N4]arginine (Arg6) and l-[2H4]lysine (Lys4), or l-[U-13C6,15N4]arginine (Arg10) and l-[U-13C6,15N2]lysine (Lys8) (Eurisotop). The labeling efficiency was determined to be greater than 97% (data not shown). Cells were starved for 3 h prior to ligand stimulation with EGF (50 ng/ml, 5 min; Peprotech) or PDGF (20 ng/ml, 5 min; Peprotech).
Cell Lysis and Anti-Tyr(P) Immunoprecipitation—
Cells were grown for 2 days in 12 × 15-cm dishes per SILAC label (2 × 106 cells/dish were seeded) and were lysed for 20 min in ice-cold lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 1 mm EDTA, 1 mm sodium orthovanadate, 1 mm PMSF, 0.1 μg/ml aprotinin, 10 mm NaF). Lysates were precleared by centrifugation at 16,500 × g for 15 min. Protein amount determination was performed using the BCA assay (Pierce). In SILAC experiments, cell lysates were mixed 1:1 (double labeling) or 1:1:1 (triple labeling) after determination of protein amounts. For immunoprecipitation, 200 μg of anti-Tyr(P) 4G10 antibody were added together with 40 μl of protein A-Sepharose (GE Healthcare) to mixed cell lysates containing up to 20 mg of total protein of each label and incubated for 4 h at 4 °C. Precipitates were washed four times with lysis buffer, and precipitated proteins were eluted twice with urea buffer (7 m urea, 2 m thiourea, 50 mm HEPES, pH 7.5, 1% n-octyl glucoside) for 10 min at 37 °C. For Western blot analysis up to 500 μg of total protein was used per immunoprecipitation, and precipitated proteins were eluted with SDS sample buffer.
In Vitro Substrate Trapping—
The catalytic domain of PTP1B (residues 1–321) was linked to GST fusion protein, and PTP1B mutants D181A and D181A/Q262A were generated as described before (18, 31). The recombinant wild-type and PTP1B mutant fusion proteins were expressed in Escherichia coli and purified with GST-Sepharose (GE Healthcare). In vitro substrate-trapping assays were performed as described previously (19). Briefly cells were grown for 2 days in eight 15-cm dishes (2 × 106 cells/plate) stimulated for 30 min with 100 μm pervanadate and lysed for 20 min in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 10% glycerol, 1 mm EDTA, 1 mm PMSF, 0.1 μg/ml aprotinin, 5 mm iodoacetic acid). DTT was added to a final concentration of 10 mm, and samples were incubated for 10 min. Lysates were precleared by centrifugation at 16,500 × g for 15 min. 10 μg of recombinant PTP1B wild-type enzyme or substrate-trapping mutant were coupled to GSH beads (GE Healthcare) and incubated for 4 h at 4 °C with lysates containing 10 mg of total protein. Trapped proteins were washed three times with HNTG buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100). For subsequent MS analysis, proteins were eluted with urea buffer (7 m urea, 2 m thiourea, 50 mm HEPES, pH 7.5, 1% n-octyl glucoside) for 10 min at 37 °C.
In-solution Protein Digestion—
After anti-Tyr(P) immunoprecipitation or substrate trapping, precipitated proteins were denatured in the urea buffer described above, and protein amount was measured using the Bradford assay (Bio-Rad). Proteins were reduced by adding 2 mm DTT (final concentration) for 45 min at 25 °C, and thiols were carboxymethylated with 5.5 mm iodoacetamide for 30 min at room temperature. Endoproteinase Lys-C (Wako) was added in an enzyme/substrate ratio of 1:100 (usually 1–3 μg of Lys-C), and the proteins were digested for 4 h at room temperature. The resulting peptide mixtures were diluted with water to reach a final urea concentration below 2 m. For double digestion, modified trypsin (sequencing grade, Promega) was added in an enzyme/substrate ratio of 1:100 (usually 1–3 μg of trypsin), and the digest was incubated at room temperature overnight. Trypsin activity was quenched by adding TFA to a final concentration of 1%.
Titansphere Enrichment of Phosphopeptides—
After trypsin digest phosphopeptides were enriched using Titansphere chromatography (TiO2) columns as described previously (28, 32). Peptide samples were diluted 1:6 with 30 g/liter 2,5-dihydroxybenzoic acid (DHB) in 80% ACN, 0.1% TFA. 5 μg of TiO2 beads (GL Sciences Inc.) were washed once with elution buffer (NH3 water in 20% ACN, pH 10.5) and equilibrated with washing buffer (50% ACN, 0.1% TFA). TiO2 beads were loaded with DHB by washing with loading buffer (6 g/liter DHB in 15% ACN). Peptide samples were loaded onto TiO2 beads for 30 min at room temperature on a rotating wheel. Subsequently beads were washed three times with washing buffer, and bound phosphopeptides were eluted twice for 10 min at room temperature with 50 μl of elution buffer. The eluates were filtered through home-made C8 STAGE Tips in 200-μl pipette tips. 30 μl of 80% ACN, 0.5% acetic acid was applied to the STAGE Tips (33) after filtering, and the flow-through was combined with the filtered sample. The pH value of the sample was adjusted with TFA to a value of approximately pH 7, and the eluates were concentrated in a vacuum concentrator. Before MS analysis, 5% ACN and 0.1% TFA (final concentrations) were added to the samples.
In-gel Protein Digestion—
Mixed protein lysates from PTP1B wild-type and KO cells (300 μg) or precipitated proteins after anti-Tyr(P) immunoprecipitation or substrate trapping were separated by SDS-PAGE using NuPAGE Novex Bis-Tris gels (Invitrogen) according to the manufacturer's instructions. The gel was stained with Coomassie Blue using the Colloidal Blue Staining kit (Invitrogen). The resulting lane was cut into 15 slices (protein expression analysis) or 10 slices (anti-Tyr(P) immunoprecipitation or substrate trapping), which were then subjected to in-gel digestion essentially performed as described previously (34). Gel slices were cut into small cubes and washed with 50 mm ammonium bicarbonate (ABC), 50% ethanol until the cubes were fully destained. Gel cubes were dehydrated with ethanol and rehydrated with 50 mm ABC containing 10 mm DTT. Proteins were reduced for 1 h at 56 °C. Resulting free thiol groups were then alkylated by adding 55 mm iodoacetamide in 50 mm ABC for 1 h at 25 °C in the dark. Gel pieces were washed twice with a 50 mm ABC, 50% ethanol solution; dehydrated with 100% ethanol; and dried in a vacuum concentrator. Each gel fraction was rehydrated in 50 mm ABC solution containing 0.4 μg of trypsin, and samples were incubated at 37 °C overnight. Supernatants were transferred to new tubes, and residual peptides were extracted out of the gel pieces by double incubation with 30% ACN in 3% TFA and double incubation with 100% ACN. All extracts were combined, and ACN was evaporated in a vacuum concentrator. Subsequently samples were desalted using home-made reverse phase (RP) C18 STAGE Tip columns (33), and the eluted peptides were used for MS analysis.
Nano-LC-MS/MS Analysis—
All peptide samples were separated by on-line RP nano-LC and analyzed by electrospray MS/MS. Using an Agilent 1100 nanoflow system (Agilent Technologies), samples were injected onto a 15-cm RP, fused silica capillary column (inner diameter, 75 μm; packed in house with 3-μm ReproSil-Pur C18-AQ, Dr. Maisch GmbH). The LC setup was connected to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems). Loaded peptides were eluted with 140-min gradients from 5 to 40% ACN in 0.5% acetic acid. Data-dependent acquisition was performed on the LTQ-Orbitrap using Xcalibur 2.0 software in the positive ion mode. The instrument was recalibrated in real time by co-injection of an internal standard from ambient air into the C-trap (“lock mass option”) (35). Survey spectra were acquired with a resolution of 60,000 in the orbitrap. In parallel, up to five of the most intense multiple charged ions per cycle were isolated, fragmented, and analyzed in the LTQ part of the instrument. To improve phosphopeptide analysis, the multistage activation option in the software was enabled, and the neutral loss species at 97.97, 48.99, or 32.66 m/z below the precursor ion were activated for 30 ms during fragmentation (pseudo-MS3) (36).
Peptide Identification Using the MASCOT Search Engine—
Raw MS data were processed using the in-house software Raw2msm (35), and peak lists were analyzed with the MASCOT search engine (Version 2.1.0, Matrix Science). All tandem mass spectra were searched against an in-house curated decoy IPI mouse protein database of the International Protein Index (Decoy IPI Version 3.18) containing forward and reversed sequences. In addition, contaminants such as human keratins, porcine trypsin, and endoproteinase Lys-C were included in this database. Using a concatenated target/decoy database allows defining a cutoff score threshold that permits a false-positive rate of peptide identification of less than 1% (p < 0.01) (37). The absolute average mass accuracy and its S.D. was 0.80/1.15 ppm for all peptides measured with the LTQ-Orbitrap. Therefore, the maximum allowed mass deviations for peptide identification on MS peaks were 5 ppm and on MS2 peaks were 0.5 Da. Carbamidomethylcysteine was set as fixed modification, and oxidized methionine, phosphorylation of Ser/Thr/Tyr, protein N-acetylation, N-pyroglutamate, and the SILAC labels (Lys4, Lys8, Arg6, and Arg10) were searched as variable modifications. Full tryptic specificity was required, and up to three “missed cleavages” were allowed. The instrument setting for the MASCOT search was specified as “ESI-Trap.”
Post-translational Modification (PTM) Scoring and Peptide Quantitation Using MSQuant—
All spectra and sequence assignments obtained from MASCOT were imported into the MSQuant software. The PTM scoring algorithm implemented into this program is a probability-based scoring system for phosphorylation site assignment within peptide sequences and was described before (28). Most of the analyzed phosphorylated peptides had a localization probability for their phosphogroups of 1. In only a few cases with p values of 0.5, a phosphogroup was assigned to two residues with the same probability. After the phosphorylation sites were assigned to peptide sequences and PTM scores were determined, phosphopeptide quantitation was performed using MSQuant. For each SILAC doublet or triplet, MSQuant calculated the corresponding extracted ion chromatogram values, and all assignments made for quantitation were displayed and manually validated (28, 38). To ensure reproducibility of the quantitation data, the comparison of PTP1B wild-type with KO cells in normal growth medium was repeated three times, and other SILAC experiments were performed twice. Final peptide quantitation ratios were determined by calculating the average value from the ratios of single measurements.
To analyze the distribution of -fold changes in tyrosine phosphorylation between PTP1B wild-type and KO cells, Tyr(P) peptides were plotted in a histogram against their -fold changes, and a Gaussian regression analysis was performed using the GraphPad Prism 5 software. The obtained values for the mean and S.D. of the Gaussian curve were used to calculate p values and to determine the significance of -fold changes across the whole data set by using the probability mass function (normal density function) of Microsoft Excel.
RESULTS
PTP1B Deficiency Leads to Alterations in the Phosphotyrosine Proteome of Mouse Embryonic Fibroblasts—
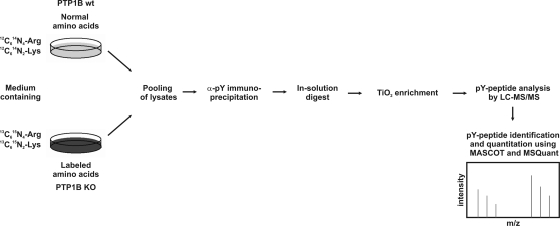
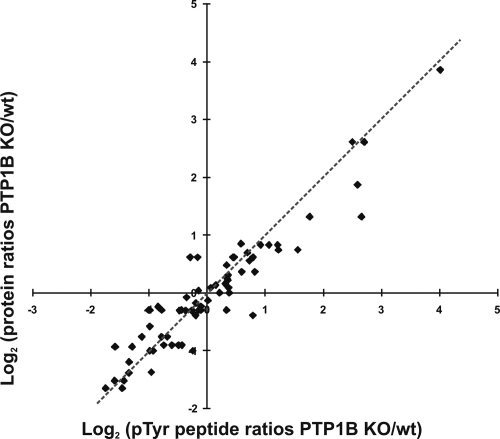
PTP1B-deficient mouse embryonic fibroblasts are a well established cell culture system to study the substrate specificity of this prototypical phosphatase. In the absence of the enzyme, tyrosine phosphorylation of potential direct substrates should be increased compared with wild-type cells. To study alterations in phosphotyrosine signaling of PTP1B-deficient cells, we used a quantitative proteomics approach based on high resolution LC-MS upon differential labeling of wild-type and KO fibroblasts with SILAC media (39) (Fig. 1). After cell lysis, cellular tyrosine phosphorylated proteins were immunoprecipitated with anti-Tyr(P) antibodies and digested in solution, and the resulting phosphopeptides were further enriched using TiO2 beads (32). This two-step procedure resulted in highly phosphopeptide-enriched samples from which more than 100 distinct tyrosine phosphorylation sites could be identified in single LC-MS experiments on a linear ion trap/orbitrap mass spectrometer (LTQ-Orbitrap) (24). In comparison with direct protein analysis after anti-Tyr(P) protein immunoprecipitation, additional phosphopeptide enrichment has potential advantages. First, because of a strong reduction of sample complexity, about 10 times shorter instrument time was required to identify Tyr(P)-containing proteins with comparable sensitivity. Second, the identification strategy focuses on Tyr(P)-containing proteins and their phosphorylation sites. In contrast, analysis of anti-Tyr(P) immunoprecipitates without additional enrichment does not effectively discriminate between tyrosine phosphorylated proteins and their interaction partners as typically only a few Tyr(P)-containing peptides are detected because of a vast excess of non-phosphorylated peptide species. To investigate how the quantitative information obtained for Tyr(P)-containing peptides after the two-step enrichment correlates with changes of protein abundance directly determined from anti-Tyr(P) immunoprecipitates, both approaches were applied to a SILAC-based, quantitative comparison of wild-type and PTP1B-deficient fibroblasts. Notably altered abundance of tyrosine phosphorylated proteins was found to correlate with at least similar or often more pronounced changes of one or more Tyr(P)-containing peptides (Fig. 2 and supplemental Table 1). Moreover in some cases levels of Tyr(P)-containing peptides were different in the absence of protein changes, indicating that regulated sites were identified in proteins that are immunoprecipitated through one or more constitutive tyrosine phosphorylation site(s). These data support the notion that phosphopeptide quantification upon consecutive enrichment represents a useful strategy to identify regulated, Tyr(P)-containing proteins. However, it is noteworthy that the quantification of individual site changes after final phosphopeptide enrichment could be complicated in certain cases, for example when distinct tyrosine phosphorylation sites are differentially regulated and exhibit different affinities for anti-Tyr(P) antibody.
Fig. 1.
Working scheme for mass spectrometric analysis of tyrosine phosphorylation alterations in PTP1B-deficient fibroblasts. pY, phosphotyrosine; wt, wild type.
Fig. 2.
Comparison of different strategies to quantitatively analyze proteins immunoprecipitated with Tyr(P)-specific antibodies. log2-transformed PTP1B KO/wild-type ratios of immunoprecipitated Tyr(P)-containing proteins were compared with the respective ratios of their Tyr(P) peptides upon further TiO2 enrichment. pTyr, phosphotyrosine; wt, wild type.
Application of the two-step enrichment protocol to quantitative phosphopeptide analysis revealed 184 phosphotyrosine sites from 81 different tyrosine phosphorylated proteins in PTP1B wild-type and KO MEFs under basal cell culture conditions (supplemental Table 2). To ensure that differences in tyrosine phosphorylation were not due to different protein expression levels, total protein extracts of wild-type and KO cells were mixed in equal amounts and then subjected to in-gel digestion prior to LC-MS analysis. All proteins found to be tyrosine phosphorylated in this study were searched in this protein expression data set, and KO/wild-type expression ratios for 56 phosphotyrosine proteins were determined by quantitation of non-phosphorylated peptides. Interestingly most of the analyzed protein expression ratios were close to 1 (supplemental Table 3).
To assess the biological reproducibility of our global phosphotyrosine analysis, the relative ratio of phosphopeptide quantifications from two independent experiments were visualized in a scatter plot and further analyzed regarding their normal distribution. This comparative analysis of biological replicates indicated that Tyr(P)-containing peptides varying by more than 2.5-fold could be considered as significantly different in wild-type versus KO cells as their ratios differed from the mean by more than three standard deviations (supplemental Fig. 1).
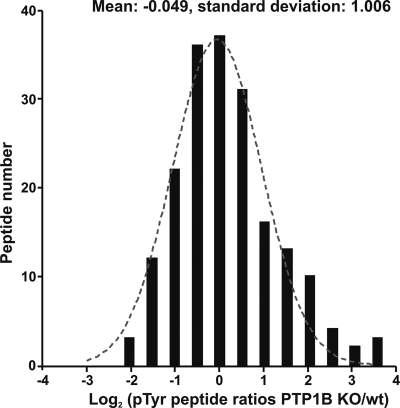
Direct cellular PTP1B substrates can be expected to exhibit increased tyrosine phosphorylation in KO cells. However, as regulated sites might either positively or negatively modulate PTP1B substrate functions, phosphatase regulation could also enhance or down-regulate phosphotyrosine-mediated signaling downstream of direct cellular substrate proteins. Alternatively compensatory mechanisms upon PTP1B knock-out, such as an up-regulation of other PTP family members, could possibly result in some alterations of phosphotyrosine-based signaling not directly related to cellular PTP1B function. These considerations provide possible reasons for the presence of Tyr(P)-containing peptides, which showed reduced abundance upon PTP1B deficiency (Fig. 3). The ratios of all quantified Tyr(P)-containing peptides gave rise to a normal distribution with a population of outliers that was more strongly phosphorylated in the KO cells. According to this normal distribution, p values were calculated for all ratios, and the threshold level for significantly regulated phosphopeptides was set to a p value of less than 0.1. Notably the ratios of all Tyr(P)-containing peptides, which were significantly up-regulated in PTP1B-defiencient cells according to this criterion, were also highly significant according to the analysis of biological replicates described above.
Fig. 3.
Distribution analysis of changes in the cellular Tyr(P)-containing proteome of PTP1B wild-type and KO cells under normal conditions. Tyr(P) peptides were binned according to the log2 values of their PTP1B KO/wild-type ratios. Values for the mean and S.D. were obtained by Gaussian regression analysis. Note the normal distribution of the whole population of Tyr(P) peptides and the outliers that were up-regulated in PTP1B-deficient cells. pTyr, phosphotyrosine; wt, wild type.
For some phosphopeptides, estimations of a minimum -fold change were made because in these cases solely the strongly induced phosphopeptide from the KO cells was detected, and quantitation was performed over background. According to these statistical criteria, 13 proteins were found with significantly hyperphosphorylated Tyr(P) peptides in PTP1B KO cells under normal cell culture conditions (Table I). Tandem MS spectra for all of these peptides are provided as supplemental data (supplemental MS/MS spectra 1). Where available, protein expression data from the parallel analysis of total cell lysates was used for the normalization of Tyr(P) peptide changes. Additional information about the main cellular functions of the respective proteins is given in the left column of Table I.
Table I.
Hyperphosphorylation of 13 proteins in PTP1B-deficient fibroblasts under basal cell culture conditions
wt, wild type; pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine; (0.5), 50% phosphorylation probability.
| Function | Protein | Phosphopeptide sequence | Tyr(P) peptide
|
Whole protein KO/wt ratio | |||
|---|---|---|---|---|---|---|---|
| Tyr(P) site | p value | KO/wt ratio | Normalized | ||||
| Cell shape and motility | LPP | SAQPSPHYMAGPSSGQIpYGPGPR | 245 | 0.05 | 4.09 | 2.62 | 1.56 |
| pY(0.5)pY(0.5)EPYpYAAGPSYGGR | 297/302 | 0.05 | 4.02 | 2.58 | 1.56 | ||
| SEGDTApYGQQVQPNTWK | 318 | 0.05 | 3.44 | 2.21 | 1.56 | ||
| EAApYAPPASGNQNHPGMYPVSGPKK | 333 | 0.04 | 4.26 | 2.73 | 1.56 | ||
| MLpYDMENPPADDYFGR | 403 | 0.01 | 6.96 | 4.47 | 1.56 | ||
| Cell motility | Vav-3 | TPIALATGIRPFPTEESINDEDIpYK | 141 | 0.00 | >10.00 | ||
| Tks5/Fish protein | VKYEEPEpYDVPAFGFDpSEPEMNEEPSGDR | 557 | 0.08 | 3.42 | |||
| VGESSEDVALEEETIpYENEGFRPYTEDTLSAR | 619 | 0.02 | 5.21 | ||||
| Fer | VQENDGKEPPPVVNpYEEDAR | 402 | 0.03 | 4.70 | 3.45 | 1.36 | |
| QEDGGVpYSSSGLK | 715 | 0.04 | 4.47 | 3.29 | 1.36 | ||
| RhoGAP12 | ApT(0.5)pT(0.5)PPNQGRPDpSPVpYANLQELK | 241 | 0.08 | 3.32 | |||
| Cell motility/vesicular transport | Cortactin | NASTFEEVVQVPSApYQK | 334 | 0.02 | 5.58 | 5.44 | 1.03 |
| KQpTPPApSPpSPQPIEDRPPpS(0.5)pS(0.5)PI pYEDAAPFK | 421 | 0.03 | 4.86 | 4.74 | 1.03 | ||
| Cell adhesion | Catenin δ-2 | DpYETYQPFPNSTR | 1176 | 0.00 | 7.73 | ||
| p120ctn | QDVpYGPQPQVR | 257 | 0.06 | 3.84 | 4.13 | 0.93 | |
| FHPEPpYGLEDDQR | 280 | 0.10 | 3.07 | 3.30 | 0.93 | ||
| Tight junction assembly | ZO-1 | TSTLRHEEQPAPApYEVHNR | 1164 | 0.06 | 3.72 | 3.71 | 1.00 |
| YRPEAQPpYSSTGPK | 1177 | 0.00 | >10.00 | >10.00 | 1.00 | ||
| IP3 and DAG signaling | PLCγ1 | IGTAEPDpYGALYEGR | 771 | 0.16 | 2.50 | 2.54 | 0.98 |
| Phagocytosis | MEGF10 protein | DSPpYAEINNSTPANR | 1061 | 0.00 | >10.00 | ||
| Proliferation | p62DOK | FSALEMLENSLpYSPTWEGSQFWVTSQK | 146 | 0.16 | 2.45 | 2.03 | 1.21 |
| GLpYDLPQEPR | 376 | 0.25 | 1.87 | 1.54 | 1.21 | ||
| Proliferation/migration | Eph receptor A3/4/5 | VLEDDPEAApYTTR | 779 | 0.06 | 3.85 | ||
| Cell growth/metabolism | Insulin-like growth factor 1 receptor/insulin receptor | DIpYETDpYYRK | 1193/1197 | 0.00 | 8.75 | ||
| Unknown | Similar to oligophrenin 1 | LWLEAMDGKEPIpYTLPAIISK | 371 | 0.06 | 3.70 | ||
Only three of these proteins, cortactin, p62DOK (docking protein 1, Dok-1), and the insulin receptor/IGF1R, have been described previously to be more strongly tyrosine phosphorylated upon cellular PTP1B inactivation and have been further characterized as direct substrates of PTP1B (12, 40–42). Although p62DOK phosphotyrosine peptides did not have p values of less than 0.1 according to our analysis, increased tyrosine phosphorylation of p62DOK has been directly linked to PTP1B deficiency in this cell system, and further analysis characterized it as a cellular PTP1B substrate protein (40). Interestingly hyperphosphorylation of p62DOK was linked to a reduced rate of proliferation of PTP1B KO cells because of decreased Ras activity by Dube et al. (40). This profound alteration in cell proliferation could also be recapitulated in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays and corresponds to our MS data (supplemental Fig. 2A). In the case of the insulin receptor/IGF1R peptide, the 8.75-fold higher tyrosine phosphorylation could not be directly linked to one of the proteins because no unique peptides of these kinases were identified.
Among the up-regulated proteins, those controlling cell motility and cell adhesion are overrepresented. Mainly adapter proteins that are involved in the regulation of the actin cytoskeleton at sites of cell-matrix and cell-cell adhesion, like lipoma-preferred partner homolog (LPP), p120ctn (Ctnnd1; catenin δ 1), ZO-1, and cortactin, are more strongly phosphorylated in KO cells. Concordantly tyrosine phosphorylation of these proteins has been linked to increased cell motility, such as for cortactin in transformed endothelial cells (43). Reduced cell adhesion has been connected to tyrosine phosphorylation of p120ctn (44) and ZO-1 (45). These results correlate well with the described increased migration rate of PTP1B KO cells compared with their wild-type counterparts (46). We confirmed this strong increase in motility in our PTP1B-deficient cells by both wounding assays and Boyden chamber assays (supplemental Fig. 2B). Other cell motility regulators were also found to be more strongly phosphorylated in KO cells, among them Fer kinase, which localizes to N-cadherin-based adherens junctions (47); the podosome regulator Tks5/Fish (48); and the Rho regulators Vav-3 and RhoGAP12.
In addition, an RTK kinase family involved in migration and proliferation was found to be activated in PTP1B KO cells: a Tyr(P)-containing peptide shared among the ephrin receptor family members A3, A4, and A5 was up-regulated. However, as no unique peptide was identified for either receptor, the exact identity of the regulated protein remained elusive.
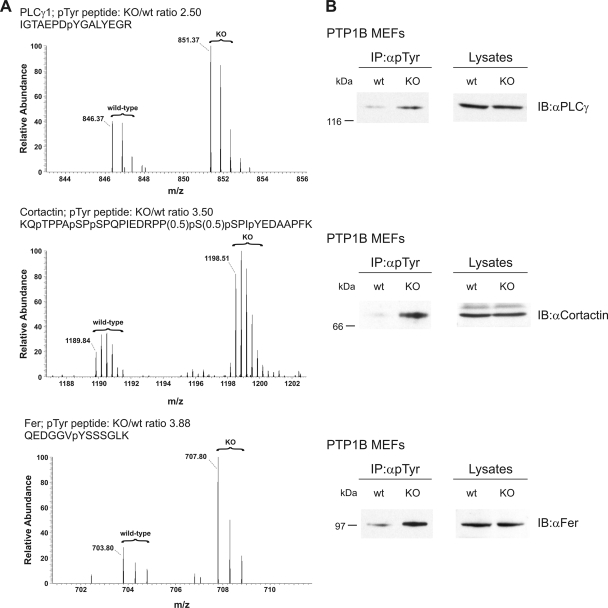
Note that all determined protein expression differences between PTP1B wild-type and KO cells in Table I are less than 2-fold, by extension indicating that most of the observed alterations in phosphotyrosine signaling do not result from altered protein expression levels. Fig. 4A shows the MS spectra of PLCγ1, cortactin, and tyrosine kinase Fer as examples for the quantification of SILAC pairs of Tyr(P)-containing peptides with the MSQuant software. The lower mass isotope clusters represent peptides derived from the normal arginine- and lysine-encoded PTP1B wild-type cells, whereas the higher mass isotope clusters were derived from the heavy isotope-labeled (Arg10 and Lys8) KO cells. To test the results of our MS analysis in a second assay, we analyzed PLCγ1, cortactin, and Fer by immunoblotting and found increased levels of these proteins upon immunoprecipitation with anti-Tyr(P) antibodies, whereas their expression in total lysates was unchanged (Fig. 4B). Thus, these results from immunoblot analyses are in excellent agreement with our quantitative MS data.
Fig. 4.
Phosphorylation analysis of selected proteins of PTP1B wild-type and KO cells using quantitative MS and Western blotting. A, selected MS spectra of Tyr(P) peptides depicted in Table I are shown here. All MS spectra were evaluated with MSQuant software. Isotope clusters of Tyr(P) peptides derived from wild-type and KO cells are indicated by braces. For quantitation, only monoisotopic peaks were used; they are marked with corresponding m/z values. B, tyrosine phosphorylated proteins of unstimulated cells were immunoprecipitated with anti-Tyr(P) 4G10 antibodies and separated using SDS-PAGE. Immunoprecipitated proteins were probed with the indicated antibodies. In addition, cellular protein expression levels were analyzed by loading whole cell lysates (right panel). IP, immunoprecipitation; pTyr, phosphotyrosine; wt, wild type; pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine; IB, immunoblot.
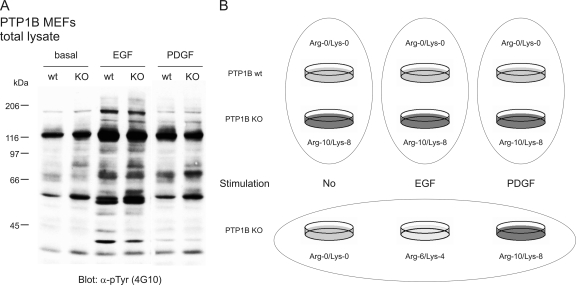
Analysis of PTP1B Function in EGF and PDGF Signaling—
PTP1B has been implicated in both EGFR and PDGFRβ signaling (9, 49), and signal transmission downstream of both RTKs is important for the growth and motility of mouse embryonic fibroblasts. To analyze how the overall cellular tyrosine phosphorylation differs between wild-type and KO cells after either EGF or PDGF stimulation, we performed a Tyr(P)-specific Western blot analysis (Fig. 5A). Detection with anti-phosphotyrosine antibody 4G10 revealed a number of differences between wild-type and KO cells under each condition. We then used quantitative MS analysis to investigate which proteins are potentially regulated by PTP1B in cellular EGFR and PDGFRβ signaling. Fig. 5B shows our strategy to detect phosphorylation differences between wild-type and KO cells in three parallel experiments under basal, EGF-, and PDGF-stimulated conditions using SILAC double labeling. In a fourth experiment, we applied SILAC triple labeling to detect phosphorylation changes in PTP1B KO cells resulting from growth factor stimulation (Fig. 5B). Using this strategy, we not only identified proteins that are functionally linked to PTP1B but also got information to what extent the respective proteins are regulated within either EGFR- or PDGFRβ-mediated signaling (supplemental Tables 4–7). In total, significant ligand-induced up-regulation of Tyr(P)-containing peptides was detected for 32 proteins upon EGF treatment and for 31 proteins upon PDGF treatment (supplemental Table 7).
Fig. 5.
Examination of PTP1B function in EGF and PDGF signaling by differential SILAC labeling of PTP1B-deficient cells. A, Western blot analysis of tyrosine phosphorylated proteins in wild-type and KO cells under basal, EGF- (50 ng/ml, 5 min) or PDGF (20 ng/ml, 5 min)-stimulated conditions after 3 h of serum starvation. B, scheme depicting the strategy to analyze phosphorylation differences with quantitative MS upon EGF (50 ng/ml, 5 min) and PDGF (20 ng/ml, 5 min) stimulation in MEFs starved for 3 h. Each oval represents one independent SILAC experiment. Thereby not only alterations in phosphorylation between wild-type and KO cells can be examined, but ligand-induced phosphorylation events are also determined. pTyr, phosphotyrosine; wt, wild type.
As PTP1B substrates are expected to be hyperphosphorylated in PTP1B KO cells under these treatment conditions, we rated all proteins as positive hits that were found significantly up-regulated (p < 0.1) with at least one Tyr(P) peptide in KO compared with wild-type cells. Applying these criteria to our EGF data set, we identified three proteins that were not only induced by EGF stimulation but also showed a higher tyrosine phosphorylation in KO cells versus wild-type cells (Table II).
Table II.
Alterations in EGF signaling in PTP1B-deficient cells
EGF stimulation led to increased phosphorylation of three proteins in KO cells compared with wild-type cells. wt, wild type; pY, phosphotyrosine.
| Function | Name | Phosphopeptide sequence | Tyr(P) peptide
|
Whole protein KO/wt ratio | Stimulation ratio EGF/basal | |||
|---|---|---|---|---|---|---|---|---|
| Tyr(P) site | p value | KO/wt ratio | Normalized | |||||
| Cell motility | Vav-3 | TPIALATGIRPFPTEESINDEDIpYK | 141 | 0.00 | 10.58 | 23.60 | ||
| IP3 and DAG signaling | PLCγ1 | IGTAEPDpYGALYEGR | 771 | 0.04 | 5.94 | 6.05 | 0.98 | 12.87 |
| Phosphoinositide signaling | SHIP2 | TLSEVDpYAPGPGR | 1136 | 0.10 | 4.06 | 6.22 | 0.65 | 48.61 |
| Proliferation | p62DOK | GFSSDTALpYSQVQK | 450 | 0.18 | 3.04 | 2.51 | 1.21 | 2.38 |
| Proliferation/migration | Epidermal growth factor receptor | pYSSDPTGAVTEDNIDDAFLPVPEpYVNQSVPK | 1069/1092 | 0.39 | 1.31 | 1.68 | 0.78 | 45.04 |
| MHLPSPTDSNFpYR | 1000 | 0.39 | 1.20 | 1.55 | 0.78 | >50.00 | ||
| GSHQMSLDNPDpYQQDFFPK | 1172 | 0.38 | 1.05 | 1.35 | 0.78 | 37.97 | ||
| GPTAENAEpYLR | 1197 | 0.37 | 1.02 | 1.32 | 0.78 | 53.28 | ||
As expected, tyrosine phosphorylation of the EGFR was 30–50-fold increased after EGF stimulation. Surprisingly although EGFR has been described as a potential PTP1B substrate, all quantified and ligand-induced EGFR Tyr(P) peptides showed a KO/wild-type ratio almost equal to 1. However, downstream of the EGFR, three proteins were found with significantly up-regulated tyrosine phosphorylation in KO cells. Among them were the phosphoinositide regulators PLCγ1 and SHIP2 as evident from 6- and 4-fold increases, respectively, of Tyr(P)-containing peptides derived from these signal transducers. In addition, the EGF-induced Rho regulator Vav-3 was hyperphosphorylated in KO cells. Similar to the basal data set, the negative regulator of proliferation p62DOK (50) was more strongly tyrosine phosphorylated upon PTP1B deficiency, although the p value of this known PTP1B substrate was not within the applied threshold of significance.
In PDGF-induced signaling, five proteins showed significantly increased KO/wild-type phosphorylation ratios (Table III). The PDGFRβ itself was about 2.5-fold more strongly phosphorylated in KO cells, although this effect was mainly due to increased protein expression. However, downstream of the PDGFRβ, both negative signaling regulators such as p62DOK and p120RasGAP (40) and the positive regulator SHP2 were up-regulated with respect to their tyrosine phosphorylation (51–53). Furthermore PLCγ1 and the diacylglycerol-binding kinase PKCδ appeared as PTP1B-regulated elements from our PDGF data. Collectively these data indicate various roles of PTP1B in modulating receptor-proximal signaling events upon EGF or PDGF treatment. All MS/MS spectra of the Tyr(P) phosphopeptides listed in Tables II and III are also available as supplemental data (supplemental MS/MS spectra 2).
Table III.
Alterations in PDGF signaling in PTP1B-deficient cells
PDGF stimulation led to hyperphosphorylation of five proteins in PTP1B KO cells. wt, wild type; pY, phosphotyrosine; (0.5), 50% phosphorylation probability.
| Function | Name | Phosphopeptide sequence | Tyr(P) peptide
|
Whole protein KO/wt ratio | Stimulation ratio PDGF/basal | |||
|---|---|---|---|---|---|---|---|---|
| Tyr(P) site | p value | KO/wt ratio | Normalized | |||||
| Phagocytosis | MEGF10 protein | DSPpYAEINNSTPANR | 1061 | 0.01 | >10.00 | 2.14 | ||
| IP3 and DAG signaling | PLCγ1 | IGTAEPDpYGALpYEGRNPGFpYVEANPMPTFK | 771/775/783 | 0.05 | 6.11 | 6.22 | 0.98 | 28.47 |
| IGTAEPDpYGALYEGR | 771 | 0.14 | 3.96 | 4.04 | 0.98 | 23.29 | ||
| Proliferation/migration | PKCδ | KLDTTESVGIpYQGFEK | 311 | 0.03 | 7.23 | 7.96 | 0.91 | |
| Ras positive regulation | SHP2 | IQNTGDpY(0.5)pY(0.5)DLYGGEK | 62 or 63 | 0.09 | 4.77 | 3.85 | 1.24 | >20.00 |
| Ras negative regulation | p120RasGAP | EIpYNTIR | 451 | 0.04 | 6.66 | 4.42 | 1.51 | |
| Proliferation | p62DOK | IPPGPSQDSVpYSDPLGSTPAGAGEGVHSK | 314 | 0.24 | 2.92 | 2.41 | 1.21 | |
| TVPPPVPQDPLGSPPALpYAEPLDSLR | 295 | 0.27 | 2.68 | 2.22 | 1.21 | |||
| Proliferation/migration | Platelet-derived growth factor receptor β | YADIESPSYMAPYDNpYVPSAPER | 778 | 0.27 | 2.63 | 1.53 | 1.72 | 18.21 |
| DESIDpYVPMLDMK | 751 | 0.31 | 2.38 | 1.39 | 1.72 | >50.00 | ||
| DIMRDSNpYISK | 857 | 0.33 | 2.18 | 1.27 | 1.72 | 21.75 | ||
| pYQQVDEEFLR | 970 | 0.36 | 2.00 | 1.16 | 1.72 | 73.45 | ||
PTP1B Substrate-trapping Approach Identifies Physical Interaction with Potential Substrates—
An increase in tyrosine phosphorylation in PTP1B-deficient cells indicates a functional linkage but is not necessarily due to direct substrate binding to and dephosphorylation by PTP1B. Therefore, we used a parallel approach to identify proteins that are potentially regulated by direct interaction with PTP1B. A well established strategy to identify direct substrates for protein-tyrosine phosphatases is the in vitro substrate-trapping method. Hereby recombinant phosphatase-dead mutants are generated by introducing substitutions in their essential catalytic residues, which are D182A and Q262A in the case of PTP1B (18, 31). These mutants have lost their ability to dephosphorylate substrate proteins and can instead form stable complexes with their substrates (supplemental Fig. 3A). In contrast, no tyrosine phosphorylated proteins are pulled down with the wild-type enzyme because all interacting proteins are dephosphorylated. To get an impression of the amount of tyrosine phosphorylated proteins interacting with recombinant PTP1B substrate-trapping mutants, Western blot analysis was performed with cellular extracts from PTP1B KO cells treated with pervanadate. As expected, no tyrosine phosphorylated proteins were pulled down with the wild-type enzyme, whereas a large number interacted similarly with both PTP1B D182A and D182A/Q262A substrate-trapping mutants (supplemental Fig. 3B).
In our experimental approach, substrate trapping was followed by elution of bound proteins with urea and a subsequent TiO2 purification of Tyr(P) peptides. Thereafter, phosphopeptides were analyzed by LC-MS/MS in an LTQ-Orbitrap mass spectrometer. This analysis of phosphopeptide-enriched fractions was done in a qualitative fashion as almost no tyrosine phosphorylated proteins were enriched with the recombinant wild-type enzyme. We chose to stimulate PTP1B KO cells with pervanadate to provide a sufficient amount of tyrosine phosphorylated proteins for substrate trapping and MS analysis as similar experiments with EGF- or PDGF-treated lysates did not result in detectable, Tyr(P)-containing peptides (data not shown). These technical limitations are most likely due to the rather low affinities of substrate-phosphatase interactions. Altogether Tyr(P)-containing peptides from 53 proteins could be identified upon in vitro association with the PTP1B D182A/Q262A substrate-trapping mutant (supplemental Table 8). Moreover eight of these proteins were also identified as significantly up-regulated in PTP1B KO cells according to our cellular analysis of the Tyr(P)-containing proteome, providing evidence that these proteins are potential direct substrates of PTP1B (Table IV). Importantly for five of these eight proteins, all Tyr(P)-containing peptides identified in the substrate-trapping experiment were also found in the cellular analyses, indicating that the pervanadate-induced tyrosine phosphorylation closely resembles the physiological states of these PTP1B substrates. Moreover there was a considerable phosphopeptide overlap for LPP and p120ctn. Only for RhoGAP12 none of the Tyr(P)-containing peptides from the substrate-trapping experiment were found in cells under physiological growth or treatment conditions.
Table IV.
In vitro substrate trapping identified eight proteins that both bound to the active site of PTP1B and were hyperphosphorylated at Tyr(P) sites in PTP1B-deficient cells
wt, wild type, pY, phosphotyrosine; pS, phosphoserine; pT, phosphothreonine; (0.5), 50% phosphorylation probability.
| Name | Qualitative Tyr(P) peptide analysisa
|
nsb (nc) | Up-regulated inc
|
Quantitative substrate trappingd protein ratio trap/wt | Already described (reference) | |||
|---|---|---|---|---|---|---|---|---|
| Tyr(P) site | Phosphopeptide sequence | Basal | EGF | PDGF | ||||
| p120ctn | 280 | FHPEPpYGLEDDQR | 9 (5) | X | 7.29 | |||
| 257 | QDVpYGPQPQVR | X | ||||||
| LPP | 245 | SAQPSPHYMAGPSSGQIpYGPGPR | 5 (3) | X | ||||
| 301/302 | YYEPpYpYAAGPSYGGR | X | ||||||
| 318 | SEGDTApYGQQVQPNTWK | X | ||||||
| Cortactin | 421 | KQpTPPApSPSPQPIEDRPPpS(0.5) pS(0.5)PIpYEDAAPFK | 2 (2) | X | 2.62 | (42) | ||
| 334 | NASTFEEVVQVPSApYQK | X | ||||||
| ZO-1 | 1164 | TSTLRHEEQPAPApYEVHNR | 1 (1) | X | 2.80 | |||
| Rho GTPase-activating protein 12 | 3 (0) | X | 21.08 | |||||
| PLCγ1 | 771 | IGTAEPDpYGALYEGR | 4 (4) | X | X | 43.96 | ||
| SHIP2 | 1136 | TLSEVDpYAPGPGR | 1 (1) | X | ||||
| p120RasGAP | 451 | EIpYNTIR | 1 (1) | X | 47.62 | |||
| p62DOK | 295 | TVPPPVPQDPLGpSPPALpYAEPLDSLR | 6 (4) | (X) | 12.66 | (40) | ||
| 376 | GLpYDLPQEPR | (X) | ||||||
| 450 | GFSSDTALpYSQVQK | (X) | ||||||
| Epidermal growth factor receptor | 1197 | GPTAENAEpYLR | 5 (5) | 7.69 | (49) | |||
| 1172 | GSHQMSLDNPDpYQQDFFPK | |||||||
| 1000 | MHLPSPTDSNFpYR | |||||||
| 1069/1092 | pYSSDPTGAVTEDNIDDAFLPVPEpYVNQSVPK | |||||||
| Platelet-derived growth factor receptor β | 751 | DESIDpYVPMLDMK | 4 (3) | 17.89 | (49) | |||
| 763/778 | pYADIESPSYMAPYDNpYVPSAPER | |||||||
Tyr(P) peptides are shown that belong to proteins that bound to PTP1B substrate-trapping mutants.
The number ns indicates how many Tyr(P) peptides were identified in the substrate-trapping experiment, and nc indicates how many of these Tyr(P) peptides were also identified under physiological cellular conditions.
Proteins found with significantly up-regulated Tyr(P) peptides (p value < 0.1) are indicated by X. Tyr(P) peptides from the described PTP1B substrate p62DOK are indicated by (X), as they were not found with p values of less than 0.1.
PTP1B substrate-trapping mutant/wild-type enzyme ratios are listed.
As shown in Table IV, the EGFR, the PDGFR, p62DOK, and cortactin have already been described to bind to PTP1B substrate-trapping mutants, thus confirming the reliability of this method (19, 42). For various other tyrosine phosphorylated proteins, we obtained evidence for their ability to directly interact with the catalytic site of PTP1B in addition to their up-regulation upon cellular PTP1B deficiency. Among these proteins are mainly regulators of cell adhesion like p120ctn or ZO-1 and regulators of motility like LPP or cortactin. Thus, PTP1B-regulated tyrosine phosphorylation correlates well with the observed increased cellular motility of phosphatase-deficient MEFs. Moreover with p120RasGAP we detected a key regulator of Ras signaling by both approaches, and our combined data further revealed a previously unknown connection of PTP1B to phosphoinositide signaling by acting on the lipid hydrolase PLCγ1 and the lipid phosphatase SHIP2.
To further confirm selective binding for the identified Tyr(P)-containing proteins, we used a quantitative SILAC approach to analyze protein binding to the substrate-trapping mutant in comparison with recombinant PTP1B wild-type enzyme (supplemental Table 9). All proteins shown in Table IV were identified in the quantitative substrate-trapping experiment except lipoma-preferred partner homolog and SHIP2. The protein binding ratios confirmed that the PTP1B substrate-trapping mutant possessed higher binding affinities to the potential substrate proteins than did the wild-type enzyme (Table IV).
DISCUSSION
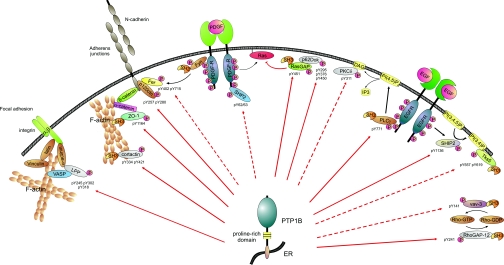
PTP1B is a well known regulator of tyrosine signaling controlling various metabolic, oncogenic, and immune functions. Previously 12 substrates of PTP1B had been described in biochemical studies (10, 42). Many of these studies took advantage of PTP1B-deficient cells where, as expected, an up-regulation of the phosphotyrosine contents of the identified substrates was observed. Several substrates such as the EGFR, the PDGFR, the insulin receptor, the IGF1R, JAK2, Tyk2, and p62DOK have been identified by analyzing PTP1B-deficient fibroblasts upon the respective cytokine and growth factor treatments. However, for most of these substrates information about Tyr(P) site-specific regulation is missing, and also little is known about the activity of PTP1B toward effector proteins acting downstream of the respective PTKs. In our study, six of these already described substrates were either found to be hyperphosphorylated in PTP1B KO cells and/ or identified in the substrate-trapping analysis (insulin receptor/IGF1R, p62DOK, cortactin, IRS1, EGFR, and PDGFR). In total, our quantitative MS analysis allowed us to compare the relative differences in tyrosine phosphorylation of 124 proteins containing 301 phosphotyrosine sites between PTP1B KO cells and their wild-type counterparts under basal and EGF- or PDGF-stimulated conditions. Therefore, this cellular phosphotyrosine analysis gives a detailed overview about global tyrosine signaling processes in this extensively used cell system and further provides new insights into the functional relevance of tyrosine phosphorylation in increased migration and decreased proliferation observed upon PTP1B deficiency. On the cellular level, regulation by PTP1B was shown for 18 of the analyzed proteins for which the absence of PTP1B led to a significant increase in tyrosine phosphorylation as evident from one or multiple Tyr(P)-containing peptides. Hyperphosphorylation of proteins in PTP1B KO cells does not necessarily implicate a direct enzyme-substrate interaction; however, it indicates that the identified protein is functionally associated with cellular PTP1B function. The analysis of direct interactions of PTP1B with its functionally associated proteins provides further information about potential enzyme-substrate relationships. A well established approach to test the binding of tyrosine phosphorylated proteins to PTP1B uses catalytically inactive substrate-trapping mutants of the phosphatase. All of the previously reported substrate proteins mentioned above have been shown to bind directly to the active center of PTP1B (19). Using this substrate-trapping approach, we demonstrated specific binding of various other tyrosine phosphorylated proteins to the enzymatically inactive mutant of the phosphatase. Interestingly eight of the 18 significantly up-regulated proteins identified in our study were found to bind to PTP1B substrate-trapping mutants: cortactin, LPP, PLCγ1, SHIP2, p120ctn, ZO-1, RhoGAP12, and p120RasGAP. Fig. 6 gives an overview of all novel putative substrates and puts these proteins into their described functional context. In addition, proteins functionally associated with PTP1B according to our global phosphotyrosine analysis data are illustrated for which tyrosine phosphorylation and function could be connected to the increased motility and decreased proliferation observed in PTP1B KO cells. It should be noted, however, that although these proteins did not show in vitro binding to substrate-trapping mutants it cannot be excluded that PTP1B directly dephosphorylates them in a rather transient cellular interaction.
Fig. 6.
Schematic drawing summarizing novel potential PTP1B enzyme-substrate interactions in a functional context. ER-localized PTP1B contains a proline-rich domain that allows interaction with substrate proteins containing SH3 domains. Red arrows indicate a direct interaction and dephosphorylation of putative substrates, whereas dashed red arrows specify a direct or indirect functional association evident from increased tyrosine phosphorylation upon PTP1B deficiency. All depicted Tyr(P) sites are hyperphosphorylated in PTP1B-deficient cells. In addition, Tyr(P) sites of potential substrates were identified as putative interaction sites in the substrate-trapping assay. This scheme was designed on the basis of the following references: focal adhesion site (73), PDGFRβ-mediated regulation of N-cadherin cell-cell adhesion (47), and p62DOK and p120RasGAP regulation of Ras signaling (40). VASP, vasodilator-stimulated phosphoprotein; pY, phosphotyrosine; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3,4,5)P2, phosphatidylinositol 3,4,5-trisphosphate; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate.
Most of the proteins identified here as potential new PTP1B substrates (Table IV) can be subdivided into two functional classes: one comprising regulators of cell migration and adhesion and the other defined by regulatory roles in cell proliferation. We verified that PTP1B-deficient cells showed a strong increase in cell motility as described by Buckley et al. (46). In accordance with this observation, ectopic expression of PTP1B in HeLa cells has been shown to reduce serum-induced cell migration (54). Intriguingly we identified as many as seven potential PTP1B substrates that have been described as modulators of cell migration and adhesion: cortactin, LPP, PLCγ1, SHIP2, p120ctn, ZO-1, and RhoGAP12. Tyrosine phosphorylation of cortactin leads to reduced actin cross-linking activity and promotes cell motility in endothelial cells (43). Using Tyr(P)-421 site-specific antibodies, Head et al. (55) showed that cortactin phosphorylated on Tyr-421 localized to lamellipodia and podosomes, whereas the function of its phosphorylation site Tyr-334 is so far unclear. Very recently, Stuible et al. (42) also identified cortactin to be a substrate of PTP1B and mapped the binding of PTP1B substrate-trapping mutants to Tyr(P)-446 of cortactin. Furthermore they showed that phosphorylation of Tyr-446 and Tyr-421 are interdependent and that interference of PTP1B function with a specific inhibitor or by expression of a dominant-negative mutant results in increased phosphorylation of both residues in HeLa cells. In our study we detected cortactin Tyr(P)-421 but not Tyr(P)-446. This may be due to technical difficulties in analyzing the corresponding peptide of Tyr(P)-446 by our MS strategy, or alternatively, it is also possible that this phosphorylation event does not occur in mouse embryonic fibroblasts. Tyrosine phosphorylation of LPP has not been studied so far, but LPP was shown to localize to focal adhesion junctions and to bind vasodilator-stimulated phosphoprotein and α-actinin (56).
Interestingly we also identified two phosphoinositide-modifying enzymes in our analysis as potential substrates of PTP1B: PLCγ1, which converts phosphatidylinositol 4,5-bisphosphate into IP3/DAG, and SHIP2, which dephosphorylates phosphatidylinositol 3,4,5-trisphosphate into phosphatidylinositol 3,4-bisphosphate. Binding of PTP1B to PLCγ1 has already been mapped to the SH3 domain of PLCγ1 (57), but no evidence for a direct dephosphorylation is currently available. Elevated activity of PLCγ1 in PTP1B KO cells leads to an increase in PKCδ phosphorylation on Tyr(P)-311 after PDGF stimulation. PKCδ tyrosine phosphorylation is positively modulated by DAG resulting in increased kinase activity that in turn leads to enhanced motility of mouse fibroblasts (58). In line with these cellular data, PTP1B was also found to dephosphorylate PKCδ in vitro (59). In the case of SHIP2, tyrosine phosphorylation on Tyr-986 or Tyr-987 is likely to control the enzymatic activity of the protein and was shown to be important for lamellipodia formation and regulation of the actin cytoskeleton (60). How increased phosphorylation of SHIP2 on residues Tyr-987 and Tyr-1136 might affect EGF signaling in PTP1B KO cells and whether there is a connection of SHIP2 to the phosphatidylinositol 3,4-bisphosphate binder Tks5/Fish (48), which is also hyperphosphorylated in KO cells, remain to be investigated.
PTP1B wild-type fibroblasts exhibited stronger cell-cell adhesion contacts than did the KO cells (data not shown). Tyrosine phosphorylation of p120ctn could explain this phenotype as it was demonstrated that p120ctn phosphorylation by Src kinases leads to loss of E-cadherin function (44). In contrast, other studies show that tyrosine phosphorylation of p120ctn is dispensable for the modulation of cadherin-based cell adhesions (61). To elucidate this discrepancy, it is important to obtain site-specific information about the regulation of p120ctn by tyrosine kinases and phosphatases (62). In our study, we found that p120ctn is likely to be regulated by PTP1B at Tyr(P)-257 and Tyr(P)-280 but not at Tyr(P)-228 and Tyr(P)-904 (supplemental Table S2). As PTP1B-deficient cells show enhanced migration and reduced cell adhesion, the regulated Tyr(P) sites can be linked to these cellular processes. Reduced cell adhesion was furthermore linked to tyrosine phosphorylation of ZO-1 (45). As no tight junctions are present in fibroblasts, ZO-1 presumably localizes to and regulates cell adherens junctions (63).
PTP1B was also found to regulate the Rho signaling regulators Vav-3 and RhoGAP12 in intact cells. Vav-3 Tyr(P)-141 is a new site, and RhoGAP12 Tyr(P)-241 has been described without further characterization of its function. Further analysis of phosphotyrosine regulation of these GTPase regulators could help to explain recent observations that PTP1B can act both upstream (64) and downstream (65) of the small GTPase RhoA.
PTP1B KO MEFs show a strong decrease in proliferation compared with wild-type cells. Dube et al. (40) correlated this reduced proliferation rate with reduced Ras activity that is a consequence of increased phosphorylation of its negative regulator p62DOK. With our quantitative MS analysis, we reproduced the reported up-regulation of p62DOK tyrosine phosphorylation in PTP1B KO cells and further showed that p120RasGAP is also hyperphosphorylated in KO cells after PDGF stimulation, whereas its expression is only slightly increased. Interestingly p120RasGAP is a p62DOK-binding protein, and it negatively regulates Ras by promoting its intrinsic GTPase activity (51).
The substrate specificity of PTP1B is dependent on its subcellular localization and organization in protein complexes. PTP1B interacts with some of its substrates via binding of its proline-rich domain to SH3 domains as demonstrated, for example, for the SH3 domain of p130cas (66). Interestingly several of the potential substrate proteins we identified here contain SH3 domains, such as cortactin, ZO-1, p120RasGAP, RhoGAP12, and PLCγ1 (Fig. 6). In addition, we found other tyrosine phosphorylated proteins with SH3 domains, like Tks5 or Vav-3, to be up-regulated in PTP1B-deficient cells for which we did not get experimental evidence for direct binding to the catalytic domain of PTP1B in vitro. However, in the cellular context, it might well be that PTP1B-SH3 domain interactions through the proline-rich domain of the phosphatase promote the direct dephosphorylation of these proteins.
According to the “zip code” model that describes non-catalytic domains in PTPs to be essential for their cellular localization (67), PTP1B localizes to the ER (68). This raises the question how PTP1B can act on substrate proteins localized at the plasma membrane and in the cytosol. The cellular functions of PTP1B can be subdivided by the localization of its substrates. It was shown to dephosphorylate precursors of receptor proteins, like the insulin receptor and other RTKs, on their biosynthetic way from the ER to the plasma membrane (9, 69).
Interestingly Haj et al. (70) also showed that PTP1B targets RTKs like the EGFR and the PDGFR after ligand stimulation when the receptors are internalized and endosomes come in close contact to the ER. This might explain the missing hyperphosphorylation of the EGFR and the PDGFR in PTP1B KO cells in our study because both receptors are not internalized after the short stimulation time points that were used here. Direct access of ER-bound PTP1B to substrates at the plasma membrane was demonstrated by Anderie et al. (71) but remains controversial. In addition, release of PTP1B into the cytosol by proteolytic calpain-mediated cleavage was demonstrated in platelets (68, 72). At which cellular location PTP1B addresses the novel potential substrates we characterized in our study will be an interesting issue for further research.
Our study provides a valuable list of potential new substrate proteins and downstream effector molecules for PTP1B in mammalian cells. In particular, our findings help to explain the involvement of PTP1B in the control of cell motility and adhesion as many of the identified potential substrate proteins localize to cell-cell adhesion sites. These observations also suggest that PTP1B acts not only on PTKs, which constitute most of its already described substrate proteins, but also on their downstream effector proteins to maintain the homeostasis of reversible cellular protein tyrosine phosphorylation. To the best of our knowledge, this study for the first time provides a global picture of the cellular functions of a member of the PTP family of enzymes. The strategies developed here for the analysis of cellular PTP1B function and substrate specificity can now be applied to other members of this important enzyme family and have the potential to define the functions of other PTPs in the context of cellular signal transmission networks.
Supplementary Material
Acknowledgments
We thank other members of the Department of Molecular Biology at the Max Planck Institute of Biochemistry for help and fruitful discussions especially Michaela Bairlein, Felix Oppermann, and Yixiang Zhang. We also thank Verena Maier for critical reading of the manuscript. Fer antibody was a kind gift from Peter A. Greer.
Footnotes
Published, MCP Papers in Press, May 31, 2008, DOI 10.1074/mcp.M800196-MCP200
The abbreviations used are: PTP, protein-tyrosine phosphatase; PTK, protein-tyrosine kinase; RPTP, receptor-like PTP; RTK, receptor tyrosine kinase; SILAC, stable isotope labeling by amino acids in cell culture; DHB, 2,5-dihydroxybenzoic acid; STAGE, stop and go extraction; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; ABC, ammonium bicarbonate; RP, reverse phase; nano, nanoscale capillary; IPI, International Protein Index; PTM, post-translational modification; MEF, mouse embryonic fibroblast; KO, knock-out; EGF, epidermal growth factor; PDGF, platelet-derived growth factor; PLCγ1, phospholipase C γ1; PKCδ, protein kinase C δ; ZO-1, zonula occludens 1 protein; LPP, lipoma-preferred partner homolog; EGFR, epidermal growth factor receptor; PDGFR, platelet-derived growth factor receptor; SH3, Src homology 3; ER, endoplasmic reticulum; GAP, GTPase-activating protein; DAG, diacylglycerol; IP3, inositol 1,4,5-trisphosphate; ERK, extracellular signal-regulated kinase.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Tiganis, T., and Bennett, A. M. ( 2007) Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 402, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lammers, R., Moller, N. P., and Ullrich, A. ( 1997) The transmembrane protein tyrosine phosphatase α dephosphorylates the insulin receptor in intact cells. FEBS Lett. 404, 37–40 [DOI] [PubMed] [Google Scholar]
- 3.Fuchs, M., Muller, T., Lerch, M. M., and Ullrich, A. ( 1996) Association of human protein-tyrosine phosphatase κ with members of the armadillo family. J. Biol. Chem. 271, 16712–16719 [DOI] [PubMed] [Google Scholar]
- 4.Anders, L., Mertins, P., Lammich, S., Murgia, M., Hartmann, D., Saftig, P., Haass, C., and Ullrich, A. ( 2006) Furin-, ADAM 10-, and γ-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of β-catenin's transcriptional activity. Mol. Cell. Biol. 26, 3917–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharitonenkov, A., Chen, Z., Sures, I., Wang, H., Schilling, J., and Ullrich, A. ( 1997) A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 386, 181–186 [DOI] [PubMed] [Google Scholar]
- 6.Gensler, M., Buschbeck, M., and Ullrich, A. ( 2004) Negative regulation of HER2 signaling by the PEST-type protein-tyrosine phosphatase BDP1. J. Biol. Chem. 279, 12110–12116 [DOI] [PubMed] [Google Scholar]
- 7.Pulido, R., Zuniga, A., and Ullrich, A. ( 1998) PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J. 17, 7337–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buschbeck, M., Eickhoff, J., Sommer, M. N., and Ullrich, A. ( 2002) Phosphotyrosine-specific phosphatase PTP-SL regulates the ERK5 signaling pathway. J. Biol. Chem. 277, 29503–29509 [DOI] [PubMed] [Google Scholar]
- 9.Lammers, R., Bossenmaier, B., Cool, D. E., Tonks, N. K., Schlessinger, J., Fischer, E. H., and Ullrich, A. ( 1993) Differential activities of protein tyrosine phosphatases in intact cells. J. Biol. Chem. 268, 22456–22462 [PubMed] [Google Scholar]
- 10.Dube, N., and Tremblay, M. L. ( 2005) Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim. Biophys. Acta 1754, 108–117 [DOI] [PubMed] [Google Scholar]
- 11.Tonks, N. K. ( 2003) PTP1B: from the sidelines to the front lines! FEBS Lett. 546, 140–148 [DOI] [PubMed] [Google Scholar]
- 12.Elchebly, M., Payette, P., Michaliszyn, E., Cromlish, W., Collins, S., Loy, A. L., Normandin, D., Cheng, A., Himms-Hagen, J., Chan, C. C., Ramachandran, C., Gresser, M. J., Tremblay, M. L., and Kennedy, B. P. ( 1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 13.Klaman, L. D., Boss, O., Peroni, O. D., Kim, J. K., Martino, J. L., Zabolotny, J. M., Moghal, N., Lubkin, M., Kim, Y. B., Sharpe, A. H., Stricker-Krongrad, A., Shulman, G. I., Neel, B. G., and Kahn, B. B. ( 2000) Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 20, 5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bence, K. K., Delibegovic, M., Xue, B., Gorgun, C. Z., Hotamisligil, G. S., Neel, B. G., and Kahn, B. B. ( 2006) Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med. 12, 917–924 [DOI] [PubMed] [Google Scholar]
- 15.Dube, N., Bourdeau, A., Heinonen, K. M., Cheng, A., Loy, A. L., and Tremblay, M. L. ( 2005) Genetic ablation of protein tyrosine phosphatase 1B accelerates lymphomagenesis of p53-null mice through the regulation of B-cell development. Cancer Res. 65, 10088–10095 [DOI] [PubMed] [Google Scholar]
- 16.Julien, S. G., Dube, N., Read, M., Penney, J., Paquet, M., Han, Y., Kennedy, B. P., Muller, W. J., and Tremblay, M. L. ( 2007) Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 17.Bentires-Alj, M., and Neel, B. G. ( 2007) Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer Res. 67, 2420–2424 [DOI] [PubMed] [Google Scholar]
- 18.Flint, A. J., Tiganis, T., Barford, D., and Tonks, N. K. ( 1997) Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 94, 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchetot, C., Chagnon, M., Dube, N., Halle, M., and Tremblay, M. L. ( 2005) Substrate-trapping techniques in the identification of cellular PTP targets. Methods 35, 44–53 [DOI] [PubMed] [Google Scholar]
- 20.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 21.Siu, R., Fladd, C., and Rotin, D. ( 2007) N-cadherin is an in vivo substrate for protein tyrosine phosphatase sigma (PTPσ) and participates in PTPσ-mediated inhibition of axon growth. Mol. Cell. Biol. 27, 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, J., Katrekar, A., Honigberg, L. A., Smith, A. M., Conn, M. T., Tang, J., Jeffery, D., Mortara, K., Sampang, J., Williams, S. R., Buggy, J., and Clark, J. M. ( 2006) Identification of substrates of human protein-tyrosine phosphatase PTPN22. J. Biol. Chem. 281, 11002–11010 [DOI] [PubMed] [Google Scholar]
- 23.Syka, J. E., Marto, J. A., Bai, D. L., Horning, S., Senko, M. W., Schwartz, J. C., Ueberheide, B., Garcia, B., Busby, S., Muratore, T., Shabanowitz, J., and Hunt, D. F. ( 2004) Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J. Proteome Res. 3, 621–626 [DOI] [PubMed] [Google Scholar]
- 24.Makarov, A., Denisov, E., Kholomeev, A., Balschun, W., Lange, O., Strupat, K., and Horning, S. ( 2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 25.Mumby, M., and Brekken, D. ( 2005) Phosphoproteomics: new insights into cellular signaling. Genome Biol. 6, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong, S. E., and Mann, M. ( 2007) Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol. Biol. 359, 37–52 [DOI] [PubMed] [Google Scholar]
- 27.Mann, M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 28.Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 29.Senis, Y. A., Craig, A. W., and Greer, P. A. ( 2003) Fps/Fes and Fer protein-tyrosine kinases play redundant roles in regulating hematopoiesis. Exp. Hematol. 31, 673–681 [DOI] [PubMed] [Google Scholar]
- 30.Cheng, A., Bal, G. S., Kennedy, B. P., and Tremblay, M. L. ( 2001) Attenuation of adhesion-dependent signaling and cell spreading in transformed fibroblasts lacking protein tyrosine phosphatase-1B. J. Biol. Chem. 276, 25848–25855 [DOI] [PubMed] [Google Scholar]
- 31.Xie, L., Zhang, Y. L., and Zhang, Z. Y. ( 2002) Design and characterization of an improved protein tyrosine phosphatase substrate-trapping mutant. Biochemistry 41, 4032–4039 [DOI] [PubMed] [Google Scholar]
- 32.Larsen, M. R., Thingholm, T. E., Jensen, O. N., Roepstorff, P., and Jorgensen, T. J. ( 2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 33.Rappsilber, J., Ishihama, Y., and Mann, M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko, A., Wilm, M., Vorm, O., Jensen, O. N., Podtelejnikov, A. V., Neubauer, G., Mortensen, P., and Mann, M. ( 1996) A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24, 893–896 [DOI] [PubMed] [Google Scholar]
- 35.Olsen, J. V., de Godoy, L. M., Li, G., Macek, B., Mortensen, P., Pesch, R., Makarov, A., Lange, O., Horning, S., and Mann, M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 36.Schroeder, M. J., Shabanowitz, J., Schwartz, J. C., Hunt, D. F., and Coon, J. J. ( 2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 37.Elias, J. E., Haas, W., Faherty, B. K., and Gygi, S. P. ( 2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 38.Schulze, W. X., and Mann, M. ( 2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 39.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 40.Dube, N., Cheng, A., and Tremblay, M. L. ( 2004) The role of protein tyrosine phosphatase 1B in Ras signaling. Proc. Natl. Acad. Sci. U. S. A. 101, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seely, B. L., Staubs, P. A., Reichart, D. R., Berhanu, P., Milarski, K. L., Saltiel, A. R., Kusari, J., and Olefsky, J. M. ( 1996) Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes 45, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 42.Stuible, M., Dube, N., and Tremblay, M. L. ( 2008) PTP1B regulates cortactin tyrosine phosphorylation by targeting Tyr446. J. Biol. Chem. 283, 15740–15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang, C., Liu, J., Haudenschild, C. C., and Zhan, X. ( 1998) The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 273, 25770–25776 [DOI] [PubMed] [Google Scholar]
- 44.Ozawa, M., and Ohkubo, T. ( 2001) Tyrosine phosphorylation of p120(ctn) in v-Src transfected L cells depends on its association with E-cadherin and reduces adhesion activity. J. Cell Sci. 114, 503–512 [DOI] [PubMed] [Google Scholar]
- 45.Takeda, H., Nagafuchi, A., Yonemura, S., Tsukita, S., Behrens, J., and Birchmeier, W. ( 1995) V-src kinase shifts the cadherin-based cell adhesion from the strong to the weak state and β catenin is not required for the shift. J. Cell Biol. 131, 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckley, D. A., Cheng, A., Kiely, P. A., Tremblay, M. L., and O'Connor, R. ( 2002) Regulation of insulin-like growth factor type I (IGF-I) receptor kinase activity by protein tyrosine phosphatase 1B (PTP-1B) and enhanced IGF-I-mediated suppression of apoptosis and motility in PTP-1B-deficient fibroblasts. Mol. Cell. Biol. 22, 1998–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greer, P. ( 2002) Closing in on the biological functions of Fps/Fes and Fer. Nat. Rev. Mol. Cell Biol. 3, 278–289 [DOI] [PubMed] [Google Scholar]
- 48.Seals, D. F., Azucena, E. F., Jr., Pass, I., Tesfay, L., Gordon, R., Woodrow, M., Resau, J. H., and Courtneidge, S. A. ( 2005) The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 7, 155–165 [DOI] [PubMed] [Google Scholar]
- 49.Haj, F. G., Markova, B., Klaman, L. D., Bohmer, F. D., and Neel, B. G. ( 2003) Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J. Biol. Chem. 278, 739–744 [DOI] [PubMed] [Google Scholar]
- 50.Zhao, M., Schmitz, A. A., Qin, Y., Di Cristofano, A., Pandolfi, P. P., and Van Aelst, L. ( 2001) Phosphoinositide 3-kinase-dependent membrane recruitment of p62(dok) is essential for its negative effect on mitogen-activated protein (MAP) kinase activation. J. Exp. Med. 194, 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donovan, S., Shannon, K. M., and Bollag, G. ( 2002) GTPase activating proteins: critical regulators of intracellular signaling. Biochim. Biophys. Acta 1602, 23–45 [DOI] [PubMed] [Google Scholar]
- 52.Neel, B. G., Gu, H., and Pao, L. ( 2003) The ‘Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28, 284–293 [DOI] [PubMed] [Google Scholar]
- 53.Agazie, Y. M., and Hayman, M. J. ( 2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol. Cell. Biol. 23, 7875–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yigzaw, Y., Poppleton, H. M., Sreejayan, N., Hassid, A., and Patel, T. B. ( 2003) Protein-tyrosine phosphatase-1B (PTP1B) mediates the anti-migratory actions of Sprouty. J. Biol. Chem. 278, 284–288 [DOI] [PubMed] [Google Scholar]
- 55.Head, J. A., Jiang, D., Li, M., Zorn, L. J., Schaefer, E. M., Parsons, J. T., and Weed, S. A. ( 2003) Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14, 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petit, M. M., Meulemans, S. M., and Van de Ven, W. J. ( 2003) The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J. Biol. Chem. 278, 2157–2168 [DOI] [PubMed] [Google Scholar]
- 57.Choi, J. H., Kim, H. S., Kim, S. H., Yang, Y. R., Bae, Y. S., Chang, J. S., Kwon, H. M., Ryu, S. H., and Suh, P. G. ( 2006) Phospholipase Cγ1 negatively regulates growth hormone signalling by forming a ternary complex with Jak2 and protein tyrosine phosphatase-1B. Nat. Cell Biol. 8, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 58.Iwabu, A., Smith, K., Allen, F. D., Lauffenburger, D. A., and Wells, A. ( 2004) Epidermal growth factor induces fibroblast contractility and motility via a protein kinase C δ-dependent pathway. J. Biol. Chem. 279, 14551–14560 [DOI] [PubMed] [Google Scholar]
- 59.Benes, C., and Soltoff, S. P. ( 2001) Modulation of PKCδ tyrosine phosphorylation and activity in salivary and PC-12 cells by Src kinases. Am. J. Physiol. 280, C1498–C1510 [DOI] [PubMed] [Google Scholar]
- 60.Prasad, N., Topping, R. S., and Decker, S. J. ( 2002) Src family tyrosine kinases regulate adhesion-dependent tyrosine phosphorylation of 5′-inositol phosphatase SHIP2 during cell attachment and spreading on collagen I. J. Cell. Sci. 115, 3807–3815 [DOI] [PubMed] [Google Scholar]
- 61.Cozzolino, M., Giovannone, B., Serafino, A., Knudsen, K., Levi, A., Alema, S., and Salvatore, A. ( 2000) Activation of TrkA tyrosine kinase in embryonal carcinoma cells promotes cell compaction, independently of tyrosine phosphorylation of catenins. J. Cell Sci. 113, 1601–1610 [DOI] [PubMed] [Google Scholar]
- 62.Alema, S., and Salvatore, A. M. ( 2007) p120 catenin and phosphorylation: mechanisms and traits of an unresolved issue. Biochim. Biophys. Acta 1773, 47–58 [DOI] [PubMed] [Google Scholar]
- 63.Itoh, M., Nagafuchi, A., Yonemura, S., Kitani-Yasuda, T., and Tsukita, S. ( 1993) The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J. Cell Biol. 121, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dadke, S., and Chernoff, J. ( 2003) Protein-tyrosine phosphatase 1B mediates the effects of insulin on the actin cytoskeleton in immortalized fibroblasts. J. Biol. Chem. 278, 40607–40611 [DOI] [PubMed] [Google Scholar]
- 65.Kabuyama, Y., Langer, S. J., Polvinen, K., Homma, Y., Resing, K. A., and Ahn, N. G. ( 2006) Functional proteomics identifies protein-tyrosine phosphatase 1B as a target of RhoA signaling. Mol. Cell. Proteomics 5, 1359–1367 [DOI] [PubMed] [Google Scholar]
- 66.Liu, F., Hill, D. E., and Chernoff, J. ( 1996) Direct binding of the proline-rich region of protein tyrosine phosphatase 1B to the Src homology 3 domain of p130(Cas). J. Biol. Chem. 271, 31290–31295 [DOI] [PubMed] [Google Scholar]
- 67.Mauro, L. J., and Dixon, J. E. ( 1994) ‘Zip codes' direct intracellular protein tyrosine phosphatases to the correct cellular ‘address'. Trends Biochem. Sci. 19, 151–155 [DOI] [PubMed] [Google Scholar]
- 68.Frangioni, J. V., Beahm, P. H., Shifrin, V., Jost, C. A., and Neel, B. G. ( 1992) The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 68, 545–560 [DOI] [PubMed] [Google Scholar]
- 69.Boute, N., Boubekeur, S., Lacasa, D., and Issad, T. ( 2003) Dynamics of the interaction between the insulin receptor and protein tyrosine-phosphatase 1B in living cells. EMBO Rep. 4, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haj, F. G., Verveer, P. J., Squire, A., Neel, B. G., and Bastiaens, P. I. ( 2002) Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 71.Anderie, I., Schulz, I., and Schmid, A. ( 2007) Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell. Signal. 19, 582–592 [DOI] [PubMed] [Google Scholar]
- 72.Kuchay, S. M., Kim, N., Grunz, E. A., Fay, W. P., and Chishti, A. H. ( 2007) Double knockouts reveal that protein tyrosine phosphatase 1B is a physiological target of calpain-1 in platelets. Mol. Cell. Biol. 27, 6038–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calderwood, D. A., Shattil, S. J., and Ginsberg, M. H. ( 2000) Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J. Biol. Chem. 275, 22607–22610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.