Abstract
Four covalent complexes between recombinant yeast cytochrome c and cytochrome c peroxidase (rCcP) were synthesized via disulfide bond formation using specifically designed protein mutants [Papa, H. S., and Poulos, T. L. (1995) Biochemistry 34, 6573–6580]. One of the complexes, designated V5C/K79C, has cysteine residues replacing valine-5 in rCcP and lysine-79 in cytochrome c with disulfide bond formation between these residues linking the two proteins. The V5C/K79C complex has the covalently-bound cytochrome c located on the ‘back-side’ of cytochrome c peroxidase, ~180° from the primary cytochrome c-binding site as defined by the crystallographic structure of the 1:1 non-covalent complex [Pelletier, H., and Kraut J. (1992) Science 258, 1748–1755]. Three other complexes have the covalently bound cytochrome c located ~90° from the primary binding site and are designated K12C/K79C, N78C/K79C, and K264C/K79C, respectively. Steady-state kinetic studies were used to investigate the catalytic properties of the covalent complexes at both 10 and 100 mM ionic strength, pH 7.5. All four covalent complexes have catalytic activities similar to those of rCcP (within a factor of two). A comprehensive study of the ionic strength dependence of the steady-state kinetic properties of the V5C/K79C complex provides evidence for significant electrostatic repulsion between the two cytochromes bound in the 2:1 complex at low ionic strength but that the electrostatic repulsion decreases as the ionic strength of the buffer increases.
Cytochrome c peroxidase (CcP)1 is a detoxification enzyme localized between the inner and outer membranes of yeast mitochondria (1). CcP detoxifies hydrogen peroxide by catalyzing its reduction to water using the reducing equivalents from two molecules of ferrocytochrome c (2). The catalytic mechanism involves oxidation of the native enzyme by hydrogen peroxide to an oxidized enzyme intermediate followed by reduction of the intermediate back to the native enzyme by ferrocytochrome c. CcP effectively couples the two-electron reduction of hydrogen peroxide to the one-electron oxidation of ferrocytochrome c by using two equivalents of cytochrome c per catalytic cycle. Since 1980, when the three-dimensional structure of CcP was first revealed (3, 4), CcP has played a significant role in elucidating the structural basis for heme protein reactivity, especially in the activation of hydrogen peroxide (5, 6), long-range electron transfer between heme proteins (7, 8), and protein-protein interactions (9, 10).
At low ionic strength, CcP can bind two molecules of cytochrome c (11) with quite different affinities. Below 30 mM ionic strength, the binding of the first yeast iso-1 cytochrome c is very strong with equilibrium dissociation constants for the 1:1 non-covalent complex, KD1, of less than 0.1 μM (12, 13) while binding of the second cytochrome c is much weaker with KD2 values in the 100 to 200 μM range (7, 14, 15). The ability of CcP to bind more than one molecule of cytochrome c had engendered a number of questions concerning the nature of the interaction between these two proteins and the role of cytochrome c bound at different locations on the surface of CcP in the catalytic mechanism. We have recently shown that only cytochrome c bound in the vicinity of the crystallographically defined (9), or primary binding site, is electron transfer active in the normal catalytic cycle of CcP (16). The role of cytochrome c binding at secondary sites is only apparent below ~100 mM ionic strength and appears to involve modulation of the rate of association and dissociation of cytochrome c from the primary site. This implies interaction between cytochrome c molecules bound at the two sites.
In this report we present the results of steady-state kinetic studies of the catalytic activity of four specifically engineered 1:1 cytochrome c/CcP covalent complexes, all of which leave the primary cytochrome c binding site open and accessible to free cytochrome c. All four covalent complexes are catalytically active with activities similar to those of free rCcP. We demonstrate that the Michaelis constants determined from steady-state kinetics studies are in good agreement with published values for the equilibrium dissociation constants of the 1:1 and 2:1 non-covalent complexes, KD1 and KD2, respectively, and can be used as a measures of binding affinity between cytochrome c and CcP. The data provide evidence for significant electrostatic repulsion between bound cytochrome c molecules on the surface of CcP at low ionic strength with the interaction energy diminishing with increasing ionic strength. Above 100 mM ionic strength, the interaction energy is effectively zero.
MATERIALS AND METHODS
Synthesis of the CcP/cytochrome c Covalent Complexes
The starting clones containing the genes for CcP and yeast iso-1 cytochrome c, mutagenesis techniques, protein expression and protein purification have been previously described (16). Both wild-type CcP and yeast iso-1 cytochrome c contain single cysteine residues, Cys-128 in CcP and Cys-102 in yeast iso-1 cytochrome c. The cysteine residues in the wild-type proteins were eliminated by conversion to Ser-128 in rCcP and Thr-102 in recombinant cytochrome c prior to constructing specifically engineered cysteine sites in the two proteins. The conversion of the wild-type cysteine residues to hydroxy-containing amino acid residues eliminates the possibility of making undesired disulfide bonds during the synthesis of the covalent complexes. Four rCcP mutants were constructed: rCcP(V5C), rCcP(K12C), rCcP(N78C), and rCcP(K264C); each of these rCcP mutants contain the C128S alteration. A single cytochrome c mutant was constructed, cytochrome c(K79C), a mutation that also contains the C102T change.
The synthesis of the covalent complexes was performed in a manner similar to that described by Papa and Poulos (17). Slight modifications to the Papa-Poulos procedure were developed to improve the yield and these modifications have been described (16). Four covalent complexes were synthesized and these are designated V5C/K79C, K12C/K79C, N78C/K79C, and K264C/K79C. The complex names give the mutational designation in rCcP followed by the mutational designation in cytochrome c.
The covalent complexes were separated from the synthetic reaction mixture by chromatography using a carboxymethyl-cellulose (Whatman CM52) column (1 × 10 cm) as previously described (16). The starting buffer was O2-saturated 25 mM ammonium acetate. Step-wise elution with increasing concentrations of ammonium acetate was used to recover fractions containing the covalent complex. Pooled fractions containing the covalent complex were dialyzed against 4 to 5 changes of deionized water at 4°C, lyophilized, and stored at −20 °C until used.
SDS-PAGE Analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a PhastSystem from Pharmacia LKB Biotechnology AB, Uppsala, Sweden. Proteins were incubated at room temperature for 15 minutes in a 10 mM Tris/HCl buffer, pH 8, containing 2.5% SDS and 1 mM EDTA. To preserve the covalent disulfide cross-links, the denaturing buffer did not contain disulfide bond reducing agents. Protein samples were loaded onto a PhastGel gradient 10–15 pre-cast gel along with standard molecular weight markers. SDS buffer strips were used during the electrophoresis. The gels were stained with Coomassie blue.
Protein Concentration Determination
Spectra of all proteins were acquired using either a Varian/Cary Model 3E spectrophotometer or a Hewlett Packard Model 8452A diode array spectrophotometer. The spectra were used to determine protein concentration using the extinction coefficients at the Soret maximum. An extinction coefficient of 98 mM−1 cm−1 at the Soret maximum (408 nm) was used to determine the concentration of rCcP and its mutants. Extinction coefficients of 118 mM−1 cm−1 (408 nm) and 150 mM−1 cm−1 (414 nm) were used to determine the concentration of oxidized and reduced recombinant yeast iso-1 cytochrome c(C102T) and its mutants, respectively (16).
Hydrogen Peroxide Concentration
Hydrogen peroxide was reagent grade 30% (v/v) purchased from Aldrich Chemical Company, Inc. Hydrogen peroxide stock solutions were standardized by titration with cerium(IV) sulfate (18).
Steady-State Kinetic Studies
Steady-state kinetic studies were performed at pH 7.5 in potassium phosphate buffers of varying ionic strength. Buffer, cytochrome c, and enzyme were thermally equilibrated at 25 °C in the spectrophotometer, initial absorbance readings made, then the reaction initiated by addition of the hydrogen peroxide. Initial velocities, v0, were determined as a function of yeast iso-1 ferrocytochrome c(C102T) concentration (generally 1 to 100 (μM) at constant hydrogen peroxide (200 (μM). Initial velocities were calculated by measuring the time-dependent change in absorbance upon oxidation of ferrocytochrome c at multiple wavelengths using a Hewlett Packard Model 8452A diode array spectrophotometer. Five different wavelengths, generally chosen from 314, 362, 418, 448, 468, 478, 548, 564, and 574 nm depending upon the substrate concentration, were used to calculate the initial velocity at each set of experimental conditions using eq 1. The symbols in eq 1
| (1) |
include the initial velocity, v0, the total enzyme concentration, e0, the change in absorbance with time, ΔA/Δt, and the difference in extinction coefficient, Δε, between oxidized and reduced yeast iso-1 cytochrome c(C102T). Samples of the substrate may contain small amounts of oxidized cytochrome c that inhibit the reaction; fox is the fraction of oxidized cytochrome c in the substrate and is used to make small corrections to the initial velocity. The factor of 2 in the denominator converts cytochrome c turnover to enzyme turnover since two cytochrome c molecules are oxidized per enzymatic cycle.
RESULTS AND DISCUSSION
Design and Synthesis of the Covalent Complexes
In a previous publication (16) we reported on the synthesis and characterization of a covalent cytochrome c/CcP complex in which the covalently-bound cytochrome c blocked the primary binding site, the cytochrome c-binding site identified in the crystallographic structure of the non-covalent complex between yeast iso-1 cytochrome c and CcP (9). This complex was completely inactive and we concluded that only cytochrome c bound at the primary binding site could transfer electrons to the oxidized CcP intermediates during catalytic turnover. The catalytic activity of CcP measures the reaction occurring at the primary binding site.
In this study, our objective is to determine the effect of cytochrome c molecules covalently bound to the surface of CcP at locations other than the primary binding site on the reactions occurring at the primary site. We designed four complexes that have the covalently bound cytochrome c sufficiently far from the primary binding site to minimize steric inhibition of free cytochrome c binding at the primary site. All of the experimental evidence indicates that free cytochrome c only binds to the primary binding site in these covalent complexes.
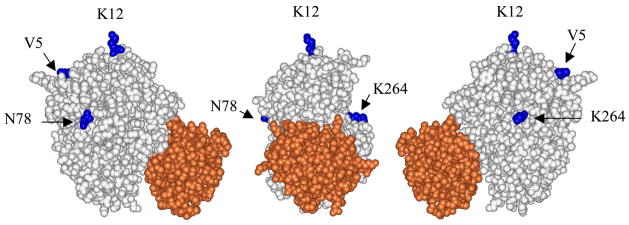
Figure 1 shows the covalent attachment sites on CcP relative to the primary binding site and it also defines the ‘front face’, the ‘right-side’, the ‘left-side’, and the ‘back-side’ of CcP. The V5C/K79C complex has a yeast cytochrome c covalently attached to the back-side of CcP, located ~180° from the primary cytochrome c binding site as far from the primary binding site as possible on the surface of CcP. The other three complexes have the covalently bound cytochrome c located ~90° from the primary binding site.
Figure 1.
The yeast iso-1 cytochrome c/CcP complex (9). Yeast cytochrome c is shown in red, CcP is shown in white and the residues in CcP that were individually converted to cysteine residues are shown in blue. Center - space-filling rendering of the complex with the cytochrome c in front of the CcP molecule. This view defines the ‘front face’ of CcP. The primary cytochrome c-binding site is located on the lower half of the front face of CcP. Three of the four CcP residues that are converted to cysteine residues can be seen in this view: Lys-12, Asn-78, and Lys-264. Right-hand side - A 90° clockwise rotation of the central figure about the vertical axis exposes the right face of the CcP molecule. Val-5, on the back side of CcP can be seen as well as the relative locations of Lys-12 and Lys-264. Left-hand side - A 90° counter-clockwise rotation of the central figure about the vertical axis exposes the left face of CcP. Val-5, Lys-12, and Asn-78 are seen in this view.
The four covalent complexes were synthesized as previously described (16, 17). Yields of the four complexes were similar to that reported for the E290C/K73C complex (16). SDS-PAGE analysis of overloaded gels of the covalent complexes showed small amounts of unreacted rCcP and cytochrome c in the purified samples. This is consistent with the results found for the E290C/K73C complex reported previously (16) where it was shown that small amounts of unreacted rCcP and recombinant yeast iso-1 cytochrome c co-purify with the covalent complexes.
Steady-State Kinetics of the Covalent Complexes at 10 mM Ionic Strength
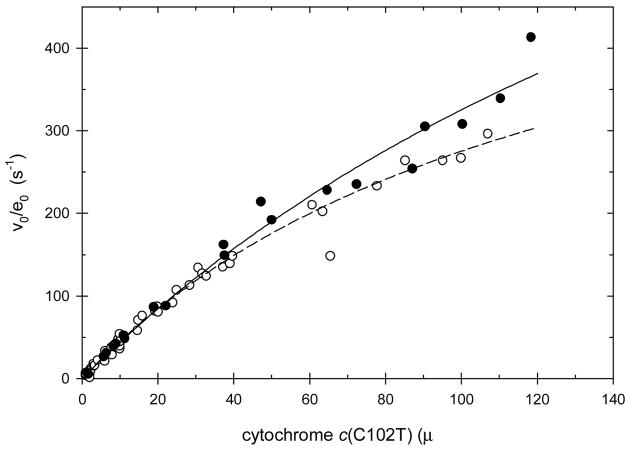
Figure 2 shows the initial velocities for the oxidation of recombinant yeast iso-1 ferrocytochrome c(C102T) by hydrogen peroxide as catalyzed by the V5C/K79C covalent complex as a function of the ferrocytochrome c concentration. Figure 2 includes similar data for rCcP catalysis for reference. It is readily apparent that the V5C/K79C covalent complex is as catalytically active as rCcP itself under these conditions. Although the data formally fit the Michaelis-Menten equation, there is insufficient curvature in the plots to obtain accurate values of both the Michaelis constant and the maximum velocity. This is a direct indication that the interaction between enzyme and substrate is very weak under these experimental conditions and that only a small fraction of the enzyme is converted to the enzyme-substrate complex during the steady-state turnover, even at the highest substrate concentration used in the experiments.
Figure 2.
Steady-state velocities for recombinant yeast iso-1 ferrocytochrome c(C102T) oxidation as a function of the cytochrome c concentration at 10 mM ionic strength, pH 7.5, [H2O2] = 200 μM. The reaction is catalyzed either by rCcP (open circles) or the V5C/K79C covalent complex (solid circles).
The most accurate steady-state parameter that can be obtained from the 10 mM ionic strength data is the limiting slope of the plot of velocity versus ferrocytochrome c concentration at low substrate concentration. A modified form of the Michaelis-Menten equation was used to fit the data, eq 2,
| (2) |
where kbi = VM/(e0·KM). kbi has units of an apparent bimolecular rate constant and is the initial slope of a plot of the normalized initial velocities versus the substrate concentration. It can be considered a lower limit to the bimolecular rate constant for the association of cytochrome c and the enzyme under these conditions. The value of the maximum turnover rate, kcat = VM/e0, can be calculated from kbi and KM. The steady-state parameters for rCcP and the V5C/K79C covalent complex are collected in Table 1. The steady-state parameters for the other three complexes are also given in Table 1. Plots of the data for the K12C/K79C, N78C/K79C, and K264C/K79C complexes are shown in the Supporting Information accompanying this article.
Table 1.
Steady-State Parameters for the Oxidation of Recombinant Yeast Iso-1 Ferrocytochrome c by rCcP and Four Cytochrome c/CcP Covalent Complexes at both 10 and 100 mM Ionic Strength.a
| 10 mM Ionic Strength
|
100 mM Ionic Strength
|
||||
|---|---|---|---|---|---|
| Catalyst | kbi (μM−1s−1) | KM (μM) | kcat (s−1) | KM (μM) | kcat (s−1) |
| rCcP | 4.8 ± 0.2 | 130 ± 20 | 630 ± 50 | 2.1 ± 0.2 | 640 ± 10 |
| V5C/K79C | 4.6 ± 0.4 | 250 ± 80 | 1100 ± 300 | 2.7 ± 0.3 | 410 ± 10 |
| K12C/K79C | 4.4 ± 0.3 | 240 ± 50 | 1000 ± 200 | 2.6 ± 0.4 | 330 ± 10 |
| N78C/K79C | 4.1 ± 0.7 | 170 ± 100 | 680 ± 290 | 2.3 ± 0.3 | 320 ± 10 |
| K264C/K79C | 4.0 ± 0.8 | 170 ± 110 | 690 ± 330 | 3.7 ± 0.6 | 470 ± 20 |
Substrate: recombinant yeast iso-1 ferrocytochrome c(C102T), [H2O2] = 200 μM, potassium phosphate buffer, pH 7.5, 25 °C.
As can be seen from the data in Table 1, the initial slopes, kbi, for all four covalent complexes are within the estimated experimental error of kbi for rCcP. The KM values vary by about a factor of two, ranging from 130 μM for rCcP to 250 μM for the V5C/K79C complex. The precision of KM is not high since all of the fitted KM values are greater that the highest substrate concentrations used in these steady-state experiments. Likewise, the calculated maximum turnover rates, kcat, vary by about a factor of two, from 630 s−1 for rCcP to about 1100 s−1 for the V5C/K79C complex. As is to be expected, there is a strong positive correlation between the KM and kcat values.
Steady-State Kinetics of the Covalent Complexes at 100 mM Ionic Strength
Figure 3 shows the initial velocities for the oxidation of recombinant yeast iso-1 ferrocytochrome c(C102T) by hydrogen peroxide as catalyzed by the V5C/K79C covalent complex as a function of the ferrocytochrome c concentration. Figure 3 includes similar data for rCcP catalysis for reference. Plots of the steady-state velocity data at 100 mM ionic strength for the other three complexes are included in the Supporting Information provided with this article. At 100 mM ionic strength, the apparent binding affinity of cytochrome c to CcP is much higher than at 10 mM ionic strength, KM values between 2 and 4 μM at 100 mM ionic strength compared to KM > 100 μM at 10 mM ionic strength. In the case of the wild-type yeast CcP, the Michaelis constants has been associated with binding of cytochrome c to the primary binding site at 100 mM ionic strength and the binding of cytochrome c to a secondary binding site at 10 mM ionic strength (19). At 100 mM ionic strength the curvature in the velocity/substrate plots is sufficiently great that accurate values for both KM and kcat can be obtained and these are included in Table 1. Both KM and kcat for the covalent complexes are within a factor of two of the corresponding parameters for rCcP.
Figure 3.
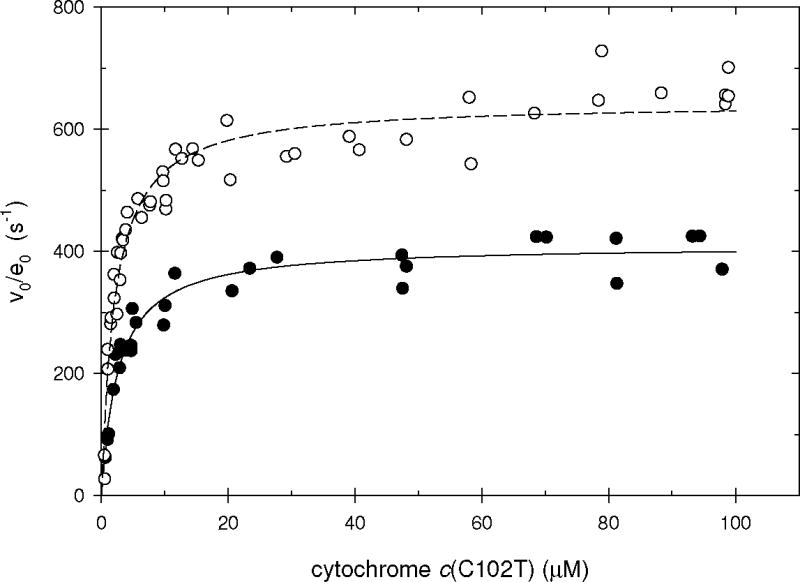
Steady-state velocities for recombinant yeast iso-1 ferrocytochrome c(C102T) oxidation as a function of the cytochrome c concentration at 100 mM ionic strength, pH 7.5, [H2O2] = 200 μM. The reaction is catalyzed either by rCcP (open circles) or the V5C/K79C covalent complex (solid circles).
All four covalent complexes have the primary cytochrome c-binding site accessible to the substrate and substrate turnover is efficient. There is a systematic trend in the data, with the kcat values for the complexes slightly lower than kcat for rCcP and the KM values for the complexes slightly larger than the KM value for rCcP. Another interesting observation is that the kbi values at 100 mM ionic strength vary much less than either KM or kcat. The kbi values range from 1.3 × 108 M−1 s−1 for both the K12C/K79C and K264/K79C complexes to 1.6 × 108 M−1 s−1 for rCcP. These apparent bimolecular rate constants are approaching the diffusion-limit for association rate constants (20) suggesting that the binding of cytochrome c to the primary binding site on CcP is very efficient at the higher ionic strength.
Ionic strength Dependence of KM for the V5C/K79C Complex
It has previously been shown (19) that plots of the steady-state initial velocities as a function of the yeast iso-1 ferrocytochrome c concentration for wild-type yeast CcP are generally biphasic and can be empirically fit to an equation that is the sum of two Michaelis-Menten terms. The high-affinity Michaelis constant, KM1, is related to substrate dissociation from the 1:1 cytochrome c/CcP complex and the low-affinity Michaelis constant, KM2, is related to dissociation from the 2:1 complex. Due to the ionic strength dependence of cytochrome c binding to both the primary and secondary binding sites on yCcP, the initial velocity plots revert to simple, single term Michaelis-Menten plots at low (≤ 20 mM) and high (≥ 100 mM) ionic strength. The low ionic strength data only monitor properties of the 2:1 complex while the high ionic strength data only reflect properties of the 1:1 complex. As shown in Figures 2, 3, and in the Supporting Information, the steady-state velocity plots for rCcP and the four covalent complexes fit the simple Michaelis-Menten equation at both 10 and 100 mM ionic strength.
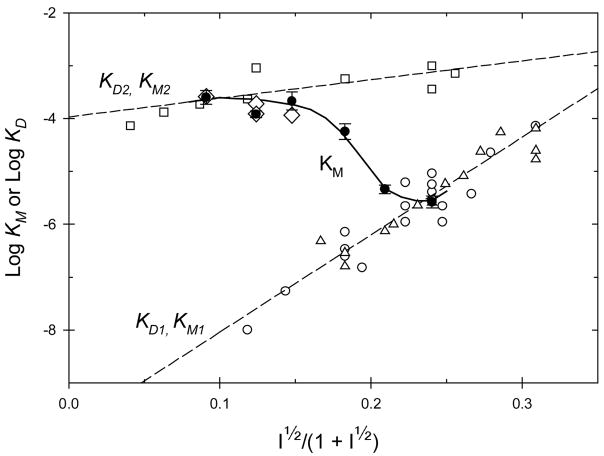
The V5C/K79C complex was chosen for a more complete study of the ionic strength dependence of the steady-state parameters. The initial velocities for the V5C/K79C complex could be fit to the simple Michaelis-Menten equation at all ionic strengths between 10 and 100 mM. The steady-state parameters, kbi, KM, and kcat, for the V5C/K79C complex at different ionic strengths are collected in Table 2. Figure 4 shows a plot of KM for the V5C/K79C complex as a function of ionic strength along with reference data from the literature (21).
Table 2.
Steady-State Parameters for the V5C/K79C Covalent Complex at Various Ionic Strengthsa
| Ionic Strength (mM) | kbi (μM−1s−1) | KM(μM) | kcat (s−1) |
|---|---|---|---|
| 10 | 4.6 ± 0.4 | 250 ± 80 | 1100 ± 300 |
| 20 | 3.2 ± 0.2 | 120 ± 20 | 390 ± 40 |
| 30 | 3.4 ± 0.3 | 210 ± 80 | 740 ± 220 |
| 50 | 6.4 ± 1.2 | 56 ± 19 | 360 ± 60 |
| 70 | 53 ± 8 | 4.6 ± 0.8 | 240 ± 10 |
| 100 | 150 ± 10 | 2.7 ± 0.3 | 410 ± 10 |
Substrate: recombinant yeast iso-1 ferrocytochrome c(C102T), [H2O2] = 200 μM, potassium phosphate buffer, pH 7.5, 25 °C.
Figure 4.
Variation of KM (solid circles) for the V5C/K79C covalent complex as a function of ionic strength. Published data are included for reference. The lower linear plot includes published values for KD1 (open circles) for the yeast iso-1 cytochrome c/CcP complex. The KD1 values include data collected between 18 and 200 mM ionic strength, between pH 5.5 and 7.0, and for complexes formed using either ferri- or ferrocytochrome c (11–13). Michaelis constants for the high-affinity phase, KM1 (open triangles), of the steady-state oxidation of yeast iso-1 ferrocytochrome c catalyzed by yCcP (19) and CcP(MI) (23) are included. The upper linear plot includes values for KD2 (open squares) obtained between 1.8 mM and 118 mM ionic strength, between pH 6 and 7, using proton uptake measurements (11) and dynamic fluorescence quenching of zinc-modified CcP (7, 14, 15). Michaelis constants for the low-affinity phase, KM2 (open diamonds), of the steady-state oxidation of yeast iso-1 ferrocytochrome c catalyzed by yCcP (19) and CcP(MI) (23) are included.
Some comment on the reference data shown in Figure 4 is useful for full understanding of the significance of the ionic strength dependence of the KM values for the V5C/K79C complex. Equilibrium dissociation constants, KD1, for the 1:1 yeast iso-1 cytochrome c/yCcP complex are shown by the open circles in the lower plot in Figure 4. These data include all published values for KD1 between pH 5.5 and 7.0 (11–13) since the pH dependence of KD1 is small relative on the logarithmic scale used in Figure 4. The most extensive data set suggests that KD1 varies by about a factor of two over the pH range 6.0 to 7.5 at 50 mM ionic strength (11). Most of the data are for the binding of the oxidized form of the yeast cytochrome but the plot includes three data points for the reduced cytochrome c/CcP complex. The binding affinity for reduced cytochrome c is about 3-fold weaker than for oxidized yeast iso-1 cytochrome c, again small relative to the range of KD1 shown in Figure 4. . The dashed line through the lower plot is generated by linear least square regression of the logarithm of KD1 as a function of the ionic strength as defined in eq 3.
| (3) |
The experimental KD1 values span a range of >7,000-fold between 18 and 200 mM ionic strength. Eq 3 predicts the values of KD1 within a factor of two in most cases with the average RMS deviation ± 60% of the mean value.
The open triangles shown in the lower plot of Figure 4 are Michaelis constants for the high-affinity phase, KM1, of the steady-state oxidation of yeast iso-1 ferrocytochrome c as catalyzed by wild-type yeast CcP (19) and by recombinant CcP(MI) (23). The data were collected at both pH 6.0 and 7.5 and between 40 and 200 mM ionic strength. As can be seen in Figure 4, the available KM1 values cluster around the KD1 values and one can assume that the Michaelis constants are reasonable measures for the binding of yeast iso-1 cytochrome c to CcP. Comparing experimental values of KM1 with values of KD1 calculated from eq 3 indicates that KM1 is only about 10% larger than KD1 on average, within experimental error of both measurements.
There are relatively few quantitative values for KD2 and KM2 in the literature. The open squares shown in the upper plot of Figure 4 are published values for KD2(7, 11, 14, 15). The open diamonds shown in the upper plot of Figure 4 are KM2 values for the low-affinity phase of the steady-state oxidation of yeast iso-1 ferrocytochrome c by wild-type CcP at pH 7.5 (19) and recombinant CcP(MI) at pH 6.0 (23). The KM2 values could only be determined between 10 and 30 mM ionic strength. Again there is a good agreement between the KD2 and KM2 values indicating that KM2 is reporting the interaction of cytochrome c at the secondary binding site. The upper dashed line in Figure 4 was calculated from eq 4, the correlation line for the logarithm of KD2 as a function of ionic strength.
| (4) |
Figure 4 shows the value of KM for the V5C/K79C as a function of ionic strength. The value of KM for the V5C/K79C complex is comparable to KD2 for CcP at low ionic strength and to KD1 for CcP at high ionic strength. As outlined above, the Michaelis constant for the V5C/K79C complex is a direct measure of the binding of yeast iso-1 cytochrome c(C102T) to the primary binding site on the surface of the CcP in the covalent complex. At high ionic strength, ≥100 mM, the covalently bound cytochrome has little influence on the binding of exogenous cytochrome c to the primary binding site. The binding affinity for exogenous cytochrome c is essentially identical for both V5C/K79C and CcP. Under these conditions there is no interaction between the two bound cytochromes, whether the cytochromes are bound covalently or non-covalently.
At low ionic strength, ≥20 mM, there is strong electrostatic repulsion between the covalently bound cytochrome c and cytochrome c binding to the primary site. In this case, the Michaelis constant is similar to KD2. Note that in the case of the V5C/K79C complex, the Michaelis constant is measuring the binding of cytochrome c at the primary site of rCcP in the presence of a cytochrome c covalently bound at a secondary site while for the non-covalent complex, KD2 is a measure of the binding of cytochrome c to a secondary site in the presence of a non-covalently bound cytochrome at the primary binding site.
Model for the Yeast cytochrome c/CcP Interactions
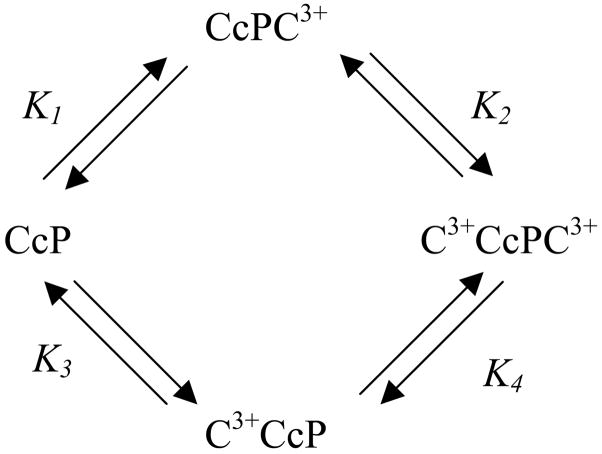
The model for cytochrome c binding to CcP that emerges from these results is one of unique, interacting sites. The primary binding site has a higher intrinsic affinity for cytochrome c than the secondary site and there are strong interactions between sites at low ionic strength. The interaction energy is modulated by ionic strength and effectively goes to zero at ≥100 mM ionic strength. The binding model is shown in Scheme 1, where binding of cytochrome c to the primary binding site is represented by placing the symbol for cytochrome c to the right-hand side of CcP and for binding at a secondary site, by placing the symbol on the left-hand side of the CcP. K1, K2, K3, and K4 represent the microscopic, or site-specific constants. Thus K1 represent the microscopic equilibrium dissociation constant for the 1:1 complex with the cytochrome c bound at the primary binding site and K3 the dissociation constant for the 1:1 complex in which the cytochrome c is bound at a secondary site. K2 represents the equilibrium dissociation constant for the 2:1 complex in which cytochrome c dissociates from the secondary site and K4 represents the dissociation of cytochrome c from the primary binding site in the 2:1 complex. For an in-depth description of this binding model see reference (24).
Scheme 1.
Microscopic model for the binding of cytochrome c to two unique sites on CcP, a primary binding site (right-hand side) and a secondary binding site (left-hand side).
The relationship between the microscopic equilibrium constants shown in Scheme 1 and the observed equilibrium dissociation constants are given by eqs 5 and 6.
| (5) |
| (6) |
The crystal structure CcP and of the 1:1 yeast cytochrome c/CcP non-covalent complex (4, 9) show no significant conformational differences in CcP upon binding of the yeast cytochrome c. This suggests that the intrinsic binding affinities of the primary and secondary binding sites are not affected by complex formation and that K2 and K4 are only affected by electrostatic repulsion between cytochromes binding at the two sites in the 2:1 complex. Eqs 7 and 8 show the relationships between K1 and K4 and between K2 and K3, respectively.
| (7) |
| (8) |
At ionic strengths of ≥ 100 mM, KM is numerically equal to KD1 indicating that the covalently bound cytochrome c in the covalent complex has essentially no effect on the binding of cytochrome c at the primary binding site. The interactions energy, ΔGi is effectively zero and K2 = K3 and K4 = K1. Under high ionic strength conditions the microscopic constants can be calculated from the observed equilibrium dissociation constants. Using eqs 3 through 8, with the stipulation that ΔGi is zero, one can determine that K1 = K4 = 3.5 μM and K2 = K3 = 750 μM at 100 mM ionic strength. The affinity for yeast iso-1 cytochrome c binding at the primary site is greater than 200 times the binding affinity at any secondary site in the formation of 1:1 complexes and less than 0.5% of the 1:1 complexes in solution have cytochrome c bound at a secondary site. The upper pathway in Scheme 1 is a good representation of binding of cytochrome c to CcP with the lower pathway making a negligible contribution.
As the ionic strength is decreased, electrostatic repulsion between cytochrome c bound at the primary binding site weakens binding at the secondary binding site relative to binding at the primary site and at 10 mM ionic strength there is a difference of about 35,000 in binding affinity, i.e., Kj ≅ KD1 = 6.2 nM and K2 = KD2 = 220 μM.
The data for the V5C/K79C complex can be used to calculate the minimum interaction energy as a function of ionic strength. KD1 ≅ K1 represents the intrinsic binding at the primary binding site as a function of ionic strength while KM for the V5C/K79C complex represents the binding of cytochrome c to the primary binding site in the presence of a covalently-bound cytochrome c, positioned as far as possible from the primary binding site on the surface of CcP. Eq 9 can be used to calculate ΔGi at any ionic strength for which KM has been determined. At 10 mM ionic strength the interaction energy is
| (9) |
+6.3 kcal/mol.
The ionic strength modulates the electrostatic repulsion between the bound cytochromes in the 2:1 complex and this provides a rational for the observation that KD2 is not as dependent upon the ionic strength as is KD1 (Figure 4 and eqs 3 and 4). The intrinsic binding constant at both the primary and secondary binding sites are determined in large part by the electrostatic attraction between the negatively charged CcP and the positively charged cytochrome c. At pH 7.5, the net charge on CcP is about −10 (25) and about +5 on cytochrome c (26) leading to a very large slope (+18.43) in the plot of log KD1 as a function of ionic strength (Figure 4 and eq 3). However, the intrinsic electrostatic attraction of the secondary binding site is largely compensated by the electrostatic repulsion from the cytochrome bound at the primary site, decreasing the ionic strength dependence of the log KD2 by a factor of 5, eq 4.
One of the initial objectives of this project was to determine if the covalent complexes could give some information concerning the location of potential secondary binding sites based on the assumption that the interaction energy between covalently bound cytochrome c and cytochrome c binding at the primary site would depend upon the location of the covalently attached cytochrome c. However, the data at 10 mM ionic strength, Table 1, indicate that the interaction between the covalently bound cytochromes and cytochrome c binding at the primary site is essentially the same for all four complexes, regardless of the location of the covalently bound cytochrome c.
Conclusions
The Michaelis constants from steady-state kinetic studies of the CcP-catalyzed oxidation of yeast iso-1 ferrocytochrome c by hydrogen peroxide agree quite well with the equilibrium dissociations constants for the 1:1 and 2:1 yeast iso-1 cytochrome c/CcP complexes and can be used as measures of the interaction between cytochrome c and CcP. The binding model for yeast cytochrome c and CcP can be described as a unique, two-site interacting model in which the primary binding site for cytochrome c on the surface of CcP has an affinity for the yeast cytochrome that is at least 200-times greater than the secondary site. The observed equilibrium dissociation constant, KD1, is essentially a specific site constant and represents dissociation of cytochrome c from the primary site at all ionic strengths. Likewise, KD2 represents dissociation of cytochrome c from a secondary binding site in the 2:1 complex. There is strong electrostatic repulsion between cytochrome c molecules bound at the two sites at low ionic strength, but the electrostatic interaction energy decreases with increasing ionic strength and is negligible above 100 mM ionic strength.
Supplementary Material
Plots of the steady-state velocities as a function of the yeast ferrocytochrome c concentration for the K12C/K769C, N78C/K79C, and K264/K79C covalent complexes at both 10 and 100 mM ionic strength. This material is available free of charge via the Internet at http://pubs.acs/org.
Acknowledgments
We thank Professor James Satterlee, Washington State University for providing the plasmid containing the gene for rCcP and Professor Gary Pielak, University of North Carolina, Chapel Hill for providing the plasmid containing the yeast iso-1 cytochrome c(C102T) and heme lyase genes.
Footnotes
This work was supported in part by a grant from the National Institutes of Health (R15 GM59740).
Abbreviations: CcP, generic abbreviation for cytochrome c peroxidase whatever the source; yCcP, authentic yeast cytochrome c peroxidase isolated from S. cervisiae; rCcP, recombinant cytochrome c peroxidase expressed in E. coli, the rCcP used in this study has an amino acid sequence identical to that of yCcP; mutations in the amino acid sequences of either CcP or cytochrome c are indicated by using the one letter code for the amino acid residue in the wild-type protein, followed by the residue number and the one letter code for the amino acid residue in the mutant protein, i.e., C102T represents a mutant in which a threonine residue replaces the cysteine residue at position 102 of the wild-type protein; the four covalent complexes investigated in this manuscript are designated V5C/K79C, K12C/K79C, N78C/K79C, and K264C/K79C where the first mutantional designation is that in rCcP and the second in recombinant yeast iso-1 cytochrome c with the disulfide bond linking the two proteins between the two cysteine residues.
References
- 1.Yonetani T, Ohnisi T. Cytochrome c peroxidase, a mitochondrial enzyme of yeast. J Biol Chem. 1966;241:2983–2984. [PubMed] [Google Scholar]
- 2.Yonetani T. Studies on cytochrome c peroxidase IV. A comparison of peroxide-induced complexes of horseradish and cytochrome c peroxidases. J Biol Chem. 1966;241:2562–2571. [PubMed] [Google Scholar]
- 3.Poulos TL, Freer ST, Alden RA, Edwards SL, Skogland U, Takio K, Eriksson B, Xuong N, Yonetani T, Kraut J. The crystal structure of cytochrome c peroxidase. J Biol Chem. 1980;255:575–580. [PubMed] [Google Scholar]
- 4.Finzel BC, Poulos TL, Kraut J. Crystal structure of yeast cytochrome c peroxidase refined at 1.7-Å resolution. J Biol Chem. 1984;259:13027–13036. [PubMed] [Google Scholar]
- 5.Poulos TL, Kraut J. The stereochemistry of peroxidase catalysis. J Biol Chem. 1980;255:8199–8205. [PubMed] [Google Scholar]
- 6.Erman JE, Vitello LB, Miller MA, Shaw A, Brown KA, Kraut J. Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I. Biochemistry. 1993;32:9798–9806. doi: 10.1021/bi00088a035. [DOI] [PubMed] [Google Scholar]
- 7.Nocek JM, Zhou JS, De Forest S, Priyadarshi S, Beratan DN, Onuchic JN, Hoffman BM. Theory and practice of electron transfer within protein-protein complexes: application to the multidomain binding of cytochrome c by cytochrome c peroxidase. Chem Rev. 1996;96:2459–2489. doi: 10.1021/cr9500444. [DOI] [PubMed] [Google Scholar]
- 8.Bendal DS, editor. Protein electron transfer. Bios Scientific Publishers; Oxford: 1996. [Google Scholar]
- 9.Pelletier H, Kraut J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 10.Mathews FS, Mauk AG, Moore GR. Protein-protein complexes formed by electron transfer proteins. In: Kleanthous C, editor. Protein-protein recognition. Oxford University Press; Oxford: 2000. pp. 60–101. [Google Scholar]
- 11.Mauk MR, Ferrer JC, Mauk AG. Proton linkage in formation of the cytochrome c-cytochrome c peroxidase complex: electrostatic properties of the high- and low-affinity cytochrome c binding sites on the peroxidase. Biochemistry. 1994;33:12609–12614. doi: 10.1021/bi00208a011. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Pielak GJ. Equilibrium thermodynamics of a physiologically-relevant heme-protein complex. Biochemistry. 1999;38:16876–16881. doi: 10.1021/bi992005i. [DOI] [PubMed] [Google Scholar]
- 13.Morar AS, Wang X, Pielak GJ. Effects of crowding by mono-, di-, and tetrasaccharides on cytochrome c peroxidase binding: Comparing experiment to theory. Biochemistry. 2001;40:281–285. doi: 10.1021/bi002296r. [DOI] [PubMed] [Google Scholar]
- 14.Stemp EDA, Hoffman BM. Cytochrome c peroxidase binds two molecules of cytochrome c: evidence for a low-affinity, electron-transfer-active site on cytochrome c peroxidase. Biochemistry. 1993;32:10848–10865. doi: 10.1021/bi00091a041. [DOI] [PubMed] [Google Scholar]
- 15.Zhou JS, Hoffman BM. Stern-Volmer in reverse: 2:1 stoichiometry of the cytochrome c-cytochrome c peroxidase electron transfer complex. Science. 1994;265:1693–1696. doi: 10.1126/science.8085152. [DOI] [PubMed] [Google Scholar]
- 16.Nakani S, Viriyakul T, Mitchell R, Vitello LB, Erman JE. Characterization of a covalently-linked yeast cytochrome c/cytochrome c peroxidase complex: Evidence for a single, catalytically-active cytochrome c binding site on cytochrome c peroxidase. Biochemistry. 2006;45:9887–9893. doi: 10.1021/bi060586n. [DOI] [PubMed] [Google Scholar]
- 17.Papa HS, Poulos TL. Site-specific crosslinking as a method for studying intramolecular electron transfer. Biochemistry. 1995;34:6573–6580. doi: 10.1021/bi00020a001. [DOI] [PubMed] [Google Scholar]
- 18.Kolthoff IM, Belcher R. Volumetric Analysis. Vol. 3. Interscience; New York: 1957. Hydrogen peroxide; pp. 75–76. [Google Scholar]
- 19.Matthis AL, Erman JE. Cytochrome c peroxidase-catalyzed oxidation of yeast iso-1 ferrocytochrome c by hydrogen peroxide: Ionic strength dependence of the steady-state parameters. Biochemistry. 1995;34:9985–9990. doi: 10.1021/bi00031a021. [DOI] [PubMed] [Google Scholar]
- 20.Hague DN. Fast reactions. Wiley-Interscience; London: 1971. [Google Scholar]
- 21.Erman JE, Vitello LB. Yeast cytochrome c peroxidase: mechanistic studies via protein engineering. Biochim Biophys Acta. 2002;1597:193–220. doi: 10.1016/s0167-4838(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 22.Yonetani T, Ray GS. Studies on cytochrome c peroxidase: HI Kinetics of the peroxidatic oxidation of ferrocytochrome c catalyzed by cytochrome c peroxidase. J Biol Chem. 1966;241:700–706. [PubMed] [Google Scholar]
- 23.Miller MA. A complete mechanism for steady-state oxidation of yeast cytochrome c by cytochrome c peroxidase. Biochemistry. 1996;35:15791–15799. doi: 10.1021/bi961488c. [DOI] [PubMed] [Google Scholar]
- 24.Klotz IM. Ligand-receptor energetics. Wiley; New York: 1997. [Google Scholar]
- 25.Conroy CW, Erman JE. pH titration study of cytochrome c peroxidase and apocytochrome c peroxidase. Biochim Biophys Acta. 1978;537:396–405. doi: 10.1016/0005-2795(78)90524-x. [DOI] [PubMed] [Google Scholar]
- 26.Theorell H, Åkesson Å. Studies on cytochrome c. III. Titration curves. J Amer Chem Soc. 1941;63:1818–1820. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plots of the steady-state velocities as a function of the yeast ferrocytochrome c concentration for the K12C/K769C, N78C/K79C, and K264/K79C covalent complexes at both 10 and 100 mM ionic strength. This material is available free of charge via the Internet at http://pubs.acs/org.