Abstract
The ubiquitin-proteasome pathway has emerged as a central player in the regulation of several diverse cellular processes. Here, we describe the important components of this complex biochemical machinery as well as several important cellular substrates targeted by this pathway and examples of human diseases resulting from defects in various components of the ubiquitin-proteasome pathway. In addition, this review covers the chemistry of synthetic and natural proteasome inhibitors, emphasizing their mode of actions toward the 20S proteasome. Given the importance of proteasome-mediated protein degradation in various intracellular processes, inhibitors of this pathway will continue to serve as both molecular probes of major cellular networks as well as potential therapeutic agents for various human diseases.
Keywords: ubiquitin-proteasome pathway, protein degradation, proteasome inhibitor, natural product
1. INTRODUCTION
The integrity of cellular processes depends upon the proper balance of different proteins. Among the possible ways to control the activities of such molecules, controlling protein half-life itself by destruction has emerged as a major cellular regulatory mechanism. In cells, there are two major destruction pathways involving either a collection of proteolytic enzymes within the lysosome or the multicatalytic proteolytic core of the ubiquitin-proteasome pathway. This review focuses this latter system.
The majority of proteins that are destined for degradation are marked by the covalent attachment of multiple ubiquitin molecules, which provide a recognition signal for the 26S proteasome. Since the ubiquitin-proteasome pathway degrades a wide range of protein substrates with exquisite specificity, nature has developed an ingenious way to divide this complex biochemical machinery into two discrete and successive steps: first, a specific recognition process, employing the ubiquitin conjugation cascade, and secondly, an indiscriminate destruction process, mediated by the proteolytic proteasome core. The ubiquitin conjugation cascade uses a highly modular approach that can be used in different combinations for different purposes. Specifically, three different sets of enzymes i.e., E1, E2, and E3, shuttle ubiquitin and eventually link ubiquitin to the protein substrate. Different combinations of E2 or E3 or both recognize each substrate’s unique degradation signal, thereby conferring exquisite specificity for ubiquitination of diverse protein substrates. Ultimately, most ubiquitinated proteins are recognized by the 26S proteasome, unfolded, and threaded into the 20S proteolytic chamber in an ATP-dependent manner. The second, indiscriminate proteolytic step provides directionality for a signaling pathway, i.e., once a protein is committed for degradation, there is no return, ensuring that partially degraded proteins do not interfere with biological processes.
This ATP- and ubiquitin-dependent process is responsible for intracellular activities as diverse as the removal of misfolded or aged housekeeping proteins, cell cycle regulation via cyclin degradation, and cellular immune response by antigenic peptide processing. Given the wide range of cellular substrates and processes that are controlled by the ubiquitin-proteasome pathway, the components of this pathway have become an attractive novel target for therapeutic intervention. Indeed, aberrations in this pathway have been implicated in the pathogenesis of several human diseases. Although the ubiquitin-proteasome pathway can be blocked at various steps, most inhibitors of this pathway directly target the proteolytic activities of the 20S proteasome. The number of such proteasome inhibitors is growing and their target and subunit specificities have improved in recent years, leading to their inclusion in several clinical trials. To widen the potential for the treatment of a wider range of disorders and to gain greater specificity, future efforts will undoubtedly focus on the development of inhibitors/activators that affect earlier steps of the ubiquitin-proteasome pathway, allowing selective inhibition of the destruction of specific proteins.
2. UBIQUITIN-PROTEASOME PATHWAY
A. Ubiquitin Cascade System
The ubiquitin-proteasome pathway degrades diverse cellular proteins with exquisite specificity. To accomplish this, a highly modular and elaborate ubiquitination machinery is employed to recognize selectively specific substrates. Ubiquitin is composed of 76 amino acid residues and its primary sequence is highly conserved from yeast to mammals, thus resulting in essentially identical tertiary structure.1 X-ray diffraction and proton NMR analyses have allowed the determination of the tertiary structure of ubiquitin,2–4 revealing its highly compact and rigid nature: only the four carboxy terminal amino acid residues, including Gly-76 that is activated in an ATP-dependent manner for subsequent ubiquitin ligation with the ε-amino group of an internal Lys residue of the protein substrate, are extended and unstructured. Another important residue, Lys-48, of ubiquitin is located on the surface of a Type III reverse turn, providing its ε-amino group for an easy access to form an additional amide isopeptide bond to generate multi ubiquitin chains.
In the first step of the ubiquitin conjugation cascade, the carboxyl group of Gly-76 of ubiquitin is activated by ubiquitin-activating enzyme (E1). This step involves the hydrolysis of ATP to PPi to generate an ubiquitinyl adenylate intermediate bound to an E1 enzyme. Subsequently, an active site Cys residue of E1 covalently links to ubiquitin via a high-energy thioester linkage, with the concomitant release of AMP. Following activation, activated ubiquitin is then transferred by transacylation reaction to a thiol group of an active site Cys residue of E2 (ubiquitin carrier protein or ubiquitin-conjugating enzyme). Finally, E2 shuttles ubiquitin either directly to a protein substrate by itself or in cooperation with ubiquitin-protein ligase (E3), to form an amide isopeptide bond between the carboxyl group of Gly-76 of ubiquitin and an ε-amino group of the protein substrate’s internal Lys residue. The last step occurs by first transferring ubiquitin from E2 to E3, which accepts ubiquitin in a similar thiol linkage and then to the protein substrate. In some cases, however, covalent linkage between E3 and ubiquitin is not observed and it appears that ubiquitin is directly transferred from E2 to the protein substrate in a ternary E2-E3-substrate complex.
Once the protein substrate is mono-ubiquitinated, a polyubiquitin chain is formed through the same ubiquitination conjugation cascade, as described above, in which the carboxyl group of the carboxy terminal Gly-76 of ubiquitin is covalently linked to an internal Lys residue of ubiquitin that is already conjugated to the protein substrate.
It should be emphasized that the specificity of ubiquitination is largely determined by a series of E3 enzymes and E3 multiprotein complexes, each of which is specific to one or a few corresponding protein substrate(s) and of E2 enzymes, each of which is dedicated to their cognate E3 enzyme(s). As a result, different combinations of E2 and E3 enzymes allow selective tagging and degradation of specific intracellular proteins. In contrast, there is a single family of E1 that is highly conserved.
1. Enzymes of the Ubiquitination Cascade System
E1 (UBIQUITIN-ACTIVATING ENZYME OR UBA)
A single E1 appears to be responsible for the activation of ubiquitin required for all modifications. In yeast, for example, only one functional UBA, UBA1 is known for its ubiquitin-activating activity.5 Its deletion is lethal in yeast, as expected, and mutation of a putative active-site Cys residue abolishes E1 activity.6
E2 (UBIQUITIN-CARRIER PROTEIN OR UBIQUITIN-CONJUGATING ENZYME OR UBC)
Many E2s have been identified, at least 13 in yeast (UBC1–13) and more than 20 in mammals,7,8 reflecting their role in conferring the specificity for ubiquitination of diverse protein substrates. All E2s contain a conserved ~130 amino acid-long catalytic core sequence i.e., UBC domain. An active-site Cys residue within the UBC domain is essential since it is required to form an ubiquitin-thioester intermediate. As a result, mutation of an active-site Cys residue abolishes UBC activity.9,10 E2s are subdivided on the basis of their distinct primary sequences, presumably reflecting their different specificities for their cognate E3s. For example, class I E2s, such as UBC4, UBC5, UBC7, and UBC9–13 consist exclusively of UBC domain and may require their cognate E3s for recognition of substrates, since they cannot alone transfer ubiquitin to substrates. Class II (UBC1, UBC2/RAD6, UBC3/CDC34, UBC6, and UBC8), class III (UBC6), and class IV E2s contain unique carboxy or amino terminal extensions or both that may mediate substrate specificity as well as intracellular localization.
Among the 13 known yeast E2s, only UBC3 and UBC9 are encoded by essential genes11,12 and both are involved in regulating cell cycle progression. UBC3/CDC34, for example, is involved in the degradation of SIC1 CDK inhibitor, which is necessary for the G1 to S phase transition, in conjunction with CDC53, SKP1, and CDC4.13,14 In addition, UBC2/RAD6 is involved in DNA repair in cooperation with its cognate E3α15 to degrade so-called N-end rule protein substrates. It is generally believed that E2s together with their cognate E3s recognize specific protein substrates. Such examples and specific E3s will be described below in more detail.
E3S (UBIQUITIN-PROTEIN LIGASES)
E3 ligases or ligase complexes recognize specific motifs in their substrate(s) and catalyze the transfer of ubiquitin directly or indirectly from a thioester intermediate from their cognate E2 to form an amide isopeptide bond between protein substrate and ubiquitin.16,17 In general, there are a few possible mechanisms for recognition of protein substrates by the different E3s. For example, some substrates carry degradation signals that are constitutively active, whereas other substrates require a covalent modification of the sequence motif, such as phosphorylation. Conversely, E3s themselves can be altered or bound with an ancillary protein to recognize a specific subset of substrates. Various examples of such regulation and different families of E3 ligases are described below.
i. N-end rule family
E3α was the first identified E3 ligase, which was originally defined as a third enzyme component, in addition to E1 and E2, of the ubiquitin cascade system.18 Yeast homolog to E3α, UBR1, was also found.19 A specific E2, UBC2/RAD6 is shown to interact with its cognate UBR1,15,20–22 and activated ubiquitin is believed to be transferred directly to the substrate in a ternary E2–E3–substrate complex.16
E3α/UBR1 is a large ~200 kDa protein with several distinct functional domains. It has two distinct and independent sites that recognize N-end rule substrates through their amino terminal residues — a type-1 site that recognizes basic residues, such as Arg, Lys, and His, and a type-2 site that recognizes bulky hydrophobic residues, such as Leu, Ile, Phe, Trp, and Tyr.23,24 It also contains a third substrate-binding site that recognizes a non-N-end rule substrate, such as GPA1 and CUP9.25,26 Interestingly, binding of type-1 or type-2 dipeptides to their respective sites allosterically stimulates association of CUP9 with UBR1,27 suggesting yet another mechanism to control the activity of E3 ligase.
Careful analysis of interaction with E3 UBR1 and its cognate E2 ligase UBC2 revealed that binding is mediated largely between the basic residue-rich (BRR) domain of UBR1 and the polyacidic carboxy terminal tail of UBC2.28 Interestingly, a RING-H2 finger motif, which was found near BRR domain, is also essential for ubiquitination and the ultimate degradation of N-end rule substrates, though this domain does not appear to involve in binding to both UBC2 and substrates. The exact nature of the role of a RING finger motif in UBR1 awaits further examination, yet it should be noted that a RING finger motif has become one of most exciting new motifs found in other E3s, as described below.
ii. Hect (homologous to E6-AP carboxy terminus) domain family
The Hect class of E3s possess an approximately 350 amino acid-long carboxy terminal sequence, which is highly homologous to that of E6-AP.29 E6-AP was initially identified as an associating protein to E6 oncoprotein produced by human papillomavirus.30 The E6-AP and E6 complex was later found to mediate ubiquitination of p53, which ultimately is degraded by the ubiquitin-proteasome pathway. Interestingly, E6-AP was found to promote ubiquitination of other cellular substrates in the absence of E6,31,32 suggesting its role as an E3 ubiquitin protein ligase. To date, at least 20 members of Hect proteins in human have been identified.33 Unlike other classes of E3s, Hect E3s form an ubiquitin-thioester intermediate via a conserved active-site Cys residue within the Hect domain and directly catalyze substrate ubiquitination.19,34
In contrast to the carboxy terminus, the amino terminal sequence of Hect E3s is highly variable and contains the primary determinants for recognition of specific substrates.29,35 For example, WW domains in the variable amino terminal extension of both yeast RSP5 and its mammalian homolog Nedd4 bind to the PPxY sequences of their cellular substrates, the yeast RNA polymerase II (Rpb1) and the human epithelial sodium channel, respectively.36–38
Analogous to a specific interaction of E3α/UBR1 with its cognate E2 ligase UBC2, Hect E3s bind to a distinct set of E2s, such as yeast UBC4 and UBC5 or human UbcH5, UbcH7, and UbcH8 via a specific interaction with the Hect domain.33,39–41 X-ray diffraction analysis of the crystal structure of the E6-AP Hect domain bound its cognate UbcH7 reveals the overall organization of this complex as well as the detailed structural determinants responsible for the E2-E3 specificity.42 Specifically, Phe-63 of the UbcH7 L1 loop was found to play a central role in determining its specificity for E6-AP. This is consistent with the fact that Phe-63 is conserved in E2s that recognize a Hect class of E3s and replacement of Phe-63 with Asn of UbcH5, another Hect class-recognizing E2, results in loss of binding to the Hect E3 RSP5.43 This suggests that the L1 loop of Hect E3-recognizing E2s is the specificity loop.
iii. Ring finger family
Although more than 200 functionally distinct proteins in eukaryotes are found to contain the RING finger protein sequence motif and its variants, until recently no specific function(s) had been identified. The RING finger motif was initially identified in the protein product of the human gene RING1 and has been defined as a consensus sequence Cys-X2-Cys-X(9–39)-Cys-X(2–3)-Y(Cys/His)-X2-Cys-X(4–48)-Cys-X2-Cys where X can be any amino acid residues and Y can be either Cys as in RING-HC or His as in RING-H2.44
One of the best characterized members of the RING finger E3 ligases, c-Cbl proto-oncoprotein attenuates signaling by activated receptor protein tyrosine kinases (RPTKs) via their ubiquitination and subsequent proteasome-mediated degradation.45–47 c-Cbl contains a variant SH2 domain that recognizes tyrosine phosphorylated substrates. Furthermore, it also contains a RING finger motif that recruits and activates allosterically its cognate E2s.46–48 An intact RING finger motif is therefore required for ubiquitination of activated RPTKs49 and its deletion or disruption aborts the function of c-CbL.50,51
A few cognate E2s, such as UBC4, UbcH5B/C, and UbcH7, are known, and recent X-ray diffraction analysis of the c-Cbl RING and SH2 domains together with a tyrosine-phosphorylated peptide and UbcH7 provided an insight into the mechanism by which the RING finger domain recognizes its cognate E2s52: structural data support the model in which the RING finger domain serves as both a binding site that recruits the substrates and the E2s and as a scaffold that positions and orients them optimally for ubiquitin transfer. A growing number of RING finger E3 ligases have been identified and other examples of E2-RING finger E3 pairs include RAD6-RAD18 and RAD5 complex,53 UBC9-PML,54 and UbcH5-MDM2.55,56
iv. Multisubunit complex family
Multisubunit E3 ubiquitin ligase complexes, e.g., APC and SCF complexes, consist of several subunits, among which a conserved accessory protein(s) is used to select a specific substrate for ubiquitination and another component binds to its cognate E2. It is not yet established, however, whether there is a direct transfer of activated ubiquitin from bound E2 to any component of this complex prior to final covalent modification of a substrate.
The best characterized E3 ligase complex, the anaphase-promoting complex (APC) also known as the cyclosome, comprises 12 subunits in yeast57–59 and at least 10 in mammals.60 The number of known substrates are relatively small, but several of these proteins including mitotic cyclins, certain anaphase inhibitors, and spindle-associated proteins are important for mitotic progression.58
Although the function of every APC component has yet to be identified, the role of several critical subunits is understood. Basic components of APC comprise, for example, a distant cullin homolog APC2, a substrate-recognizing Cdc20, and a recently identified RING finger-containing APC11. The similarity of basic architecture of the APC and SCF complexes has recently been appreciated, suggesting a common mechanism for recognition and ubiquitination of substrates by the ubiquitin ligase complexes.61,62
In order to ubiquitinate selective cell cycle inhibitors, the APC complex associates chronologically with two different coactivator proteins, i.e., Cdc20/fizzy/Slp1 and Cdh1/Hct1/fizzy-related.63–66 Cdc20 and Cdh1 both contain WD40 repeats that presumably recognize a “destruction box”, i.e., a conserved 9-amino acid residue motif found in most APC substrates. As expected, for example, deletion or disruption of a destruction box in cyclin A leads to a metaphase delay.67 It should be noted that the mode of regulation of APC by the above-mentioned selective association differs in the case of Cdc20, whose expression level itself is regulated. In contrast, Cdh1/Hct1 is activated by the phosphatase Cdc14 only at specific points of cell cycle but normally phosphorylated by Cdk during most of cell cycle.59
A RING finger-containing APC11 is a recently identified APC component, which is required for ubiquitination of Clb2.68 It also mediates its own ubiquitination as well as APC substrates, such as cyclin B and securin.69,70 More importantly, APC11 is found to interact with APC2, which is a cullin homolog and an essential APC component, in a similar manner as RBX1, an essential RING finger-containing SCF component, that is also homologous to APC11.71,72 It is likely that APC2 and APC11 together are involved in recruiting the cognate E2, though the nature of their interactions has yet to be determined.
The SCF complex is another well-characterized E3 ligase complex that targets a number of important cell cycle regulators, such as G1 cyclins, CDK inhibitors, and other short-lived proteins.73 In general, the SCF complex is composed of two evolutionary conserved factors, i.e., Skp1 and a member of the Cullin/Cdc53 family of proteins, together with an F-box protein.14,74–76 Substrate-recognizing F-box proteins contain WD40 repeats or leucine-rich domains that are mainly responsible for selecting the diverse SCF substrates in a phosphorylation-dependent manner. For example, phosphorylated Sic1 is recognized by the WD40 repeats of the F-box protein Cdc4 and ubiquitinated by its cognate E2 Cdc34. Similarly, the phosphorylated form of Cln is specifically bound via the leucine-rich domain of the F-box protein Grr1. In addition to regulation by phosphorylation of substrates, F-box proteins themselves are regulated transcriptionally77,78 and post-translationally.79 In contrast to the WD40 repeats or leucine-rich domains, the F-box motifs of these proteins appear to be responsible for interacting with other SCF components, particularly with Skp1.
In addition to the three traditionally defined subunits of the SCF complex (Skp1, cullin/Cdc53, and F-box proteins), as described above, a small RING finger-containing protein Rbx1/ROC1/HRT1 is also identified as an additional essential component of the SCF complex62,71,80–82: Rbx1 interacts with different cullins and together enhances activity of the SCF complex apparently by recruiting cognate E2 Cdc34 into the SCF complex. Interestingly, Rbx1 also stimulates auto-ubiquitination of Cdc34, suggesting that Rbx1 acts as an allosteric activator of Cdc34. Interestingly, RING finger-containing Rbx1 is also an essential component of another multisubunit E3 ligase complex, VCB-CUL2 that consists of the von Hippel Lindau (VHL) tumor suppressor protein, a Skp1 homolog Elongin C, and a Cdc53 homolog Cul2. In this complex, VHL protein contains a SOCS box, a distant homolog of the F-box83 and can be replaced with other proteins that also contain a SOCS box.
Taken all together, a core complex of cullins and RING finger proteins appears to form the basis for a multisubunit complex E3 ligase complex84,85: While this core structure binds and activates cognate E2, Skp1 in SCF and an as of yet unidentified component in APC mediates recruitment of the substrate-recognizing component, which confers substrate specificity to this complex.
B. 20S and 26S Proteasome: Converging Points of Ubiquitinated Substrates
1. Overall Organization of the Proteolytic System
The structure of the 20S proteasome is well conserved in virtually all organisms ranging from archaebacteria86 to yeast87 to humans88: a hollow and barrel shape consists of four stacked heptameric rings, forming a central chamber that runs through the stack from top to bottom. Each ring is composed of seven subunits, which may be classified into two groups, i.e., α-subunits that comprise the outer two rings and β-subunits the inner two rings. Structural and mutagenesis studies revealed that the amino terminal threonines of the β-subunits serve as the catalytic nucleophiles. This is in contrast to other cellular proteases, such as serine and cysteine proteases, that utilize an internal amino acid as a nucleophile.
In contrast to the ancestral proteasome found in archabacteria, the eukaryotic proteasome contains an additional 19S regulatory complex, consisting of a lid and a base89 that binds to the 20S particle to form the 26S proteasome holoenzyme. While the lid recognizes ubiquitinated protein substrates with high fidelity, the base, which contains six ATPase and caps the end of the 20S proteasome core, unfolds protein substrates and threads them into the catalytic chamber of the 20S particle, in an ATP-dependent manner.90 It should be noted that the eukaryotic 20S proteasome alone shows only modest proteolytic activities that can be stimulated by treatment of SDS91–93 and by association with 19S and 11S regulatory complexes.94–96 As has been long expected, the 19S regulatory complex was recently shown to open a gated channel into the 20S proteasome core,97 in which an access to the central chamber is blocked in the free 20S proteasome.87 Similarly, the 11S regulatory complex (PA26)98 was also shown to activate the 20S proteasome by opening the gate separating the sequestered proteasome internal chamber from the intracellular environment.99
Unlike typical proteases, the 20S proteasome ensures that virtually all peptide bonds within a protein substrate are susceptible to cleavage, by possessing multiple proteolytic activities in one proteolytic internal chamber. Traditionally, the proteolytic activity of the 20S proteasome is expressed by its preference to cleave a peptide bond immediately after a particular amino acid, i.e., a P1 amino acid residue of model peptides and natural polypeptide substrates.100 For example, the 20S proteasome hydrolyzes a peptide bond after bulky hydrophobic amino acid residues, reminiscent of chymotrypsin. Similarly, it also cleaves a peptide bond after basic residues, analogous to trypsin, and after acidic residues.101 Hence, these activities are referred to as the chymotrypsin-like (CT-L), trypsin-like (T-L), and post-glutamyl peptide hydrolyzing (PGPH) activities, respectively.102 In addition to these well-characterized proteolytic activities, the 20S proteasome is also known to possess two other proteolytic activities, i.e., branched-chain amino acid-preferring (BrAAP) and small neutral amino acid-preferring (SNAAP) activities.103 It awaits, however, to determine whether these activities simply reflect a broad specificity of the catalytic subunits that are responsible for the above-mentioned three classical activities104–106 or a novel as of yet unidentified catalytic mechanism is responsible for BrAAP and SNAAP activities.
Attempts to map the active sites of the 20S proteasome catalytic subunits responsible for these proteolytic activities revealed that different subunits can be assigned to corresponding activities. For example, extensive mutations in the yeast proteasome provide evidence that β1 (PRE3) is responsible for the PGPH activity,107,108 β5 (PRE2) provides for the CT-L activity, and β2 (PUB1) is necessary for the T-L activity.104,109,110 Based on the high sequence homology of the catalytic subunits between the yeast and mammalian proteasomes, X (mammalian counterpart to yeast β5) is assigned for the CT-L activity, Y (mammalian counterpart to yeast β1) for the PGPH activity, and Z (mammalian counterpart to β2) for the T-L activity. In general, these assignments agree with extensive inhibitor studies.111
2. Catalytic Mechanism for the 20S Proteasome-Mediated Proteolysis
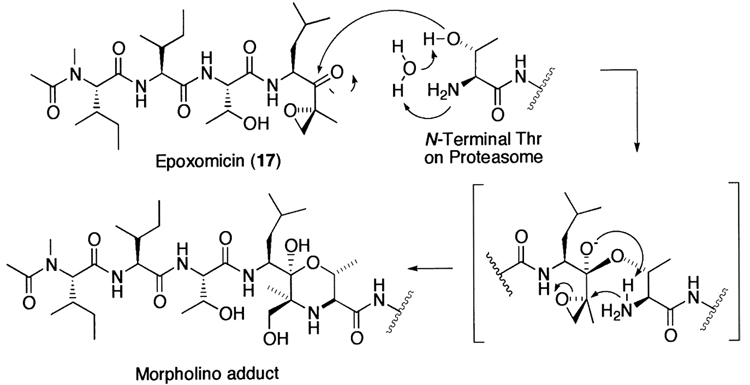
After many years of uncertainty as to whether the 20S proteasome belongs to the classical serine or cysteine proteases, a combination of genetic, structural, and inhibitor studies have revealed the importance of the amino terminal Thr as a nucleophile catalyst in the 20S proteasome-mediated proteolysis. The first line of evidence comes from genetic studies. Mutation of the amino terminal Thr in the catalytic subunits of the 20S proteasome from Thermoplasma acidophilum was found to abolish the proteasome activity,112 suggesting a role of this residue in the proteolysis. In contrast, other amino acid residues, such as Ser, Cys, and Asp, which potentially act as a nucleophile catalyst, did not greatly affect the proteolysis. Next, X-ray diffraction analysis showed that the proteasome inhibitor, N-acetyl-Leu-Leu-norleucinal (2) is located in proximity to the hydroxyl group of the amino terminal Thr of the catalytic subunits from Thermoplasma proteasome,86 forming a hemiacetal intermediate between the aldehyde group of the inhibitor and the hydroxyl group of the amino terminal Thr, reminiscent of the tetrahedral intermediate of a protease reaction. Later, it was reported that all of the six catalytic β-subunits in the yeast proteasome covalently bind to the same inhibitor N-acetyl-Leu-Leu-norleucinal in an analogous manner.87 Finally, mechanistic insight into the role of the amino terminal Thr in the proteasome proteolysis has come from several inhibitor studies. Mode-of-action studies revealed that natural product proteasome inhibitor lactacystin (15) targets the 20S proteasome and inhibits irreversibly the CT-L and T-L activities of the 20S proteasome, by covalent modification of the amino terminal Thr of the catalytic β-subunit.113 Recently, another natural product proteasome inhibitor epoxomicin (17) was also found to target the 20S proteasome,114,115 with concomitant modification of the amino terminal catalytic Thr of the 20S proteasome by its α′,β′-epoxyketone pharmacophore.116
From these lines of evidence, the 20S proteasome is classified as an amino terminal nucleophile (Ntn) hydrolase that contains the amino terminal Thr acting as the nucleophile catalyst. Other examples of such Ntn hydrolases include penicillin acylase with an amino terminal Ser, glutamine PRPP amidotransferase with an amino terminal Cys,117,118 and aspartylglucosaminidase with the amino terminal Thr.119 All Ntn hydrolases share a common Ntn hydrolase fold.
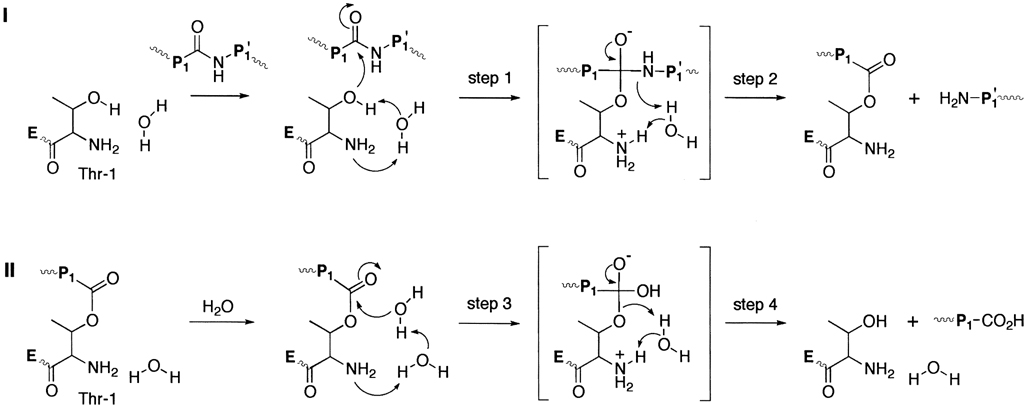
Considering the general mechanisms for the proteolysis by other Ntn hydrolases and genetic, structural, and inhibitor studies of the 20S proteasome, the following mechanism was proposed for the proteolysis catalyzed by the 20S proteasome (Fig. 1). In step 1, as the hydroxyl group of the amino terminal Thr-1 attacks the peptide carbonyl group of the scissile bond, a proton is transferred from the hydroxyl group of the nucleophile to the uncharged α-amine group of the same Thr-1, most likely through an intermediate water molecule. Evidence of the involvement of a water molecule comes from its proximity to the amino terminal Thr-1 in the active site of the catalytic subunit, as observed in the crystal structure of both penicillin acylase and the 20S proteasome.87,118 It should be noted that potential involvement of Glu-17 or Lys-33 as a general acid-base catalyst is also suggested.120 The tetrahedral addition intermediate then breaks down to result in the formation of the covalent acyl enzyme intermediate, with a proton transfer to the leaving group (step 2). Hydrolysis of the acyl enzyme intermediate is proposed to occur by a similar mechanism as described above for its formation. The free α-amine group of the amino terminal Thr-1 removes a proton from the attacking water molecule again through an intermediate water molecule (step 3). A proton is finally transferred to the oxygen atom of Thr-1 as the product is expelled from the tetrahedral addition intermediate (step 4).
Figure 1.
The proposed mechanism for the proteolysis catalyzed by the 20S proteasome.
Just as chymotrypsin, trypsin, and elastase are synthesized as inactive proenzymes, e.g., chymotrypsinogen and trypsinogen, the catalytic β-subunits of the 20S proteasome are synthesized in an inactive form containing an amino terminal prosequence that is removed upon assembly into the 20S proteasome core. This prevents inadvertent proteolysis of cellular proteins by the newly synthesized β-subunits of the 20S proteasome.121,122 The β-subunits of the 20S proteasome from Thermoplasma contain 8 amino acid-long prosequences and all 14 subunits are processed and become catalytically active. In eukaryotes, however, prosequences are longer and more heterogeneous123,124 and only three subunits (PRE2, PRE3, and PUB1) of the seven distinct β-subunits of the yeast proteasome are processed and become catalytically active.107–109,125 Similarly, X, Y, and Z β-subunits of mammalian proteasome are catalytically active. The processing event appears to be autocatalytic as in other Ntn hydrolases117 and coupled to proteasome assembly.
3. REPRESENTATIVE CELLULAR SUBSTRATES AND PROCESSES TARGETED BY THE UBIQUITIN-PROTEASOME PATHWAY
Orderly progression through the cell cycle requires programmed and periodic expression of cell cycle proteins. Cyclins were first identified as proteins whose levels oscillate during the cell cycle.126,127 This oscillation of cyclins was later found to be due to the regulated degradation mediated by the ubiquitin-proteasome pathway128 and the proteasome-mediated proteolysis has now emerged as a major regulatory mechanism of the cell division cycle. Among many, mammalian cyclins A, B, D, and E, CDK inhibitor p27, p21, transcription factor E2F, Rb, the tumor suppressor p53 proteins73,129 and yeast Clns, Clbs, Sic1, and Far1130 are best characterized. Given the fact that the inhibition of the ubiquitin-proteasome pathway results in blockage of cell cycle progression at many different phases, this antiproliferative activity of the proteasome inhibitors has been suggested to be used against various forms of tumors.
The transcription factor p53 plays a role of the key integrator in an intricate network of proteins of diverse functions that sense a variety of stress signals.131 The activation of p53 in response to such signals results in apoptosis or cell-cycle arrest, thereby maintaining the integrity of healthy cells and thus preventing the transformation to tumor cells. Not surprisingly, the tumor repressor p53 has been found as one of the most frequently mutated genes in many types of cancers. Its levels are normally quite low mainly due to its very rapid turnover and the ubiquitin-proteasome pathway is responsible for regulating its level.132,133 Proteasome inhibitors indeed can inhibit its degradation, resulting in accumulation of p53-ubiquitin aggregates. One of the major regulators of p53 is MDM2,134,135 which is inducible by its cellular substrate p53. MDM2 binds to the amino terminal trans-activation domain of p53, thereby resulting in inhibition of p53 transcriptional activity. Moreover, MDM2 binding to p53 also allows ubiquitination of both p53 and itself, eventually leading to destruction by the proteasome-mediated proteolysis.134,136
MDM2 transfers activated ubiquitin to both p53 and itself with the help of its cognate E2, UbcH5.55,56 Recognition of p53 is mainly mediated by binding of MDM2 via its carboxy terminal RING finger domain to the trans-activation domain of p53.136,137 It should be noted that phosphorylation of Ser in the amino terminus of p53 stabilizes p53 as a result of decreased binding to MDM2.138–141 Conversely, phosphorylation of MDM2 also plays a role in regulating p53 stability.142,143
The transcriptional factor NF-κB (nuclear factor κB) is another regulatory protein, whose mode of activation is controlled by the ubiquitin-proteasome pathway. Originally thought to be a B cell-specific transcription factor,144 NF-κB is an ubiquitous and central player in a variety of cellular processes, including immune and inflammatory responses, apoptosis, and cellular proliferation.145–150 Different forms of NF-κB exist, each composed of various members of the Rel family, reflecting the complexity of its response to diverse stimuli. In resting cells without extracellular stimuli, NF-κB is held latent in the cytoplasm as a complex bound with unphosphorylated IκB, thereby blocking the nuclear translocation of NF-κB. In response to the proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1 (IL-1), for example, a series of signal transduction cascades are initiated, eventually leading to activation of a multisubunit IκB kinase (IKK). Activated IKK in turn phosphorylates the critical Ser residues in the regulatory domain of a member of the inhibitor κB (IκB) family. Phosphorylation of IκB by IKK, however, leads to degradation of IκB. As a result, free NF-κB is allowed to translocate into the nucleus and activates transcription of κB sequence-specific genes.
Extensive study of such signal-induced destruction of IκBα (the best characterized member of the IκB family) has revealed that the ubiquitin-proteasome pathway plays a crucial role in NF-κB activation. Specifically, it is found that a variety of inducing signals converges on IKK α and β, which eventually phosphorylate highly conserved Ser-32 and -36 of IκBα‥151,152 These two highly conserved Ser residues reside in the amino terminal regulatory domain that precedes a well-conserved ankyrin core and a PEST domain.147,149 Upon phosphorylation, IκBα is recognized by the SCF complex containing the F-box protein β-TrCP153–155 and the RING finger protein ROC1.81 The resulting E2-E3 complex in turn covalently attaches polyubiquitination chain to IκBα that is eventually targeted for degradation by the 26S proteasome.156–158 It should be noted that other members of IκBs, such as Cactus in Drosophila and IκBβ and ε in mammals, also share a similar amino terminal regulatory domain that contains two invariant Ser residues within a defined consensus sequence and are believed to undergo a similar signal-induced degradation mediated by the ubiquitin-proteasome pathway.
Interestingly, the ubiquitin-proteasome pathway is also implicated in controlling the activity of NF-κB pathway through a limited proteolysis of p105.156,159–162 Full length p105 forms a stable complex with p65,163–165 in a similar manner as a ternary complex formation of p50/p65 dimer bound with IκB. Analogous to recognition of IκBα, the SCF complex containing the F-box protein β-TrCP recognizes p105, albeit two different recognition motifs are used for IκBα and p105.166 While an exact mechanism for p105 processing in response to NF-κB inducing stimuli is yet to be determined, results are beginning to shed light on the importance of IKK-mediated phosphorylation of p105 that leads to accelerated processing of p105.166,167 In contrast, pathways involving cotranslational degradation by the 26S proteasome of newly synthesized p105 appear to provide an alternative mechanism to control the activity of NF-κB via a balanced production of p50 and p105 in the cells.168,169
The importance of the ubiquitin-proteasome machinery in regulating critical biological processes culminates as this system is shown to be used to generate antigenic peptides from cytosolic proteins for presentation on MHC class I molecules.170,171 As to the definitive role of ubiquitination reaction for the generation of the antigenic peptides, conflicting results are reported.172–174 In contrast, as to the role of the proteolysis mediated by the 26S proteasome, ingenious use of proteasome aldehyde inhibitors clearly demonstrated that effective presentation of the antigenic peptides is inhibited in proteasome inhibitor-treated cells,175,176 because of the accumulation of MHC class I molecules continuously bound with TAP in the ER due to the lack of proper antigenic peptides.177 Later, the more specific proteasome inhibitor lactacystin (15) was also found to reduce antigen presentation in a similar manner.178,179 Interestingly, after stimulation of antigen-presenting cells with interferon-γ (INF-γ), three constitutive β-subunits, X, Y, and Z, are exchanged by LMP7, LMP2, and MECL1, respectively. These new subunits possess different profiles for proteolytic activities, most likely allowing a more efficient generation of antigenic peptides recognized by the MHC class 1 molecules. Moreover, INF-γ also induces proteasome activator PA28, which is thought to play an important role in MHC class 1 antigen presentation. Exact nature of roles of such INF-γ-inducible subunits and activators is currently under intense scrutiny.
It is well known that viruses exploit host cells’ machinery for their own replication. The ubiquitin-proteasome pathway is one of these intracellular processes co-opted by viruses. For example, the ubiquitin-proteasome pathway has been implicated in retrovirus assembly through the finding that an unusually high concentration of free ubiquitin is present in avian leukosis virus (ALV).180 Recently human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency virus (SIV), and Moloney murine leukemia virus (Mo-MuLV)181 have also been shown to contain high level of free ubiquitin inside of mature virus. Moreover, it was found that about 30% of the mature HIV-1 and Mo-MuLV Gag proteins are ubiquitinated181 and up to 50% of Gag proteins are conjugated with ubiquitins in the released virus-like particles (VLPs).182 Although it is yet to decipher a detailed mechanism, the ubiquitin-proteasome pathway was recently found to be an important player in retrovirus assembly.182–184
The involvement of the ubiquitin-proteasome pathway in retrovirus assembly, therefore, provides an excellent opportunity to develop a new line of anti-HIV therapeutic strategies. Of particular note, it was found that retrovirus budding or proteolytic maturation is inhibited upon treatment of infected cells with proteasome-specific inhibitors, e.g., lactacystin (15) or epoxomicin (17) at concentrations well below their cytotoxicity levels.183,184 This should provide a starting point in the development of potential anti-HIV drugs.
Human cytomegalovirus (CMV) represents yet another example how viruses exploit the ubiquitin-proteasome machinery to persist infection. Among a number of genes that CMV carries to inhibit the surface expression of MHC class I molecules, US2 and US11 encode ER-resident proteins, which bind to newly synthesized MHC class I molecules in the ER and target them to degradation by the ubiquitin-proteasome pathway.185,186 As a result, the presentation of antigenic viral proteins is inhibited, allowing viruses to elude the immune surveillance.
4. HUMAN DISEASES DUE TO THE DEFECTS OF THE UBIQUITIN-PROTEASOME PATHWAY
Reflecting the fact that the regulation of diverse short-lived proteins and regulatory proteins is mediated by the ubiquitin-proteasome pathway, it is not surprising that defects of various components of this complex biochemical machinery result in a range of human diseases. As a result, these components would provide an attractive novel target for therapeutic intervention. A few examples of inherited and acquired human diseases due to the defects of the ubiquitin-proteasome pathway are described below, emphasizing the molecular mechanism of pathogenesis.
A. Inherited Disorders
1. Angelman’s Syndrome
Angelman’s syndrome is a complex neurological disorder187 due to various genetic mechanisms that map to human chromosome 15q11–q13.188 Clinical characteristics of this disorder include developmental delay, speech impairment, movement or balance disorder, and behavioral uniqueness.189 Further genetic analysis revealed that truncated mutants of the UBE3A gene on chromosome 15q were identified in some patients with Angelman’s syndrome.190,191 UBE3A gene encodes a protein called E6-AP protein ligase (also known as ubiquitin-protein ligase or E3). Although the true substrate(s) of this E6-AP in AS patients has not yet been identified, the ubiquitin-proteasome pathway plays an important role during brain development and perturbation of this pathway causes a neurolopathological disorder.
2. Liddle Syndrome
Liddle et al.192 recognized this rare hereditary disorder in a family who were unable to maintain the proper balance of salt and water in the body, resulting in abnormally high blood pressure. The molecular mechanism of Liddle syndrome was later understood by Yale researchers193 who observed mutations in the carboxy terminus of β and γ subunit of the epithelial sodium channel in the kidney. Among many different mutations, deletion or mutation of the proline-rich (PPxY) region leads to constitutive activation of the kidney sodium channel, resulting in retention of excessive amounts of salt and water. Using a yeast two-hybrid screen and in vitro binding studies,37 WW domain of an ubiquitin ligase, Nedd4, is found to bind specifically to the PPxY sequences of the above-mentioned kidney sodium channel. Taken together, these findings suggests a novel mechanism regulating sodium reabsorption, in which failure of proper ubiquitination of the sodium channel results in increased retention of the sodium channel at the cell surface and thus excessive reabsorption of sodium and water leading to hypertension. It should be noted, however, that this short-lived sodium channel, after ubiquitination, is targeted and degraded in the lysosome, but not by the 26S proteasome machinery.
B. Acquired Disorders
1. Cervical Cancer
The most important risk factor for cervical cancer is human papillomavirus (HPV) infection. In cases of cervical cancer caused by the high-risk strains of HPV, levels of the tumor suppressor protein p53 were found to be unusually low due to the presence of the E6 proteins encoded by the oncogenic HPV.194 This E6-promoted degradation of p53 involves the ubiquitin-proteasome pathway and represents a major mechanism, by which virus transforms the host cells. The E6 oncoprotein was later found to modify the functions of E6-AP31,195 whose absence prevents ubiquitination of p53 by E6-AP.196 Specifically, E6 appears to function as an adapter protein to maintain a ternary complex of E6-AP ubiquitin-protein ligase (E3), its cognate UbcH8 ubiquitin-conjugating enzyme (E2), and a target substrate to ensure specific ubiquitination of substrate molecules.41,197 Interestingly, E6 also promotes the ubiquitination and degradation of E6-AP.198
2. Colorectal Cancer
Colorectal cancer, which arises in the epithelium of the lumen of the colon and rectum, is the second most common cause of cancer death in the United States. Generally speaking, the transformation of the normal epithelial cells into cancer cells occurs through a multistage process as a result of many different defects. One of most well-characterized genetic defects is found in the APC (adenomatous polyposis coli) gene located on chromosome 5q21. Mutations of the APC gene have been documented in familial adenomatous polyposis and in more than 80% of the sporadic colorectal cancer patients.199
Among its many functions, APC is known to control the cellular level of β-catenin by forming a complex with Axin, which recruits and facilitates ubiquitination of β-catenin for subsequent degradation by the 26S proteasome.200,201 This process appears to be dependent on the phosphorylation of both APC and β-catenin by glycogen synthase kinase-3 (GSK-3).202 In cancer cells expressing mutant APC, β-catenin accumulates,203,204 and interacts with T-cell factor as well as activates transcription of the proto-oncogenes, such as cyclin D1 and c-myc.205–207 Mutations of Ser to amino acid residues that cannot be phosphorylated stabilize β-catenin.208 It appears that ubiquitination of β-catenin is catalyzed by the SCF complex and its cognate E2. A receptor component of the SCF E3 ligase complex, i.e., F box- and WD domain-containing β-TrCP binds to the phosphorylated form of β-catenin, and over-production of a defective β-TrCP blocks β-catenin degradation.155,209,210
3. Alzheimer’s Disease
Alzheimer’s disease is a chronic brain disorder due to a progressive degeneration of brain tissue, leading to loss of mental functions, such as memory and learning. Alzheimer’s disease is marked by abnormal aggregates (senile plaques) and irregular knots (neurofibrillary tangles) of brain matter. By immunohistochemical analysis of protein contents in intracellular inclusion bodies, ubiquitin is found in both neurofibrillary tangles and senile plaques.211,212 Although it is not yet clear whether accumulation of ubiquitin conjugates is primary or secondary, a growing number of evidence indicate the involvement of the ubiquitin-proteasome pathway in Alzheimer’s disease. For example, findings of a simultaneous frameshift mutation of both β amyloid precursor protein (βAPP) and ubiquitin have implicated the ubiquitin-dependent pathway in the pathogenesis of Alzheimer’s disease.213 In addition, treatment of cells expressing presenilins 1 and 2 (PS1 and PS2) with proteasome inhibitors was shown to increase the PS1/PS2-mediated production of both APPα and Aβ40, which are βAPP maturation products.214 Senile plaques are thought to derive from the abnormal level of Aβ40 production.215 PS1 and PS2 are also found to be the proteasome substrates.216–218 Taken altogether, the ubiquitin-proteasome pathway appears to control the level of PS1 and PS2, thereby regulating the βAPP maturation pathway.
5. PROTEASOME INHIBITORS
Most currently available inhibitors of the ubiquitin-proteasome pathway directly target and inhibit the 20S proteasome, the core of the proteolysis machinery, rather than the upstream ubiquitination and recognition of ubiquitinated protein substrates. These proteasome inhibitors are broadly categorized into two groups: synthetic analogs and natural products. Synthetic inhibitors are peptide-based compounds with diverse pharmacophores. These include peptide benzamides, peptide α-ketoamides, peptide aldehydes, peptide α-ketoaldehydes, peptide vinyl sulfones, and peptide boronic acids. In contrast, natural product proteasome inhibitors display a variety of scaffolds of core structures and pharmacophores. Known natural product proteasome inhibitors include linear peptide epoxyketones, peptide macrocycles, γ-lactam thiol ester, and epipolythio-dioxopiperazine toxin. The inhibitory mechanisms and the limits and potentials of proteasome inhibitors are discussed in the following.
A. Synthetic Proteasome Inhibitors
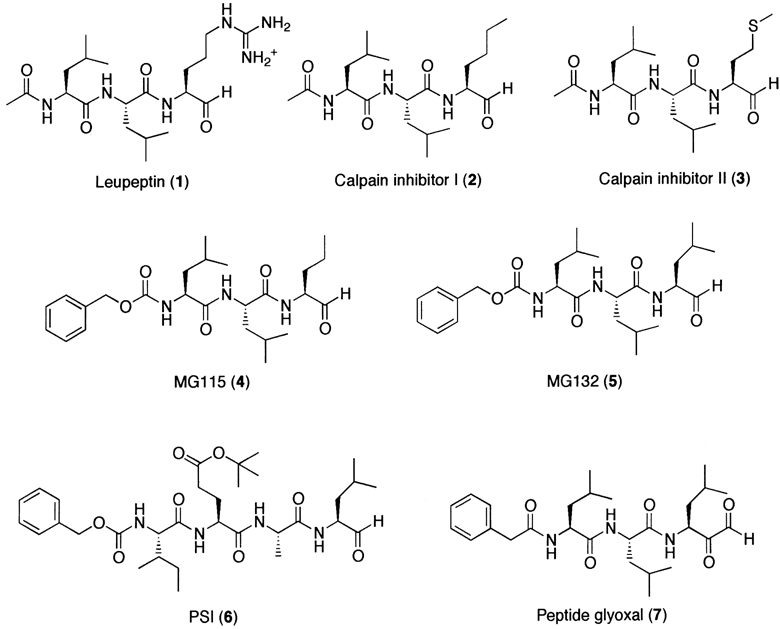
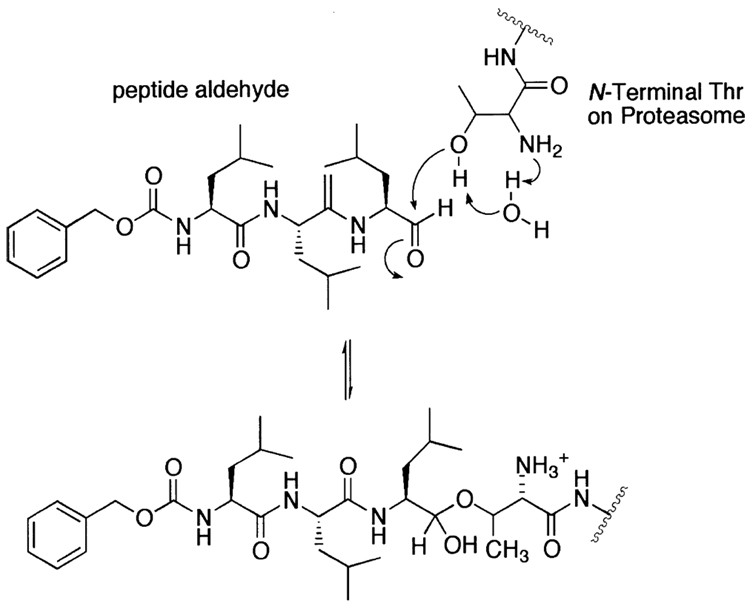
Peptide aldehydes have long been used as inhibitors for both serine and cysteine proteases. Given that the aldehyde functional group is readily subject to a nucleophilic attack by hydroxyl or thiol groups and that the proteasome uses the hydroxyl group of the amino terminal threonine as a nucleophile, it is not surprising that the commercially available peptide aldehydes were the first proteasome inhibitor family identified (Fig. 2). In the early 1980s, leupeptin (1), a standard serine (trypsin, plasmin) and cysteine (papain, cathepsin B) protease inhibitor, was already recognized as a T-L activity blocking agent of the 20S proteasome.101 Later, calpain inhibitors I (2) and II (3) were also shown to inhibit the CT-L activity of the 20S proteasome.219,220 Upon binding to the active site of the 20S proteasome, a peptide aldehyde forms a covalent hemiacetal adduct that is reversible under physiological conditions (Fig. 3). Since peptide aldehydes can be easily prepared and optimized, researchers have developed myriad of aldehyde inhibitors with higher potency as well as increased selectivity toward the CT-L activity of the 20S proteasome (Fig. 2). Such examples include MG115 (4), MG132 (5)175 and PSI (6),221 which are all potent and CT-L selective inhibitors that are widely used in studying the role of the proteasome in various cellular processes. Similarly, other peptide aldehyde inhibitors have been synthesized to study other proteasomal activities, such as BrAAP activity.222 Meanwhile, researchers have also attempted to improve peptide aldehydes’ potency by adding an additional ketone moiety at α position, yielding a glyoxal (7).223
Figure 2.
Proteasome inhibitor family of synthetic peptide aldehydes.
Figure 3.
The proposed mechanism for the inhibition of the 20S proteasome by the peptide aldehyde.
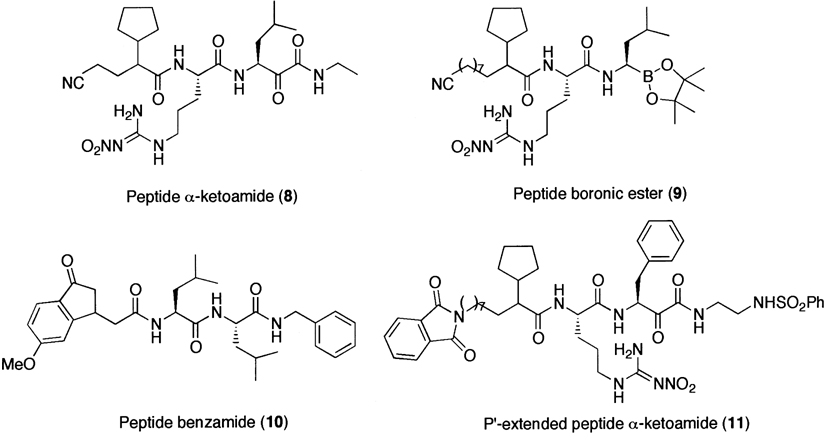
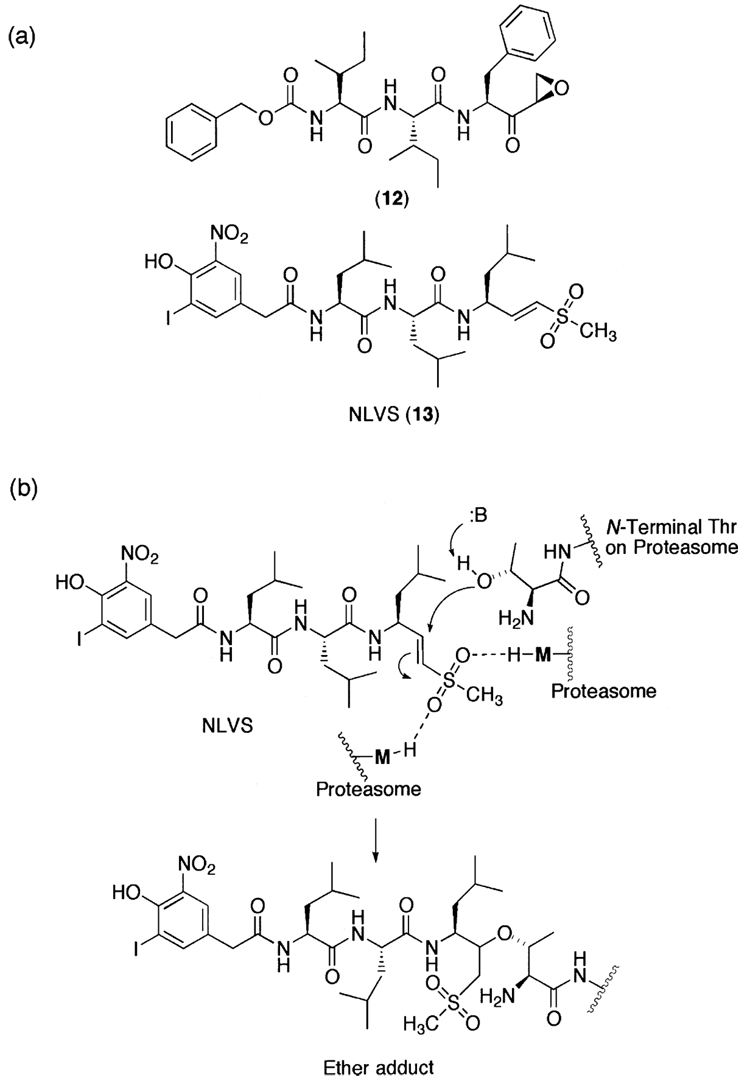
Although these peptide aldehydes, in general, are cell-permeable, potent inhibitors of the 20S proteasome, and are still widely used in biological studies, the lack of specificity due in part to the presence of highly reactive aldehyde functional group is a major limitation for their use as potential therapeutic agents or as molecular probes in dissecting complex signaling pathways. To overcome such limitations, researchers have attempted to develop non-reactive peptide inhibitors (Fig. 4), such as α-ketocarbonyl and boronic ester derivatives,224 indanone dipeptide benzamides,225 and P′- extended α-ketoamides.226 Non-aldehyde irreversible proteasome inhibitors have also been developed (Fig. 5), such as tripeptides α′,β′-epoxyketones227 and vinyl sulfones.228
Figure 4.
A family of reversible synthetic proteasome inhibitors.
Figure 5.
(a) A family of irreversible synthetic proteasome inhibitors. (b) The proposedmechanism for the inhibition of the 20S proteasome by NLVS.
Vinyl sulfones were first introduced as cysteine protease inhibitors.229,230 Although vinyl sulfones are sufficiently inert in the absence of a target, it is well characterized that they are capable of forming a hydrogen bond in the active site of enzymes, thereby becoming more electrophilic (Fig. 5). Despite being more widely known as inhibitors of cysteine proteases, peptide vinyl sulfones have also been developed as potent inhibitors toward the 20S proteasome and successfully employed for many biological studies.228,231 Peptide vinyl sulfones have also been shown to be irreversible and active site-directed proteasome inhibitors by modification of the hydroxyl group of the amino terminal threonine (Fig. 5).185,228,232,233 Nevertheless, the lack of specificity is a major concern for this class of inhibitors, since the peptide vinyl sulfones inhibit both the 20S proteasome and other cysteine proteases.229,232
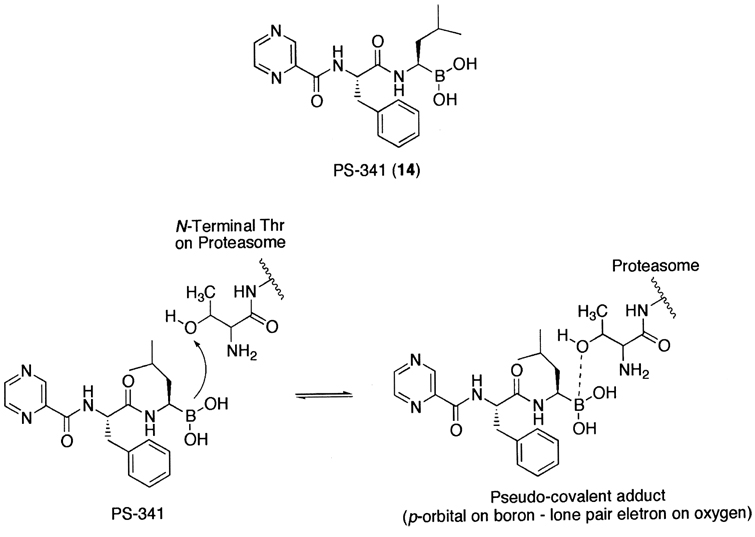
Another class of peptide-based inhibitors exploits the boronic acid functional group.234 It should be noted that this class of inhibitors is thought to exhibit proteasome inhibition via a non-covalent complex formation. Considering no difference in the potency of the boronic ester derivatives as compared to that of the corresponding aldehyde derivatives,224 the inhibitory potency and the target selectivity towards the 20S proteasome of boronic acid-based inhibitors are quite remarkable. This is presumably due to the fact that an empty p-orbital on a boron atom is positioned to accept the oxygen lone pair of the amino terminal threonine residue of the 20S proteasome to form a stable tetrahedral intermediate (Fig. 6). The stable tetrahedral borane complex allows the peptide chain length of the boronic acid-based inhibitor to be truncated to a dipeptide with good retention of inhibitory activity. This provides practical advantages, such as solubility and membrane permeability, as a potential therapeutic agent. It should be noted that, for other class of proteasome inhibitors, at least three amino acid chains are required to moderately inhibit the proteasomal activity.223,228,235
Figure 6.
A synthetic peptide boronic acid inhibitor (PS-341) and its proposed inhibition mechanism.
The most well characterized compound of this class is a dipeptide boronic acid PS-341 (14).234,236 Analogous to many other proteasome inhibitors that block cell cycle progression, PS-341 treatment also results in accumulation of cells in the G2-M phase,237 eventually leading to apoptosis after prolonged treatments of cells.238–240 Moreover, PS-341 inhibits the proteasome mediated-IκBα degradation and NFκB-dependent gene expression in vivo236 and reduces tumor growth in both murine and human xenograft models.237,241 To exploit such effects, PS-341 is currently under clinical evaluation for advanced breast and prostate cancer.242
B. Natural Product Proteasome Inhibitors
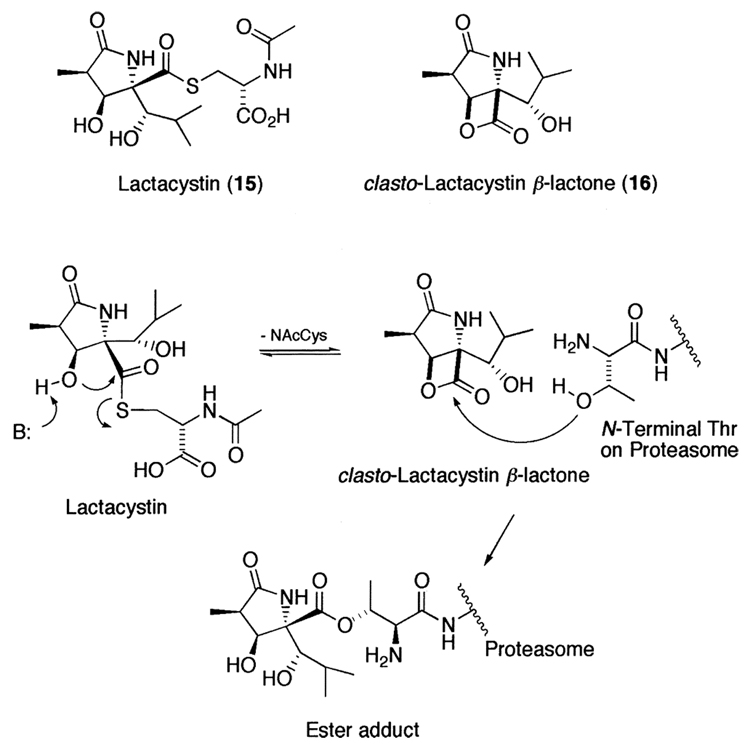
While the synthetic inhibitors mentioned above have been rationally designed, synthesized, and optimized, nature has also provided selective and potent proteasome inhibitors. For example, lactacystin (15) is a Streptomyces lactacystinaeus metabolite that was discovered on the basis of its ability to induce neurite outgrowth in the murine neuroblastoma cell line Neuro-2a.243,244 Subsequently, others showed that lactacystin also inhibited cell cycle progression.245,246 Later, Schreiber et al.113 showed that lactacystin targets the 20S proteasome by an irreversible modification of the amino terminal threonine of β-subunits. It should be noted that the active component of lactacystin is the clasto-lactacystin β-lactone (16)247 that is a rearrangement product of lactacystin in aqueous condition and reacts with the hydroxyl group of the amino terminal threonine to make an ester adduct (Fig. 7). Despite initial reports of high proteasome specificity and its wide use in biological studies, there have been reports that lactacystin also inhibits other cellular proteases.248–250 Moreover, while many elegant synthetic strategies have been achieved over the years,251 its complex synthesis has hampered the development of lactacystin-based therapeutic agents.
Figure 7.
Natural product Lactacystin, its active component, clasto-lactacystin β-lactone, and its proposed inhibition mechanism.
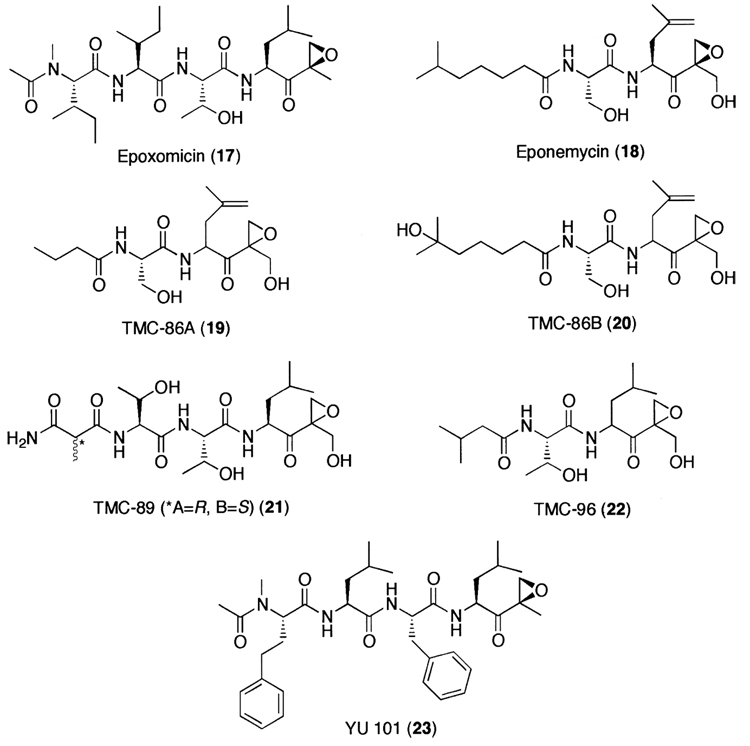
Meanwhile, in an effort to search for antitumor agents displaying specific activity against B16 murine melanoma, Hanada et al.252 found epoxomicin (17) from an unidentified actinomycete strain No. Q996-17. Epoxomicin (17) is a member of growing family of linear peptide α′,β′-epoxyketone natural products (Fig. 8). Recently, it has been shown that epoxomicin specifically targets the 20S proteasome.114,115 Unlike other classes of proteasome inhibitors that show non-target specificity, epoxomicin (17) is highly specific for the 20S proteasome. As revealed by the crystal structure of the yeast 20S proteasome complexed with epoxomicin, this unique specificity of epoxomicin is a result of a six-membered morpholino ring formation between the amino terminal catalytic Thr-1 of the 20S proteasome and the α′,β′-epoxyketone pharmacophore of epoxomicin (Fig. 9).116 In addition, eponemycin (18), an anti-angiogenic linear peptide α′,β′-epoxyketone isolated from Streptomyces Hygroscopicus No. P247–271 on the basis of its specific activity against B16 melanoma,253 has been shown to target the 20S proteasome as well.254 More recently, other linear peptide α′,β′-epoxyketone natural products have been isolated directly on the basis of proteasome inhibition screening from microbial metabolites (Fig. 8). Examples include TMC-86A (19) and B (20) from Streptomyces sp. TC 1084,255 TMC-89A and B (21) from Streptomyces sp. TC 1087256 and TMC-96 (22) from Saccharothrix sp. TC 1094.255 The facile syntheses of linear peptide α′,β′-epoxyketones has prompted the development of more potent peptide α′,β′-epoxyketones, such as YU101 (23) that was developed by the optimization of the amino acids of the P2–P4 positions to maximize its potency toward the CT-L activity.235
Figure 8.
A family of natural product peptide α′,β′-epoxyketone-based proteasome inhibitors.
Figure 9.
The proposed morpholino adduct formation mechanism by epoxomicin.
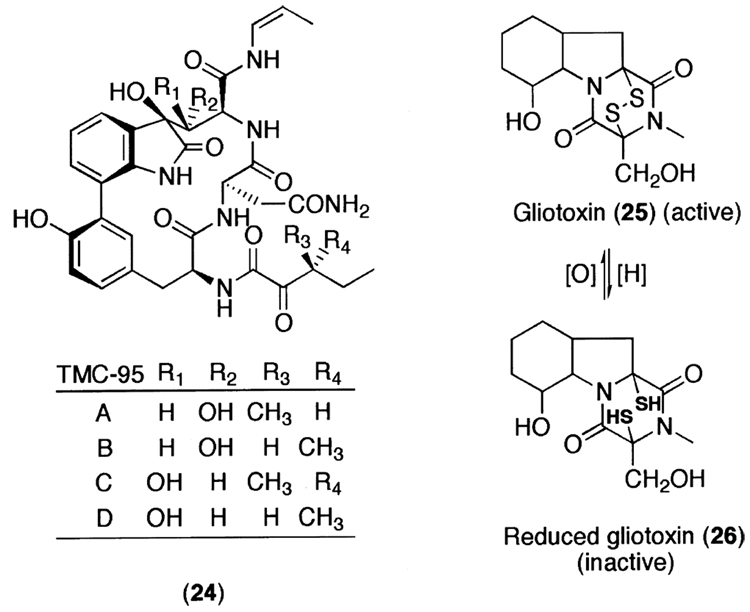
While lactacystin and epoxomicin are the best known natural product inhibitors of the 20S proteasome, there have been other potent natural product proteasome inhibitors with novel structures. For example, in the course of proteasome inhibitor screening procedure, Kohno et al.257 found a series of TMC-95 (24) from the fermentation broth of Apiospora montagnei Sacc. TC 1093 (Fig. 10). Despite their unusual macrocyclic peptide architecture, they possess potent inhibitory activity toward the CT-L activity of the 20S proteasome.258 Given that most well known potent proteasome inhibitors are the linear peptide backbone-based compounds, it will be interesting to see how the active site of the 20S proteasome can accommodate the macrocyclic ring of TMC-95s. Another unexpected class of natural product proteasome inhibitors includes gliotoxin (25), a member of the fungal epipolythiodioxopiperazine toxins that are characterized by a heterobicyclic core containing a disulfide bridge(s) (Fig. 10). Gliotoxin was originally identified as a potent inhibitor of NF-κB activation in T and B cells.259 Recently, Kroll et al.260 showed that gliotoxin noncompetitively targets the CT-L activity of the 20S proteasome and that the disulfide bridge is responsible for its inhibitory action.
Figure 10.
Natural product proteasome inhibitors with unusual scaffolds.
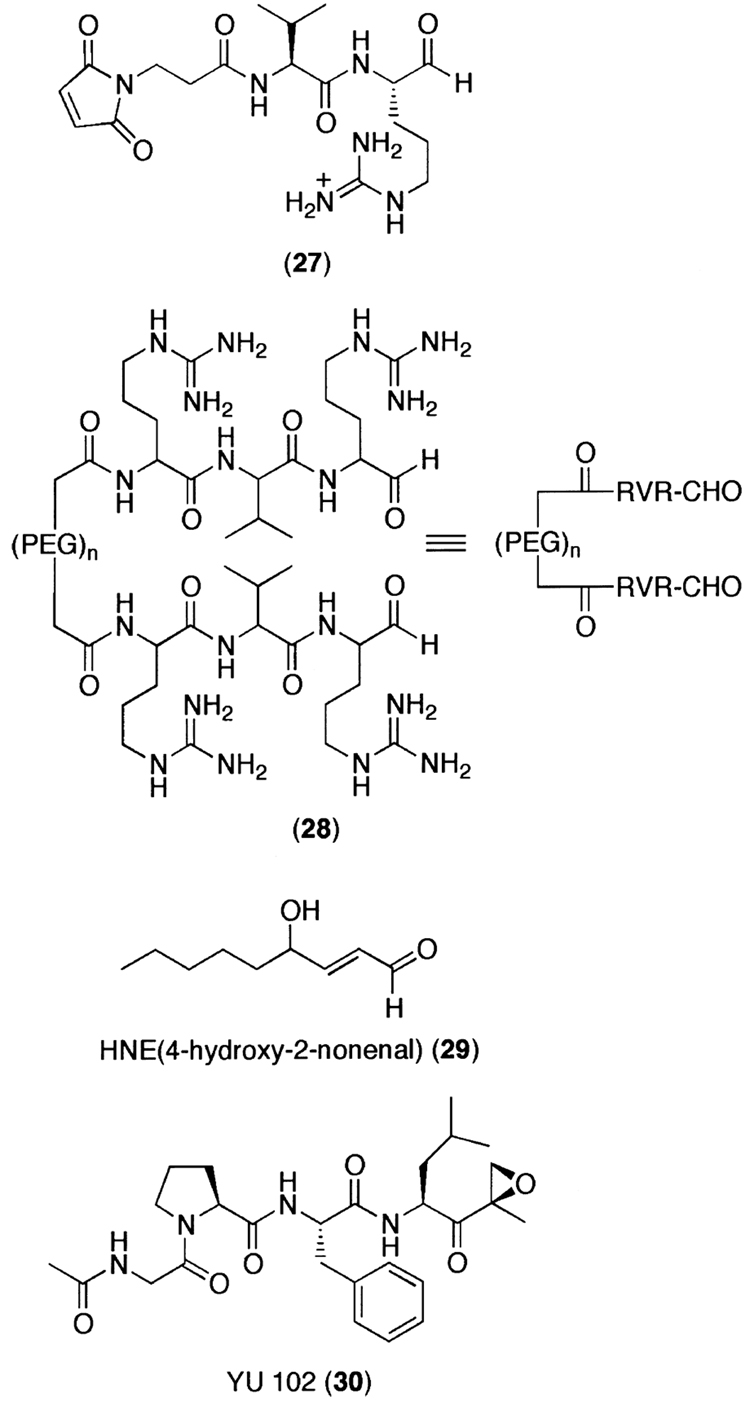
Although biochemical basis of catalytic activity generated by each subunit has been extensively analyzed by classical genetic mutation and X-ray crystal structure of the 20S proteasome, it is not yet clear how each catalytic activity contributes to the overall biological processes. To further separate the role(s) of the proteasome catalytic subunits, inhibition of each proteolytic activity is required. Most most of the synthetic and natural proteasome inhibitors described above are, however, CT-L activity-directed compounds. Moreover, only a limited number of compounds have been shown to inhibit the PGPH or T-L activities of the 20S proteasome. Therefore, a new generation of compounds that specifically inhibit the PGPH or T-L activities may be required to perform a careful analysis of proteasome subunit function and to gain insights into the possibility for potential therapeutic intervention. For example, Loidl et al.261 developed a T-L activity specific aldehyde inhibitor (27) by structure-based rational design and a bivalent T-L specific proteasome inhibitor (28), in which two tripeptide aldehydes were linked through a polymeric spacer that is appropriate for simultaneous binding at two active sites.262 Another interesting but not fully understood proteasome inhibitor 4-hydroxy-2-nonenal (HNE) (29) is a major end product of lipid peroxidation (Fig. 11) and is associated with the decrease of both the T-L and the PGPH but not the CT-L activities during in vivo oxidative stress.263 Further investigation of HNE’s mode(s) of action may prove to be useful for the design of non-peptidic PGPH or T-L specific inhibitors.
Figure 11.
Subunit-specific proteasome inhibitors.
To probe the role of different catalytic subunits in in vivo and in vitro proteasomal degradation, we have recently developed a series of PGPH-specific peptide α′,β′-epoxyketones by optimizing the amino acids at the P1-P4 positions.264 Successful application of these compounds including YU102 (30) in cell-based protein degradation assays in living cells revealed that selective PGPH inhibition was not sufficient to inhibit protein degradation, indicating that the CT-L and PGPH sites function independently.
6. CONCLUSION
Given that the ubiquitin-proteasome pathway is involved in so many critical cellular processes, proteasome inhibitors are of great interest as potential therapeutics. Indeed, a boronate inhibitor and a lactacystin inhibitor are currently in clinical trials for cancer and for stroke-associated ischemia reperfusion injury, respectively. In order to improve the safety of proteasome-inhibitor drugs and enable more specific inhibition of biological processes, however, attempts to target earlier steps of ubiquitin-proteasome pathway are desirable. Although such inhibitors are not yet available, the growing amount of information about cellular substrates and components of ubiquitin conjugation cascade may aid the design of potential E2 or E3 inhibitors.
Biographies
Jayhyuk Myung received his PhD in biochemistry in 1994 after doctoral training in the laboratory of Dr William P. Jencks at Brandeis University. After postdoctoral training with Dr Ira S. Mellman at Yale School of Medicine, studying the molecular mechanism of polarized vesicle traffic in the MDCK cells, he is currently involved in examining the mechanism of the 20S proteasome using a “chemical knockout” approach in the laboratory of Dr Craig M. Crews at Yale University. He received a National Research Service Award (NIH) and an Anderson Fellowship (Yale University). His research interests focus on understanding complex intracellular signaling networks by exploring the nature of biologically active natural products.
Kyung Bo Kim graduated from Yonsei University in 1988 and obtained his MS with Dr Dong Han Kim from the Pohang University of Science and Technology in 1991. After completing his PhD in the field of sugar nucleotide chemistry under the supersivion of Dr Edward Behrman at the Ohio State University in 1997, he is currently conducting postdoctoral studies in the laboratory of Dr Craig Crews. His research interests focus on the development of novel molecular probes to unravel complex signaling pathways and the identification novel potential drug targets.
Craig M. Crews graduated from the University of Virginia in 1986 and then studied for a year in Tübingen, Germany, as a DAAD Fellow. He received his PhD from Harvard University in 1993. After completing his postdoctoral training with Stuart Schreiber at Harvard University, he joined the faculty at Yale University where he is currently Associate Professor of Molecular, Cellular, and Developmental Biology as well as of Pharmacology. His research interests lie at the interface of chemistry and biology. In particular, his laboratory uses biologically active natural products to identify and validate novel drug targets.
REFERENCES
- 1.Vijay-Kumar S, Bugg CE, Wilkinson KD, Vierstra RD, Hatfield PM, Cook WJ. J Biol Chem. 1987;262:6396–6399. [PubMed] [Google Scholar]
- 2.Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Proc Natl Acad Sci USA. 1985;82:3582–3585. doi: 10.1073/pnas.82.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijay-Kumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 4.Weber PL, Brown SC, Mueller L. Biochemistry. 1987;26:7282–7290. doi: 10.1021/bi00397a013. [DOI] [PubMed] [Google Scholar]
- 5.McGrath JP, Jentsch S, Varshavsky A. Embo J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatfield PM, Vierstra RD. J Biol Chem. 1992;267:14799–14803. [PubMed] [Google Scholar]
- 7.Jentsch S. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 8.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 9.Sung P, Prakash S, Prakash L. J Mol Biol. 1991;221:745–749. doi: 10.1016/0022-2836(91)80169-u. [DOI] [PubMed] [Google Scholar]
- 10.Sommer T, Jentsch S. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 11.Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 12.Seufert W, Futcher B, Jentsch S. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 13.Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 14.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen RJ, Madura K, Bartel B, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 17.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 18.Ponzio G, Rossi B, Lazdunski M. J Biol Chem. 1983;258:8201–8205. [PubMed] [Google Scholar]
- 19.Bartel B, Wunning I, Varshavsky A. Embo J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. Embo J. 1991;10:2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madura K, Dohmen RJ, Varshavsky A. J Biol Chem. 1993;268:12046–12054. [PubMed] [Google Scholar]
- 22.Watkins JF, Sung P, Prakash S, Prakash L. Genes Dev. 1993;7:250–261. doi: 10.1101/gad.7.2.250. [DOI] [PubMed] [Google Scholar]
- 23.Reiss Y, Kaim D, Hershko A. J Biol Chem. 1988;263:2693–2698. [PubMed] [Google Scholar]
- 24.Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky A. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 25.Madura K, Varshavsky A. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 26.Byrd C, Turner GC, Varshavsky A. Embo J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner GC, Du F, Varshavsky A. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 28.Xie Y, Varshavsky A. Embo J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibregtse JM, Scheffner M, Howley PM. Embo J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 32.Oda H, Kumar S, Howley PM. Proc Natl Acad Sci USA. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz SE, Rosa JL, Scheffner M. J Biol Chem. 1998;273:12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 34.Scheffner M, Nuber U, Huibregtse JM. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 35.Huibregtse JM, Yang JC, Beaudenon SL. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshavsky A. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 37.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. Embo J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 38.Goulet CC, Volk KA, Adams CM, Prince LS, Stokes JB, Snyder PM. J Biol Chem. 1998;273:30012–30017. doi: 10.1074/jbc.273.45.30012. [DOI] [PubMed] [Google Scholar]
- 39.Scheffner M, Huibregtse JM, Howley `PM. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. J Biol Chem. 1996;271:2795–2800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Kao WH, Howley PM. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 42.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 43.Nuber U, Scheffner M. J Biol Chem. 1999;274:7576–7582. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- 44.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 45.Miyake S, Lupher ML, Jr, Druker B, Band H. Proc Natl Acad Sci USA. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 47.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 48.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 49.Waterman H, Levkowitz G, Alroy I, Yarden Y. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 50.Blake TJ, Shapiro M, Morse HCd, Langdon WY. Oncogene. 1991;6:653–657. [PubMed] [Google Scholar]
- 51.Andoniou CE, Thien CB, Langdon WY. Embo J. 1994;13:4515–4523. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 53.Ulrich HD, Jentsch S. Embo J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de The H, Hay RT, Freemont PS. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 55.Honda R, Tanaka H, Yasuda H H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 56.Honda R, Yasuda H. Embo J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasmyth K. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 58.King RW, Deshaies RJ, Peters J-M, Kirschner MW. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 59.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 60.Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 61.Peters JM. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 62.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 63.Schwab M, Lutum AS, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 64.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 65.Fang G, Yu H, Kirschner MW. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 66.Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 67.Sigrist S, Jacobs H, Stratmann R, Lehner CF. Embo J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 69.Leverson JD, Joazeiro CA, Page AM, Huang H, Hieter P, Hunter T. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. Proc Natl Acad Sci USA. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 72.Zachariae W, Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 73.Koepp DM, Harper JW, Elledge SJ. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 74.Schwob E, Bohm T, Mendenhall MD, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 75.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 76.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H, Kobayashi R, Galaktionov K, Beach D. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 78.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 79.Zhou P, Howley PM. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- 80.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 81.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 82.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 84.Joazeiro CA, Weissman AM. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 85.Tyers M, Jorgensen P. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 86.Loöwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 87.Groll M, Ditzel L, Loöwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 88.Kopp F, Hendil KB, Dahlmann B, Kristensen P, Sobek A, Uerkvitz W. Proc Natl Acad Sci USA. 1997;94:2939–2944. doi: 10.1073/pnas.94.7.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 90.Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 91.Orlowski M, Cardozo C, Hidalgo MC, Michaud C. Biochemistry. 1991;30:5999–6005. doi: 10.1021/bi00238a025. [DOI] [PubMed] [Google Scholar]
- 92.Dahlmann B, Becher B, Sobek A, Ehlers C, Kopp F, Kuehn L. Enzyme Protein. 1993;47:274–284. doi: 10.1159/000468685. [DOI] [PubMed] [Google Scholar]
- 93.Shibatani T, Ward WF. Arch Biochem Biophys. 1995;321:160–166. doi: 10.1006/abbi.1995.1381. [DOI] [PubMed] [Google Scholar]
- 94.Ma CP, Slaughter CA, DeMartino GN. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 95.Chu-Ping M, Vu JH, Proske RJ, Slaughter CA, DeMartino GN. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 96.Hoffman L, Rechsteiner M. J Biol Chem. 1994;269:16890–16895. [PubMed] [Google Scholar]
- 97.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 98.Yao Y, Huang L, Krutchinsky A, Wong ML, Standing KG, Burlingame AL, Wang CC. J Biol Chem. 1999;274:33921–33930. doi: 10.1074/jbc.274.48.33921. [DOI] [PubMed] [Google Scholar]
- 99.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 100.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 101.Wilk S, Orlowski M. J Neurochem. 1983;40:842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x. [DOI] [PubMed] [Google Scholar]
- 102.Cardozo C. Enzyme Protein. 1993;47:296–305. doi: 10.1159/000468687. [DOI] [PubMed] [Google Scholar]
- 103.Orlowski M, Cardozo C, Michaud C. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 104.Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, Keilholz W, Stevanovic S, Wolf DH, Huber R, Rammensee HG, Schild H. J Biol Chem. 1998;273:25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- 105.McCormack TA, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Plamondon L, Stein RL, Dick LR. Biochemistry. 1998;37:7792–7800. doi: 10.1021/bi980097q. [DOI] [PubMed] [Google Scholar]
- 106.Cardozo C, Michaud C, Orlowski M. Biochemistry. 1999;38:9768–9777. doi: 10.1021/bi990735k. [DOI] [PubMed] [Google Scholar]
- 107.Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- 108.Enenkel C, Lehmann H, Kipper J, Guckel R, Hilt W, Wolf DH. FEBS Lett. 1994;341:193–196. doi: 10.1016/0014-5793(94)80455-9. [DOI] [PubMed] [Google Scholar]
- 109.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 110.Salzmann U, Kral S, Braun B, Standera S, Schmidt M, Kloetzel PM, Sijts A. FEBS Lett. 1999;454:11–15. doi: 10.1016/s0014-5793(99)00768-1. [DOI] [PubMed] [Google Scholar]
- 111.Orlowski M, Wilk S. Arch Biochem Biophys. 2000;383:1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- 112.Seemüller E, Lupas A, Zuhl F, Zwickl P, Baumeister W. FEBS Lett. 1995;359:173–178. doi: 10.1016/0014-5793(95)00036-9. [DOI] [PubMed] [Google Scholar]
- 113.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 114.Sin N, Kim K, Elofsson M, Meng L, Auth H, Kwok BHB, Crews CM. Bioorg Med Chem Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- 115.Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Groll M, Kim KB, Huber R, Crews CM. J Am Chem Soc. 2000;122:1237–1238. [Google Scholar]
- 117.Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 118.Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, Moody PC. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 119.Oinonen C, Tikkanen R, Rouvinen J, Peltonen L. Nat Struct Biol. 1995;2:1102–1108. doi: 10.1038/nsb1295-1102. [DOI] [PubMed] [Google Scholar]
- 120.Seemuüller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 121.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel PM. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 122.Yang Y, Fruh K, Ahn K, Peterson PA. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 123.Tamura T, Tanaka K, Kumatori A, Yamada F, Tsurumi C, Fujiwara T, Ichihara A, Tokunaga F, Aruga R, Iwanaga S. FEBS Lett. 1990;264:91–94. doi: 10.1016/0014-5793(90)80773-c. [DOI] [PubMed] [Google Scholar]
- 124.Tamura T, Lee DH, Osaka F, Fujiwara T, Shin S, Chung CH, Tanaka K, Ichihara A. Biochim Biophys Acta. 1991;1089:95–102. doi: 10.1016/0167-4781(91)90090-9. [DOI] [PubMed] [Google Scholar]
- 125.Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Embo J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 127.Swenson KI, Farrell KM, Ruderman JV. Cell. 1986;47:861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- 128.Glotzer M, Murray AW, Kirschner MW. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 129.Lohrum MA, Vousden KH. Trends Cell Biol. 2000;10:197–202. doi: 10.1016/s0962-8924(00)01736-0. [DOI] [PubMed] [Google Scholar]
- 130.Deshaies RJ. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 131.Caspari T. Curr Biol. 2000;10:R315–R317. doi: 10.1016/s0960-9822(00)00439-5. [DOI] [PubMed] [Google Scholar]
- 132.Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL, Brodeur GM. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maki CG, Huibregtse JM, Howley PM. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 134.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 135.Kubbutat MH, Jones SN, Vousden KH. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 136.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 137.Honda R, Yasuda H. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 138.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 139.Shieh SY, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 140.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Embo J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ashcroft M, Vousden KH. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 142.Mayo LD, Turchi JJ, Berberich SJ. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 143.Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sen R, Baltimore D. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 145.Baeuerle PA, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 146.Baeuerle PA, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 147.Baldwin AS., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 148.Barnes PJ, Karin M. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 149.Ghosh S, May MJ, Kopp EB. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 150.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 151.Stancovski I, Baltimore D. Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 152.Maniatis T. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 153.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 154.Spencer E, Jiang J, Chen ZJ. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 157.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 159.Fan CM, Maniatis T. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 160.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz AL, Ciechanover A. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 161.Sears C, Olesen J, Rubin D, Finley D, Maniatis T. J Biol Chem. 1998;273:1409–1419. doi: 10.1074/jbc.273.3.1409. [DOI] [PubMed] [Google Scholar]
- 162.Coux O, Goldberg AL. J Biol Chem. 1998;273:8820–8828. doi: 10.1074/jbc.273.15.8820. [DOI] [PubMed] [Google Scholar]
- 163.Rice NR, MacKichan ML, Israel A. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- 164.Mercurio F, DiDonato JA, Rosette C, Karin M. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 165.Naumann M, Wulczyn FG, Scheidereit C. Embo J. 1993;12:213–222. doi: 10.1002/j.1460-2075.1993.tb05647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Orian A, Gonen H, Bercovich B, Fajerman I, Eytan E, Israel A, Mercurio F, Iwai K, Schwartz AL, Ciechanover A. Embo J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Heissmeyer V, Krappmann D, Wulczyn FG, Scheidereit C. Embo J. 1999;18:4766–4778. doi: 10.1093/emboj/18.17.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lin L, DeMartino GN, Greene WC. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 169.Lin L, DeMartino GN, Greene WC. Embo J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pamer E, Cresswell P. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 171.Rock KL, Goldberg AL. Ann Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 172.Michalek MT, Grant EP, Gramm C, Goldberg AL, Rock KL. Nature. 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 173.Cox JH, Galardy P, Bennink JR, Yewdell JW. J Immunol. 1995;154:511–519. [PubMed] [Google Scholar]
- 174.Michalek MT, Grant EP, Rock KL. J Immunol. 1996;157:617–624. [PubMed] [Google Scholar]
- 175.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 176.Harding CV, France J, Song R, Farah JM, Chatterjee S, Iqbal M, Siman R. J Immunol. 1995;155:1767–1775. [PubMed] [Google Scholar]
- 177.Hughes EA, Ortmann B, Surman M, Cresswell P. J Exp Med. 1996;183:1569–1578. doi: 10.1084/jem.183.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 179.Cerundolo V, Benham A, Braud V, Mukherjee S, Gould K, Macino B, Neefjes J, Townsend A. Eur J Immunol. 1997;27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]
- 180.Putterman D, Pepinsky RB, Vogt VM. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 181.Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, 2nd, Yoshinaka Y, Oroszlan S, Arthur LO, Henderson LE. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patnaik A, Chau V, Wills JW. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 186.Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 187.Angelman H. Dev Med Child Neurol. 1965;7:681–688. [Google Scholar]
- 188.Mann MR, Bartolomei MS. Hum Mol Genet. 1999;8:1867–1873. doi: 10.1093/hmg/8.10.1867. [DOI] [PubMed] [Google Scholar]
- 189.Williams CA, Angelman H, Clayton-Smith J, Driscoll DJ, Hendrickson JE, Knoll JH, Magenis RE, Schinzel A, Wagstaff J, Whidden EM, et al. Am J Med Genet. 1995;56:237–238. doi: 10.1002/ajmg.1320560224. [DOI] [PubMed] [Google Scholar]
- 190.Kishino T, Lalande M, Wagstaff J. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 191.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 192.Liddle G, Bledsoe T, Coppage W. Trans Assoc Am Phys. 1963;76:199–213. [Google Scholar]
- 193.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Jr, Ulick S, Milora RV, Findling JW, et al. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 194.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 195.Huibregtse JM, Scheffner M, Howley PM. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Talis AL, Huibregtse JM, Howley PM. J Biol Chem. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 197.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias KE, Schwartz AL, Kahana C, Ciechanover A. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kao WH, Beaudenon SL, Talis AL, Huibregtse JM, Howley PM. J Virol. 2000;74:6408–6417. doi: 10.1128/jvi.74.14.6408-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kinzler KW, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 200.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Embo J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 202.Ikeda S, Kishida M, Matsuura Y, Usui H, Kikuchi A. Oncogene. 2000;19:537–545. doi: 10.1038/sj.onc.1203359. [DOI] [PubMed] [Google Scholar]
- 203.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 205.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 206.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 207.Tetsu O, McCormick F. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 208.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 209.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 210.Latres E, Chiaur DS, Pagano M. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 211.Mori H, Kondo J, Ihara Y. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- 212.Perry G, Friedman R, Shaw G, Chau V. Proc Natl Acad Sci USA. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RDJ, Salehi A, Martens GJM, Grosveld FG, Peter J, Burbach H, Hol EM. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 214.Checler F, da Costa CA, Ancolio K, Chevallier N, Lopez-Perez E, Marambaud P. Biochim Biophys Acta. 2000;1502:133–138. doi: 10.1016/s0925-4439(00)00039-9. [DOI] [PubMed] [Google Scholar]
- 215.Checler FJ. Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 216.Marambaud P, Ancolio K, Lopez-Perez E, Checler F. Mol Med. 1998;4:147–157. [PMC free article] [PubMed] [Google Scholar]
- 217.Kim TW, Pettingell WH, Hallmark OG, Moir RD, Wasco W, Tanzi RE. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 218.Marambaud P, Alves da Costa C, Ancolio K, Checler F. Biochem Biophys Res Commun. 1998;252:134–138. doi: 10.1006/bbrc.1998.9619. [DOI] [PubMed] [Google Scholar]
- 219.Orlowski M. Biochemistry. 1990;29:10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- 220.Figueiredo-Pereira ME, Banik N, Wilk S. J Neurochem. 1994;62:1989–1994. doi: 10.1046/j.1471-4159.1994.62051989.x. [DOI] [PubMed] [Google Scholar]
- 221.Wilk S, Figueiredo-Pereira ME. Enzyme Protein. 1993;47:306–313. doi: 10.1159/000468688. [DOI] [PubMed] [Google Scholar]
- 222.Vinitsky A, Cardozo C, Sepp-Lorenzino L, Michaud C, Orlowski M. J Biol Chem. 1994;269:29860–29866. [PubMed] [Google Scholar]
- 223.Lynas JF, Harriott P, Healy A, McKervey MA, Walker B. Bioorg Med Chem Lett. 1998;8:373–378. doi: 10.1016/s0960-894x(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 224.Iqbal M, Chatterjee S, Kauer JC, Mallamo JP, Messina PA, Reiboldt A, Siman R. Bioorg Med Chem Lett. 1996;6:287–290. [Google Scholar]
- 225.Lum RT, Nelson MG, Joly A, Horsma AG, Lee G, Meyer SM, Wick MM, Schow SR. Bioorg Med Chem Lett. 1998;8:209–214. doi: 10.1016/s0960-894x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 226.Chatterjee S, Dunn D, Mallya S, Ator MA. Bioorg Med Chem Lett. 1999;9:2603–2606. doi: 10.1016/s0960-894x(99)00443-6. [DOI] [PubMed] [Google Scholar]
- 227.Spaltenstein A, Leban JJ, Huang JJ, Reinhardt KR, Viveros OH, Sigafoos J, Crouch R. Tet Lett. 1996;37:1343–1346. [Google Scholar]
- 228.Bogyo M, Shin S, McMaster JS, Ploegh HL. Chem Biol. 1998;5:307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- 229.Palmer JT, Rasnick D, Klaus JL, Bromme D. J Med Chem. 1995;38:3193–3196. doi: 10.1021/jm00017a002. [DOI] [PubMed] [Google Scholar]
- 230.Bromme D, Klaus JL, Okamoto K, Rasnick D, Palmer JT. Biochem J. 1996;315:85–89. doi: 10.1042/bj3150085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Glas R, Bogyo M, McMaster JS, Gaczynska M, Ploegh HL. Nature. 1998;392:618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- 232.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.McCormack T, Baumeister W, Grenier L, Moomaw C, Plamondon L, Pramanik B, Slaughter C, Soucy F, Stein R, Zuhl F, Dick L. J Biol Chem. 1997;272:26103–26109. doi: 10.1074/jbc.272.42.26103. [DOI] [PubMed] [Google Scholar]
- 234.Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Bioorg Med. Chem Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 235.Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Chem Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- 236.Palombella VJ, Conner EM, Fuseler JW, Destree A, Davis JM, Laroux FS, Wolf RE, Huang J, Brand S, Elliott PJ, Lazarus D, McCormack T, Parent L, Stein R, Adams J, Grisham MB. Proc Natl Acad Sci USA. 1998;95:15671–15676. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- 238.Drexler HC. Proc Natl Acad Sci USA. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lopes UG, Erhardt P, Yao R, Cooper GM. J Biol Chem. 1997;272:12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- 240.Chandra J, Niemer I, Gilbreath J, Kliche KO, Andreeff M, Freireich EJ, Keating M, McConkey DJ. Blood. 1998;92:4220–4229. [PubMed] [Google Scholar]
- 241.Teicher BA, Ara G, Herbst R, Palombella VJ, Adams J. Clin Cancer Res. 1999;5:2638–2645. [PubMed] [Google Scholar]
- 242.Adams J, Palombella VJ, Elliott PJ. Invest New Drugs. 2000;18:109–121. doi: 10.1023/a:1006321828515. [DOI] [PubMed] [Google Scholar]
- 243.Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. J Antibiot (Tokyo) 1991;44:113–116. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- 244.Omura S, Matsuzaki K, Fujimoto T, Kosuge K, Furuya T, Fujita S, Nakagawa A. J Antibiot (Tokyo) 1991;44:117–118. doi: 10.7164/antibiotics.44.117. [DOI] [PubMed] [Google Scholar]
- 245.Fenteany G, Standaert RF, Reichard GA, Corey EJ, Schreiber SL. Proc Natl Acad Sci USA. 1994;91:3358–3362. doi: 10.1073/pnas.91.8.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Katagiri M, Hayashi M, Matsuzaki K, Tanaka H, Omura S. J Antibiot (Tokyo) 1995;48:344–346. doi: 10.7164/antibiotics.48.344. [DOI] [PubMed] [Google Scholar]
- 247.Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. J Biol Chem. 1996;271:7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- 248.Ostrowska H, Wojcik C, Omura S, Worowski K. Biochem Biophys Res Commun. 1997;234:729–732. doi: 10.1006/bbrc.1997.6434. [DOI] [PubMed] [Google Scholar]
- 249.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 250.Ostrowska H, Wojcik C, Wilk S, Omura S, Kozlowski L, Stoklosa T, Worowski K, Radziwon P. Int J Biochem Cell Biol. 2000;32:747–757. doi: 10.1016/s1357-2725(00)00021-2. [DOI] [PubMed] [Google Scholar]
- 251.Corey EJ, Li W. Chem Pharm Bull. 1999;47:1–10. doi: 10.1248/cpb.47.1. [DOI] [PubMed] [Google Scholar]
- 252.Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T. J. Antibiotics. 1992;45:1746–1752. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- 253.Sugawara K, Hatori M, Nishiyama Y, Tomita K, Kamei H, Konishi M, Oki T. J. Antibiotics. 1990;43:8–18. doi: 10.7164/antibiotics.43.8. [DOI] [PubMed] [Google Scholar]
- 254.Meng L, Kwok BHB, Sin N, Crews CM. Cancer Research. 1999;59:2798–2801. [PubMed] [Google Scholar]
- 255.Koguchi Y, Kohno J, Suzuki S, Nishio M, Takahashi K, Ohnuki T, Komatsubara S. J Antibiot (Tokyo) 2000;53:63–65. doi: 10.7164/antibiotics.53.63. [DOI] [PubMed] [Google Scholar]
- 256.Koguchi Y, Nishio M, Suzuki S, Takahashi K, Ohnuki T, Komatsubara S. J Antibiot (Tokyo) 2000;53:967–972. doi: 10.7164/antibiotics.53.967. [DOI] [PubMed] [Google Scholar]
- 257.Kohno J, Koguchi Y, Nishio M, Nakao K, Kuroda M, Shimizu R, Ohnuki T, Komatsubara S. J Org Chem. 2000;65:990–995. doi: 10.1021/jo991375+. [DOI] [PubMed] [Google Scholar]
- 258.Koguchi Y, Kohno J, Nishio M, Takahashi K, Okuda T, Ohnuki T, Komatsubara S. J Antibiot (Tokyo) 2000;53:105–109. doi: 10.7164/antibiotics.53.105. [DOI] [PubMed] [Google Scholar]
- 259.Pahl HL, Krauss B, Schulze-Osthoff K, Decker T, Traenckner EB, Vogt M, Myers C, Parks T, Warring P, Muhlbacher A, Czernilofsky AP, Baeuerle PA. J Exp Med. 1996;183:1829–1840. doi: 10.1084/jem.183.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kroll M, Arenzana-Seisdedos F, Bachelerie F, Thomas D, Friguet B, Conconi M. Chem Biol. 1999;6:689–698. doi: 10.1016/s1074-5521(00)80016-2. [DOI] [PubMed] [Google Scholar]
- 261.Loidl G, Groll M, Musiol H-J, Ditzel L, Huber R, Moroder L. Chem Biol. 1999;6:197–204. doi: 10.1016/S1074-5521(99)80036-2. [DOI] [PubMed] [Google Scholar]
- 262.Loidl G, Groll M, Musiol H-J, Huber R, Moroder L. Proc Natl Acad Sci USA. 1999;96:5418–5422. doi: 10.1073/pnas.96.10.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Okada K, Wangpoengtrakul C, Osawa T, Toyokuni S, Tanaka K, Uchida K. J Biol Chem. 1999;274:23787–23793. doi: 10.1074/jbc.274.34.23787. [DOI] [PubMed] [Google Scholar]
- 264.Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Molecular Cell. 2001;7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]