Abstract
Lipopolysaccharide (LPS) is a major component of the outer membrane of gram-negative bacteria. The lipid A region of LPS stimulates the immune system in a structure dependent manner. We have previously identified the two major lipid A species from Francisella tularensis as asymmetric tetraacylated structures containing four long acyl chains (16 and 18 carbons) and a single phosphate group that is partially modified by galactosamine (Phillips, N.J.; Schilling B.; McLendon, M.K.; Apicella, M.A.; Gibson, B.W. Infect. Immun. 2004, 72, 5340–5348). In this current study, we used matrix-assisted laser desorption / ionization on a vacuum source (vMALDI) coupled to a linear ion trap (LIT) mass spectrometer in multiple-stage mass fragmentation mode (MSn) to determine the structures of several minor and low abundant lipid A species present in F. tularensis, F. novicida, and F. philomiragia LPS that have not been previously characterized. Comprehensive vMALDI-MSn fragmentation studies allowed us to deduce the composition and the position of the fatty acid substituents within the lipid A moieties. Unexpectedly, most of these minor lipid A species consisted of multiple isobaric species with acyl chains of various lengths. Moreover, we found that a small portion of these lipid A’s may be modified by the addition of a hexose or hexosamine sugar, in addition to the galactosamine that was previously identified. Overall, we found that MSn analysis on the vMALDI-LIT MS platform was highly efficient and sensitive, allowing for thorough analysis of very minor lipid A species.
INTRODUCTION
Francisella tularensis is a gram-negative, non-sporulating, encapsulated, facultative intracellular coccobacillus that causes tularemia, a zoonotic disease of humans and mammals. Tularemia is characterized by acute febrile illness and, commonly, ulceration at the site of entry1. F. tularensis is transmitted to humans and other mammals by the bite of infected insects or animals, direct inoculation of the mucosa, ingestion of contaminated water or food and, most seriously, by inhalation of aerosolized particles2, 3. Inhalation of as few as 10 bacteria can cause serious disease in humans4, 5. Because of its high infectivity, ease of aerosol dissemination and severe morbidity and mortality, F. tularensis is considered a possible biological warfare agent6, 7.
The lipopolysaccharide (LPS) of F. tularensis is structurally and biologically distinct from the LPS of other gram-negative bacteria. The lipid A moiety functions as a hydrophobic anchor of the LPS and it has been shown that the degree of acylation and other modifications of lipid A influence toxicity. As depicted in Figure 1A, we have previously reported8 that the major lipid A form from F. tularensis is asymmetric and contains four long chain fatty acids (16 and 18 carbons), lacks a 4′-phosphate residue, and is partially substituted with phosphate or phosphogalactosamine at the 1-position of the diglucosamine backbone. The structures of lipid A moieties isolated from F. tularensis and F. novicida strains have also been reported by Vinogradov et al.9, 10. In their studies, a very similar structure was found to that reported by Phillips et al.8, but differed in that the major lipid A species contained one or more fatty acids of shorter length as well as a free reducing terminus with no phosphate or sugar substitutions. We have previously suggested that these structural differences may have been due to the use of positive-ionization MALDI-MS by Vinogradov and colleagues9, 10 for lipid A profiling, thus favoring the ionization of non-phosphate containing isoforms, and/or differences in the isolation and purification steps that may have effected the extraction efficiencies of the different lipid A forms. The presence of lipid A variants with two methylene groups shorter or longer relative to the major species was indicated by both groups, but was not investigated further. Recent studies by Wang et al.11, 12 used complementation in Escherichia coli with Francisella genes to demonstrate functions of the F. novicida lipid A biosynthesis genes lpxE and lpxF. In addition, using complementation and mutational analysis our group identified an F. tularensis gene encoding a homolog of the E. coli laurate transferase, LpxL (McLendon et al., unpublished results).
Figure 1.
Negative-ion vMALDI-LIT mass spectra of partially purified lipid A preparations. (A) Francisella major tetraacyl lipid A structure consists of a diglucosamine backbone bearing four fatty acids: three C18:0(3-OH) fatty acids (in the 2-, 3-, and 2′-position) and one C16:0 fatty acid (linked to the C18:0(3-OH) acyl chain in the 2′-position), plus additional phosphate and galactosamine groups. Negative-ion vMALDI-LIT MS of lipid A isolated from (B) Francisella tularensis, (C) Francisella novicida, and (D) Francisella philomiragia. Major tetraacyl lipid A species were observed at m/z 1665.2 and m/z 1504.2 with a composition of C16:0, 3x C18:0(3-OH), phosphate, GalNn=1/0 and are colored in blue. Additional minor and low abundant lipid A species were observed with masses 28 Da higher (colored in red) and 28 Da lower (colored in green) suggesting lipid A species containing acyl chains that are two methylene groups longer or shorter than the major species. For further detail of lipid A compositions see Supplementary Table S1.
In this study, we have systematically investigated low abundant lipid A species from LPS obtained from F. tularensis, F. novicida, and F. philomiragia grown in vitro using a new linear ion trap (LIT) mass spectrometer equipped with a vacuum MALDI ion source (vMALDI-LTQ). We have developed a sensitive mass spectrometric strategy using multiple-stage MS (MSn) to characterize these novel low abundant lipid A structures relative to their acylation patterns and the possible presence of unusual sugar modifications. Here, we report the identification of several minor lipid A structures containing very long acyl chains (up to 20 carbons) and show them to consist of multiple isobaric species exhibiting a complex and variable acylation pattern (involving the 2-, 3-, and 2′-positions) as well as being further modified with a second hexosamine and/or hexose sugar.
EXPERIMENTAL SECTION
Strains and Culture Conditions
F. tularensis subsp. holarctica strain 1547-578 (referred to as F. tularensis in this manuscript) was cultured on chocolate agar medium (Remel, Lenexa, KS). Because of F. tularensis’ high infectivity, ease of aerosol dissemination and severe morbidity and mortality, all work involving F. tularensis required appropriate safety measures and was carried out in a CDC-approved BSL-3 laboratory with select agent approval. F. philomiragia strain 25015 and F. novicida strain U112 bacteria were cultured on cysteine heart agar medium (Difco Co., Sparks, MD) supplemented with 9% defibrinated sheep’s blood (Colorado Serum Co., Denver, CO). Bacteria were incubated at 37°C with 5% CO2 for 48 h.
LPS Isolation
Bacteria were collected by flooding plates with PBS and scraping colonies from the surface. LPS was isolated using a standard phenol extraction protocol as previously described in detail8.
Acid Hydrolysis of LPS-lipid A Preparation
LPS from the various Francisella species (1.0–1.5 mg) were hydrolyzed in 200 μl 1% acetic acid at 100°C for 2 h. During incubation samples were agitated/vortexed every 30 min to ensure completeness of reaction. Samples were then centrifuged at 4°C for 40 min, and the lipid A pellets were washed 2 times with water and dried under a stream of nitrogen. For further purification, the lipid A pellets were partitioned in 525 μl CHCl3/CH3OH/H2O (10/5/6, v/v/v) and the bottom organic layers and interfaces were saved and evaporated to dryness under a stream of nitrogen.
Multiple-Stage Mass Spectrometry (MSn)
Data were acquired on a LTQ linear ion trap (LIT) fitted to a vMALDI™ ion-source (Thermo Electron, San Jose, CA). The intermediate-vacuum (170 mTorr) MALDI ion source uses a 337.7 nm SI nitrogen laser with a frequency of 20 Hz and energy of ~250 μjoules per pulse. Lipid A samples were dissolved in 3:1 CHCl3/CH3OH (v/v); l μl of analyte was mixed with l μl saturated CMBT matrix solution (6-chloro-3-mercaptobenzothiazole) in 3:1 CHCl3/CH3OH (v/v), and then spotted on 96- or 384-position stainless steel target plates. Data were acquired in both negative- and positive-ion mode using XCalibur 1.4 software scanning a mass range from m/z 700–4000 (in MS mode). To record multiple-stage mass spectra (MSn, n=2–5) the following parameters were used: precursor ion isolation width of 3 m/z, normalized collision energy of 40% (percentage of RF amplitude used to fragment ions), activation Q of 0.25 and activation time of 30 msec. Spectra were recorded using automatic gain control to control the number of laser shots and the automatic spectrum filter tool. Lipid A isoform ratios were calculated from fragment ion abundances (ion peak abundances) derived from MSn fragmentation patterns.
RESULTS AND DISCUSSION
To investigate the lipid A profiles and corresponding structures in several Francisella species, LPS was isolated from F. tularensis, F. novicida, and F. philomiragia strains using a classical phenol extraction protocol. Crude lipid A fractions were then prepared from these LPS by acetic acid hydrolysis, followed by partial purification using organic extraction. To characterize these lipid A preparations, we first profiled the purified fractions by MALDI-MS to identify the major and minor species, and then compared these profiles to what has been previously reported for the major species from F. tularensis8, 9. Lastly, an in-depth mass spectrometric investigation of the uncharacterized lipid A components was carried out using multiple-stage mass fragmentation (MSn) on a linear ion trap (LIT) mass spectrometer equipped with a vacuum MALDI ionization source.
MS Profiles of Francisella lipid A’s
The partially purified lipid A fractions from F. tularensis, F. novicida, and F. philomiragia were initially analyzed by vMALDI-LIT MS in the negative-ion mode to determine the monoisotopic masses of the major and minor species. Overall, the masses and the relative abundances of the major (and even minor) lipid A species among the three Francisella species were very similar (see Figure 1), and all contained the major two species at m/z 1504 and 1665 that we had previously identified from F. tularensis8.
As shown in Figure 1A, the two major lipid A species from F. tularensis consist of a diglucosamine backbone bearing four fatty acids; three 3-hydroxy stearic acids (C18:0(3-OH)) in the 2-, 3-, and 2′-positions and one palmitic acid (C16:0) linked to the hydroxy group of 3-hydroxy stearic acid at the 2′-position8. These non-symmetric tetraacyl lipids A’s are further modified with a phosphate or a galactosamine moiety in the 1-position, yielding the two major deprotonated molecular ions (M-H)- at m/z 1504.2 and m/z 1665.2, respectively. While the lipid A species at m/z 1665 has been fully characterized for F. tularensis, to the best of our knowledge, a lipid A species containing three 3-hydroxy stearic acids and one palmitic acid has not been previously characterized for the F. novicida and F. philomiragia strains. However, it is clear from MS profiles that they exhibit the two major lipid A species at the same masses at m/z 1665 and m/z 1504. A non-phoshorylated lipid A species described by Vinogradov et al. for F. novicida and F. tularensis9, 10 appears in positive-ion mode only (at m/z 1392) and contains shorter fatty acids.
Upon closer inspection of the vMALDI-MS profiles shown in Fig. 1B-D, several additional minor lipid A species were present with mass shifts of +/−28 Da relative to the two major lipid A species at m/z 1504 and 1665. These mass shifts most likely result from incorporation of acyl chains that are either longer or shorter by two carbons. In fact, similar lipid A mass shifts of +/−28 Da have been previously observed in several other bacterial species that have undergone thorough mass spectrometric analysis, such as Salmonella minnesota Re59513 and Pseudomonas aeruginosa14. However, unlike the Francisella species examined here, these additional minor lipid A isoforms were attributed to fatty acid alkane chain length heterogeneity that typically involved only a single fatty acyl chain substitution, such as lauroyl to myristoyl13, or 3-hydroxycaproyl to 3-hydroxylauroyl14.
For F. philomiragia the lipid A MS profile data suggests a shift towards lower masses (i.e., shorter fatty acid chains) compared to the two other Francisella strains investigated.
Additional low abundant lipid A species were observed for F. philomiragia at m/z 1609.4 and 1448.3, indicating the presence of even shorter (− 4 carbons, −56 Da) fatty acid chains. Although not indicated in the MS profile data, the m/z 1609 species was also detected in trace amounts in F. tularensis and F. novicida.
Lastly, a higher mass peak was observed for F. novicida at m/z 1827, that corresponds to a mass shift of 162 Da relative to the major conserved lipid A peak at m/z 1665 (see Fig. 1C). After close inspection of the lipid A spectra of F. tularensis and F. philomiragia (Fig. 1B and D), a similar peak at m/z 1827 was seen for F. philomiragia, but at considerably lower abundance. The MS profile spectrum for F. tularensis, however, did not contain this additional component, although a minor peak at m/z 1826 was detected. Nominally, these peaks would correspond to the addition of a hexose (+162 Da) to the galactosamine-containing lipid A isoform at m/z 1665 for F. novicida and F. philomiragia, and possibly the addition of a second amino sugar (+161 Da) to the corresponding lipid A species in F. tularensis.
As described in detail below, to characterize and identify the additional low abundant lipid A species mentioned above, a series of MSn experiments were carried out on these lipid A species from F. tularensis and F. novicida, and in some limited cases, from F. philomiragia (see Supplementary Table S1 for overview). To confirm the identities, we systematically investigated these lower abundant lipid A species by MSn, with special attention to the species at m/z 1693 and 1637 (m/z 1665 +/−28 Da), and at m/z 1532 and 1476 (m/z 1504 +/−28 Da). Moreover, the major lipid A forms for F. novicida were also examined, as these have not been reported before.
MSn Analysis of m/z 1665 lipid A from F. novicida
To verify the identity of the major lipid A species from F. novicida, a vMALDI-LIT MSn fragmentation study was carried out on the (M-H)− precursor ion at m/z 1665.3, whose structure has not been previously reported. A comparison of the resulting MSn spectra with the same spectral data set obtained from the analogous precursor ion in F. tularensis (whose structure is known8) showed essentially identical fragment ions, suggesting that the lipid A composition and structure is indeed the same (see Supplementary Fig. S1). It is worth noting that the vMALDI-MSn fragmentation pattern of the m/z 1665 species from both the F. novicida and F. tularensis strains show a close correspondence to the fragment ion distribution reported in the previous ESI-MSn studies used to establish the F. tularensis lipid A structure8. Briefly, the major fragments can be attributed to a loss of hexosamine (−161 Da, HexN) in combination with subsequent losses of O-linked acyl chains, i.e., loss of C16:0 (−256 Da) and/or loss of O-linked C18:0(3-OH) (−300 Da), (see Supplementary Fig. S1). In addition, other fragments result from the loss of N-linked C18:0(3-OH) as ketene (−282 Da), and a specific loss of a 240 Da moiety was previously assigned to a McLafferty type rearrangement resulting from N-linked 3-hydroxy stearic acid8. The deciphering of acyl chain positioning was supported by a glycosidic fragment ion pair Y1/Z1 at m/z 522.3 and 504.2, according to the nomenclature described by Costello and Vath15.
MSn Analysis of minor lipid A species at m/z 1693 and m/z 1532 from F. tularensis
As previously noted, the major lipid A isoforms at m/z 1665 and 1504 in all three Francisella species show additional satellite peaks at m/z 1693 and 1532, respectively, that were 28 Da higher in mass. The shift in mass alone suggest 2 carbon extensions in one or more of the four acyl chains, perhaps substituting one of the three C18:0(3-OH) chains with 3-hydroxy arachidic acid (C20:0(3-OH)). To explore the source of this fatty acid heterogeneity, an in-depth MSn analysis was carried out on the deprotonated molecular ion for this minor lipid A species at m/z 1693.0 for F. tularensis. Although the m/z 1693 peak appears as a very minor component in the overall lipid A profile (see Fig. 1B), the high sensitivity of the vMALDI-LIT in MSn mode (see Fig. 2) allowed for relatively straightforward deciphering of the lipid A structure and acylation pattern including the position of individual acyl groups as described below.
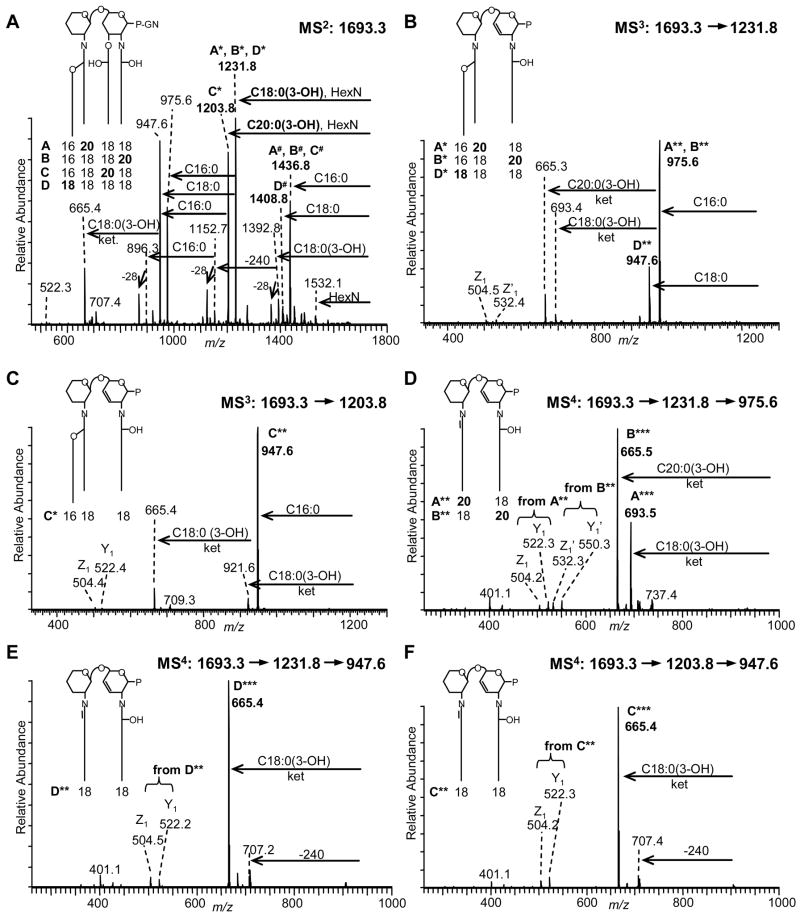
Figure 2.
Multiple-stage negative-ion vMALDI-MSn spectra of F. tularensis minor lipid A species at m/z 1693.3. (A) MS2 of m/z 1693.3; (B) MS3 of m/z 1231.8 (from MS2 1693.3); (C) MS3 of m/z 1203.8 (from MS2 1693.3); (D) MS4 of m/z 975.6 (from MS3 1231.8, from MS2 1693.3); (E) MS4 of m/z 947.6 (from MS3 1231.8, from MS2 1693.3); and (F) MS4 of m/z 947.6 (from MS3 1203.8, from MS2 1693.3). At least 4 different isoforms “A-D” were characterized (for more details see Suppl. Figs. S2 and S3). P: phosphate, GN: galactosamine.
MSn analysis of the (M-H) − precursor ion at m/z 1693 from F. tularensis gave a very distinctive and definitive fragmentation pathway (Fig. 2). As seen in Fig. 2A, the two most abundant fragments ions in the MS2 spectrum appear at m/z 1231.8 and 1203.8, with a relative abundance ratio of 1.2:1.0. These two fragment ions can be readily assigned as arising from the combined loss of hexosamine and either an O-linked C18:0(3-OH) or C20:0(3-OH) fatty acid (−461 and 489 Da, respectively). In the MS2 spectra of m/z 1665 from F. novicida (see Supplementary Fig. S1) as well as from the previously published F. tularensis ESI-MSn study8, this abundant fragmentation process was assigned to the loss of the unsubstituted O-linked fatty acid located on the 3-position, as this O-acyl chain is considerably more labile than the N-linked chains. Thus, the two fragment ions at m/z 1231.8 and 1203.8 provide the first evidence of multiple isobaric lipid A structures at m/z 1693.2. Additionally, the m/z 1693.2 precursor ion can lose an O-linked non-hydroxylated fatty acid, either C16:0 or C18:0, that substitutes the 3-hydroxy group of the 2′-N-linked fatty acid (referred to as 2′-O) to produce fragment ions at m/z 1436.8 and 1408.0, respectively (Fig. 2A). These latter fragment ions, which arise from the heterogeneous loss of the fatty acid in acyloxyacyl linkage on the N-linked 2′-position, provide further support for fatty acid heterogeneity. As discussed above, the initial analysis of the MS2 spectrum of the precursor ion at m/z 1693 clearly indicates acyl chain heterogeneity, in this case involving two carbon chain extensions at multiple lipid A backbone positions, i.e., positions 3 and 2′-O.
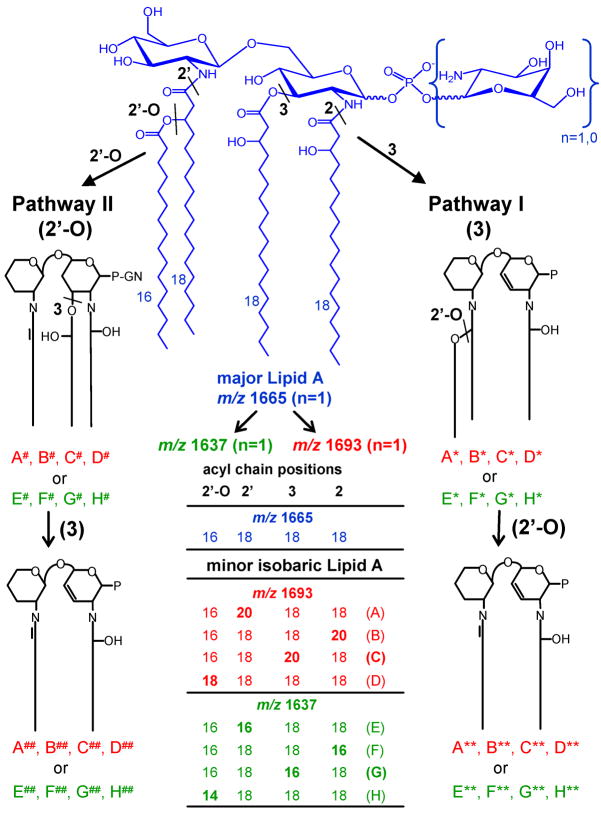
As shown in Fig. 3, the major difference between these two primary fragmentation pathways can be attributed to which O-linked acyl group, the most labile of fatty acid linkages, is cleaved first. O-acyl cleavage can occur on the O-linked 3-position (Pathway I, “3-cleavage”) or the secondary acyl chain attached to the hydroxy group of the primary N-linked acyl group on the 2′-position (Pathway II, “2′-O-cleavage”). Given this nomenclature (see Fig. 3, and for more details Suppl. Fig. S2), Pathway I gives rise to the two most abundant fragment ions in the MS2 spectrum of precursor ion m/z 1693.2 (Fig. 2A) at m/z 1231.8 and 1203.8, loss of galactosamine and either C16:0(3-OH) or C18:0(3-OH), and Pathway II gives rise to the fragment ions at m/z 1436.8 and 1408.8 (loss of C16:0 or C18:0). To follow specific fragmentation pathways through multiple stages of MSn, an annotation system was devised as follows. Fragment ions associated with Pathway I are marked with asterisks (*), and fragmentation Pathway II uses a cross-hatch annotation (#). Specific lipid A isoforms are given a letter designation, i.e., A-D, and the fragment ions derived from a specific lipid A isoform are annotated with one or more asterisks or cross-hatches to denote the MS cycle from which it was derived, i.e., isoform A fragment ions are annotated as A* (MS2), A** (MS3) or A*** (MS4) within Pathway I, and as A# (MS2), A## (MS3) or A### (MS4) within Pathway II.
Figure 3.
MSn fragmentation scheme defining the major fragmentation Pathways I and II, of Francisella lipid A species and isobaric lipid A species featuring heterogeneous acyl chain distribution. Francisella major tetraacyl lipid A at m/z 1665 consists of one isoform only (blue). Further minor isobaric lipid A species featuring acyl chain distribution on the disaccharide backbone are indicated in red for m/z 1693 (heterogeneous isoforms A-D) and in green for m/z 1637 (heterogeneous isoforms E-G). Major MSn lipid A fragmentation pathways are attributed to which O-linked acyl group is cleaved first, i.e., either loosing the O-linked acyl group on the 3-position (pathway I, “3-cleavage”) or the O-linked secondary acyl chain attached the primary N-linked acyl group on the 2′-position (pathway II, “2′-O-cleavage”). Fragment ions are annotated with one or more asterisks or cross-hatch to denote the MS cycle and the fragmentation pathway it was derived from.
A surprisingly complex fragmentation pattern was uncovered for the species at m/z 1693.2 even at the MS2 stage, suggesting that the m/z 1693 peak consists of a series of isobaric lipid A species with different acyl chains located at several positions on the lipid A backbone. This observation is in contrast to the two major lipid A species at m/z 1665 and 15048, that feature one major isoform each. At least four isobaric lipid A species could ultimately be identified for the peak at m/z 1693.2 as indicated in the inset of Fig. 2 and in Supplementary Fig. S2 (also see Fig. 3 showing heterogeneous acyl chain distribution for m/z 1693 marked in red). Isobaric structures were generated by substitution of one 3-hydroxy stearic acid with 3-hydroxy arachidic acid, i.e., at the 2′-, 2- or 3-position, yielding “isoforms A-C” respectively (see Supplementary Table S1). Alternatively, substitution of the palmitic acid linked to the 3-hydroxy stearic acid in the 2′-position with stearic acid would yield a fourth “isoform D” (all isoforms discussed above also contain one galactosamine and phosphate).
To further deconvolute the isomeric contributions to the MS2 fragment ions and assign the underlying isobaric lipid A isoforms A-D, additional MS3 and MS4 fragmentation cycles were performed. For MS3 analysis, fragment ions at m/z 1231.8 (A*, B*, and/or D*) and m/z 1203.8 (C*) were selected and these spectra are shown in Fig. 2B and 2C, respectively (see Suppl. Fig. S2). As shown in Fig. 2B, fragmentation of m/z 1231.8 led to a loss of C16:0 to yield an ion at m/z 975.6 (A**, B**), and simultaneously a loss of C18:0 to yield an ion at m/z 947.6 (D**). To confirm the latter fragment ion structures, MS4 analyses were carried out on m/z 975.6 and m/z 947.6 (Figs. 2D and 2E). Fragmentation of the peak at m/z 975.6 showed losses of the more stable N-linked 3-hydroxy fatty acids at the 2-position as ketenes to yield MS4 fragment ions m/z 693.5 (A**− C18:0(3-OH)ketene = A***) and m/z 665.5 (B** − C20:0(3-OH)ketene = B***). Additional evidence was provided from the coexistence of specific glycosidic fragment ions Y1/Z1 at m/z 522.3 and 504.2 that are generated from isoform A** (with C18:0(3-OH) in the 2-position), and Y′1/Z′1 at m/z 550.3 and 532.3 that are generated from isoform B** (with C20:0(3-OH) in the 2-position). The fragment ion at m/z 947.6 from the MS3 spectrum (observed in Fig. 2B) was then selected for MS4 analysis as shown in Fig. 2E. This latter spectrum confirmed the structure of isoform fragment D** based on the loss of the N-linked C18:0(3-OH) fatty acid in the 2-position as ketene yielding m/z 665.4 (D***). Additional structural evidence was provided by the observation of glycosidic ions Y1/Z1 at m/z 522.2 and 504.5.
To investigate the second branch of fragmentation Pathway I, the peak at m/z 1203.8 (C*) was selected for MS3 and its spectrum is shown in Fig. 2C. As expected, a loss of a C16:0 fatty acid yielded an ion at m/z 947.6 (C**). Subsequent selection of the m/z 947.6 for MS4 analysis is shown in Fig. 2F, and clearly shows the anticipated loss of an N-linked C18:0(3-OH) fatty acid as a ketene from the 2-position to yield an ion at m/z 665.4 (C***). Glycosidic cleavage ions Y1 and Z1 at m/z 522.3 and 504.2 also indicated a C18:0(3-OH) fatty acid in the 2-position. Overall, fragmentation Pathway I (see Suppl. Fig. S2) clearly identifies four different isobaric lipid A structures A-D.
Independent evidence for the presence of these four isomers (A-D) in the molecular ion m/z 1693.3 was provided by the alternative fragmentation pathway (Pathway II) that investigated the MS2 fragment ions at m/z 1436.8 (A#-C#) and 1408.8 (D#). The m/z 1436.8 and 1408.8 ion fragment pair (abundance ratio 7.0:1.0) reflects the initial loss from the m/z 1693.3 precursor ion of C16:0 and C18:0 fatty acids, respectively, which were attached to the hydroxyl group of the 3-hydroxy acyl group in the 2′-position (see Fig. 3 and Suppl. Fig. S2). Further selection of the ion m/z 1436.8 (A#, B# & C#) for MS3 fragmentation (see Suppl. Fig. S3A), resulted in a loss of hexosamine and an O-linked 3-hydroxy acyl chain at the 3-position, which was either C18:0(3-OH) (A##, B##) or C20:0(3-OH) (C##) yielding m/z 975.7 and m/z 947.6, respectively. In fact, these lipid A fragments A##, B##, and C## contain one N-linked 3-hydroxyl acyl chain in the 2-position and one N-linked monounsaturated acyl chain in the 2′-position and were already investigated as part of fragmentation Pathway I as described previously (A**, B**, and C**). Nevertheless, for completion, MS4 fragmentation was performed on these latter two ions as part of Pathway II which is described more fully in the supplement (Suppl. Figs. S3C and S3D). Similarly, the low abundant fragment ion at m/z 1408.8 (D#, see Suppl. Fig. S3B) was selected for MS3 analysis, and the resulting ion at m/z 947.6 (D##) was subsequently selected for MS4 analysis (see Suppl. Fig. S3E). While we are aware of the intrinsic difficulties of extracting quantitative data regarding the relative proportions of the four lipid A isoforms (A-D; M =1694.01 Da), a ratio among the different isoforms was calculated by comparing peak abundances of corresponding fragments in the distinct pathways, leading to an estimated of 1.0:2.0:3.7:1.0 (A:B:C:D) for F. tularensis lipid A isoforms that arise from the minor peak at m/z 1693.3 (see Supplementary Table S1).
An analogous set of lipid A isoforms were also observed for the related, low abundant lipid A species at m/z 1532.1, which differs from the m/z 1693.3 peak in that it lacks galactosamine (GalN). The vMALDI-MSn fragmentation spectra of the (M-H) − precursor at m/z 1532.1 (Suppl. Fig. S4A) revealed a mixture of four isobaric species (A–D) with similar acyl chain compositions as discussed above for the corresponding m/z 1693 lipid A species. Longer acyl chains such as 3-hydroxy arachidic acid or stearic acid are incorporated into the lipid A structures (for more details and varying backbone positions of acyl groups, see Supplementary Table S1 and Suppl. Figs. S4 and S5). The relative abundances of the four lipid A isoforms from F. tularensis determined from the fragmentation studies of the m/z 1532 peak were estimated to be 1.0:1.8:3.2:1.2 (A:B:C:D), and were very similar to the relative ratios calculated for the m/z 1693 lipid A peak. The relative abundances of the four lipid A isoforms from F. tularensis determined from the fragmentation studies of the m/z 1532 peak were estimated to be 1.0:1.8:3.2:1.2 (A:B:C:D), and were very similar to the relative ratios calculated for the m/z 1693 lipid A peak. As already stated, this method of estimating lipid A isoform ratios is based on the assumption that ionization efficiencies are similar amongst the different acylation isoforms. While this is not an unreasonable assumption, this correlation has not been independently validated and therefore should only be considered a rough estimate of actual molar ratios.
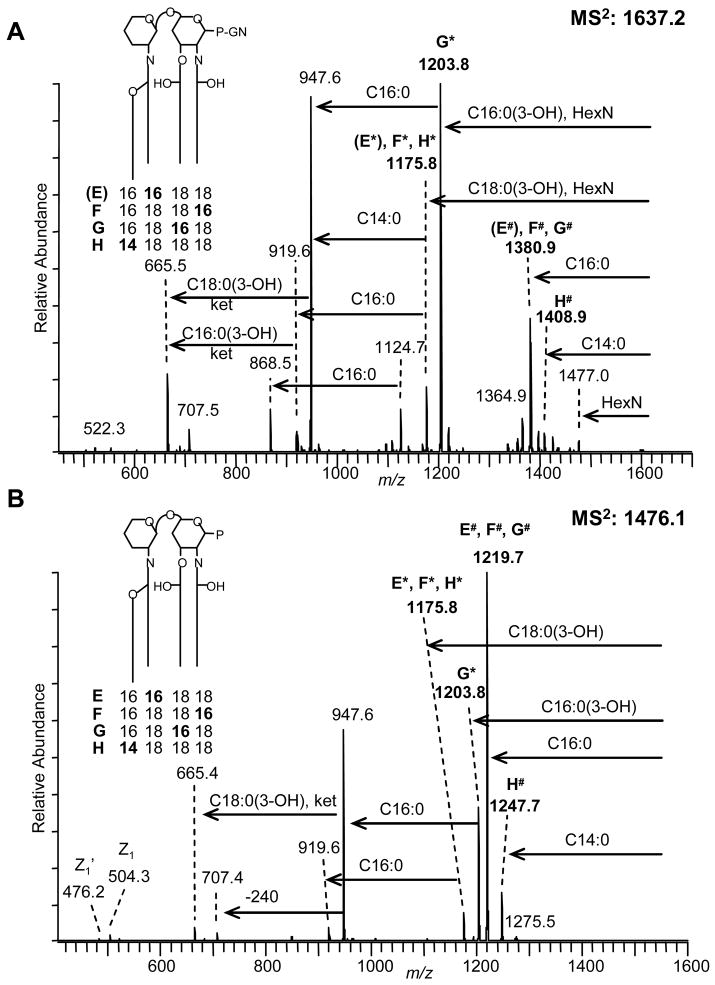
MSn Analysis of lipid A species at m/z 1637 and m/z 1476 from F. tularensis
In addition to the lipid A species with two carbon chain extensions, peaks in the MS profile data (Fig. 1) were observed at m/z 1637.2 and 1476.2 that were 28 Da lower in mass relative to the two major species, suggesting lipid A isoforms containing shorter fatty acids, such as 3-hydroxy palmitic acid and myristic acid. These latter species have been previously reported8 and appear at higher abundance than the corresponding +28 Da isoforms at m/z 1693 and 1532 as discussed in the previous section. These lower mass species, however, have not been characterized relative to their fatty acid substituents. Therefore, to investigate these latter lipid A species, multiple-stage fragmentation was carried out on both peaks from F. tularensis.
Fragmentation of the (M-H) − precursor ion at m/z 1637.2 (see Fig. 4A and Suppl. Fig. S6) indicated multiple isobaric lipid A species, annotated as isoforms “E-H” (see Fig. 3), with varying fatty acid compositions, i.e., either 2 × C18:0(3-OH), C16:0(3-OH), C16:0 or 3× C18:0(3-OH), C14:0, but all containing phosphate and galactosamine. Distribution of the acyl groups in different backbone positions was further investigated by detailed MSn studies that are shown in Suppl. Fig. S6 and described in Suppl. Fig. S7. In addition, their overall features are listed in Supplementary Table S1.
Figure 4.
Negative-ion vMALDI-MS2 spectra of F. tularensis lipid A species at m/z 1637.2 and m/z 1476.1. (A) MS2 of m/z 1637.2; and (B) MS2 of m/z 1476.1. Four different isoforms “E-G” were characterized for lipid A species at m/z 1637.2 and m/z 1476.1. However, for the 1637 species, isoform “(E)” only showed minor contribution (for more details see Suppl. Figs. S6-S9). P: phosphate, GN: galactosamine.
Multiple-stage mass spectrometry was also carried out on the analogous lipid A species lacking the galactosamine with an (M-H) − at m/z 1476.2 as shown in Fig. 4B (also see Suppl. Figs. S8 and S9). Fragmentation patterns were observed for these two lipid A molecular ions (m/z 1637 and 1476) that were analogous to those discussed in the previous section for the +28 Da lipid A species at m/z 1693 and 1532. The MS2 spectra of these latter lower mass species showed comparable levels of fatty acid heterogeneity (Figs. 4A and 4B). For example, both spectra show ion pairs at m/z 1203.8 and m/z 1175.8 that reflect the heterogeneous loss of O-linked C16:0(3-OH) and O-linked C18:0(3-OH), respectively, emanating from the 3-position. In the case of the m/z 1637.2 precursor ion (Fig. 4A), a simultaneous loss of hexosamine was required to yield the above fragment ions. Losses of C14:0 and C16:0, respectively, reveal heterogeneity of the acyl chain connected to the hydroxyl group of the N-linked acyl group attached at the 2′-position of the lipid A backbone, giving rise to characteristic fragment ion pairs at m/z 1408.9/1380.9 (from m/z 1637.2) and at m/z 1247.7/1219.7 (from m/z 1476.2). All major MS2 fragment ions mentioned here were further analyzed by MS3 and MS4 fragmentation studies to decipher the isobaric lipid A’s (see Suppl. Fig. S6 and S8). Lipid A isoform structures E-H are schematically drawn in Fig. 4A and 4B (see insets) and could clearly be identified for precursor ions m/z 1637.2 and m/z 1476.2 (for details see Suppl. Figs. S6-S9 and Suppl. Table S1). However, for the m/z 1637 species, the lipid A isoform E bearing a C16:0(3-OH) fatty acid in the 2′-position showed minor contribution to the isoform mixture. The corresponding lipid A structures and estimated isoform abundance ratios are summarized in Supplementary Table S1.
MSn Analysis of low abundant lipid A species at m/z 1609 from F. philomiragia
One of the least abundant lipid A species observed for F. philomiragia at m/z 1609.4 indicates the presence of even shorter fatty acid chains than seen in the other species, i.e., −56 Da or 4 carbon chain reductions. To determine the makeup of this lipid A, a similar set of MSn experiments was conducted (see Suppl. Fig. S10). The complex fragmentation pathways of the lipid A (M-H) − precursor ion observed at m/z 1609 could be readily assigned as a mixture of isobaric lipid A species (see Suppl. Fig. S11) where the most abundant isoform (so called ”I-1”) could be assigned a fatty acid composition; of 2x C18:0(3-OH)[2, 2′], C16:0(3-OH)[3], and C14:0. Several more heterogeneous lipid A isoforms were also observed containing fatty acids, such as C18:0(3-OH), C16:0(3-OH), C14:0, C16:0, and C12:0, with different distribution of the acyl groups within the diglucosamine backbone positions 2, 3, and 2′ (for details see Supplementary Table S1).
Non-Phosphorylated lipid A at m/z 1392 from F. tularensis
As part of this extensive lipid A fragmentation study, we also investigated the major F. tularensis lipid A detected in positive-ion mode using vMALDI-LIT MSn, a non-phosphorylated lipid A observed with (M+Na)+ at m/z 1392.0 (see Suppl. Fig. S12). As expected, only one major isoform was determined with the specific acyl group distribution o 2x C18:0(3-OH)[2′ and 2], C16:0(3-OH)[3], and C14:0. These observations correspond to structural data for the m/z 1392 lipid A previously reported by Vinogradov et al.9. Analyzing the fragmentation pattern of the m/z 1392 species, it was interesting to note that the non-phosphorylated lipid A’s in positive-ion mode showed quite abundant cross-ring cleavages, such as 0,4A2 and 2,4 A1 fragment ions (see Suppl. Fig. S12). These cross-ring fragment ions are typically seen in lipid A’s with a free (non-phosphorylated) reducing terminus16, 17 which is the case here in these non-phosphorylated lipid A species.
Possible Sugar Modifications in lipid A species at m/z 1827 and 1826
As stated earlier, in addition to the lipid A species that originate from fatty acid heterogeneity, two lipid A species at m/z 1827.0 and 1826.2 were observed in F. novicida and F. tularensis that suggest the presence of hexose (+162 Da) and hexosamine (+161 Da), respectively. The negative-ion fragmentation MS2 spectra for (M-H) − at m/z 1827 and 1826 are shown in Fig. 5A and 5B, and display distinct mass shifts of 1 Da comparing the resulting fragment ions, e.g., 1570.6/1569.9; 1365.5/1364.8; and 1109.5/1108.6. The F. novicida lipid A peak at m/z 1827.0 (see Fig. 1C and Suppl. Fig. S13A) has been recently described by Shaffer et al.18 and features an additional hexose (+162 Da) that was previously determined by the same group to be glucose added to the 4′-position of the major tetraacyl lipid A at m/z 1665. However, the corresponding peak in the F. tularensis lipid A was detected at even lower abundance, but with a distinct and isotopically resolved ion at m/z 1826.2 (compare MS trace in Fig. 1B, and zoom in MS trace Suppl. Fig. S13B). Presumably this latter lipid A displays an additional hexosamine instead of the hexose observed for F. novicida. Further details of the fragmentation MSn spectra (n=2–4) of precursor ions 1827.0 and 1826.2 are shown in Suppl. Figs. S14 and S15. Due to the extremely low abundance of the F. tularensis precursor ion at m/z 1826.2, we were not yet able to determine the exact position of the additional hexosamine group on this lipid A. The 4′-position of the distal glucosamine of the major lipid A represents the most likely location based on the observations made for the extra hexose found in F. novicida18.
Figure 5.
Negative-ion vMALDI-MS2 spectra of lipid A species at m/z 1827.1 (A) and m/z 1826.3 (B) isolated from F. novicida and F. tularensis, respectively. The F. novicida lipid A at m/z 1827.1 was previously described by Shaffer et al.18 to feature an additional hexose (+162 Da) in the 4′-position of the lipid A backbone compared to the major species at m/z 1665 with a composition of C16:0, 3x C18:0(3-OH), phosphate, HexN (see Fig. 5A). However, the novel corresponding F. tularensis lipid A species at m/z 1826.3 displays an additional hexosamine (HexN: +161 Da) instead (see Fig. 5B). The asterisk indicates that the position of the additional hexosamine on the backbone has not yet been determined (it possibly is located in the 4′-position as is the case for F. novicida). MS2 fragment ions from precursor ions m/z 1827.1 (A) and 1826.3 (B) marked bold differ in mass by 1 Da, indicating the difference between the fragment ion bearing a hexose (Hex) or hexosamine (HexN) moiety. For more details see Suppl. Figs. S14 and S15.
CONCLUSION
The investigation of lipid A fragmentation, primarily from F. tularensis but also from F. novicida and F. philomiragia, led to the unexpected discovery that these lipid A’s consist of multiple isobaric species. Using multiple-stage fragmentation (MSn) on a vMALDI-LIT platform, we were able to decipher and characterize novel, multiple isobaric lipid A structures differing primarily in their acyl chain lengths and acyl chain backbone positions (2-, 3-, and 2′-positions). The fatty acid heterogeneity and presence of lipid A isoforms was observed primarily in the minor lipid A species at m/z 1693.0 and 1532.1 containing longer acyl chains, such as C20:0(3-OH) or C18:0, and also in low to medium abundant lipid A species at m/z 1637.2 and 1476.2 containing shorter acyl chains, such as C16:0(3-OH) or C14:0, compared to the major lipid A species.
From the individual lipid A fragmentation data and their MSn fragmentation pathways, we were able to estimate isoform abundance ratios (Supplementary Table S1). Interestingly, for F. tularensis, the 3-position of the lipid A backbone featuring an O-linked fatty acid possibly appears as a preferred position to add in longer, i.e., C20:0(3-OH), or shorter acyl chains, i.e., C16:0(3-OH) as investigated for these novel low abundant lipid A structures. In fact, as described by Vinogradov et al.9 and as also found by our group, the 3-position of the lipid A backbone can carry a C16:0(3-OH) fatty acid in the abundant non-phosphorylated lipid A species observed at m/z 1392 in positive-ion mode with a composition of 2xC18:0(3-OH)[2,2′], C16:0(3-OH)[3], and C14:0.
In addition to fatty acid heterogeneity, we provided structural evidence for a F. novicida lipid A species at m/z 1827.0 that appears to contain an additional hexose sugar (+162 Da) relative to the most abundant species at m/z 1665.2. Such a novel extension has been recently presented by Shaffer and colleagues using Fourier Transform mass spectrometry18, 19. F. tularensis lipid A, however, does not appear to show this additional hexose extension, and rather shows a very minor component at m/z 1826.2 that most likely contains an additional hexosamine sugar (+161 Da). Further studies will be needed to confirm this second hexosamine extension, as well as to define a biological role for this modification and growth conditions that increase its relative expression.
F. tularensis subspecies holarctica, F. novicida and F. philomiragia are zoonotic pathogens which live in the environment and are found primarily throughout the Northern Hemisphere1. These organisms infect mammals, arthropods, and amoeba, which may serve as environmental reservoirs1, 20. The lipid A portion of the LPS of the family Francisellaceae has the interesting characteristic of elaborating an unusual tetraacyl structure which does not bind or inhibit binding to the lipid A receptor, TLR47. The observation that the lipid A can show such a diverse repertoire of acyl structures was unexpected. The mechanisms which contribute to the control of this diversity and the advantage this would give these microbes in there different environmental niches is unclear. The lipid A of gram-negative bacteria is a major structural component of the bacterial outer membrane. It is interesting to speculate that this heterogeneity in acylation contributes to membrane integrity and insuring survival in the wide environmental range in which these organisms are found (insects, amoeba, aqueous environments, mammals) and the wide temperature range over which it is capable of surviving.
In summary, the combination of vMALDI ionization and multiple-stage fragmentation on a linear ion trap has proven particularly useful for analysis of low abundant lipid A’s. Although previous studies have successfully employed electrospray ionization on various mass spectrometry platforms, the methodology we described here provides efficient, sensitive and fast analysis of lipid A. This vMALDI-LIT platform allows for stable data acquisition from a single spot sufficient for multiple MSn experiments per sample, even for components that represent less than 1% relative intensity of the major peak. Such a robust and highly sensitive vMALDI-LIT platform should greatly advance our ability to study complicated, heterogenous lipid A mixtures and provide the analytical and structural framework for ongoing biological studies.
Supplementary Material
Additional experimental data, such as multiple-stage fragmentation spectra (MSn) and MSn fragmentation schemes. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by a Program Project Grant from National Institutes of Health grant #AI44642-05. We would also like to acknowledge Thermo Electron Corporation for providing a vMALDI-LTQ mass spectrometer for evaluation.
References
- 1.Ellis J, Oyston PC, Green M, Titball RW. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohara Y, Sato T, Homma M. FEMS Immunol Med Microbiol. 1996;13:185–189. doi: 10.1111/j.1574-695X.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 3.Sanford JP. JAMA. 1983;250:3225–3226. doi: 10.1001/jama.250.23.3225. [DOI] [PubMed] [Google Scholar]
- 4.Saslaw S, Eigelsbach HT, Wilson HE, Prior JA, Carhart S. Arch Intern Med. 1961;107:689–701. doi: 10.1001/archinte.1961.03620050055006. [DOI] [PubMed] [Google Scholar]
- 5.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. Arch Intern Med. 1961;107:702–714. doi: 10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- 6.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 7.McLendon MK, Apicella MA, Allen LA. Ann Rev Microbiol. 2006 doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Infect Immun. 2004;72:5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinogradov E, Perry MB, Conlan JW. Eur J Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 10.Vinogradov E, Perry MB. Carbohydr Res. 2004;339:1643–1648. doi: 10.1016/j.carres.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, McGrath SC, Cotter RJ, Raetz CR. J Biol Chem. 2006 doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CR. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan S, Reinhold VN. Anal Biochem. 1994;218:63–73. doi: 10.1006/abio.1994.1141. [DOI] [PubMed] [Google Scholar]
- 14.Bedoux G, Vallee-Rehel K, Kooistra O, Zahringer U, Haras D. J Mass Spectrom. 2004;39:505–513. doi: 10.1002/jms.611. [DOI] [PubMed] [Google Scholar]
- 15.Costello CE, Vath JE. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- 16.Kussak A, Weintraub A. Anal Biochem. 2002;307:131–137. doi: 10.1016/s0003-2697(02)00004-0. [DOI] [PubMed] [Google Scholar]
- 17.Post DM, Phillips NJ, Shao JQ, Entz DD, Gibson BW, Apicella MA. Infect Immun. 2002;70:909–920. doi: 10.1128/iai.70.2.909-920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaffer SA, Kalhorn TF, Harvey MD, Ernst RK, Goodlett DR. Proceedings of the 54th Annual Meeting of the American Society for Mass Spectrometry. Seattle; Washington: 2006. [Google Scholar]
- 19.Shaffer SA, Goodlett DR, Ernst RK. Proceedings of the 53rd Annual Meeting of the American Society for Mass Spectrometry. San Antonio, Texas: 2005. [Google Scholar]
- 20.Abd H, Johansson T, Golovliov I, Sandstrom G, Forsman M. Appl Environ Microbiol. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental data, such as multiple-stage fragmentation spectra (MSn) and MSn fragmentation schemes. This material is available free of charge via the Internet at http://pubs.acs.org.