Abstract
Green fluorescent proteins (GFP) have become powerful markers for numerous biological studies due to their robust fluorescence properties, site-specific labeling, pH-sensitivity, and mutations for multiple-site labeling. Fluorescence correlation spectroscopy (FCS) studies have indicated that fluorescence blinking of anionic GFP mutants take place on a time scale of 45-300 μs, depending on pH, and have been attributed to external proton transfer. Here we present experimental evidence indicating that conformational change of the protein β-barrel is a determining step for such external protonation of GFP-S65T (at low pH) using time-resolved fluorescence and polarization anisotropy measurements. While the average anionic fluorescence lifetime of GFP-S65T is reduced by ∼18% over pH 3.6-10.0 range, the fluorescence polarization anisotropy decays mostly as a single-exponential with a rotational time, φ=17±1 ns, that indicates an intact β-barrel with a hydrodynamic volume of 78±5 nm3. In contrast, the total fluorescence (525±50 nm) of the excited neutral state of S65T reveals a strong correlation between the fluorescence lifetime, structural conformation, and pH. The average fluorescence lifetime of the excited neutral state of S65T as a function of pH yields a pKa≈5.9 in agreement with literature values using steady-state techniques. In contrast to the intact β-barrel at high pH, the anisotropy of neutral S65T (at pH≤pKa) decays as a biexponential (e.g., at pH 5.8, φ1=1.86 ns, β1=0.03, φ2=17.5 ns, and β2=0.25), which suggests a segmental mobility of the chromophore associated with conformational changes of the protein. The segmental motion of the S65T chromophore becomes faster with an enhanced amplitude ratio as pH is reduced. For comparative purposes, we also provide complementary FCS results on fluorescence blinking of the neutral-state of EGFP mutant (F64L/S65T), on a much slower time scale. Our results indicate that conformational rearrangement of the β-barrel and the amino acids surrounding the embedded chromophore is a rate determining step for external proton transfer and possibly cis/trans isomerization as non-radiative pathways that underlie fluorescence blinking of GFP mutants in acidic environment. In addition, the neutral-state transition is likely to be involved in the blinking process previously observed for the anionic-state transition in several GFP mutants.
Keywords: Green fluorescent protein, GFP-S65T, fluorescence blinking, fluorescence anisotropy, fluorescence correlation spectroscopy, structural conformation
1. Introduction
Green fluorescent proteins (GFP), from the jellyfish Aequorea victoria, have been widely used as a fluorescence marker in a wide range of cell studies.1,2 The chromophore of GFP (located in the center of a β-sheet barrel with an α-helix) is formed by an intermolecular autocatalytic reaction between amino acids Ser65, Tyr66, and Gly67.3,4 The β-barrel shields the chromophore from external quenchers5,6 and provides steric hindrance that prevents the cis/trans isomerization pathway,7,8 which explains the superb fluorescence properties of GFP mutants, even when fused with other proteins.5,6 Recent theoretical calculations on GFP-like chromoprotein protein (asFP595, isolated from Anemonia sulcata) have suggested that the non-fluorescent (dark) state is associated with a trans configuration of the embedded chromophore.7,8 In contrast, molecular dynamics calculations of GFP chromophore indicate that the trans and cis conformations of GFP chromophore exhibit undistinguishable energy minima.44 As a result, GFP mutants provide interesting model systems for studying the underlying mechanisms of their unique photophysical properties, which can be exploited for quantitative biological studies.
There have been recent efforts to develop new GFP variants with minimized sensitivity to pH, chloride ion concentration [Cl-] and photobleaching.9,10 Yellow fluorescent protein (YFP) mutant, T203Y, has remarkable pH-resistance and may find use in the labeling of acidic organelles.9 Citrine, from the third generation of YFP derivatives, exhibits a reduced sensitivity to [Cl-], a larger pKa-value suited to physiological pH, and improved fluorescence properties.11 There is a strong demand for red fluorescent proteins for (i) multi-color protein-tracking applications, (ii) new donor-acceptor pairs for fluorescence resonance energy transfer (FRET) studies of molecule-molecule interactions, and (iii) distinctive emission from the underlying cellular autofluorescence.12 Applications of DsRed (a red fluorescent protein isolated from a coral of the Discosoma genus)12,13 to cell and protein studies are limited due to its tetramerization14,15 and long maturation time.9 Tsien and co-workers engineered a monomeric DsRed variant (mRFP1) that matures quickly.9,10 Recently, a new generation of intrinsically red-fluorescent protein (RFP) mutants (e.g., mRFP1, mCherry, mOrange and mStrawberry) has been reported16 with superb spectroscopic properties such as large extinction coefficient, high fluorescence quantum yield, and long absorption and emission wavelengths. An alternative approach has been proposed to generate long-wavelength fluorescence emission using a photoactivation mechanism.17 Stepanenko et al. have provided a systematic comparison between green and red fluorescent proteins, concerning their structural properties and conformational stabilities, using steady-state, Stern-Volmer quenching, and circular dichroism (CD) spectroscopy.18
GFP mutants exhibit both neutral and anionic state conformers2,3,6 with electronic transitions at 396 nm and 488 nm (or longer), respectively, whereas the fluorescence quantum yield of the latter is much higher19,20 than the former. As a result, numerous GFP mutations have been created to enhance the anionic state population with variable emission peaks (from blue to yellow) for multiple-site labeling and FRET studies. Dickson and co-workers reported fluorescence blinking of T203Y and T203F mutants upon 488 nm (anionic) excitation and a subsequent recovery following 405 nm (neutral) illumination.21 Despite the two different absorption bands, GFP emits mainly around 505 nm due to the excited-state deprotonation of the hydroxyl group of Tyr-66.22,23 A minor emission shoulder around 457 nm of EGFP was assigned to the neutral-state emission.3,24 The population of the neutral state of GFP increases as the proton concentration increases in the surrounding bulk solution. Such interconversion between the anionic (bright) and neutral (relatively dark) states of GFP was assigned as the underlying mechanism for fluorescence blinking observed in fluorescence correlation spectroscopy (FCS) studies.15,25,26 Anionic GFP mutants (e.g., GFP-S65T, EGFP, Citrine, T203Y, and T203F) exhibit fluorescence blinking that takes place on a timescale of ∼ 45-340 μs due to external proton transfer from the buffer solution to the protein interior.15,25,26 However, it remains unclear whether structural changes in the β-barrel of GFP are necessary for such proton exchange between low-pH buffer and the protein interior. Furthermore, it is not yet clear whether the underlying mechanisms for GFP blinking in FCS studies are the same as those observed in single-molecule studies, which reveal a slow (seconds) blinking process.21,27
Enoki et al. have investigated the acid denaturation and folding/unfolding kinetics of a GFP mutant (Cycle3: F99S/M153T/V163A) by measuring the chromophore and tryptophan fluorescence.28 Using far-UV CD with a stopped-flow system, they have demonstrated that the refolding kinetics of the acid-denatured (at pH 1.5 and 2.0) state of Cycle3 involve five kinetic phases.28 In addition, the slow phase was fully rate-limited by slow isomerization in the denatured and intermediate states. Molecular dynamics (MD) simulations of GFP have also been carried out to investigate the conformational rearrangements induced by deprotonation.29 In addition to MD studies, the authors also used quantum mechanics/molecular mechanics (QM/MM) to investigate the β-barrel effects on the internal rotation of the chromophore, while providing a quantitative estimate of the torsional rotation barrier around the bridging bond.29 Real-time experimental measurements of acid-induced, ultrafast conformational changes of GFP remain elusive.
Here, we use time-resolved fluorescence lifetime and polarization anisotropy to address the correlation between the ultrafast conformational changes of GFP-S65T mutant (as a model system) and the loss of its fluorescence (i.e., blinking) in acidic environments. We also investigate the neutral state blinking kinetics of EGFP using FCS under 396-nm excitation. These results will be discussed in terms of a three electronic state transition model30 to explain the blinking pathways in GFP and the excited-state dynamics of neutral and anionic GFP-S65T.
2. Materials and Methods
GFP-S65T mutant (S65T hereafter) was a generous gift from the group of Prof. Stephen Benkovic (Chemistry, Penn State University). The stock solution of the protein (30 μM) was then diluted in buffer solutions of varying pH (pH 3.6 - 10). Potassium phosphate (0.1 mM) buffers (for pH 6 - pH 8 range) were prepared from potassium monobasic and dibasic solutions. 0.1 mM glacial acetic acid and sodium borate solutions were used to prepare pH 3.0 to 5.5 and pH 8.5 to 10 buffer solutions, respectively. The desired pH was adjusted with a small amount of NaOH (0.5 mM). Absorption spectra were measured with a single beam, diode array spectrophotometer (DU800, Beckman Coulter), whereas the fluorescence spectra were recorded using a PTI-QM-1 spectrofluorimeter (FL3-21, Fluorolog). The second harmonic (SHG4500, Coherent) of Ti:sapphire femtosecond laser pulses (∼120 fs at 76 MHz, Mira900F, Coherent) was used for 1P-excitation of both the neutral (396 nm) and anionic (488 nm) state-transition of S65T. The pulse repetition rate was reduced to 4.22 MHz using a pulse picker (Mira900, Coherent) before steering the laser pulses to the sample via the back entrance port of an inverted microscope (IX81, Olympus) and a dichroic mirror (488LP, Chroma). The filtered (using 525±50, 525±25, or 475±25 filters from Chroma) epi-fluorescence was split into two channels using a 50/50 beam splitter and the detection polarization of each was selected independently using Glan-Thompson polarizers for time- and polarization-resolved fluorescence measurements. The polarized fluorescence was detected by microchannel plate (MCP) (R3809U-50, Hamamatsu), amplified, and then fed to a time-correlated single-photon counting module (SPC-830, Becker & Hickl, Germany). The back aperture of a 1.2 NA objective (60× water immersion, Olympus) was underfilled to minimize depolarization effects.31
2.1 Time-Resolved Fluorescence and Polarization Anisotropy
A non-linear least-square fitting routine (SPCM, Becker & Hickl, Germany) was used to analyze the measured magic-angle (54.7°) fluorescence decays, F(t), that are convoluted with the system response function, R(t), (full-width half maximum, FWHM=50±5 ps). Generally, the deconvoluted fluorescence decay is best described as a sum of exponentials with time constants (τi) and amplitudes (αi) that are dependent on pH and electronic-state transitions (i.e., anionic versus neutral):3
| (1) |
The average fluorescence lifetime is calculated as: . The excited-state fluorescence decay rate (kfl = 1/τi) of a given fluorophore is dependent on the radiative (kr) and non-radiative (knr) rates as follows:32kfl= 1/τi = kr+ knr. While the radiative rate is inherent in the molecular structure of the chromophore, the non-radiative processes may include isomerization, charge transfer, and ionization pathways that compete with fluorescence. As a result, the reduction of excited-state lifetime of a fluorophore can be attributed to the enhanced non-radiative processes under certain conditions. However, the identification of the underlying non-radiative processes is not straightforward and requires systematic studies of the excited-state dynamics in different environments and with targeted structural manipulations.
For time-resolved fluorescence polarization anisotropy, the measured parallel (I//(t)) and perpendicular (I⊥(t)) fluorescence decays were used to calculate the anisotropy decay as follows:32
| (2) |
The G-factor, which accounts for the polarization sensitivity of the detection channels, was estimated using a tail matching32 approach on fluorescein emission under the same excitation and detection conditions. Based on the complexity of the molecular system, the anisotropy decays were either described as a single exponential (e.g., GFP in high pH buffer) or a complex multi-exponential (e.g., GFP in low pH buffer), which can be generally described as:32
| (3) |
where the sum of pre-exponential factor (βi) equals to the initial anisotropy (r0). When the fluorophore undergoes a segmental motion while tethered with a large biomolecule, the corresponding rotational time (φi) becomes too slow to observe within the excited-state lifetime. As a result, the corresponding pre-exponential factor becomes assigned as a residual anisotropy (r∞). The rotational time (φ) is related to the hydrodynamic volume (V) of a chromophore by the following relationship:32φ = ηV/KBT, where η is the viscosity of the buffer (∼ 0.89 cP) at a given temperature (T∼293 K), and KB is the Boltzmann constant.
2.2 Fluorescence Correlation Spectroscopy
The autocorrelation function of rhodamine green (lateral diffusion coefficient DRhG ∼2.8×10-6 cm2/s)15 was measured to calibrate the FCS setup that includes femtosecond laser pulses (at 82 MHz), an underfilled 1.2NA, 60×, water immersion objective and a fiber with 50-μm diameter, which serves as a confocal pinhole. The autocorrelation function G(τ) is defined as:15,25,26GD(τ) = [δF(t).δF(t+τ)]/ < F(t) >2, where F(t) is the fluorescence intensity at a given time t, and δF(t) is the corresponding fluorescence fluctuation. The autocorrelation function of rhodamine green is best described by diffusion such that:15,25,26
| (4) |
where N is the average number of molecules with a residence diffusion time of τD (for RhG, τD=440±10 μs) in the open three-dimensional Gaussian observation volume with an axial-to-lateral dimension, ω (∼ 5.5 for this setup). In addition to diffusion, however, it is known that GFP exhibits fluorescence blinking processes (i.e., light and pH-dependent), both of which are much faster than the overall diffusion of GFP molecules. The autocorrelation function due to these two blinking processes can be expressed as:15,25,26
| (5) |
where fBi is the amplitude fraction of the GFP population that undergoes blinking (i = 1, 2 for light- and pH-driven processes, respectively)25 over a time constant τBi. Such a relationship is only valid when the time scales of these blinking processes are significantly different, which might not be the case at very low pH or high excitation intensity.
3. Results and Discussion
3.1 Steady-State Spectroscopy of the Anionic and Neutral-State Transitions in S65T
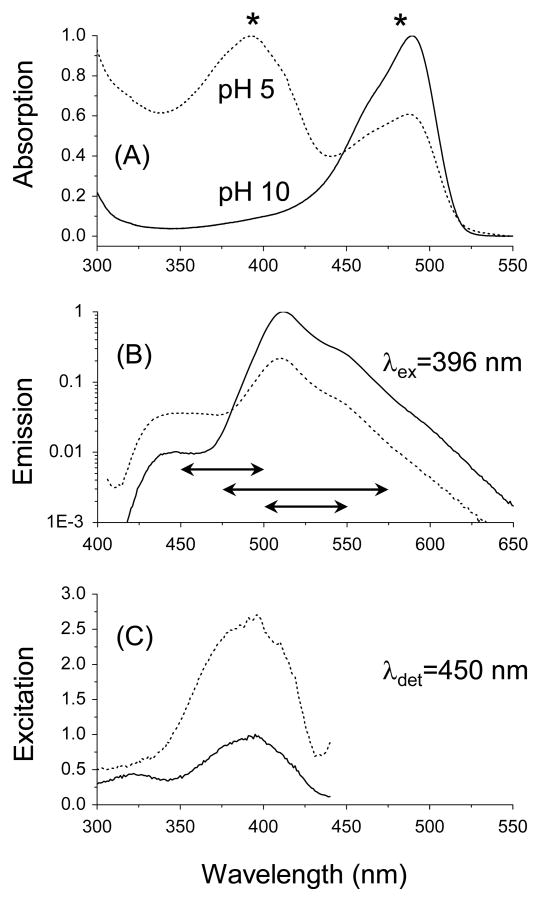
The absorption, emission, and excitation spectra of S65T were measured at pH 10 and pH 5 (Figure 1). The observed general trend here is similar to other literature studies,3,25 where the absorption band of anionic S65T at 488 nm (ε488∼56,000 M-1cm-1)1,2 is reduced as pH decreases, with a concomitant increase in the neutral state absorption at ∼392 nm and an isobestic point at ∼445 nm (Figure 1A)25. In addition to the main emission band at ∼509 nm (pH 10), a new blue emission shoulder appears around 450 nm as the pH decreases (Figure 1B). To assign the transition associated with the observed blue emission shoulder at 450±25 nm, we have measured the excitation spectra of S65T as a function of pH (5 and 10) and detection wavelength (450 nm) as shown in Figure 1C. At pH 10.0, the absorption and excitation spectra are basically congruent when the emitted fluorescence was detected at 510 nm. However, an excitation shoulder can be identified around 396 nm at pH 5.0, even with 510-nm detection (data not shown). Interestingly, when the fluorescence was detected at 450 nm, the excitation band at 396 nm was identical to the neutral absorption at low pH (Figure 1C). These results enable us to assign the blue emission shoulder around 450 nm (Figure 1B) to the excited neutral-state emission with a low quantum yield based on the time-resolved fluorescence measurements (see below).3,4 Furthermore, a small portion of the neutral state emission of S65T (excited at 396 nm) can be detected around 510 nm. Such spectral overlap between the neutral and anionic transitions might also explain the observed pH-dependent blinking in FCS studies15,25,26 under 488 nm excitation. The absorption and emission bands associated with anionic S65T disappear at pH 4, while the neutral state transition of the denatured protein dominates with absorption at 382 nm (with a roughly estimated extinction coefficient, ε382∼44,600 M-1cm-1) and emission shoulder around ∼450 nm (data not shown). The reduced extinction coefficient of the neutral-state transition of S65T at pH 4 is in agreement with Haupts et al.,25 but it is not yet clear if such reduction can explain the loss of the neutral emission as was previously proposed (see below).25
Figure 1.
Steady-state spectroscopy of GFP-S65T is sensitive to the environmental pH. (A) In addition to the main absorption band of S65T (pH 10.0, solid line), a new absorption band appears around 392 nm (pH 5.0), which is assigned to the neutral state transition (dotted line). The asterisks (*) correspond to the excitation wavelengths used in this work. (B) The anionic-state emission of S65T peaks around 509 nm (pH 10) and is reduced significantly at a lower pH (5.0). Depending on the excitation wavelengths (480 or 396 nm), the detection-filter bandwidths used here were varied as represented by the horizontal arrows (525±25, 525±50, and 475±25 nm) shown above for guidance. In addition, an enhanced emission shoulder around 450 nm can be identified at pH 5.0, which is assigned to the neutral state transition as shown in the excitation spectrum (C) under 396-nm excitation.
It has been shown that fluorescence emission of anionic GFP mutants (e.g., S65T, EGFP, citrine, pHfluorin),15,25,26 is very sensitive to pH due to external proton transfer from low-pH buffer to the protein interior in a single-protonation step,33,34 with a standard free reaction energy ΔG0=33±1 kJ/mol for S65T at room temperature.25 Such pH-sensitivity of GFP fluorescence has been exploited in various biological applications.35,36 It is not clear, however, whether the external protonation that underlies the fluorescence blinking of GFP mutants15,25,26 at low pH is correlated with conformational changes of the β-barrel. To address this question, we conducted time-resolved fluorescence lifetime and polarization anisotropy measurements on S65T, as a function of pH and excitation wavelengths. Furthermore, we used the absorption and emission spectra for qualitative analysis of the radiative rate constant of both anionic and neutral state transitions (see below) using Strickler-Berg equation.32
3.2 The Excited Anionic-State Dynamics of S65T is less Sensitive to pH
3.2.a pH-Effects on Excited Anionic-State Lifetime of S65T
At high pH, the excited (λex=480 nm) anionic state fluorescence (525±25 nm) decay of S65T can be described satisfactorily as a single exponential with a time constant of 2.95±0.01 ns (Table 1, Figure 2D). These results are consistent with previous studies using the single-photon counting technique.3,4 Assuming a fluorescence quantum yield of 0.661, the estimated radiative rate constant (kr) is ∼2.20×108 s-1, using the measured fluorescence decay rate (kfl =1/τfl=3.38×108 s-1). In addition, the calculated non-radiative rate constant (knr) of S65T (high pH) is ∼1.18×108 s-1 at 480 nm excitation. Following Strickler-Berg approach,32 we also used the absorption and emission spectra of anionic S65T (pH10) to calculate the radiative rate constant (∼2.88×108 s-1), which agree well with the above value.
Table (1).
pH-dependence of the time-resolved fluorescence of anionic S65T, when excited at 480 nm. The fluorescence is detected at 525±25 nm using magic angle (54.7°) polarization with 24.4 ps/channel.
| pH | τ1 (ps) | α1 | τ2 (ns) | α2 | τ3 (ns) | α3 | 〈τfl〉 (ns) |
|---|---|---|---|---|---|---|---|
| 8.0 | - | - | - | - | 2.95(1)a | 1.0 | 2.95(1) |
| 5.0 | - | - | 2.4(1) | 0.46(8) | 3.04(2) | 0.54(8) | 2.75(1) |
| 4.5 | - | - | 2.32(2) | 0.50(4) | 3.03(7) | 0.51(4) | 2.68(3) |
| 4.0 | - | - | 2.16(3) | 0.35(2) | 3.05(6) | 0.47(1) | 2.58(2) |
| 3.6 | 87(11) | 0.38(2) | 1.29(6) | 0.34(2) | 3.82(2) | 0.28(1) | 1.54(6) |
The number in parentheses represents the standard deviation (uncertainty) of the last digit.
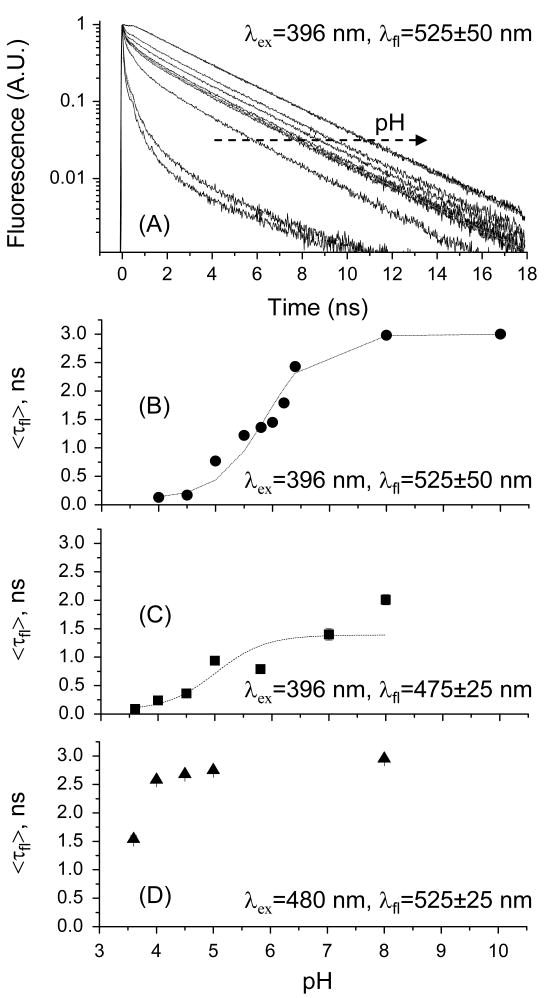
Figure 2.
The pH-sensitivity of the excited-state fluorescence dynamics of S65T depends on the excitation and detection wavelengths. (A) Representative fluorescence decays of S65T as a function of pH (10.0, 8.0, 6.4, 6.2, 6.0, 5.8, 5.5, 5.0, 4.5, and 4.0), excited at 396 nm with magic-angle (54.7°) detection of 525±50 nm emission. (B) The average fluorescence (525 ± 50 nm) average lifetimes are fit with the following equation:25 < F > α < τfl >= a + b[1 + 10(pKa−pH)]−1, where a=0.4±0.1, b=2.6±0.1, and pKa=5.8±0.1, which yields ΔG0 =33.61 kJ/mol. (C) The estimated pKa-value is reduced (pKa=4.94) when the fluorescence is detected at 475±25 nm. By comparison, the average fluorescence lifetime of anionic S65T exhibits a relatively less pronounced pH-dependence (D).
In an acidic environment (pH<pKa), the excited anionic state fluorescence decays as a biexponential (e.g., τ1=2.16 ns, α1=0.35, τ2=3.05 ns, and α2=0.47 at pH 4.0) with a significantly faster average lifetime of 2.58±0.02 ns. At pH 3.6, the fluorescence decays as triple exponential (τ1=87 ps, α1=0.38, τ2=1.29 ns, α2=0.34, τ3=3.82 ns, and α3=0.28) with an average lifetime of 1.54±0.06 ns (Table 1, Figure 2D). Under similar conditions, however, FCS studies on anionic S65T revealed fast (up to 350 μs) fluorescence blinking that was attributed to a combination of internal (light-driven) and external (pH-driven) proton transfers.3,25,37 Since the external proton transfer (from buffer to the protein interior) takes place on a slow time scale (10-6 to 10-4 s), it is unlikely to occur on the potential energy surface of excited anionic state during the 2.95-ns lifetime.3,4 Instead, the partitioning of the ground state population between anionic and neutral S65T can be excited at 488 nm at low pH (Figure 1), albeit with a different excitation probabilities due to Franck-Condon principle. We have measured time-resolved fluorescence polarization anisotropy of S65T, as a function of pH, to examine possible correlation between confirmation changes43 and fluorescence blinking due to external proton transfer in FCS studies25.
3.2.b pH-Effects on the Rotational Diffusion and Structural Conformation of Anionic S65T
The time-resolved fluorescence (525±25 nm) polarization anisotropy of anionic S65T decays as a single exponential with a rotational time φ=17±1 ns and an initial anisotropy of 0.33±0.01, over the range of pH 4.5 to pH 8.0. The single-exponential anisotropy decay indicates that the embedded anionic chromophore is restricted by the protein environment with an overall hydrodynamic volume of ∼ 78.2 nm3. In contrast, the fluorescence anisotropy of S65T decays as a biexponential as pH is reduced below the pKa-value (e.g., φ1=5.1 ns, β1=0.11, φ2=28 ns and β2=0.22 at pH 3.6). The observed biexponential anisotropy decay indicates a segmental mobility of the chromophore in a denatured protein at pH 3.6.
These results suggest that the chromophore of anionic S65T is protected by the intact β-barrel, which explain the absence of fluorescence pH-blinking in S65T using FCS at high pH 15,25,26 At very low pH, the ground-state population of neutral S65T increases, with concurrent reduction of the anionic population as well as denaturation of the protein. Consequently, the blinking process in FCS studies of S65T and other GFP mutants, 15,25,26 under similar conditions, is likely due to the conversion of the anionic (i.e., bright) to the neutral (i.e., presumed dark or non-fluorescent) state on the ground state potential energy surface. However, there are not thorough FCS and conformational studies on the neutral state transition of GFP mutants as a function of pH. Here, we conducted fluorescence polarization anisotropy and FCS measurements of neutral S65T and EGFP, respectively, as a function of pH.
3.3 The Neutral-State Transition is Sensitive to both pH and Detection Wavelength
3.3.a pH-Effects on the Neutral State Fluorescence (475±50 nm)
Based on the steady-state spectroscopy (Figure 1) and the energy diagram of the electronic state transitions of S65T (shown below in Figure 6), it is possible to probe the excited neutral state dynamics of S65T using a combination of excitation (396 nm) and detection (450±25 nm) selectivity. In addition, excited-state proton transfer and/or direct excitation of high vibronic levels may also lead to populating the excited-state of anionic GFP mutants. Under 396-nm excitation, the excited state fluorescence (475±25 nm) of S65T (pH 8.0) decays as a triple exponential with τ1=100±16 ps, α1=0.29±0.01, τ2=1.2±0.2 ns, α2=0.32±0.06, τ3=4.1±0.3 ns, α3=0.39±0.05 (Table 2). In addition to the multiexponential decay, the estimated average lifetime is 2.01±0.09 ns, as compared to the 2.95 ns measured for the anionic state transition (Figure 2C and 2D). The fast time constants of the measured triple exponential decays dominate with pH-reduction. At pH 4.0 (for example), the fluorescence of neutral S65T decays as a triple exponential with τ1=60±5 ps, α1=0.64±0.01, τ2=0.35±0.02 ns, α2=0.31±0.01, τ3=1.90±0.05 ns, α3=0.05±0.02 and an average lifetime of 240±10 ps (i.e., ∼4.17×109 s-1). Figure 2C depicts the average fluorescence lifetime of S65T as a function of pH with an estimated pKa=4.9±0.9. We also used Strickler-Berg equation32 with the absorption and emission spectra of neutral S65T (pH 4) to calculate the radiative rate constant (kr∼6.21×108 s-1). Consequently, the fluorescence quantum yield of the neutral state transition is S65T can be roughly estimated ( ∼0.15), which is significantly lower than 0.65 for the anionic state.1 As a result, we conclude that the neutral state transition is relatively non-fluorescent (or dark) due to efficient non-radiative processes that may include proton transfer as well as isomerization pathways. In addition, it is likely that the neutral state transition is comprised of multiple states (〈SN〉, Figure 6) to account for the observed multi-exponential fluorescence decays.
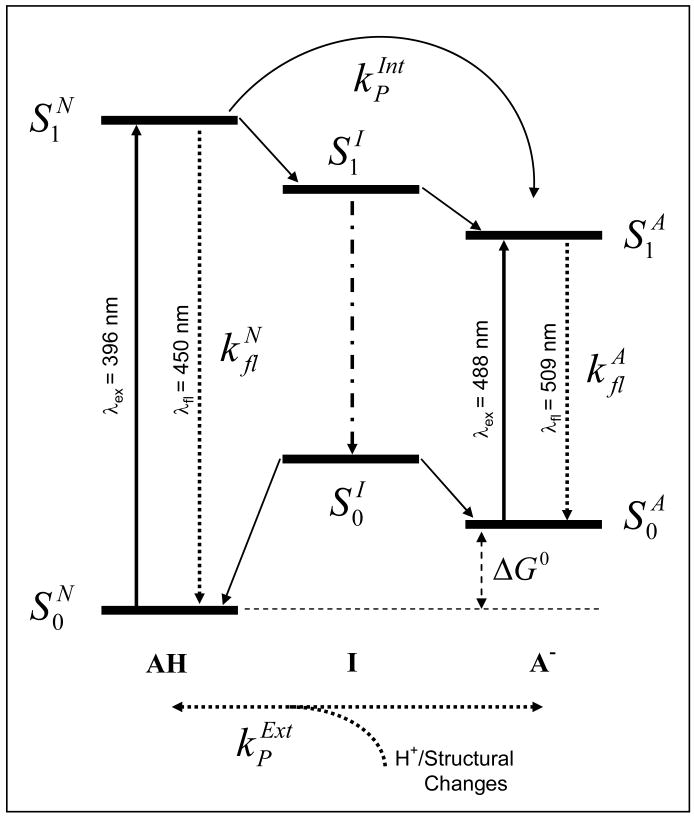
Figure 6.
Schematic energy diagram of the neutral (AH), anionic (A-) and hypothesized intermediate (I) electronic state transitions of GFP-S65T (and possibly other GFP mutants). The electronic energy between the corresponding ground and first-excited states for each transition can be calculated using steady-state spectroscopy (Figure 1), except the intermediate state transition . The proposed scheme also accounts for excited-state relaxation dynamics (e.g., neutral and anionic fluorescence decay rates), as well as photoconversion via either external ( , pH-driven) or internal ( , light-driven) proton transfer, that are observed using time-resolved fluorescence and FCS measurements as a function of pH and excitation/detection wavelength. The observed reduction of the excited-state fluorescence lifetime and segmental mobility of GFP chromophore, at low pH, indicates a correlation between ultrafast conformational changes of the protein structure and external-proton transfer (from low-pH buffer to the protein interior) in acidic environments.
Table (2).
pH-dependence of the time-resolved fluorescence of neutral S65T, when excited at 396 nm. The fluorescence is detected at 475±25 nm using magic angle (54.7°) polarization with 4.88 ps/channel.
| pH | τ1 (ps) | α1 | τ2 (ns) | α2 | τ3 (ns) | α3 | 〈τfl〉 (ns) |
|---|---|---|---|---|---|---|---|
| 8.0 | 100(16)a | 0.29(1) | 1.2(2) | 0.32(6) | 4.1(3) | 0.39(5) | 2.01(9) |
| 7.0 | 90(21) | 0.34(2) | 0.6(2) | 0.28(2) | 3.2(4) | 0.38(4) | 1.4(1) |
| 5.8 | 85(2) | 0.40(2) | 0.40(3) | 0.32(6) | 2.75(3) | 0.22(1) | 0.79(2) |
| 5.0 | 84(5) | 0.39(1) | 0.41(1) | 0.34(1) | 2.81(2) | 0.27(2) | 0.94(4) |
| 4.5 | 77(7) | 0.52(2) | 0.32(2) | 0.40(1) | 2.26(3) | 0.08(1) | 0.36(1) |
| 4.0 | 60(5) | 0.64(1) | 0.35(2) | 0.31(1) | 1.90(5) | 0.05(2) | 0.24(1) |
| 3.6 | 36(8) | 0.86(1) | 0.31(5) | 0.13(2) | 1.2(2) | 0.013(6) | 0.09(2) |
The number in parentheses represents the standard deviation (uncertainty) of the last digit.
3.3.b pH-Effects on the Neutral Plus Anionic State Fluorescence (525±50 nm)
The partitioning of ground state population of anionic, neutral and intermediate species of GFP is pH-dependent. As a result, it is more informative to assemble most of the ground state population(s) to examine how the corresponding excited-state dynamics correlates with steady-state thermodynamics. Such collective probing of different S65T populations can be achieved using 396-nm excitation with wide-band emission detection (e.g., 525±50 nm filter). In addition to the direct excitation of neutral-state transition of S65T, the excited anionic state can also be populated either directly (but with much weaker probability according to the Franck-Condon principle) or indirectly (via excited state proton transfer or intramolecular energy transfer), as shown in Figure 1 and 6.
When excited at 396 nm, the total fluorescence (525±50 nm) of S65T (pH 10) decays as a biexponential, where τ1=2.7±0.1 ns, α1=0.43±0.03, τ2=3.25±0.05 ns, and α2=0.55±0.05 with an estimated average lifetime of 3.00±0.01 ns (Table 3). At low pH (pKa=5.9±0.1),25 the fluorescence decays become triple exponentials with significantly reduced average fluorescence lifetime (Figure 2A and 2B) as the environment becomes more acidic. At pH 4.0 (for example), the fluorescence decay is best described with τ1=45±5 ps, α1=0.84±0.08, τ2=450±20 ps, α2=0.12±0.02, τ3=3.6±0.5 ns, α3=0.01±0.01 and average lifetime of 130±20 ps (Table 3). The pH-dependence of the average fluorescence lifetime yields a pKa of 5.8±0.1 (Figure 2B), which is in a agreement with literature values obtained by FCS (5.9±0.1),25 despite the difference between the time scales of slow blinking (≥μs) and fast excited-state dynamics (ps-ns). As a result, we conclude that pH alters the partitioning of the ground state populations of neutral and anionic states of GFP mutants by modifying the ground state potential energy surfaces of the anionic and neutral state as well as their barriers.
Table (3).
pH-dependence of the time-resolved fluorescence of S65T, when excited at 396 nm. The total fluorescence (525±50 nm) is detected at magic-angle (54.7°) polarization with time/channel of 24.4 ps.
| pH | τ1 (ps) | α1 | τ2 (ns) | α2 | τ3 (ns) | α3 | 〈τfl〉 (ns) |
|---|---|---|---|---|---|---|---|
| 10 | - | - | 2.7(1)a | 0.43(3) | 3.25(5) | 0.55(5) | 3.00(1) |
| 8.0 | - | - | 2.6(1) | 0.54(8) | 3.4(1) | 0.5(1) | 2.98(1) |
| 6.4 | 403(54) | 0.21(5) | 2.4(2) | 0.3(2) | 3.3(2) | 0.5(2) | 2.43(6) |
| 6.2 | 102(25) | 0.30(1) | 0.84(2) | 0.17(1) | 2.99(3) | 0.54(2) | 1.79(6) |
| 6.0 | 98(17) | 0.36(1) | 0.69(9) | 0.21(1) | 2.96(1) | 0.43(1) | 1.45(4) |
| 5.8 | 94(11) | 0.38(1) | 0.68(3) | 0.23(1) | 2.93(2) | 0.40(1) | 1.36(2) |
| 5.5 | 87(2) | 0.42(4) | 0.65(6) | 0.26(8) | 2.51(1) | 0.36(2) | 1.22(1) |
| 5.0 | 88(2) | 0.52(4) | 0.55(3) | 0.28(2) | 2.77(1) | 0.21(2) | 0.77(6) |
| 4.5 | 48(6) | 0.82(3) | 0.48(3) | 0.16(3) | 3.4(2) | 0.018(4) | 0.169(7) |
| 4.0 | 45(5) | 0.84(8) | 0.45(2) | 0.12(2) | 3.6(5) | 0.01(1) | 0.13(2) |
The number in parentheses represents the standard deviation (uncertainty) of the last digit.
The results are consistent with the hypothesis that the neutral state(s) of S65T (and possibly other GFP mutants) dominate at low pH with relatively dark (i.e., weakly fluorescent) with lower fluorescence quantum yield ( ∼0.15, see above) than the anionic state transition ( ∼0.651). In an acidic environment, the β-barrel of S65T is partially denatured, altering the net ionic charges by electrostatic repulsion and hydrogen bond disruption due to excess proton concentration.38 In addition to the protonation of the chromophore in acidic environment,25,39 the observed reduction of fluorescence lifetime (i.e., quantum yield) also suggests that the interruption of the hydrogen-bonding network surrounding the chromophore may lead to cis/trans isomerization,42,43 as an additional non-radiative mechanism. To examine this hypothesis, we carried out time-resolved fluorescence polarization anisotropy measurements of S65T as a function of pH.
3.3.c Segmental Mobility and Conformation Changes of S65T in Acidic Environment
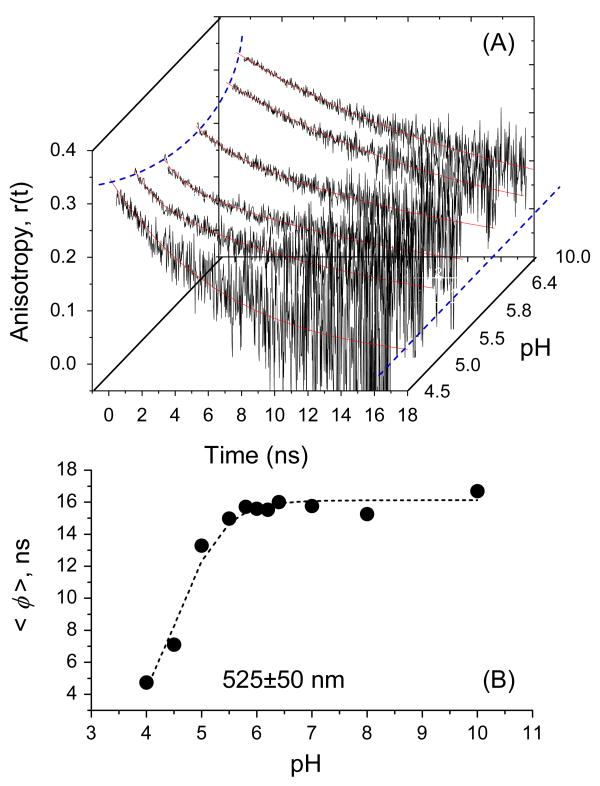
Figure 3 shows fluorescence polarization anisotropy decays of S65T as a function of pH and the fitting parameters are summarized in Table 4. Under 396-nm excitation, the time-resolved fluorescence (525±50 nm) anisotropy of S65T (high pH) decays as a single exponential with an estimated rotational time φ=15.9±0.3 ns, which yields a hydrodynamic volume of ∼72.6 nm3, consistent with its molecular weight (29±1 kDa) and x-ray crystal structure.39 The observed single exponential decay of polarization anisotropy also implies a rigid S65T protein with an intact β-barrel, which might explain the lack of pH-driven fluorescence blinking in FCS studies at high pH.15,25,37
Figure 3.
Rotational diffusion and segmental mobility of S65T is sensitive to pH as revealed by time-resolved fluorescence polarization anisotropy. In this experiment, parallel and perpendicular fluorescence polarizations were detected simultaneously, using a wideband filter (bandwidth of 525 ± 50 nm), prior to calculating the anisotropy decays. (A) A single exponential describes the anisotropy decays with ∼16 ns rotational time over the pH 6.4 – pH 10 range, but a slight reduction of the initial anisotropy can be identified. The observed biexponential at pH 5.8 and 5.5 suggests a segmental mobility of the embodied chromophore in S65T due to partial denaturation (i.e., conformational changes) of the β-barrel. The rotational time of the embodied chromophore of S65T becomes faster as the β-barrel denatures in the acidic environment below the pKa-value. The dotted lines serve as a guide through the variation of initial and residual anisotropy with pH. (B) The estimated average rotational time of S65T was calculated from the measured anisotropy decays (shown in Figure 3A) as a function of pH. The estimated pKa-value (4.9 ± 0.2) using these conformational changes of S65T are low compared with the pKa calculated using the average fluorescence lifetime (Figure 2B).
Table (4).
Time-resolved fluorescence polarization anisotropy of GFP-S65T as a function of pH. The protein is excited at 396 nm while the total fluorescence emission is detected at 525±50 nm. Similar measurements are also conducted while detecting 475/50 nm as well as the anionic transition (data not shown).
| pH | φ1 (ns) | β1 | φ2 (ns) | β2 | r∞ | r0 | 〈φ〉 (ns) |
|---|---|---|---|---|---|---|---|
| 10 | - | - | 16.7 | 0.33 | - | 0.33 | 16.7 |
| 8.0 | - | - | 15.3 | 0.33 | - | 0.33 | 15.3 |
| 7.0 | - | - | 15.8 | 0.33 | - | 0.33 | 15.8 |
| 6.4 | - | - | 16.1 | 0.32 | - | 0.32 | 16.1 |
| 6.2 | - | - | 15.5 | 0.31 | - | 0.31 | 15.5 |
| 6.0 | 1.4 | 0.01 | 15.8 | 0.31 | - | 0.31(a) | 12.0 |
| 5.8 | 1.86 | 0.03 | 17.5 | 0.25 | - | 0.28 | 9.20 |
| 5.5 | 1.1 | 0.04 | 17.5 | 0.23 | - | 0.27 | 5.45 |
| 5.0 | 1.29 | 0.06 | 15.9 | 0.25 | - | 0.31 | 5.00 |
| 4.5 | - | - | 7.1 | 0.35 | - | 0.35 | 7.1 |
| 4.0 | - | - | 5.6 | 0.34 | 0.06 | 0.40 | 5.6 |
| 3.6 | - | - | 4.5 | 0.33 | 0.04 | 0.37 | 4.5 |
The sum of the pre-exponential terms is the initial anisotropy at zero-time.
Near the pKa (∼5.94), the fluorescence polarization anisotropy of S65T decays as a bi-exponential with φ1=1.4 ns, β1=0.01, φ2=15.8 ns, and β2=0.31, which indicates a segmental mobility of the chromophore due to a slight denaturation of the protein and, therefore, less constraint inside the β-barrel. The second slow component is the overall rotational time of the protein. As the pH decreases further below the pKa, the amplitude of the fast decay component increases, suggesting a larger degree of freedom for the segmental mobility (i.e., tumbling) of the chromophore. In addition, the initial anisotropy (r0) decreases slightly (but consistently) with pH reduction, possibly due to (i) the enhanced rotational mobility of the less-restricted chromophore, and/or (ii) intramolecular energy transfer between the excited neutral and anionic states of S65T. Interestingly, further denaturation of the protein at pH≤4.0 leads to a single exponential anisotropy decay with a rotational time of ∼5.6 ns, which is much faster than the rotation of intact S65T protein (e.g., at pH>pKa). The measured fast rotational time (∼5.6 ns), associated with the segmental mobility, of denatured S65T (pH≤4.0) is also too slow to be assigned as a free, fully isolated chromophore. As a result, we conclude that the chromophore is still tethered to the adjacent amino acid backbone via a hydrogen-bond network and remains as an integral part of the denatured protein.
These results suggest that the β-barrel of S65T (and possibly other GFP mutants)1,2 is denatured in an acidic environment and that the segmental mobility of the embodied chromophore increases with buffer acidity at pH≤pKa. Under these conditions, the chromophore experiences more rotational degrees of freedom (or segmental mobility), even while remaining attached to the amino acid backbone of the denatured protein (based on the residual anisotropy observed under acidic environments (r∞∼0.06 at pH 4.0). These real-time ultrafast dynamics indicate that structural changes of S65T in an acidic environment correlate with (i) the external proton transfer from the buffer to the interior moiety of the protein and, therefore, (ii) the loss (or blinking) of GFP fluorescence. The observed segmental mobility also suggest an interruption of the hydrogen-bonding network42,43 between the chromophore and the neighboring amino acid residues upon the denaturation of the protein β- barrel at low pH.3,4 In the absence of such steric hindrance, therefore, the chromophore becomes less restricted and likely undergoes cis/trans isomerization, which may explain the observed reduction of (i) initial anisotropy, (ii) the time scale of the segmental mobility, and (ii) fluorescence lifetime as an additional non-radiative pathway, besides proton transfer. This conclusion is also in agreement with molecular dynamics simulations of GFP deprotonation.29
Due to the combined influence of pH on the overall rotational diffusion and segmental mobility of the chromophore of S65T, the average rotational time (weighted by the amplitude fractions) can be used to qualitatively assess the pKa associated with conformational changes as a function of pH. The estimated pKa-value appears dependent on the detection wavelength, where pKa=5.2±0.2 and 4.9±0.2 (Figure 3B) using filters with bandwidths of 525±25 nm and 525±50 nm (Figure 3B), respectively. To correlate these ultrafast structural changes of GFP at low pH with fluorescence blinking on the microsecond time scale, we compare FCS results of neutral EGFP at 405-nm excitation, which is almost non-existent.30 Furthermore, these measurements will also enable us to examine the role of the neutral state transition (which is considered dark)25 in fluorescence blinking mechanism.
3.3.d Detection-Wavelength Dependence of Neutral S65T (pH 5.0) Rotational Diffusion
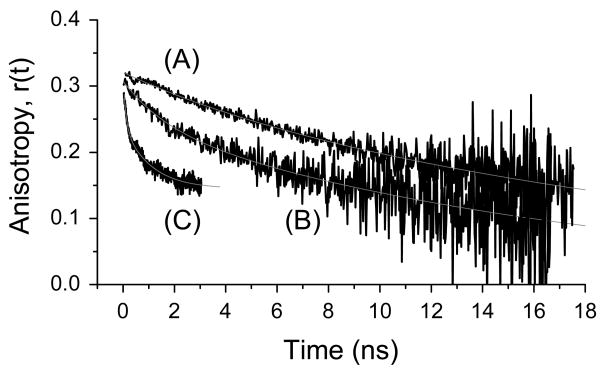
Time-resolved fluorescence polarization anisotropy of S65T (pH 5.0) depends on the detection wavelength (Figure 4). Ultimately, these results may enable us to assign the different electronic state transition to specific structural conformations of the protein. The fluorescence anisotropy of S65T (pH5.0), excited at 396 nm, decays as a biexponential with φ1=1.7 ns, β1=0.07, φ2=17.9 ns, and β2=0.24 (B), compared with the single-exponential decay φ=20.3 ns, r0=0.32) of the anionic state under 480-nm excitation (A). Further, the anisotropy associated with 475±50 nm emission is best described as a triple exponential with φ1=100 ps, β1=0.05, φ2=763 ps, β2=0.08, φ3=24.9 ns, and β3=0.167 over a short observation window that is dictated by the corresponding excited-state lifetime (C). Due to the bandwidth of the detection filter used here, contributions of different electronic states can be expected. Further, the multi-exponential anisotropy decays also indicate pronounce dependence of the segmental mobility of S65T (pH 5.0) as a function of the detection wavelength.
Figure 4.
Fluorescence polarization anisotropy of S65T (pH 5.0) reveals wavelength-dependent access to different protein states. The fluorescence anisotropy of S65T (pH5.0) excited at 396 nm decays as a biexponential with φ1=1.7 ns, β1=0.065, φ2=17.9 ns, and β2=0.24 (B), compared with a single-exponential decay (φ=20.3 ns, r0=0.32) of the anionic state under 480-nm excitation (A). Further, the anisotropy associated with 475±50 nm emission is best described as a triple exponential with φ1=100 ps, β1=0.05, φ2=763 ps, β2=0.076, φ3=24.9 ns, and β3=0.167 over a short observation window that is dictated by the corresponding excited-state lifetime (C).
3.4 Complementary Studies of Fluorescence Blinking of EGFP using FCS
3.4.a Fluorescence blinking of Anionic EGFP
At pH 11, the correlation curve of anionic EGFP (F64L/S65T), excited at 488 nm, is described mainly by diffusion with τD =5.85±0.09 ms (i.e., the diffusion coefficient, D≈6.9×10-7 cm2/s) and a minor (amplitude fraction, fB1∼0.11) light-driven blinking time τB1 =450±50 μs, in agreement with Haupts et al.25 The measured diffusion time of EGFP is basically the same at pH 5 under low-intensity excitation. At pH 5, the autocorrelation of anionic EGFP additionally reveals two blinking processes with time constants (amplitude fraction) of 85 μs (0.77) and ∼1.1 ms (0.24) that are attributed to external and internal proton transfer processes, respectively.25 The observed fast blinking component is attributed to external proton exchange between the buffer and the protein interior.3,25,26,37 Similar measurements have been conducted on other anionic GFP mutants, such as S65T,25 citrine,15 pHlurine and sapphire (H9),30 and yellow fluorescent proteins (namely, T203Y and T203F)40 using FCS. The light and pH driven conversion between bright and dark states in GFP mutants has been modeled using the ground and excited-state potential energy surface of three electronic transitions including a hypothesized intermediate state.21,30
In contrast to the anionic states of EGFP, FCS studies of the neutral state of GFPs are almost non-existent,41 even though such studies might address the role of neutral state transitions in the blinking mechanism of GFPs. Here, we are present fluorescence fluctuation dynamics studies of neutral EGFP (excited at 405 nm) using FCS as a function of pH (4.9 - 11). These measurements enable us to correlate the fast structural changes of GFP reported above with the slow blinking dynamics of the neutral and anionic states.
3.4.b Fluorescence Blinking of Excited Neutral-state Transition of EGFP
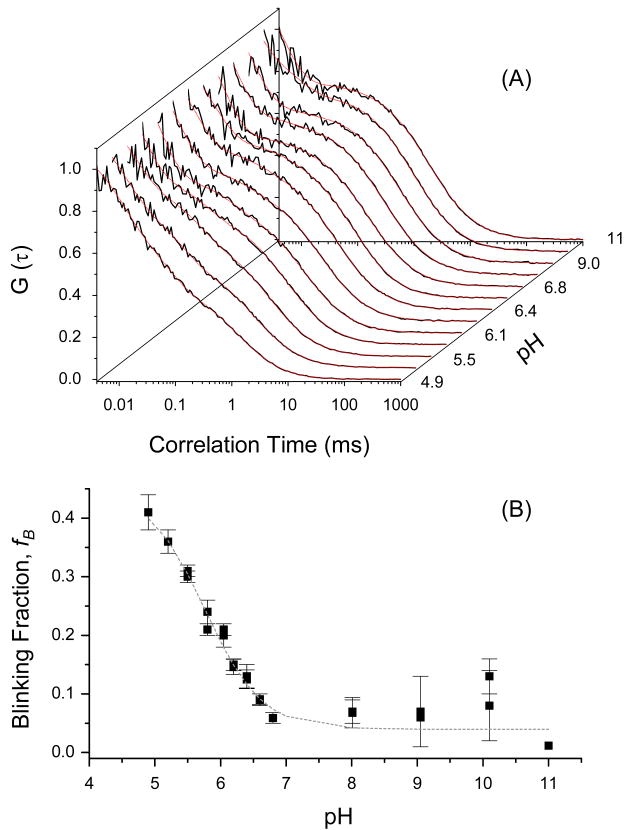
The overall autocorrelation function can be described adequately using the product of the diffusion and blinking functions (equation 5): G(τ) = GD(τ) · GB(τ).25 The autocorrelation curve of neutral EGFP (pH 11) is best described by diffusion (τD =1.78±0.08 ms) with a blinking component (τB1=7±3 μs, fB1∼0.48), which is significantly faster than the anionic EGFP light-driven blinking (τB1=450±50 μs, fB1∼0.11). The calculated diffusion coefficient of neutral EGFP (pH 11) is ∼ 2.2 ×10-7 cm2/s, which is slightly smaller than the anionic EGFP diffusion (∼6.9×10-7 cm2/s) and likely due to different photophysics of both EGFP and the reference (rhodamine green) under 396 and 488 nm excitation. The fast blinking component, which seems pH-independent (Figure 5A), is tentatively attributed to light-driven blinking between the bright and dark state of the protein.3,25,26,37 The significance of this fast decay component is likely to be compromised by the low signal-to-noise (Figure 5A) and the so called “after pulsing” inherent in the avalanche photodiodes.16 As the acidity of the buffer is increased (pH≤pKa), the fractional amplitude of an additional blinking component (fB2) increases and the pH-dependent time constant (τB2) decreases with an estimated pKa=5.77±0.08 (Figure 5B). The observed differences of the time constants and the amplitude fractions between neutral and anionic EGFP can be attributed to the nature of the potential energy surfaces and barrier heights of the corresponding electronic state transitions.21,15,30
Figure 5.
Complementary FCS studies of neutral EGFP (F64L/S65T), which was excited at 405 nm, as a function of pH. Haupts et al. have demonstrated that the pH-dependence of EGFP and GFP-S65T mutants exhibits comparable photoconversion kinetics under 488 nm excitation (i.e., the anionic state transition).25 The autocorrelation curves (A) of neutral EGFP are best described by diffusion and blinking terms (see equation 5). The amplitude fraction of the blinking population of EGFP, as a function of pH, yields pKa∼5.76 (B), which is slightly smaller than that of the anionic state transition (pKa∼5.9) of EGFP.25
3.5 Modeling the Electronic Transitions and Photoconversion in GFP-S65T
Based on our studies of both fast excited-state dynamics3 (using time-resolved fluorescence and polarization anisotropy) and slow fluorescence blinking processes25 (using FCS) of GFP-S65T and EGFP, we propose an energy diagram3,4 of different state transitions (Figure 6) and the following conclusions emerge. First, the β-barrel of anionic GFP (at high pH and 488-nm excitation) is intact and exerting a constraining force on the embedded chromophore leading to negligible non-radiative pathways such as cis/trans isomerization and external proton exchange with the buffer. Second, the excited neutral state of S65T can be excited at 396 nm and detected selectively around 450±25 nm. In addition, the anionic state transition of GFP can be excited at 396 nm via either direct excitation (but with low Frank-Condon factor) or excited-state internal proton transfer , as observed in some GFP mutants. Third, our FCS studies of the neutral EGFP, for the first time, suggest that the neutral-state transition is involved of GFP fluorescence blinking. The observed kinetics also confirm the hypothetical intermediate state as the dark-state invoked in previous studies in GFP fluorescence. Fourth, the time scales associated with the excited state dynamics (ps-ns) and fluorescence blinking (μs-ms) are distinctly different, which indicates that the observed pH-driven conversion is likely to occur on the ground-state potential energy surface. Despite these differences, the estimated pKa values using ultrafast time-resolved fluorescence agree well with FCS results. As a result, the observed multiexponential fluorescence decays of neutral GFP (low pH) are assigned to the excitation of multiple, ground-state GFP conformations (or states) that exist at equilibrium. Fifth, the observed conformational changes of the β-barrel and the amino acids surrounding the chromophore are likely to facilitate external-proton transfer from the buffer to the protein interior, which underlies fluorescence blinking of GFPs. In an acidic environment, these conformational changes of the β-barrel and the disruption of hydrogen-bonding network reduce the restriction on the GFP chromophore and, therefore, may lead to isomerization pathway.42,43 Using the experimental results provided here, however, it is difficult to be more specific concerning the isomerization mechanism involved (e.g., whether it involves Thr203)42,43 or the exact configuration (trans, cis, or hula-twist)44 associated with neutral and anionic states of these GFP mutants. While these conclusions may be generalized to other GFP mutants, the rates and amplitude fractions associated with blinking and conformational changes are likely to depend on pH and mutation site/type.15 We are currently applying the same experimental approach to determined whether these findings are still valid in the new generation of monomeric red-fluorescent proteins that have been engineered recently by Roger Tsien and co-workers.16
4. Conclusion
Time-resolved fluorescence and polarization anisotropy measurements of GFP-S65T are carried out as a function of pH (3.6–10.0), as well as excitation and detection wavelengths. At high pH, the β-barrel of anionic GFP-S65T is intact with a rotational time consistent with its molecular weight, which explains the absence of pH-dependent fluorescence blinking in FCS studies. As pH reduced below the pKa-value (∼5.9), the average fluorescence lifetime decreases (by ∼18%) with minor conformational changes (except at pH 3.6). The pH effects on the total S65T fluorescence dynamics and conformational changes become more pronounced upon assembling different ground-state populations under 396-nm excitation. The observed segmental mobility in acidic environment indicates that the embodied chromophore becomes less restricted by the surrounding hydrogen-bonding network and the amino acids. Such conformational changes of the protein coincide with pH-driven fluorescence blinking due to external proton transfer using fluorescence correlation spectroscopy, which occurs on much slower time scales. As a result, we conclude that structural changes of the β-barrel at low pH facilitate external proton transfer from the low-pH buffer to the protein moiety. Furthermore, the observed changes in S65T structure may also trigger cis/trans isomerization of the chromophore as an additional non-radiative pathway. Due to these conformational changes and fluorescence sensitivity as a function of pH, GFP mutants can be used as efficient non-invasive, pH sensors for biological studies.
Acknowledgments
We would like to thank Prof. Stephen Benkovic (PSU, Chemistry) for the gift of purified GFP-S65T. The FCS measurements on EGFP were carried out in the laboratory of Prof. Watt Webb (DRBIO, Cornell University). We are also thankful for the comments and critiques of Angel Davey and Dr. Michael E. Webb (PSU, Chemistry). This work was supported by the Penn State Materials Research Institute and the Penn State MRSEC (supported by the National Science Foundation, Grant number: DMR 0213623) and the Lehigh-Penn State Center for Optical Technologies (supported by the Commonwealth of Pennsylvania). Additional support was also provided by the Huck Institutes of the Life Sciences (PSU). The help of Chieh-Hsin Kuan, at the early stage of this project, is deeply appreciated. HK also acknowledges the support of BBSI (an NIH and NSF undergraduate program; Bioengineering, PSU). Finally, we are also grateful to Coherent Lasers, Inc. for their generous loan of a pulse picker (MIRA9200, Coherent).
References
- 1.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Chalfie M, Kain SR. Green Fluorescent Protein: Properties, Applications, and Protocols. 2nd. Vol. 47. Wiley-Liss; New York: 2005. [Google Scholar]
- 3.Heikal AA, Hess ST, Webb WW. Chem Phys. 2001;274:37–55. [Google Scholar]
- 4.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Biochemistry. 1993;32:1212. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 5.Zimmer M. Chem Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher DC. Science. 1994;263:802. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 8.Andresen M, Wahl MC, Stiel AC, Grater F, Schafer LV, Trowitzsch S, Weber G, Eggeling C, Grubmuller H, Hell SW, Jakobs S. Proc Natl Acad Sci USA. 2005;102:13070–13074. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Campbell RE, Ting AY, Tsien RY. Nature Reviews. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 10.Tsien RY. Federation of European Biochemical Societies. 2005;579:927–932. [Google Scholar]
- 11.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 12.Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Nature Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 14.Tsien RY. Nature Biotechnol. 1999;17:956–957. doi: 10.1038/13648. [DOI] [PubMed] [Google Scholar]
- 15.Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Nature Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 17.Elowitz MB, Surette MG, Wolf PE, Stock J, Leibler S. Current Biology. 1997;7:809–812. doi: 10.1016/s0960-9822(06)00342-3. [DOI] [PubMed] [Google Scholar]
- 18.Stepanenko OV, Verkhusha VV, Kazakov VI, Shavlovsky MM, Kuznetsova IM, Uversky VN, Turoverov KK. Biochemstry. 2004;43:14913–14923. doi: 10.1021/bi048725t. [DOI] [PubMed] [Google Scholar]
- 19.Heim R, Prasher DC, Tsien RY. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt AB, Tsien RY. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 22.Chattoraj M, Kong BA, Bublitz GU, Boxer SG. Proc Natl Acad Sci USA. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossau H, Kummer A, Heinecke R, Pöllinger-Dammer F, Kompa C, Bieser G, Jonsson T, Silva CM, Yang MM, Youvan DC, Michel-Beyerle ME. Chem Phys. 1996;213:1–16. [Google Scholar]
- 24.Volkmer A, Subramaniam V, Birch DJS, Jovin TM. Biophys J. 2000;78:1589–1598. doi: 10.1016/S0006-3495(00)76711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haupts U, Maiti S, Schwille P, Webb WW. Proc Natl Acad Sci USA. 1998;95:13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess ST, Huang S, Heikal AA, Webb WW. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 27.Baldini G, Cannone F, Chirico G. Science. 2005;309:1096–1100. doi: 10.1126/science.1115001. [DOI] [PubMed] [Google Scholar]
- 28.Enoki S, Saeki K, Maki K, Kuwajima K. Biochemstry. 2004;43:14238–14248. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 29.Patnaik SS, Trohalaki S, Pachter R. Biopolymers. 2004;75:441–452. doi: 10.1002/bip.20156. [DOI] [PubMed] [Google Scholar]
- 30.Hess ST, Heikal AA, Webb WW. J Phys Chem B. 2004;108:10138–10148. [Google Scholar]
- 31.Axelrod D. Methods Cell Biol. 1989;30:333–352. [PubMed] [Google Scholar]
- 32.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd. Kluwer Academic; New York: 1999. [Google Scholar]
- 33.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward WW, Bokman SH. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 35.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Proc Natl Acad Sci USA. 1998;95:6803. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miesenbock G, DeAngelis DA, Rothman JE. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 37.Schwille P, Kummer S, Heikal AA, Moerner WE, Webb WW. Proc Natl Acad Sci USA. 2000;97:151–156. doi: 10.1073/pnas.97.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson DL, Cox MM. Lehninger's Principles of Biochemistry. 3rd. New York: 2000. [Google Scholar]
- 39.Brejc J, Sixma T, Kitts PA, Kain SR, Tsien RY, Ormö M, Remington JS. Proc Natl Acad Sci USA. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwille P, Kummer S, Heikal AA, Moerner WE, Webb WW. Proc Natl Acad Sci USA. 2000;97:151–156. doi: 10.1073/pnas.97.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heikal AA, Webb WW. Proceedings of SPIE-The International Society for Optical Engineering. 2002;4812:1–14. [Google Scholar]
- 42.Warren A, Zimmer M. Journal of Molecular Graphics and Modelling. 2001;19:297–303. doi: 10.1016/s1093-3263(00)00057-7. [DOI] [PubMed] [Google Scholar]
- 43.Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 44.Weber W, Helms V, McCammon JA, Langhoff PW. Proc Natl Acad Sci USA. 1999;96:6177–6182. doi: 10.1073/pnas.96.11.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]