Abstract
Cysteine (Cys) synthesis in plants is carried out by two sequential reactions catalyzed by the rate-limiting enzyme serine acetyltransferase (SAT) and excess amounts of O-acetylserine(thiol)lyase. Why these reactions occur in plastids, mitochondria, and cytosol of plants remained unclear. Expression of artificial microRNA (amiRNA) against Sat3 encoding mitochondrial SAT3 in transgenic Arabidopsis (Arabidopsis thaliana) plants demonstrates that mitochondria are the most important compartment for the synthesis of O-acetylserine (OAS), the precursor of Cys. Reduction of RNA levels, protein contents, SAT enzymatic activity, and phenotype strongly correlate in independent amiSAT3 lines and cause significantly retarded growth. The expression of the other four Sat genes in the Arabidopsis genome are not affected by amiRNA-SAT3 according to quantitative real-time polymerase chain reaction and microarray analyses. Application of radiolabeled serine to leaf pieces revealed severely reduced incorporation rates into Cys and even more so into glutathione. Accordingly, steady-state levels of OAS are 4-fold reduced. Decrease of sulfate reduction-related genes is accompanied by an accumulation of sulfate in amiSAT3 lines. These results unequivocally show that mitochondria provide the bulk of OAS in the plant cell and are the likely site of flux regulation. Together with recent data, the cytosol appears to be a major site of Cys synthesis, while plastids contribute reduced sulfur as sulfide. Thus, Cys synthesis in plants is significantly different from that in nonphotosynthetic eukaryotes at the cellular level.
Cys synthesis in plants is a fundamental process for protein biosynthesis and all anabolic pathways that require reduced sulfur. Bacteria and fungi are able to reduce sulfate by assimilatory sulfate reduction to sulfide and to integrate it into Cys in the cytosol. In contrast, mammals need to take up reduced sulfur as Met and can synthesize Cys via trans-sulfurylation in the cytosol. Protein biosynthesis in the mitochondria in animals and fungi receives Cys, therefore, from the cytosol. In plants, the assimilatory sulfate reduction pathway is localized to plastids, whereas Cys synthesis is found in the cytosol, plastids, and mitochondria. Why Cys synthesis in plants is localized in three compartments has long been speculated.
Serine acetyltransferase (SAT; also termed Serat; EC 2.3.1.30) catalyzes the activation of Ser by acetyl-CoA to O-acetylserine (OAS). In Arabidopsis (Arabidopsis thaliana), five genes encoding SAT1 to SAT5 are localized on the five chromosomes (Hell et al., 2002). SAT1 is plastid localized, SAT3 is found in mitochondria, and SAT5 is cytosolic (Kawashima et al., 2005). SAT2 and SAT4 are also cytosolic proteins, but they differ substantially from the other SATs in their amino acid sequences and are much less expressed (Kawashima et al., 2005). The Arabidopsis Sat genes are almost constitutively expressed (www.genevestigator.ethz.ch), and the encoded proteins are of low abundance in comparison with O-acetylserine(thiol)lyase (OAS-TL; EC 2.5.1.47), since the activity of the latter is 300-fold higher than SAT activity in leaves (Ruffet et al., 1994). SAT activity is generally regarded as the rate-limiting step of Cys synthesis (Wirtz and Droux, 2005; Wirtz and Hell, 2006). Fractionation of pea (Pisum sativum) leaf cells showed that about 80% of extractable SAT activity under substrate-saturating conditions resides in mitochondria, while about 10% each is found in plastids and cytosol (Ruffet et al., 1995; Droux, 2003). The second step of Cys synthesis consists of the β-substitution of the acetyl moiety in OAS by sulfide and is catalyzed by OAS-TL. OAS-TLs are members of the pyridoxal phosphate-dependent β-substituted Ala synthase family (Hatzfeld et al., 2000b). Of the nine OAS-TL-like genes in Arabidopsis, one is a pseudogene, three encode the cytosolic OAS-TL A, plastid OAS-TL B, and mitochondrial OAS-TL C enzymes, and one is a β-cyanoalanine synthase that localizes to mitochondria (Hatzfeld et al., 2000b; Jost et al., 2000; Hell et al., 2002; Watanabe et al., 2008). According to cell fractionation of spinach (Spinacia oleracea), cauliflower (Brassica oleracea), and Datura innoxia, 40% to 45% of maximal OAS-TL activity is localized in plastids and cytosol, while about 5% is attributed to mitochondria (Lunn et al., 1990; Rolland et al., 1992; Kuske et al., 1996). As a result of the low mitochondrial OAS-TL activity, this activity is believed to be a side activity of β-cyanoalanine synthase in spinach (Warrilow and Hawkesford, 2000). The other four OAS-TL-like genes in Arabidopsis are only weakly expressed and characterized and show little or no Cys synthase activities (Wirtz and Hell, 2006; Heeg et al., 2008).
The three major SATs, SAT5, SAT1, and SAT3, form the Cys synthase protein complex with the major OAS-TLs, A, B, and C, in the cytosol, plastids, and mitochondria, according to interaction studies of the Arabidopsis proteins (Bogdanova and Hell, 1997; Jost et al., 2000). The Cys synthase complex was hypothesized to function in the cellular control of Cys synthesis, because SAT requires the presence of OAS-TL to gain full activity (Droux et al., 1998). At the same time, complex formation inactivates OAS-TL, and thus the reaction intermediate OAS dissociates from the complex and is freely available. OAS is known from feeding experiments to activate the expression of numerous genes, including genes related to sulfate uptake and reduction (Hirai et al., 2003; Maruyama-Nakashita et al., 2005). At the same time, OAS is able to dissociate the Cys synthase complex, at least in vitro, which is antagonized through stabilization by sulfide (Droux et al., 1998; Berkowitz et al., 2002; Wirtz et al., 2004; Wirtz and Hell, 2006). This effector-driven, reversible protein-protein interaction supposedly functions in cytosol, plastids, and mitochondria, although cell fractionations suggest considerably different ratios of SAT and OAS-TL activities in these compartments (Droux, 2003; Wirtz and Droux, 2005).
The role of OAS-TL in the three compartments with independent protein biosynthesis was recently addressed using Arabidopsis T-DNA insertion lines. Knockout of cytosolic OAS-TL A apparently caused oxidative stress and lesions as well as sensitivity to heavy metals (Lopez-Martin et al., 2008). While this phenotype was not reported by Watanabe et al. (2008) and Heeg et al. (2008), the latter studies confirmed the major roles of OAS-TL A, B, and C. OAS-TL A and B together contribute 95% of OAS-TL activity in leaves, and OAS-TL C contributes only about 5%. Despite this small contribution, a growth rate retardation of approximately 20% was observed for the oastlC mutant line. However, an oastlAB double mutant with OAS-TL C as the only Cys-forming activity was fully viable, with only a small reduction in growth rate (Heeg et al., 2008). OAS contents in the oastlC mutant were decreased, but in the oastlAB mutant they were found to be elevated, while labeling studies with [35S]sulfate revealed lowered incorporation of 35S into Cys only in oastlA mutants. From these findings and the high specific SAT activity in isolated mitochondria, it was concluded that mitochondria are the major site of OAS synthesis and that the cytosol, but not chloroplasts, provides most of the cellular Cys (Heeg et al., 2008). Despite the partial redundancy of Cys synthesis between these compartments, the results point toward a specific role of SAT in mitochondria as a pacemaker for the rate of cellular Cys synthesis. The investigations presented here use specific down-regulation of the gene encoding mitochondrial SAT3 by artificial microRNA (amiRNA) expression in Arabidopsis. Direct evidence for the dominant role of mitochondrial OAS formation for cellular Cys synthesis and its significance for plant growth is provided.
RESULTS
Expression of amiRNA against Sat3 Causes a Severe Growth Phenotype
According to in vitro analyses, the majority of SAT activity resides in mitochondria in pea leaves and heterotrophic Arabidopsis cell cultures (Ruffet et al., 1995; Heeg et al., 2008). The expression of the gene that encodes the mitochondrial isoform SAT3 (Sat3, equivalent to Serat2;2; At3g13110) was down-regulated by RNA interference to determine (1) if mitochondria indeed provide most of the OAS in vivo and (2) if this step limits Cys biosynthesis in the plant cell. Highest specificity of the designed amiRNA construct to Sat3 mRNA compared with the other four Sat messages was achieved using the WMD 2 - Web MicroRNA Designer (www.weigelworld.org). Furthermore, the sequence homology and hybridization energy of amiSAT3 were carefully analyzed and revealed specific targeting of amiSAT3 to Sat3 mRNA compared with RNAs of Sat1, Sat2, Sat4, and Sat5 genes as well as the rest of the Arabidopsis genome (Supplemental Fig. S1). The amiSAT3 construct was expressed under the control of the cauliflower mosaic virus 35S promoter and used to transform Arabidopsis ecotype Columbia (Col-0; amiSAT3 lines).
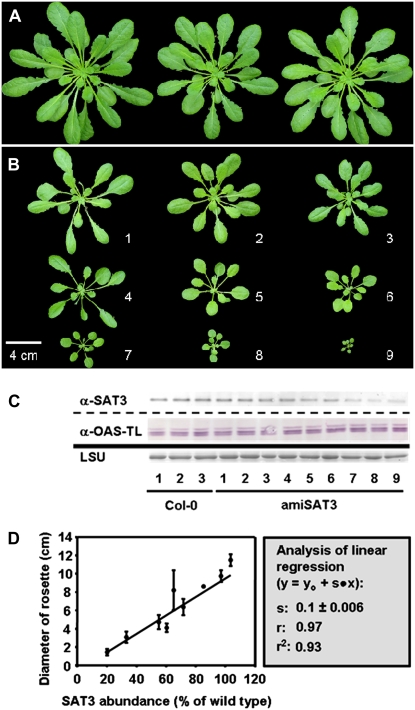
Several independent transformants (T1) were identified using kanamycin as a selectable marker. Nine plants of quite variable size survived (Fig. 1, A and B), while numerous others failed to develop any further and died at the early rosette stage before production of flowers (data not shown). Determination of SAT3 protein levels in the surviving amiSAT3 lines by specific antibodies (Wirtz and Hell, 2003) revealed decreasing amounts with declining plant size. In comparison, protein contents of the major OAS-TL forms A, B, and C, which can be simultaneously detected by an OAS-TL-specific antibody (Fig. 1C; Heeg et al., 2008), were not affected at all. Indeed, SAT3 protein contents and rosette sizes of amiSAT3 lines showed a strong linear correlation (r = 0.97; Fig. 1D).
Figure 1.
Impact of reduced mitochondrial SAT levels on the growth rate of Arabidopsis. A, Wild-type Arabidopsis plants were transformed with pBinAR-amiSAT3 and selected on kanamycin-containing medium. B, After 2 weeks, surviving transgenic plants were transferred to soil for an additional 6 weeks. C, The abundance of mitochondrial SAT3 was measured by immunological detection from soluble Arabidopsis protein extracts (15 μg) using a SAT3-specific antibody (Wirtz and Hell, 2007). The large subunit of ribulose-1,5-bisphosphate carboxylase (LSU) served as a loading control after staining with Coomassie Brilliant Blue. D, At the same time of harvesting the sample for protein extraction, the rosette diameters of the transgenic plants were measured three times for each individual and plotted against the SAT abundance of the corresponding plant. The data were fitted using a linear equation. Error bars indicate sd. [See online article for color version of this figure.]
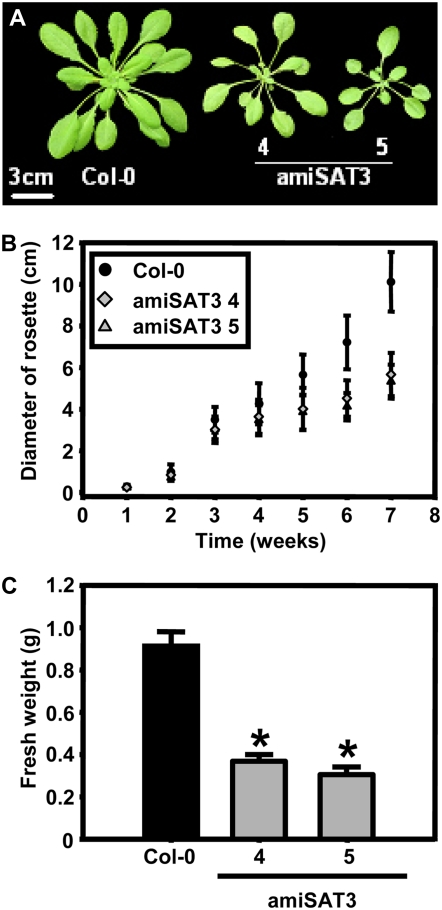
Lines 4 and 5 showed intermediate growth retardation and were selected for further analyses. The T2 generation of amiSAT3 lines 4 and 5 confirmed the growth phenotypes of the T1 generation (Fig. 2). Growth rates of lines 4 and 5 differed significantly from those of the wild type from week 4 on after germination, and fresh weights of rosettes were 40% ± 4% and 33% ± 4%, respectively, of wild-type levels after 8 weeks.
Figure 2.
Comparison of growth between wild-type and transgenic Arabidopsis plants with intermediate reduction of mitochondrial SAT. A, Phenotypes of a 7-week-old wild-type plant (Col-0) and two independent transgenic plant lines (amiSAT3 lines 4 and 5) with intermediate reduction of mitochondrial SAT. B, The growth of eight individuals from each genotype was monitored for 7 weeks by determining the diameter of the rosette three times for each individual (n = 24). C, The fresh weights of 8-week-old wild-type plants and the two amiSAT3 lines (n = 4) were determined. Error bars indicate sd, and asterisks mark significant differences using the unpaired t test. [See online article for color version of this figure.]
Specific Down-Regulation of Mitochondrial SAT Causes Growth Retardation
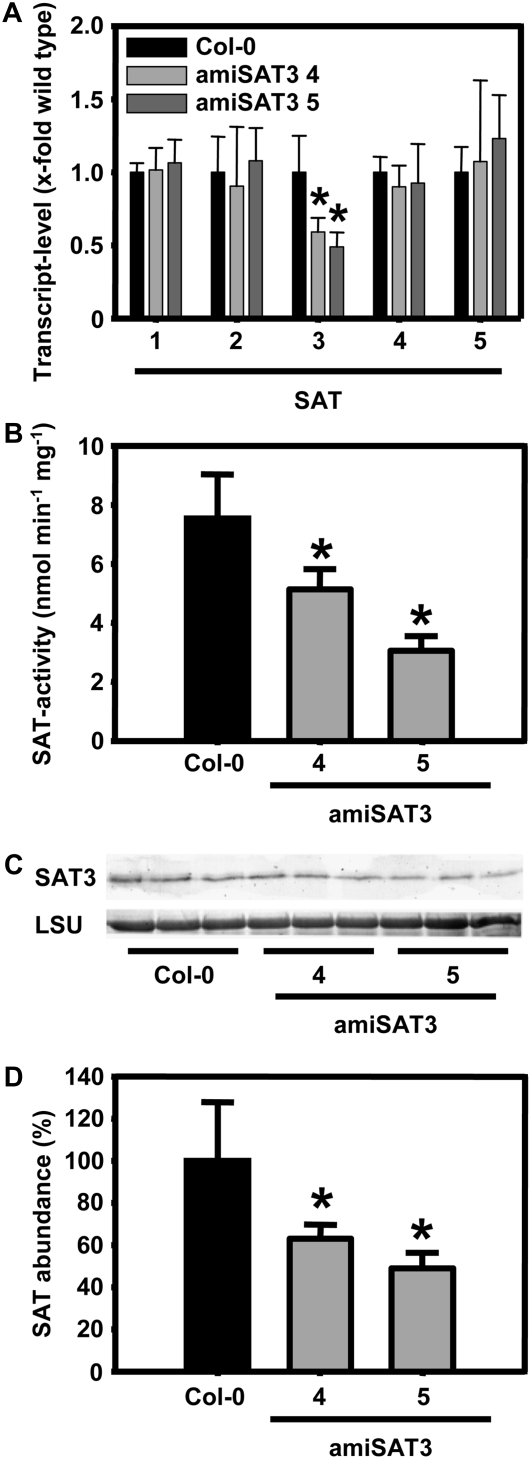
The amiSAT3 lines 4 and 5 were characterized in detail. First, the expression pattern of the Sat gene family was compared with the pattern in wild-type plants by quantitative real-time PCR. Lines 4 and 5 showed approximately 50% of wild-type Sat3 mRNA levels but no changes in abundance of Sat1, Sat2, Sat4, and Sat5 mRNA (Fig. 3A). This pattern was independently confirmed by microarray analyses of amiSAT3 line 5 (see below). These findings confirmed the specificity of the amiRNA approach and demonstrated that no compensatory up-regulation of other Sat genes occurred in the transgenic lines. Second, SAT enzyme activities were determined from whole leaf protein extracts (Fig. 3B). The down-regulation of Sat3 mRNA was mirrored by decreases of SAT enzyme activity in the two lines. Finally, the same extent of decrease in lines 4 and 5 was observed with respect to protein levels using a SAT3-specific antiserum for immunoblotting and subsequent densitometric quantification (Fig. 3, C and D). Thus, the phenotype of growth retardation is caused by gene-specific down-regulation of the gene encoding mitochondrial SAT.
Figure 3.
Molecular and biochemical characterization of transgenic Arabidopsis amiSAT3 plants. A, Transcript levels for all members of the SAT gene family in Arabidopsis were determined by quantitative real-time PCR after extraction of total mRNA from leaves of 8-week-old plants (n > 5). Proteins were extracted from three individuals of wild-type plants (black bars) and amiSAT3 plants of lines 4 and 5 (gray bars) of the same age. B, The crude protein extracts (60–100 μg) were tested three times for SAT activity (n = 9). C and D, The abundance of mitochondrial SAT3 was analyzed (n = 9) by immunological detection (C) and quantified by densitometry (D). The large subunit of ribulose-1,5-bisphosphate carboxylase (LSU) served as a loading control after staining with Coomassie Brilliant Blue. Error bars indicate sd, and asterisks mark significant differences using the unpaired t test.
Flux of Ser into Cys and Glutathione Is Reduced in amiSAT3 Lines
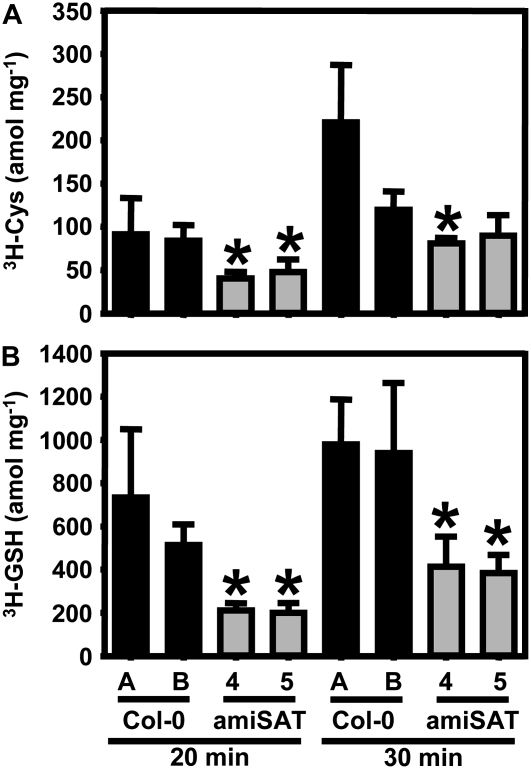
The causal relationship between reduced growth and specific down-regulation of the gene encoding mitochondrial SAT3 implies a reduced rate of OAS formation and ultimately of Cys synthesis, unless other SAT isoforms compensated for the decreased activity of the major SAT location in the cell in other ways than transcriptional increases. This assumption was tested by incubation of leaf discs in the light with radiolabeled [3H]Ser and quantification of label in Cys and glutathione (Fig. 4). Application of labeled Ser rather than sulfate was chosen to monitor flux via OAS but not of the assimilatory sulfate reduction pathway. [3H]Ser uptake of wild-type plants and amiSAT3 lines 4 and 5 was compared after 20 and 30 min, respectively. Radioactive labeling of Cys formed from [3H]OAS corresponded to half of the flux rate compared with the wild type at both time points due to down-regulation of mitochondrial SAT3 activity. An even stronger reduction of incorporation, down to 30% to 40%, was observed for glutathione. This difference suggests that, after uptake into the leaf cells, [3H]Ser was transported into the mitochondria to be used for the synthesis of [3H]OAS. This entered the cytosol to serve for the synthesis of Cys, which finally moved into the chloroplasts as substrate for the initial step of glutathione formation.
Figure 4.
Incorporation of Ser into thiols in wild-type and transgenic plants with reduced mitochondrial SAT activity. Eight-week-old transgenic plants with reduced mitochondrial SAT activity (amiSAT3 lines 4 and 5; gray bars) were tested for incorporation of [3H]Ser into Cys and glutathione (GSH). Wild-type plants of the same age (Col-0 A) and of the same developmental stage (Col-0 B; 6 weeks old) as the analyzed amiSAT lines served as controls (black bars). All plants were grown on soil under the same conditions as plants used for analysis of metabolites, transcript levels, and protein abundance. Samples of three individuals of wild-type and transgenic plants were taken and analyzed for incorporation of the radioactive label from [3H]Ser into thiols after 20 and 30 min according to Heeg et al. (2008). Error bars indicate sd, and asterisks mark significant differences from both wild types using the unpaired t test.
It is concluded that decreased mitochondrial SAT activity has a profound effect on flux into Cys and glutathione due to reduced overall OAS synthesis. As expected from the constant expression levels of the four unaffected SAT genes in Arabidopsis, no significant flux compensation could be observed, since the reductions in Sat3 mRNA and SAT3 protein levels were in the same range as the reductions of incorporation rates into Cys. Furthermore, these matching quantitative molecular and metabolic changes due to amiSAT3 expression imply that the contributions of the potentially important plastid SAT1 and cytosolic SAT5 to cellular OAS synthesis are rather small in vivo and by far exceeded by mitochondrial SAT3.
Steady-State Levels of Sulfur-Related Metabolites
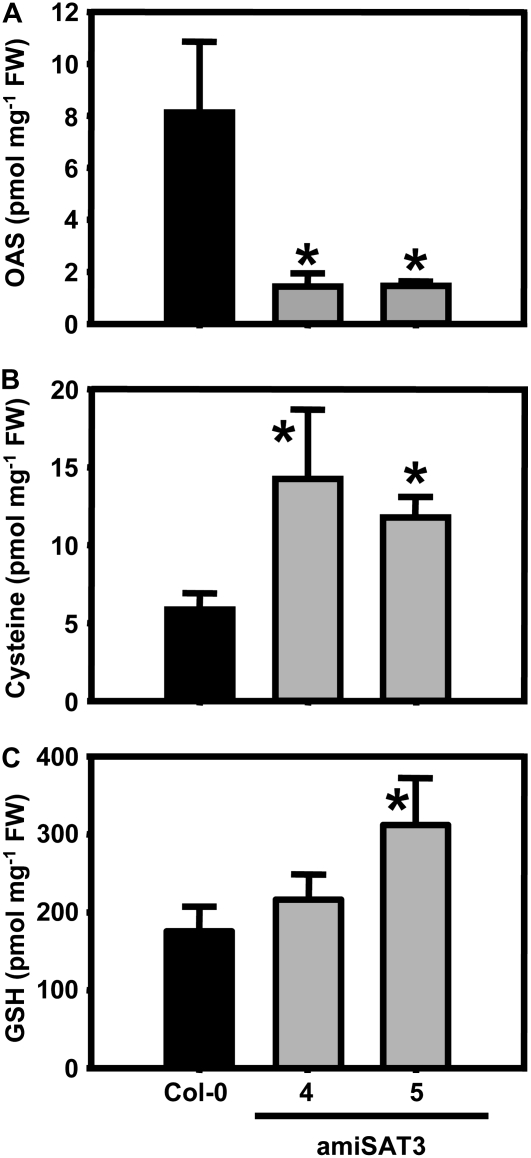
The overall cellular contents of major metabolites of primary sulfur metabolism were determined in leaves. OAS concentrations in lines 4 and 5 were decreased by 82% ± 2% and 83% ± 6% compared with the wild type and confirmed the down-regulation of the flux rate (Fig. 5A). Remarkably, steady-state levels of Cys were doubled and those of glutathione increased up to 1.5-fold compared with wild-type levels (Fig. 5, B and C). Therefore, despite strongly reduced rates of synthesis, the free concentration of both thiols was maintained, at least at the whole cell level.
Figure 5.
OAS and thiol contents in leaves of wild-type and transgenic Arabidopsis plants. Rosette leaves of three Arabidopsis wild-type plants (black bars) and three individuals of each independent transgenic plant line with reduced mitochondrial SAT activity (gray bars) were analyzed for OAS (A), Cys (B), and glutathione (GSH; C) after extraction of metabolites with 0.1 m hydrochloric acid. Error bars indicate sd, and asterisks mark significant differences using the unpaired t test. FW, Fresh weight.
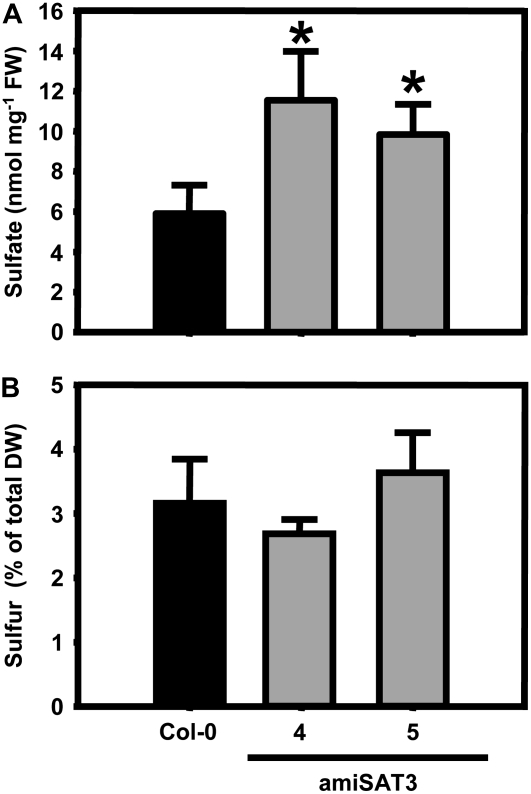
Also, the content of sulfate was significantly increased in the amiSAT3 lines (Fig. 6A). It should be noted that phosphate contents increased 1.5- and 2-fold, respectively, in amiSAT lines 4 and 5, but not the levels of nitrate (Supplemental Fig. S2). Despite elevated sulfate levels, the total sulfur contents of these lines were not affected (Fig. 6B), as were the total carbon and nitrogen contents (Supplemental Fig. S3). The significant increase of sulfate in combination with unchanged levels of total sulfur levels point toward a deregulation of the ratio between oxidized and reduced sulfur in amiSAT3 lines.
Figure 6.
Sulfate and total sulfur contents in leaves of wild-type and transgenic Arabidopsis plants. A, Rosette leaves of three Arabidopsis wild-type plants (black bars) and three individuals of each independent transgenic plant line with reduced mitochondrial SAT activity (gray bars) were analyzed for sulfate contents. FW, Fresh weight. B, The total sulfur content in dried rosette leaves of 8-week-old wild-type plants (black bars) and the amiSAT3 lines 4 and 5 (gray bars) was analyzed using an elemental analyzer (n = 4). Error bars indicate sd, and asterisks mark significant differences using the unpaired t test. DW, Dry weight.
Impact of Reduced OAS Synthesis on Expression of Sulfur Metabolism-Related Genes
The transcriptome response of amiSAT3 lines was investigated with an array containing 912 genes designed for the detection of even small changes of mRNA abundance, as is often observed with genes of primary metabolism. Sixty genes with different expression intensity by robust constitutive patterns according to Czechowski et al. (2005) were included to optimize the evaluation of all genes of sulfur metabolism and key regulatory genes of other metabolic pathways, as well as stress response, redox homeostasis, hormone metabolism, and membrane transport. In addition, 152 candidate genes were included, which are reported to be regulated in response to sulfur availability (Hirai et al., 2003; Higashi et al., 2006; Maruyama-Nakashita et al., 2006). The whole list of investigated genes is shown in Supplemental Table S1.
Based on three biological repetitions of the wild type and amiSAT3 line 5 with four technical replicates, each including dye swaps for each set, 26 genes were found to be significantly up- or down-regulated according to P values lower than 0.05 (Table I). Most of the significantly regulated genes (eight) belong to the category of sulfur metabolism, while a lower number of redox-related genes (six) and genes involved in hormone function (five), primary metabolism (four), and stress response (three) were also found to be significantly altered in abundance.
Table I.
Genes regulated in response to reduced mitochondrial SAT activity
The list comprises all genes that were regulated significantly (P < 0.05) in response to the down-regulation of mitochondrial SAT. The list of all tested genes is deposited as Supplemental Table S1. The annotation of genes and grouping by functional category is according to The Arabidopsis Information Resource (www.arabidopsis.org).
| Gene ID | Annotation | Change | P | Category |
|---|---|---|---|---|
| At5g07000 | ST2b, sulfotransferase | +1.45 | 0.022 | Sulfur metabolism |
| At5g27380 | GSH2, glutathione synthetase | +1.28 | 0.017 | Sulfur metabolism |
| At5g43780 | APS4, ATP-sulfurylase 4 | −1.20 | 0.041 | Sulfur metabolism |
| At5g43850 | ARD4, acireductone dioxygenase | −1.23 | 0.048 | Sulfur metabolism |
| At5g61420 | Myb28, transcription factor | −1.25 | 0.028 | Sulfur metabolism |
| At2g47730 | GSTF8, φ-class glutathione transferase | −1.45 | 0.028 | Sulfur metabolism |
| At1G62180 | APR2, APS reductase 2 | −1.78 | 0.028 | Sulfur metabolism |
| At3G13110 | SAT3, Ser acetyltransferase 3 | −2.01 | 0.008 | Sulfur metabolism |
| At2g32880 | TRAF homology domain-containing protein | +1.33 | 0.006 | Redox related |
| At3g24170 | GR1, cytosolic GSH reductase | −1.27 | 0.047 | Redox related |
| At1g75300 | Isoflavone reductase | −1.34 | 0.027 | Redox related |
| At5g21100 | l-Ascorbate oxidase | −1.55 | 0.004 | Redox related |
| At1g10960 | FD1, ferredoxin | −2.03 | 0.037 | Redox related |
| At1g64500 | Glutaredoxin family protein | −2.08 | 0.005 | Redox related |
| At5g07010 | ST2a, hydroxyjasmonate sulfotransferase | +2.66 | 0.001 | Hormone function |
| At1g75750 | GASA1, GA regulated | +1.87 | 0.003 | Hormone function |
| At5g05690 | CPD, cytochrome P450 monooxygenase | +1.34 | 0.022 | Hormone function |
| At5g15230 | GASA4, GA regulated | +1.27 | 0.035 | Hormone function |
| At4g39800 | MIPS, myoinositol-1-P synthase | −2.32 | 0.006 | Hormone function |
| At2g18700 | TPS11, trehalose-6-P synthase | +1.67 | 0.006 | Primary metabolism |
| At4g39330 | CAD9, cinnamyl alcohol dehydrogenase | −1.30 | 0.027 | Primary metabolism |
| At3g19450 | CAD4, cinnamyl alcohol dehydrogenase | −1.39 | 0.022 | Primary metabolism |
| At1g17745 | PGDH, 3-phosphoglycerate dehydrogenase | −1.57 | 0.009 | Primary metabolism |
| At3g44480 | RPP1, TIR-NB-LRR R protein | −1.43 | 0.033 | Stress response |
| At1g33960 | AIG1, avirulence-responsive protein | −1.51 | 0.041 | Stress response |
| At2g14610 | PR1, pathogen-related protein 1 | −1.74 | 0.048 | Stress response |
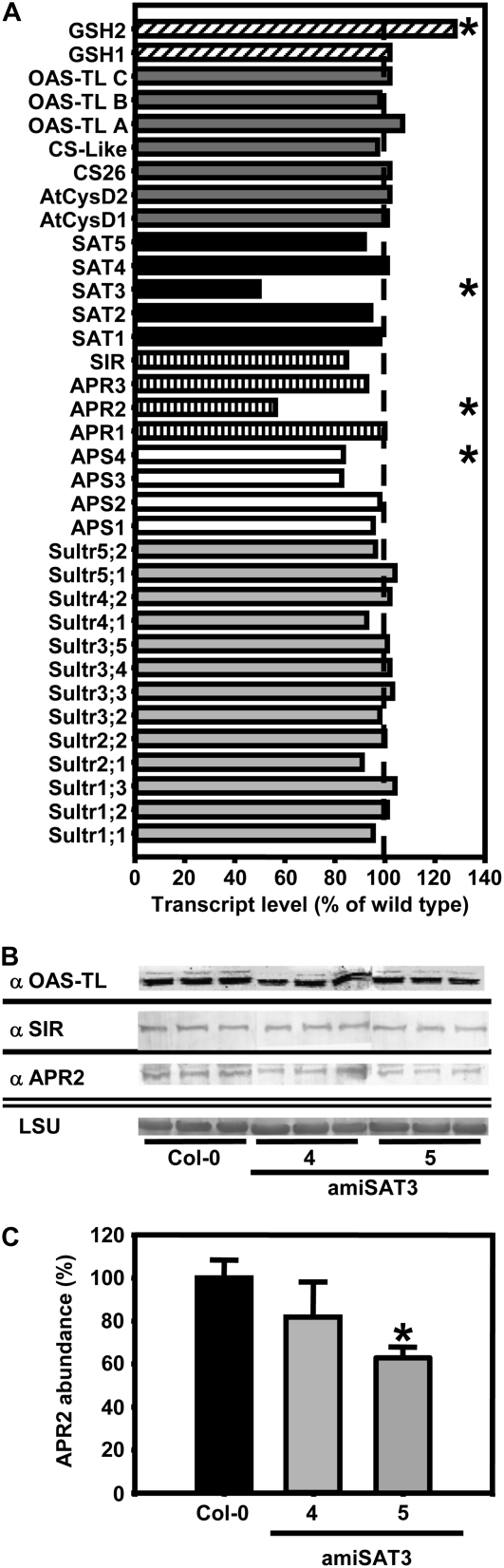
Microarray analyses of amiSAT3 line 5 confirmed the specificity and extent of down-regulation of Sat3 expression and the unchanged mRNA levels of the other four Sat genes that had been observed by real-time PCR (Fig. 3). Most interestingly, two genes upstream of SAT in assimilatory sulfate reduction were down-regulated: Aps4, encoding a ubiquitously expressed plastidic isoform of ATP-sulfurylase, which is responsible for the activation of sulfate prior to reduction (Hatzfeld et al., 2000a); and Apr2, encoding adenosine 5′-phosphosulfate reductase (APR), the major plastid isoform for the first reduction step. None of the other genes of primary sulfur metabolism was significantly affected (Fig. 7A). The mRNA data of selected genes were complemented by immunoblotting of leaf proteins. Protein contents of sulfite reductase (SIR) and OAS-TL A, B, and C were unchanged, while APR protein was decreased by 40% in amiSAT3 line 5 (Fig. 7B). Thus, all three protein species precisely reflected the mRNA contents determined by microarray analysis, although it should be noted that the APR2 antiserum can probably cross-react with all three APR isoforms (Koprivova et al., 2000).
Figure 7.
Protein abundances and transcript levels of sulfur metabolism-related genes in leaves of transgenic Arabidopsis plants. A, The transcript levels of sulfur metabolism-related genes in leaves of amiSAT3 line 5 and wild-type Arabidopsis plants were compared using a targeted microarray approach. Total mRNA was extracted form three individuals of each plant line, labeled independently two times with Cy3 and Cy5, and cohybridized with the microarray two times (n = 11). The transcript levels of sulfate transporters (gray bars), ATP-sulfurylases (white bars), sulfate-reducing genes (dashed bars), SATs (black bars), OAS-TLs (dark gray bars), and genes participating in glutathione (GSH) synthesis (inclined dashed bars) in amiSAT3 line 5 are shown as percentages of wild-type levels. B, Proteins were extracted from three individuals of wild-type plants and amiSAT3 lines 4 and 5 of the same age. The abundances of OAS-TL A, B, and C, SIR, and APR2 were determined by immunoblotting using specific antibodies from crude extracts (15 μg). The large subunit of ribulose-1,5-bisphosphate carboxylase (LSU) served as a loading control after staining with Coomassie Brilliant Blue. C, The reduction of APR2 protein in amiSAT3 lines 4 and 5 (gray bars) in comparison with wild-type plants (black bars) was quantified densitometrically. Error bars indicate sd, and asterisks mark significant differences using the unpaired t test.
Only two genes out of the 152 candidate genes known for sulfur-dependent regulation (Hirai et al., 2003) were found to be significantly regulated in amiSAT3 line 5. Both genes, At1g33960 (avirulence-responsive protein) and At1g75300 (putative isoflavone reductase), are known to be transcriptionally regulated also by other environmental factors or hormone stimuli. Especially At1g75300 is regulated more strongly by methyl jasmonate (MJ) than by sulfur availability (www.genevestigator.ethz.ch). In this respect, it is noteworthy that the gene with the highest up-regulation (2.6-fold) is ST2a, a hydroxyjasmonate sulfotransferase (At5g0710), which is supposed to inactivate MJ by sulfonation (Gidda et al., 2003). Reponses to disease and pathogen attack have been known for a long time to be coordinated by MJ. All significantly regulated genes of the category stress response in this experiment (PR1, AIG1, and PRP1) are involved in responses to diseases or pathogens. In agreement with the idea of inactivation of MJ by induction of ST2a, all of them are down-regulated. Besides genes responding to MJ, two genes (GASA1 and GASA4) that were known as markers for GA stimulus are induced. Taken together, the expression data point toward a down-regulation of the sulfate assimilation pathway in amiSAT3 lines in order to coordinate plastid sulfate reduction with the reduced synthesis of OAS in mitochondria. Additionally, an inactivation of stress-responsive genes was observed, which could be a result of perturbed hormone metabolism.
DISCUSSION
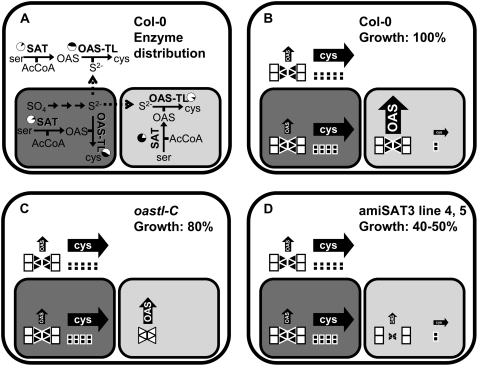
The subcellular organization of Cys synthesis in plants differs strongly from that in other eukaryotes. Why plant SAT and OAS-TL form Cys in the cytosol, plastids, and mitochondria, the reasons for their strongly different activity ratios, and whether this distribution reflects redundancy or specific functions remained enigmatic (Fig. 8, A and B). With respect to the second step of Cys synthesis, recent analyses of the OAS-TL protein family members in Arabidopsis using T-DNA insertion lines indicated redundancy as well as compartment-specific functions (Heeg et al., 2008; Lopez-Martin et al., 2008; Watanabe et al., 2008). In nonstressed plants, cytosolic OAS-TL turned out to be the most important isoform in terms of enzymatic activity (Heeg et al., 2008; Watanabe et al., 2008). However, a mitochondrial OAS-TL C mutant showed a mild growth phenotype despite a measurable OAS-TL C contribution of less than 5% of total activity. On the other hand, this little OAS-TL C activity alone was sufficient for comfortable growth under nonstress conditions in a double mutant without cytosolic OAS-TL A and plastid OAS-TL B (Heeg et al., 2008). At the same time, these mutants allowed us to conclude that Cys must be exchangeable between cytosol and organelles, at least in Arabidopsis, making an earlier hypothesis of a lack of such exchange as reason for the three sites of Cys very unlikely (Lunn et al., 1990). Feeding experiments with [35S]sulfate to oastl mutants furthermore confirmed the importance of the second step of Cys synthesis in the cytosol. The explanation for these findings was that the rate of formation of the reaction intermediate OAS in mitochondria dominates the overall rate of Cys synthesis in the cell (Fig. 8C; Heeg et al., 2008).
Figure 8.
Organization of Cys biosynthesis pathways in plant cells and transgenic Arabidopsis plants with reduced mitochondrial SAT activity. A, Schematic overview of enzymatic activities and metabolites involved in synthesis of Cys in the cytosol (white panel), the mitochondrion (light gray panel), and the chloroplast (dark gray panel). Enzymes are indicated in boldface letters. Black fields of pie charts show the ratio of enzymatic activities present inside the respective compartment as percentages of total extractable activity calculated from subcellular fractionation experiments with pea and spinach leaves (Droux, 2004). Full arrows represent enzymatic reactions, while dashed arrows indicate transport of metabolites across membranes. B, Relative fluxes of OAS and Cys (indicated by the sizes of filled arrows) in different subcellular compartments as deduced from the enzymatic activities of SAT and OAS-TL. Most likely, hexameric SAT (triangles) is associated with two OAS-TL dimers (cubes) to form the CSC. Black filling of triangles and cubes indicates the active form of enzyme, while white filling indicates inactivated or less active enzyme. The impacts on growth rate and subcellular fluxes of metabolites as a consequence of disturbed CSC formation in the mitochondria of OAS-TL C knockout plants (oastlC) and specific down-regulation of mitochondrial SAT activity in amiSAT3 plants are shown in C and D, respectively. The sizes of triangles in C and D represent the amount of SAT with respect to the wild-type level (B).
If this assumption was correct, one has to conclude that plastids and mitochondria provide most of the substrates sulfide and OAS, respectively, while the majority of Cys synthesis actually takes place in the cytosol. The experimental down-regulation of mitochondrial SAT activity in vivo, therefore, should result in reduced formation of OAS and Cys. Indeed, targeting of mitochondrial SAT3 by amiRNA resulted in reduced OAS contents, reduced rates of Cys synthesis, and reduced growth rates. Importantly, the degree of down-regulation of Sat3 mRNA, SAT3 protein levels, and overall SAT enzyme activity quantitatively matched the lowered OAS levels, Cys synthesis rates, and growth reduction. The amiRNA approach was shown by two independent methods to be specific with respect to the expression levels of the other four Sat genes. This demonstrates that mitochondrial SAT3 activity is indeed responsible for the bulk of OAS production under these conditions, and since SATs in the other compartments showed no compensation for the reduction, this isoform limited Cys production, as shown by retarded growth (Fig. 8D).
This strong dependence of total cellular Cys production on the pacemaking role of mitochondrial synthesis of OAS is unexpected, because until now cytosol and plastids were believed to be the major production sites of both OAS and Cys (Urano et al., 2000; Droux, 2003). In this respect, numerous transgenic approaches aimed to enhance Cys synthesis in these two compartments by overexpression of SAT or OAS-TL indeed achieved moderate 1.5- to 3-fold increases of total Cys and glutathione contents (Blaszczyk et al., 1999; Harms et al., 2000; Noji and Saito, 2002; Wirtz and Hell, 2003, 2007; Sirko et al., 2004). The mitochondria have not been a target for overexpression of SAT or OAS-TL until now.
The cytosol was assumed to be the major site for regulation of OAS synthesis as a consequence of the strong feedback inhibition of cytosolic SAT isoforms by Cys (50% inhibition of initial activity at 2–10 μm Cys; Noji et al., 1998; Saito, 2000; Urano et al., 2000). Organelle-localized SAT isoforms showed much less feedback inhibition by Cys in several higher plants species (Saito et al., 1995; Noji et al., 1998; Saito, 1998). Indeed, overexpression of less feedback-inhibited SAT isoforms compared with stronger feedback-inhibited SATs in the cytosol of transgenic plants resulted in a higher increase of Cys steady-state levels in Arabidopsis and tobacco (Nicotiana tabacum; Blaszczyk et al., 1999; Wirtz and Hell, 2007). On the other hand, the estimated cytosolic Cys steady-state levels in wild-type plants led to the question: is cytosolic feedback-inhibited SAT active at in vivo concentrations of Cys (Saito, 1998; Wirtz and Hell, 2006)? The SAT of the enterobacterium Escherichia coli has similar inhibition constant values for Cys (1 μm) compared with strongly feedback-inhibited plant SATs, but it is enzymatically almost completely inactive during normal growth (Kredich and Becker, 1971). In the case of E. coli, such a strict down-regulation makes sense due to the lifestyle of E. coli, which supports the organism with exogenous Cys. It remains unclear why cytosolic SAT, which contributes less than 10% of the total SAT activity (Ruffet et al., 1995), would be potently inhibited by Cys. Possible explanations for the presence of this SAT could be the production of OAS in this compartment during a stress situation when cytosolic Cys pools are depleted, or perhaps its role is not the synthesis of OAS but instead the formation of the CSC complex. A regulatory model proposes the reversible dissociation of the CSC in response to OAS and sulfide as a sensor of the actual sulfur supply in a subcellular compartment and allows the regulation of SAT activity in response to the actual demand for OAS for the production of Cys (Wirtz and Hell, 2006). In this model, the function of the feedback-inhibited cytosolic SAT would be signal recognition of OAS and sulfide supply of the cytosol rather than production of OAS. The latter scenario would provide an elegant explanation for the presence of SAT in all three compartments, although total Cys synthesis depends almost entirely on the synthesis of OAS in mitochondria, according to the results presented here.
It is remarkable that radiolabel from [3H]Ser feeding appears in glutathione. This implies an uptake of Ser into mitochondria, formation of OAS, transport to the cytosol for Cys synthesis, and import into plastids to serve as substrate of γ-glutamylcysteine, the precursor of glutathione that is exclusively formed in plastids in Arabidopsis (Wachter et al., 2005). The possibility of growth retardation in amiSAT3 lines due to deficiency of Cys in the mitochondria themselves can be excluded, since Cys can be imported into mitochondria from the cytosol (Heeg et al., 2008). Consequently, the retarded growth phenotype must be a consequence of reduced total Cys synthesis rates in the cell, although cytosolic SAT5 and plastid SAT1 are still present.
OAS contents were decreased in Arabidopsis amiSAT3 lines 4 and 5, but surprisingly, steady-state levels of Cys and glutathione were increased despite lowered flux into these thiols. Maintenance of Cys and glutathione contents was also observed in the Arabidopsis oastlC T-DNA mutant line, which lacked mitochondrial OAS-TL C and showed reduced growth (Heeg et al., 2008). In this respect, the different functions of OAS and thiols for cell metabolism have to be considered: OAS functions as reaction intermediate and potential signal for gene expression during sulfate starvation (Kim et al., 1999; Hirai et al., 2003), Cys is essential for protein biosynthesis and as a donor of reduced sulfur in numerous reactions (Hell and Bergmann, 1990; Pilon-Smits et al., 2002; Picciocchi et al., 2003; Wirtz and Droux, 2005), and glutathione is an integral component of redox homeostasis (Meyer and Hell, 2005; Meyer et al., 2007). It is conceivable that the maintenance of thiol levels for these reasons is a prerequisite for the proper function of cell metabolism. Nonetheless, this would not fully explain the elevation of Cys steady-state levels. More likely, in amiSAT3 lines, Cys steady-state levels are up-regulated as a signal to serve as a signal to adjust cell metabolism to reduced fluxes of OAS.
The microarray analysis of leaves of the Arabidopsis amiSAT3 line 5 in comparison with wild-type plants revealed that none of the OAS-TL genes and none of the genes of sulfate uptake and primary sulfur metabolism were changed in expression, with the exception of one member of the Aps and Apr gene family each. Down-regulation of APR2 protein was confirmed by immunoblotting. Reduced activation and reduction of sulfate would explain the increased contents of sulfate in amiSAT3 lines. Especially the strong down-regulation of APR2 could be of particular importance, since lower APR2 activity in leaves was shown to be responsible for the higher sulfate content of the Arabidopsis ecotype Shahdara in comparison with ecotype Bay-0 (Loudet et al., 2007). High levels of Cys as well as glutathione were shown to down-regulate genes for sulfate activation and reduction in roots of several higher plants (Koprivova et al., 2000; Vauclare et al., 2002). It seems conceivable, therefore, that the high steady-state levels of Cys and glutathione in amiSAT3 lines 4 and 5 are needed to efficiently down-regulate sulfate reduction in order to avoid the accumulation of toxic sulfide, which may not be entirely scavenged by Cys synthesis in the transgenic lines. In agreement with this hypothesis, sulfate accumulates 2-fold in amiSAT3 lines 4 and 5 in comparison with the wild type.
Further putative signals for the down-regulation of APR2 are OAS and MJ. It has been repeatedly observed that OAS, possibly due to the dissociation of the CSC, acts as a signal for the entire sulfur assimilation pathway. Feeding of OAS regulates more than 650 and 850 genes in leaves and roots of Arabidopsis, respectively (Hirai et al., 2003). At least in roots of Arabidopsis, all APR isoforms are up-regulated in response to exogenous application of OAS (Koprivova et al., 2000). Induction of APR by application of OAS was also shown in Lemna minor, indicating that regulation of APR by OAS is a general mechanism in higher plants (Kopriva et al., 2002). Thus, the 4-fold lowered OAS and doubled thiol steady-state levels in amiSAT3 lines may, independently or in combination, act as signals for the repression of Apr2 in amiSAT3 lines. MJ has been shown to modulate expression levels of sulfate assimilation-related genes (Jost et al., 2005; Sasaki-Sekimoto et al., 2005). According to microarray analysis of the amiSAT3 line 5 ST2a, a hydroxyjasmonate sulfotransferase, which is supposed to inactivate jasmonic acid signals (Gidda et al., 2003), is one of the strongest responding genes. In agreement with such a possible mechanism, several genes of the stress response to pathogens and diseases were down-regulated and none was up-regulated on the microarray in this study. Therefore, a diminished octadecanoic acid signal may have contributed to the reduced Aps4 and Apr2 expression.
CONCLUSION
Specific down-regulation of mitochondrial SAT3 activity in Arabidopsis by amiRNA expression demonstrated that mitochondria, but not chloroplasts or the cytosol, are the dominant source of OAS in vivo. Despite strongly reduced flux into Cys in amiSAT3 plants, the steady-state levels of Cys and glutathione were elevated and presumably contributed to the repression of assimilatory sulfate reduction and accumulation of free sulfate. This, together with previous evidence, strongly suggests that OAS from mitochondria and sulfide from chloroplasts serve as substrates for the bulk of Cys synthesis in the cytosol.
MATERIALS AND METHODS
General Cloning
Standard molecular biology technologies like growth of bacteria, plasmid isolation, and PCR were performed as described by Sambrook et al. (1989) according to GLP standards. The amiRNA-SAT3-specific sequences were selected with the WMD 2 - Web MicroRNA Designer (www.weigelworld.org). The amiRNA of SAT3 was constructed using an overlapping PCR approach with primers P1 (5′-gaTAAAATACAAGTCCCAGCCCCtctctcttttgtattcc-3′), P2 (5′-gaGGGGCTGGGACTTGTATTTTAtcaaagagaatcaatga-3′), P3 (5′-gaGGAGCTGGGACTTCTATTTTTtcacaggtcgtgatatg-3′), P4 (5′-gaAAAAATAGAAGTCCCAGCTCCtctacatatatattcct-3′), pRS300-A (5′-CTGCAAGGCGATTAAGTTCGGTAAC-3′), and pRS300-B (5′-GCGGATAACAATTTCACACAGGAAACAG-3′) according to Schwab et al. (2006), which resulted in the amplification of the amiRNA loop structure, the SAT-specific sequence, and terminal restriction sites for KpnI and BamHI at the 5′ and 3′ ends, respectively. The resulting PCR product was digested with KpnI and BamHI and cloned in the plant transformation vector pBINAR under the control of the cauliflower mosaic virus 35S promoter, resulting in pBINAR-amiSAT3 (Supplemental Fig. S1). The following cDNA was employed in this study: AtSAT3 (X80938; Bogdanova et al., 1995).
Plant Growth and Transformation
Transformation of Agrobacterium tumefaciens C58 with binary vectors and subsequent transformation and selection of Arabidopsis (Arabidopsis thaliana Col-0) were carried out as described by Clough and Bent (1998). Wild-type plants and plants expressing amiRNA-SAT3 (F1 or F2 generation) were grown in climate chambers in growth medium containing one-half soil and one-half substrate 2 (Klasmann-Deilmann) under controlled conditions: 8.5 h of light, 100 μE light photon flux density, 24°C and 18°C at day and night, respectively, and 50% humidity. Rosette leaves of each 8-week-old plant were pooled and used to analyze metabolites as well as the expression of sulfur-related genes at the transcript and protein levels.
Determination of Metabolites
Hydrophilic metabolites were extracted from leaves of Arabidopsis plants according to Wirtz and Hell (2003). Thiols, OAS, anions, and total contents of carbon, nitrogen, and sulfur were quantified as described by Heeg et al. (2008) in order to ensure the comparability of data sets.
Determination of Enzyme Activities
Total soluble proteins were isolated with 0.5 mL of 50 mm HEPES, pH 7.4, 10 mm KCl, 1 mm EDTA, 10% glycerin, 30 mm dithiothreitol, and 0.5% phenylmethylsulfonyl fluoride from 0.2 g of leaf material that was ground to a fine powder in liquid nitrogen. Cell debris was removed by centrifugation at 16,000g and 4°C for 10 min. Proteins were quantified as described by Bradford (1976) using bovine serum albumin as a standard. The enzymatic activities of SAT and OAS-TL were determined according to Nakamura et al. (1987): SAT activity was assayed by coupling to the OAS-TL reaction. All SAT activity determinations were supplemented with 2 units of purified recombinant OAS-TL (Wirtz et al., 2004) to ensure high excess of OAS-TL activity during coupling of both reactions.
Immunological Detection of Proteins
Total proteins from leaves were separated according to Laemmli (1970) by discontinuous SDS-PAGE in Mini-Protean II cells (Bio-Rad). Transfer of proteins, washing, and development of signals were performed as described by Heeg et al. (2008) using the alkaline phosphatase-anti-rabbit IgG conjugate (Promega; 1:10,000) detection of SAT and OAS-TL proteins. APR and SIR were detected with primary antibodies (1:2,000; incubation at 4°C overnight) after blocking of the nitrocellulose membrane with 3% milk powder. The primary antibodies were detected with a 1:5,000 dilution of the above-mentioned secondary antibody. The resulting signals were quantified by densitometry using the INTAS gel documentation system in combination with Gel-Pro Express 4.0 software (INTAS).
Determination of Incorporation Rates Using 3H-Labeled Ser
Twelve hours prior to the labeling experiment, soil-grown plants were enclosed in transparent plastic bags to ensure opening of the stomata. Leaf pieces (30 mg) were cut out from the interveinal fields of leaves and floated on the labeling solution (half-strength Hoagland medium containing 2.5 μm [3H]Ser [185–925 GBq mmol−1; Hartmann Analytic]) on a horizontal shaker with 60 rpm in the light (17 μE). Samples were taken after 20 and 30 min, washed twice with half-strength Hoagland medium [2.5 mm Ca(NO3)2, 2.5 mm KNO3, 0.5 mm MgSO4, 0.5 mm KH2PO4, 40 mm Fe-EDTA, 25 mmH3BO3, 2.25 mm MnCl2, 1.9 mm ZnSO4, 0.15 mm, CuSO4, and 0.05 mm (NH4)6MO7O24, pH 5.9], and frozen in liquid nitrogen. A negative control for contamination of the leaf surface with [3H]Ser was taken by dipping leaf pieces into the labeling solution for 1 s, prior to washing and harvesting like the samples. The samples were powdered using the Bio101 ThermoSavant Fast Prep system (Qbiogene) according to the manufacturer's instructions. The metabolites were extracted and separated prior to quantification of the incorporated 3H label as described by Heeg et al. (2008).
Determination of Transcript Levels Using a Custom-Made Microarray
Design and Production of the Microarray
50mer oligonucleotides of 912 selected genes were synthesized by Ocimum Biosolutions with an N-terminal amino linker and checked for cross-hybridization with the Smith-Waterman alignment. Oligonucleotides were diluted to a concentration of 0.01 to 1 mm in 5× SSC (0.75 M sodium chloride and 0.075 M sodium citrate) containing 2.5 m betaine and spotted on epoxy-coated NexterionE slides (Peqlab) with a MicroGRid robot (Biorobotics) using SMP3 pins (TeleChem International). A microarray contains all probes in four replicates, while the assembly of replicates on the microarray was randomized to avoid positional effects.
Sample Preparation, Hybridization, and Evaluation
Fluorescently labeled cDNA samples were prepared from 10 μg of total RNA that had been isolated from Arabidopsis leaf tissue with the RNeasy Plant Kit (Qiagen) according to the manufacturer's protocol. Routinely, Cy3/Cy5-dCTP was directly incorporated during first-strand synthesis according to The Institute for Genomic Research direct labeling protocol (www.tigr.org). The labeled cDNA probe was purified using the QIAquick Purification Kit (Qiagen) according to the manufacturer's instructions, and the incorporation rate of label was determined by photometric measurement. A Cy3-labeled wild-type cDNA sample and a Cy5-labeled amiSAT3 cDNA sample were mixed after concentration of each sample in a vacuum concentrator to a final volume of 20 μL. The resulting sample was immediately denatured by the addition of 20 μL of prewarmed hybridization buffer h (5× SSC, 0.1% [v/v] SDS, 0.06% [w/v] DNA from fish sperm, MB grade, and 40% [v/v] formamide) followed by incubation at 100°C for 5 min. The microarray was prehybridized with buffer p (5× SSC, 0.2% SDS, and 0.5% bovine serum albumin) for 45 min and washed three times with 250 mL of double-distilled water, before the denatured sample was applied and incubated for 20 h at 50°C in a Slide Booster (Advalytix). The hybridized microarray was washed at 30°C by subsequently applying buffer w1 (2× SSC and 0.5% SDS), w2 (0.2% SSC and 0.5% SDS), and w3 (0.1% SSC) for 10 min each. Signals were detected using the ScanArray5000 confocal laser scanner (Packard BioChip Technologies) and quantified with the GenePix Pro 4.1 analysis software (Axon Instruments). Data quality assessment, normalization, and correspondence cluster analysis were performed with the analysis and data warehouse software M-CHiPS (DKFZ) according to Fellenberg et al. (2001; www.mchips.org). Signal intensities of repeated hybridizations were normalized by the majority of spots, and the minimal signal intensity was at least above twice the sd of the background signal.
Determination of Transcript Levels by Real-Time PCR
Total RNA from leaf tissue was extracted with the RNeasy Plant Kit (Qiagen) according to the manufacturer's protocol. Total RNA was transcribed into cDNA and analyzed by quantitative real-time PCR as described by Talke et al. (2006). The following primers were used for specific amplification of actin7 and all members of the SAT gene family from Arabidopsis: SAT1-f (5′-CACATGCCGAACCGGTAATAC-3′), SAT1-r (5′-GGTGAATCTTCCGGTTTACAGAGA-3′), SAT2-f (5′-ACGCTAAGGGAACTCATAAGTCAGA-3′), SAT2-r (5′-TCTTCTCTTATAGCATCCCAAATAGGA-3′), SAT3-f (5′-AATGGAACCCAGACCAAAACC-3′), SAT3-r (5′-GCCCAAACATCATCGACTTCA-3′), SAT4-f (5′-CTCTTCCAATGATTGTCTCCCG-3′), SAT4-r (5′-CCTCTCGAAAGGAAACTCGTCA-3′), SAT5-f (5′-TGGACACAGATCAAGGCGG-3′), SAT5-r (5′-ATGAGAAAGAATCGTCGAATATAGATAGC-3′), actin7-f (5′-CAACCGGTATTGTGCTCGATTC-3′), and actin7-r (5′-GAGTGAGTCTGTGAGATCCCG-3′).
Statistical Analysis
Regression analyses of data sets were performed with SigmaPlot 8.0 that uses the Marquardt-Levenberg algorithm for determination of independent variables. Comparison of means from different sets of data was analyzed for statistical significance with the unpaired t test. Constant variance and normal distribution of data were carefully checked with SigmaStat 3.0 prior to statistical analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Specificity of the amiRNA-SAT3 approach.
Supplemental Figure S2. Nitrate and phosphate contents in leaves of wild-type and transgenic Arabidopsis plants.
Supplemental Figure S3. Carbon and nitrogen contents in leaves of wild-type and transgenic Arabidopsis plants.
Supplemental Table S1. List of genes that were analyzed by the targeted microarray approach.
Supplementary Material
Acknowledgments
We are indebted to Dr. S. Kopriva (John Innes Institute, Norwich, UK) and Prof. Dr. D. Weigel (Max Planck Institute, Tübingen, Germany) for the kind gifts of the APR2 antiserum and the pRS300 vector, respectively. We thank S. Hassel (University of Heidelberg) for excellent technical assistance, Dr. Maria Bernal (University of Heidelberg) for support of quantitative real-time PCR analysis of the SAT gene family, and Dr. A. Meyer (University of Heidelberg) for critically reading the manuscript.
This work was supported by the Stiftung der deutschen Wirtschaft and the Schmeil-Foundation, Heidelberg (grants to F.F.H.), and by the Bioquant Landesgraduiertenkolleg Baden-Württemberg (grant to C.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rüdiger Hell (rhell@hip.uni-heidelberg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R (2002) Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem 277 30629–30634 [DOI] [PubMed] [Google Scholar]
- Blaszczyk A, Brodzik R, Sirko A (1999) Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J 20 237–243 [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Bork C, Hell R (1995) Cysteine biosynthesis in plants: isolation and functional identification of a cDNA encoding a serine acetyltransferase from Arabidopsis thaliana. FEBS Lett 358 43–47 [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Hell R (1997) Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11 251–262 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M (2003) Plant serine acetyltransferase: new insights for regulation of sulphur metabolism in plant cells. Plant Physiol Biochem 41 619–627 [Google Scholar]
- Droux M (2004) Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynth Res 79 331–348 [DOI] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D (1998) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem 255 235–245 [DOI] [PubMed] [Google Scholar]
- Fellenberg K, Hauser NC, Brors B, Neutzner A, Hoheisel JD, Vingron M (2001) Correspondence analysis applied to microarray data. Proc Natl Acad Sci USA 98 10781–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem 278 17895–17900 [DOI] [PubMed] [Google Scholar]
- Harms K, von Ballmoos P, Brunold C, Hofgen R, Hesse H (2000) Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of cysteine and glutathione. Plant J 22 335–343 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Lee S, Lee M, Leustek T, Saito K (2000. a) Functional characterization of a gene encoding a fourth ATP sulfurylase isoform from Arabidopsis thaliana. Gene 248 51–58 [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K (2000. b) β-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol 123 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R (2008) Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R, Bergmann L (1990) γ-Glutamylcysteine synthetase in higher plants: catalytic properties and subcellular localization. Planta 180 603–612 [DOI] [PubMed] [Google Scholar]
- Hell R, Jost R, Berkowitz O, Wirtz M (2002) Molecular and biochemical analysis of the enzymes of cysteine biosynthesis in the plant Arabidopsis thaliana. Amino Acids 22 245–257 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Hirai MY, Fujiwara T, Naito S, Noji M, Saito K (2006) Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J 48 557–571 [DOI] [PubMed] [Google Scholar]
- Hirai M, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33 651–663 [DOI] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, Bogs J, Gershenzon J, Hähnel U, Hänsch R, Hartmann T, Kopriva S, Kruse C, et al (2005) Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res 86 491–508 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Wirtz M, Hopkins L, Hawkesford MJ, Hell R (2000) Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene 253 237–247 [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K (2005) Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol 137 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T (1999) Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 209 282–289 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Suter M, von Ballmoos P, Hesse H, Krahenbuhl U, Rennenberg H, Brunold C (2002) Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiol 130 1406–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S (2000) Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol 122 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich NM, Becker MA (1971) Cysteine biosynthesis: serine transacetylase and O-acetylserine sulfhydrylase (Salmonella typhimurium). Methods Enzymol 197: 459–471
- Kuske CR, Hill KK, Guzman E, Jackson PJ (1996) Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia Mill. Plant Physiol 112 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lopez-Martin MC, Becana M, Romero LC, Gotor C (2008) Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Plant Physiol 147 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North K, Kopriva S, Daniel-Vedele F (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39 896–900 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R (1990) Localization of ATP-sulfurylase and O-acetylserine(thiol)lyase in spinach leaves. Plant Physiol 94 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42 305–314 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Hell R (2005) Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth Res 86 435–457 [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hayama A, Masada M, Fukushima K, Tamura G (1987) Measurement of serine acetyltransferase activity in crude plant extracts by a coupled assay system using cysteine synthase. Plant Cell Physiol 28 885–891 [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K (1998) Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem 273 32739–32745 [DOI] [PubMed] [Google Scholar]
- Noji M, Saito K (2002) Molecular and biochemical analysis of serine acetyltransferase and cysteine synthase towards sulfur metabolic engineering in plants. Amino Acids 22 231–243 [DOI] [PubMed] [Google Scholar]
- Picciocchi A, Douce R, Alban C (2003) The plant biotin synthase reaction: identification and characterization of essential mitochondrial accessory protein components. J Biol Chem 278 24966–24975 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EA, Garifullina GF, Abdel-Ghany S, Kato S, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M (2002) Characterization of a NifS-like chloroplast protein from Arabidopsis: implications for its role in sulfur and selenium metabolism. Plant Physiol 130 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Droux M, Douce R (1992) Subcellular distribution of O-acetylserine(thiol)lyase in cauliflower (Brassica oleracea L.) inflorescence. Plant Physiol 98 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet ML, Droux M, Douce R (1994) Purification and kinetic properties of serine acetyltransferase free of O-acetylserine(thiol)lyase from spinach chloroplasts. Plant Physiol 104 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffet ML, Lebrun M, Droux M, Douce R (1995) Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. Eur J Biochem 227 500–509 [DOI] [PubMed] [Google Scholar]
- Saito K (1998) Molecular Aspects of Sulfur Assimilation and Acclimation to Sulfur Supply in Plants. Elsevier Science, Amsterdam
- Saito K (2000) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3 188–195 [PubMed] [Google Scholar]
- Saito K, Yokoyama H, Noji M, Murakoshi I (1995) Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J Biol Chem 270 16321–16326 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44 653–668 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko A, Blaszczyk A, Liszewska F (2004) Overproduction of SAT and/or OASTL in transgenic plants: a survey of effects. J Exp Bot 55 1881–1888 [DOI] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Kramer U (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y, Manabe T, Noji M, Saito K (2000) Molecular cloning and functional characterization of cDNAs encoding cysteine synthase and serine acetyltransferase that may be responsible for high cellular cysteine content in Allium tuberosum. Gene 257 269–277 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krahenbuhl U, den Camp RO, Brunold C (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31 729–740 [DOI] [PubMed] [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T (2005) Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J 41 15–30 [DOI] [PubMed] [Google Scholar]
- Warrilow AG, Hawkesford MJ (2000) Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. J Exp Bot 51 985–993 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji M, Saito K (2008) Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol 146 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M (2005) Synthesis of the sulfur amino acids: cysteine and methionine. Photosynth Res 86 345–362 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R (2004) O-Acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2003) Production of cysteine for bacterial and plant biotechnology: application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24 195–203 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2006) Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J Plant Physiol 163 273–286 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2007) Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 19 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.