Abstract
Pathogen-inducible antimicrobial defense-related proteins have emerged as key antibiotic peptides and enzymes involved in disease resistance in plants. A novel antimicrobial protein gene, CaAMP1 (for Capsicum annuum ANTIMICROBIAL PROTEIN1), was isolated from pepper (C. annuum) leaves infected with Xanthomonas campestris pv vesicatoria. Expression of the CaAMP1 gene was strongly induced in pepper leaves not only during pathogen infection but also after exposure to abiotic elicitors. The purified recombinant CaAMP1 protein possessed broad-spectrum antimicrobial activity against phytopathogenic bacteria and fungi. CaAMP1:smGFP fusion protein was localized mainly in the external and intercellular regions of onion (Allium cepa) epidermal cells. The virus-induced gene silencing technique and gain-of-function transgenic plants were used to determine the CaAMP1 gene function in plant defense. Silencing of CaAMP1 led to enhanced susceptibility to X. campestris pv vesicatoria and Colletotrichum coccodes infection, accompanied by reduced PATHOGENESIS-RELATED (PR) gene expression. In contrast, overexpression of CaAMP1 in Arabidopsis (Arabidopsis thaliana) conferred broad-spectrum resistance to the hemibiotrophic bacterial pathogen Pseudomonas syringae pv tomato, the biotrophic oomycete Hyaloperonospora parasitica, and the fungal necrotrophic pathogens Fusarium oxysporum f. sp. matthiolae and Alternaria brassicicola. CaAMP1 overexpression induced the salicylic acid pathway-dependent genes PR1 and PR5 but not the jasmonic acid-dependent defense gene PDF1.2 during P. syringae pv tomato infection. Together, these results suggest that the antimicrobial CaAMP1 protein is involved in broad-spectrum resistance to bacterial and fungal pathogen infection.
In response to microbial attack, plants activate a complex series of responses that lead to the local and systemic induction of a broad spectrum of antimicrobial defenses (Kunkel and Brooks, 2002; Kim and Martin, 2004). When induced defense responses are rapidly and coordinately triggered during a given plant-pathogen interaction, plants become resistant to diseases. These defense responses include the strengthening of mechanical barriers, the oxidative burst, and the production of antimicrobial compounds (Hammond-Kosack and Parker, 2003; Park, 2005).
Much research has focused on proteins that are specifically induced in resistant plants during infection. These proteins are referred to as PATHOGENESIS-RELATED (PR) proteins, some of which have enzymatic activities, while others have no clearly defined functions. The induced resistance response of plants to diseases correlates intimately with the accumulation of PR proteins. A number of signaling molecules, including salicylic acid (SA), ethylene, and jasmonic acid (JA), have been shown to amplify and regulate defense responses in plants during initial activation events, such as oxidative bursts or the expression of PR genes (Johnson et al., 2003; Schenk et al., 2003; Lee and Hwang, 2005).
The past decade has seen advances in understanding the activation of antimicrobial proteins in plant defense and their roles in determining the outcome of many plant-pathogen interactions (Selitrennikoff, 2001). Antimicrobial peptides and proteins have been isolated from numerous plants (Selitrennikoff, 2001) and appear to be involved in constitutive or induced resistance to various pathogens. Their modes of action are varied and include fungal cell wall degradation, membrane channel and pore formation, inhibition of DNA synthesis, damage to cellular ribosomes, and cell cycle inhibition (DeGray et al., 2001; Selitrennikoff, 2001; Shatters et al., 2006).
Virus-induced gene silencing (VIGS), a method that utilizes the RNA interference pathway to induce transient knockdown expression of endogenous plant genes, involves a homology-based degradation mechanism triggered by double-stranded RNA molecules (Dinesh-Kumar et al., 2003; Robertson, 2004; Hein et al., 2005; Scofield et al., 2005). VIGS has been especially powerful for dissecting signaling events involved in disease resistance (Liu et al., 2002; Peart et al., 2002). The tobacco rattle virus (TRV)-based VIGS has been performed for functional genetics in various solanaceous plant species, such as tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), pepper (Capsicum annuum), potato (Solanum tuberosum), and petunia (Petunia hybrida; Chung et al., 2004; Robertson, 2004; Senthil-Kumar et al., 2007). More recently, defense-related genes encoding pepper peroxidase CaPO2 and lipase CaGLIP1 were functionally characterized in pepper plants using VIGS (Choi et al., 2007; Hong et al., 2008).
Strategies to enhance plant disease resistance based on transgenic approaches have used genes associated with plant defense pathways (Makandar et al., 2006; Zhang et al., 2007), genes encoding plant or fungal hydrolytic enzymes (Lorito et al., 1998; Bieri et al., 2003), and genes encoding defense-related transcription factors (Chen and Chen, 2002; Sohn et al., 2006) and antimicrobial proteins (DeGray et al., 2001; Li et al., 2001). The accumulation of PR proteins represents a major quantitative change in protein composition that occurs during the hypersensitive response (HR), leading to the synthesis of low molecular weight compounds, proteins, and peptides with antimicrobial activity (Fritig et al., 1998). Many plant proteins are known to possess antimicrobial activity in vitro (Spelbrink et al., 2004; Oh et al., 2005; Prost et al., 2005), and they are characterized by low molecular weight, globular structures with disulfide bonds and domains rich in Cys (Garcia-Olmedo et al., 1998; Spelbrink et al., 2004). To date, several proteins with antibacterial and/or antifungal properties have been isolated and characterized from different plant species (Selitrennikoff, 2001). Some of these have been classified as thionins, lipid transfer proteins, plant defensins, chitinases, and ribosome-inactivating proteins (Lee et al., 2000; Wang et al., 2001; Soares-Costa et al., 2002; Gonorazky et al., 2005). One of the best known examples of protection conferred by transgenic expression of plant antifungal genes is the overexpression of chitinase and β-1,3-glucanase (Brogue et al., 1991; Sareena et al., 2006). Transformation of plants with genes encoding antimicrobial proteins is a promising approach for conferring resistance to a wide range of bacteria, fungi, and viruses (Zasloff, 2002). A plant defensin gene, alfAFP from Medicago sativa, was successfully expressed in tobacco and potato (Gao et al., 2000), and transgenic potato plants overexpressing a synthetic cecropin-melittin chimeric peptide gene exhibited field-level resistance to Verticillium dahliae (Osusky et al., 2000). More recently, overexpression of the pepper SAR8.2 gene in Arabidopsis (Arabidopsis thaliana) conferred enhanced resistance to infection by Pseudomonas syringae pv tomato DC3000 (Pst DC3000), Fusarium oxysporum f. sp. matthiolae, and Botrytis cinerea (Lee and Hwang, 2006).
In this study, the antimicrobial CaAMP1 (for C. annuum ANTIMICROBIAL PROTEIN1) gene was isolated and functionally characterized from pepper leaves infected with the avirulent strain Bv5-4a of Xanthomonas campestris pv vesicatoria (Xcv). The antimicrobial activity of the CaAMP1 protein toward several fungi and bacteria was examined to better understand the function of the pepper CaAMP1 gene. We also used the VIGS technique with a TRV vector to determine the CaAMP1 loss-of-function phenotype in pepper plants. CaAMP1-overexpression (OX) transgenic Arabidopsis plants were further evaluated for their resistance to infection by plant pathogenic bacteria, oomycetes, and fungi.
RESULTS
Isolation of the CaAMP1 Gene
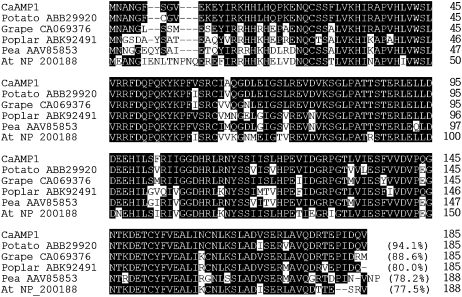
The putative pepper pathogen-induced protein gene CaAMP1 was isolated from a pepper cDNA library made from HR lesions of leaves inoculated with the Xcv avirulent strain Bv5-4a, using the differential hybridization technique (Jung and Hwang, 2000). The CaAMP1 cDNA comprises an open reading frame of 185 amino acids, and the mature protein has a molecular mass of 21,152 D and a pI of 5.9. The putative protein encoded by CaAMP1 (accession no. AY548741) was 94.1% identical to the potato (accession no. ABB29920), 88.6% identical to the grape (Vitis vinifera; accession no. CAO69376), 80.0% identical to the poplar (Populus spp.; accession no. ABK92491), 78.2% identical to the pea (Pisum sativum; accession no. AAV85853), and 77.5% identical to the Arabidopsis (accession no. NP_200128) putative AT-rich binding protein (Fig. 1). The functions of these proteins have not been elucidated.
Figure 1.
Alignment of deduced amino acid sequences of the pepper CaAMP1 protein with related sequences of potato (accession no. ABB29920), grape (accession no. CA069376), poplar (accession no. ABK92491), pea (accession no. AAV85853), and Arabidopsis (accession no. NP_200188) proteins. Identical amino acids are represented by white letters on black. Dashes indicate gaps introduced to maximize the alignment of homologous regions.
Induction of the CaAMP1 Gene in Pepper Tissues by Biotic and Abiotic Stresses
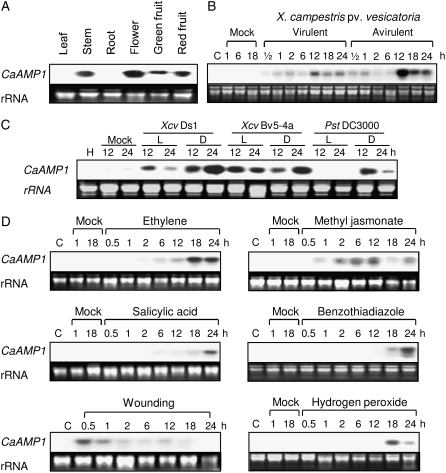
RNA-blot analysis was performed to determine whether the CaAMP1 gene is constitutively expressed in different organ tissues of pepper plants (Fig. 2A). CaAMP1 transcripts were not detected in leaf and root tissues, but high levels of transcripts were found in stem, flower, green fruit, and red fruit tissues.
Figure 2.
RNA-blot analysis of the expression of the CaAMP1 gene in pepper plants. A, Constitutive expression of CaAMP1 in various pepper plant organs. B, Expression of CaAMP1 in pepper leaves at various time points after inoculation with the virulent strain Ds1 and the avirulent strain Bv5-4a of Xcv at the six-leaf stage. C, Expression of CaAMP1 in pepper leaves at 12 and 24 h after inoculation with live (L) and dead (D) bacterial cells of Xcv virulent strain Ds1 and avirulent strain Bv5-4 and nonpathogenic Pst DC3000. D, Expression of CaAMP1 in pepper leaves at various time points after treatment with ethylene (10 μL L−1), MeJA (100 μm), SA (5 mm), BTH (10 μm), wounding, and H2O2 (100 μm).
The transcription levels of CaAMP1 were analyzed in compatible and incompatible interactions of pepper with Xcv (Fig. 2B). Pepper leaves inoculated with the virulent strain Ds1 did not exhibit any symptoms at 24 h after inoculation, while typical chlorotic and necrotic symptoms developed 6 d later. In contrast, pepper leaves infected with the avirulent strain Bv5-4a exhibited the HR at 18 h after inoculation (Lee and Hwang, 1996). In incompatible interactions, CaAMP1 transcripts accumulated rapidly and extensively in infected leaves, compared with those in compatible interactions. The CaAMP1 gene was strongly expressed at 12 to 24 h after inoculation with the avirulent strain Bv5-4a.
To determine whether CaAMP1 expression is triggered by pathogenicity factors of living pathogens, including effector proteins, live or dead cells of virulent Ds1 and avirulent Bv5-4a of Xcv and nonpathogenic Pst DC3000 were inoculated on the abaxial side of pepper leaves as described (Jung et al., 2004; Gust et al., 2007). Consistent with the results shown in Figure 2B, the CaAMP1 gene was more strongly induced in the avirulent Xcv-infected leaves compared to the virulent Xcv-infected leaves (Fig. 2C). However, infection with the nonpathogenic bacterium Pst DC3000 did not induce CaAMP1 expression. Interestingly, dead bacterial cells of virulent and avirulent Xcv or nonpathogenic Pst DC3000 strongly induced CaAMP1 expression. These results suggest that expression of CaAMP1 is strongly induced by pathogen-associated molecular patterns (PAMPs) of virulent and avirulent Xcv and nonpathogenic Pst.
Plant signal molecules, including ethylene, methyl jasmonate (MeJA), and SA, which may accumulate upon pathogen infection, are involved in the signal transduction pathways that mediate defense responses (Glazebrook, 2001). To assess the involvement of ethylene in CaAMP1 expression, pepper plants were treated with ethylene (Fig. 2D). CaAMP1 transcripts were first detected at 6 h after ethylene treatment, with maximal levels at 18 and 24 h. The three abiotic elicitors were evaluated for their ability to induce CaAMP1 gene expression. In MeJA-treated pepper leaves, CaAMP1 transcripts started to accumulate at 1 h after treatment and reached a peak at 12 h. SA has been implicated as a key component in the signal transduction pathway leading to plant resistance to various pathogens (Ryals et al., 1996). Benzothiadiazole (BTH), an analog of SA, is an inducer of systemic acquired resistance (Sticher et al., 1997). CaAMP1 transcription started to be induced at 24 h after SA treatment. As shown in Figure 2D, treatment with BTH slightly induced CaAMP1 transcription during an 18- to 24-h exposure. Hydrogen peroxide (H2O2) acts not only as a local signal for HR but also as a diffusible signal for the induction of defense genes in adjacent cells (Alvarez et al., 1998). CaAMP1 expression was evaluated in pepper leaves following H2O2 treatment. Transcripts were first detected in leaves at 18 h after treatment and declined thereafter. In response to mechanical wounding, CaAMP1 expression was induced in pepper plants. CaAMP1 expression was observed in leaves at 0.5 h after wounding, but transcripts gradually disappeared thereafter (Fig. 2D).
In Vitro Antimicrobial Activity of the CaAMP1 Protein
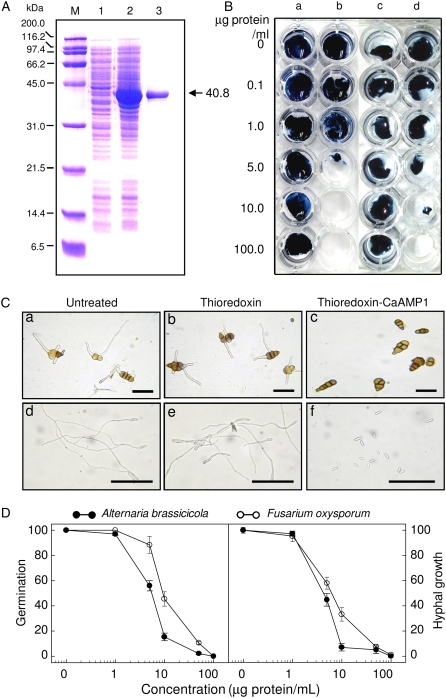
To determine whether the CaAMP1 protein has a direct antimicrobial effect, the CaAMP1 protein was purified from CaAMP1-expressing Escherichia coli (Fig. 3A). The in vitro antimicrobial activity of the CaAMP1 protein was evaluated against plant pathogenic fungi, oomycetes, yeast, and bacteria (Table I). The CaAMP1 protein completely inhibited the growth of B. cinerea, Cladosporium cucumerinum, Phytophthora capsici, Candida albicans, Saccharomyces cerevisiae, and Bacillus subtilis at levels below 30 μg mL−1. However, the growth of Rhizoctonia solani and Micrococcus luteus was unaffected, even at levels above 100 μg mL−1. The antifungal activity of the purified recombinant CaAMP1 protein at various concentrations was tested against Alternaria brassicicola and F. oxysporum f. sp. matthiolae using a microtiter broth dilution assay (Fig. 3B). The purified recombinant CaAMP1 protein completely inhibited the growth of F. oxysporum f. sp. matthiolae at 100 μg mL−1 and of A. brassicicola at 50 μg mL−1 (Fig. 3, B and D). Fungal spore germination was inhibited by recombinant thioredoxin-CaAMP1 protein but not by thioredoxin alone (Fig. 3C). Treatment with 100 μg mL−1 purified recombinant CaAMP1 protein completely inhibited spore germination (Fig. 3D). CaAMP1 protein at 1 μg mL−1 did not suppress the conidial germination of A. brassicicola and F. oxysporum f. sp. matthiolae. However, the germination of A. brassicicola was strongly inhibited at 5 and 10 μg mL−1. The hyphal growth of A. brassicicola and F. oxysporum f. sp. matthiolae was completely inhibited at 50 and 100 μg mL−1, respectively. These results indicate that CaAMP1 directly affects the growth of bacterial or fungal pathogens.
Figure 3.
Antifungal activity of the CaAMP1 protein against A. brassicicola and F. oxysporum f. sp. matthiolae at various concentrations. A, SDS-PAGE of the recombinant thioredoxin-CaAMP1 fusion protein purified after expression in E. coli. CaAMP1 expression from pET32a in E. coli BL21 cells containing the gene insert was examined in Luria-Bertani medium with or without 1 mm IPTG. Lane M, Molecular markers (kD); lane 1, soluble crude E. coli/CaAMP1 extract, noninduced; lane 2, soluble crude E. coli/CaAMP1 extract, induced with 1 mm IPTG; lane 3, the recombinant thioredoxin-CaAMP1 fusion protein purified on a nickel-nitrilotriacetic acid agarose column. B, Inhibition of hyphal growth of A. brassicicola (lanes a and b) and F. oxysporum f. sp. matthiolae (lanes c and d) by the CaAMP1 protein. Lanes a and c, Treatment with thioredoxin; lanes b and d, treatment with the thioredoxin-CaAMP1 fusion protein. C, Inhibitory effect of recombinant proteins (100 μg mL−1) on the germination of fungal spores as observed by light microscopy. Bars=50 μm. Sections a to c, A. brassicicola; sections d to f, F. oxysporum f. sp. matthiolae. D, Inhibitory effect of the CaAMP1 protein on conidial germination and hyphal growth of A. brassicicola and F. oxysporum f. sp. matthiolae at various concentrations. All results are expressed as relative values compared with those (100) of untreated controls. The data are means ± sd from three independent experiments. The absence of an error bar indicates a low error. [See online article for color version of this figure.]
Table I.
Minimum inhibitory concentrations of the CaAMP1 protein against microorganisms, including plant pathogenic bacteria, fungi, and oomycetes, as determined by the microtiter broth dilution method
| Microorganism | Minimum Inhibitory Concentrationa |
|---|---|
| μg mL−1 | |
| Alternaria brassicicola | 50 |
| Alternaria mali | 70 |
| Botrytis cinerea | 20 |
| Cladosporium cucumerinum | 5 |
| Colletotrichum coccodes | 50 |
| Fusarium oxysporum f. sp. matthiolae | 50 |
| Magnaporthe grisea | 50 |
| Phytophthora capsici | 10 |
| Rhizoctonia solani | >100b |
| Sclerotinia sclerotiorum | 50 |
| Candida albicans | 30 |
| Saccharomyces cerevisiae | 5 |
| Bacillus subtilis | 10 |
| Micrococcus luteus | >100 |
| Pseudomonas syringae pv tomato | 100 |
| Ralstonia solanacearum | 50 |
| Xanthomonas campestris pv vesicatoria | 100 |
The lowest concentration of the CaAMP1 protein required for complete inhibition of microbial growth.
Values of >100 indicate that growth of the microorganisms was not completely inhibited by the CaAMP1 protein at 100 μg mL−1.
Subcellular Localization of CaAMP1 in Onion Epidermal Cells
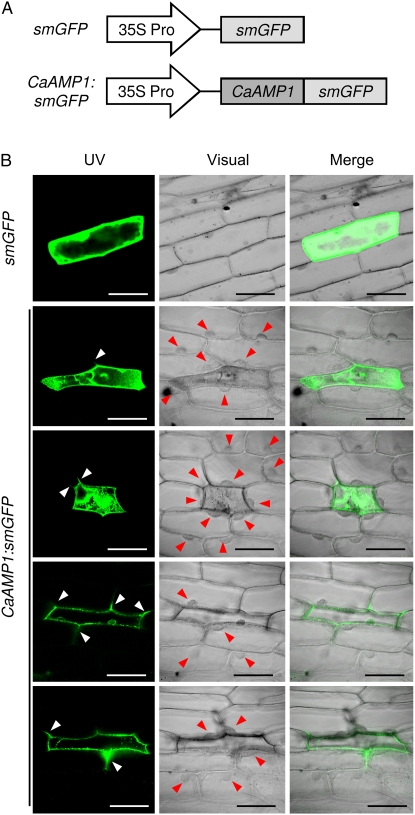
To examine the subcellular localization of CaAMP1 protein, we used the biolistic transformation system in onion (Allium cepa) epidermal cells. Computational analyses of the predicted protein sequence revealed that CaAMP1 may be a cytoplasmic protein (PSORT; 65%), but it has no known signal peptide sequence (http://www.expasy.org). The soluble-modified GFP gene (smGFP) was fused to the C-terminal region of CaAMP1 under the control of the cauliflower mosaic virus 35S promoter (Fig. 4A). The 35S:CaAMP1:smGFP fusion construct and a 35S:smGFP control were introduced into onion epidermal cells by particle bombardment. As shown in Figure 4B, the control smGFP was uniformly distributed throughout the cell. However, CaAMP1:smGFP fusion protein was mainly localized in external and intercellular regions of the cell. Intriguingly, the onion epidermal cells expressing the CaAMP1:smGFP construct were shrunken in an abnormal cell shape, compared with its neighboring cells. Furthermore, cell nuclei were localized in the periphery of the CaAMP1-expressing cell.
Figure 4.
Analysis of the subcellular localization of the CaAMP1 protein using transient expression of the CaAMP1:smGFP construct in onion epidermal cells. A, Schematic maps of the smGFP-tagged CaAMP1 construct and of a control construct. The smGFP gene was fused to the 3′ region of the CaAMP1 gene. B, Transient expression of smGFP or CaAMP1:smGFP in onion epidermal cells, as detected by confocal laser scanning microscopy at 24 h after biolistic transformation. White arrowheads indicate CaAMP1 protein localized in the intercellular spaces. Red arrowheads indicate nuclei of cells neighboring the CaAMP1-expressing cells. Bars=100 μm.
Enhanced Susceptibility of CaAMP1-Silenced Pepper Plants to Xcv Infection
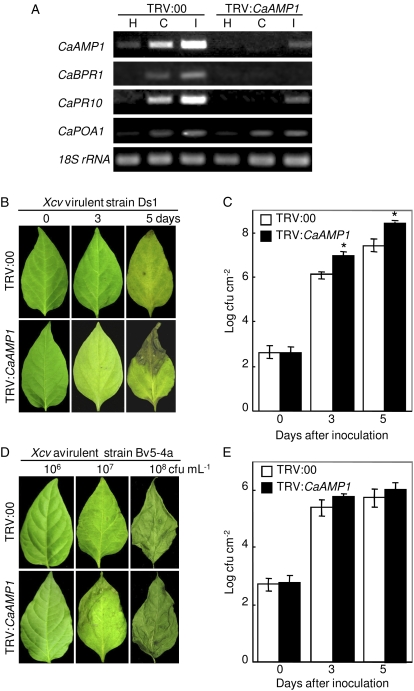
The VIGS technique was used to further investigate the roles of CaAMP1 in pepper plant defense responses. Pepper seedlings were inoculated with recombinant TRV silencing constructs. The efficiency of VIGS was tested by reverse transcription (RT)-PCR in empty vector control (TRV:00) and CaAMP1-silenced (TRV:CaAMP1) pepper leaves at 18 h after inoculation with virulent and avirulent strains of Xcv (Fig. 5A). CaAMP1 expression was compromised in CaAMP1-silenced pepper leaves; however, it remained at a slightly visible level in silenced leaves infected with the avirulent Xcv strain. To determine whether CaAMP1 silencing affects the expression of genes in the defense response pathway, the expression levels of several defense-related genes were further analyzed by RT-PCR (Fig. 5A). Silencing of CaAMP1 remarkably reduced the induction of CaBPR1 and CaPR10 but not of CaPOA1 compared with empty vector control plants (TRV:00).
Figure 5.
Enhanced susceptibility of CaAMP1 gene-silenced pepper plants to Xcv infection. A, RT-PCR analysis of the expression of CaAMP1 and of pepper defense-related genes in empty vector control (TRV:00) and CaAMP1 gene-silenced (TRV:CaAMP1) pepper plants at 12 h after inoculation with the Xcv virulent strain Ds1 (C, compatible) and the avirulent strain Bv5-4a (I, incompatible). 18S rRNA served as a control. H, Uninoculated healthy leaves; CaBPR1, pepper basic PR1; CaPR10, pepper PR10; CaPOA1, pepper ascorbate peroxidase. B, Disease symptoms developed on empty vector control and CaAMP1-silenced leaves at various time points after inoculation with the Xcv virulent strain Ds1 (5 × 106 cfu mL−1). C, Bacterial growth in empty vector control and CaAMP1 gene-silenced pepper leaves at 0, 3, and 5 d after inoculation with the Xcv virulent strain Ds1 (104 cfu mL−1). D, Disease symptoms developed on empty vector control and CaAMP1-silenced leaves at 24 h after inoculation with various concentrations of the Xcv avirulent strain Bv5-4a (106, 107, and 108 cfu mL−1). E, Bacterial growth in empty vector control and CaAMP1 gene-silenced pepper leaves at 0, 3, and 5 d after inoculation with the Xcv avirulent strain Bv5-4a (104 cfu mL−1). Experiments were repeated three times with similar results. Data are means ± sd from three independent experiments. Asterisks indicate significant differences as determined by Student's t test (P < 0.05). [See online article for color version of this figure.]
Silencing of CaAMP1 not only compromised the expression of some PR genes but also led to a highly susceptible response to infection by the Xcv virulent strain Ds1, accompanied by more severe disease symptoms at 5 d after inoculation (Fig. 5B). Consistent with these results, CaAMP1-silenced leaves exhibited significantly higher levels of bacterial growth compared with empty vector control plants (Fig. 5C). These results indicate that CaAMP1 expression is required for the PAMP-triggered immunity of pepper plants to Xcv infection (Jones and Dangl, 2006). To investigate the role of CaAMP1 during HR-based immunity, empty vector control (TRV:00) and CaAMP1-silenced (TRV:CaAMP1) plants were inoculated with various concentrations of the Xcv avirulent strain Bv5-4a (108, 107, and 106 colony forming units [cfu] mL−1; Fig. 5D). CaAMP1-silenced pepper plants did not show distinctive changes in the HR phenotype compared with empty vector control plants. Bacterial growth was only slightly and insignificantly enhanced in CaAMP1-silenced plants compared with empty vector control plants (Fig. 5E). Overall, these findings indicate that CaAMP1 expression plays a crucial role in basal defense of pepper plants against Xcv infection rather than in HR-based immunity induced by effector proteins via the type III secretion system (Jones and Dangl, 2006).
Enhanced Susceptibility of CaAMP1-Silenced Pepper Plants to Colletotrichum coccodes Infection
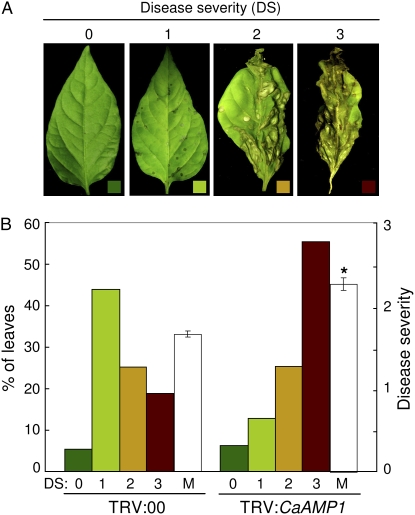
Purified recombinant CaAMP1 exhibited strong in vitro antifungal activities against diverse phytopathogenic fungi (Table I; Fig. 3). To examine the role of CaAMP1 in the defense response of pepper plants to fungal pathogens, we inoculated empty vector control (TRV:00) and CaAMP1-silenced (TRV:CaAMP1) pepper leaves with a conidial suspension of virulent Colletotrichum coccodes isolate 2-25 (105 conidia mL−1), the causal agent of pepper anthracnose. CaAMP1-silenced pepper leaves exhibited remarkably enhanced susceptibility to C. coccodes (Fig. 6). Disease severity was rated at 5 d after inoculation based on the extent of brown or dark brown areas (Fig. 6A). Approximately 55% of CaAMP1-silenced leaves exhibited severe disease symptoms (class 3: enlarged dark brown lesions with severe chlorosis) compared with 20% of empty vector control plants (Fig. 6B). CaAMP1-silenced plants showed remarkable differences in average disease severity compared with empty vector control plants. These data demonstrate that CaAMP1 expression is important for the PAMP-triggered resistance of pepper plants to the fungal pathogen C. coccodes.
Figure 6.
Enhanced susceptibility of CaAMP1-silenced (TRV:CaAMP1) pepper plants to virulent C. coccodes isolate 2-25 infection. A, Disease severity rated on a 0 to 3 scale (0, no symptoms; 1, weakly visible symptoms; 2, dark brown lesions with mild chlorosis; 3, enlarged dark brown lesions with severe chlorosis). B, Percentage of empty vector control and CaAMP1 gene-silenced pepper leaves exhibiting the indicated extent of disease at 5 d after inoculation with virulent C. coccodes isolate 2-25 (105 conidia mL−1). Disease severity was monitored on more than 50 leaves. Asterisk indicates significant difference between means as determined by Student's t test (P < 0.05). M, Means of disease severities. [See online article for color version of this figure.]
Enhanced Resistance of CaAMP1-OX Transgenic Plants to Pathogen Infection
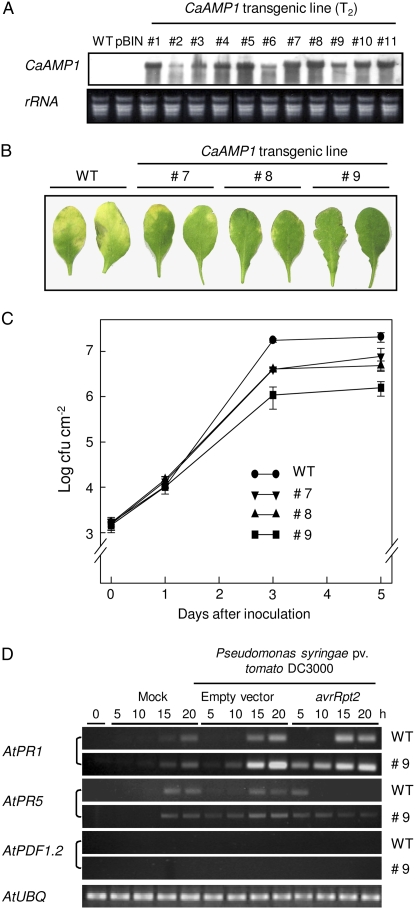
Arabidopsis transgenic plants overexpressing the entire CaAMP1 precursor were generated to determine the in vivo effect of a gain of function of the CaAMP1 gene. To verify 35S:CaAMP1 transgene expression in independent T2 lines, northern-blot analysis was performed using the CaAMP1 cDNA as a probe. CaAMP1-OX transcription was confirmed in these lines (Fig. 7A).
Figure 7.
Enhanced resistance of CaAMP1-OX transgenic Arabidopsis plants to Pst DC3000 infection. A, RNA-blot analysis of the expression of the CaAMP1 gene in wild-type and CaAMP1-OX transgenic Arabidopsis plants (T2). Total RNA (10 μg) from leaf samples was loaded. The EcoRI/XhoI fragment of the putative pepper CaAMP1 cDNA insert in pBluescript SK− was used as a probe. A duplicate gel was stained with ethidium bromide as a control for RNA loading. B, Disease symptoms developed on leaves of wild-type or transgenic plants at 5 d after infiltration with Pst DC3000 (105 cfu mL−1). C, Bacterial growth in leaves of wild-type and transgenic plants at different time points after infiltration with Pst DC3000 (105 cfu mL−1). Data are means ± sd from three independent experiments. D, RT-PCR analysis of the expression of the Arabidopsis AtPR1a, AtPR5, and AtPDF1.2 genes in wild-type and transgenic plants (T2) inoculated with Pst DC3000. Ubiquitin (UBQ) expression level was analyzed as a quantification control. Mock, 10 mm MgCl2 infiltrated; WT, Col-0 plants. [See online article for color version of this figure.]
The bacterial pathogen Pst DC3000 is virulent to Arabidopsis ecotype Columbia (Col-0) plants and causes symptoms similar to bacterial speck disease of tomato (Whalen et al., 1991). To determine whether CaAMP1 overexpression affects disease responses to this pathogen, plants were inoculated with Pst DC3000 and the progress of infection was observed over 7 d (Fig. 7B). CaAMP1-OX plants did not exhibit macroscopic signs of infection, as shown in Figure 7B. In contrast, wild-type plants displayed chlorotic symptoms compared with CaAMP1-OX plants, indicating a high susceptibility to Pst DC3000. To confirm that these macroscopic disease symptoms reflect bacterial proliferation inside the plants, bacterial growth was measured in wild-type and CaAMP1-OX plants upon inoculation and at 1, 3, and 5 d thereafter (Fig. 7C). The bacterial population at the inoculation site of the wild-type leaves infected with Pst DC3000 was approximately 4- to 15-fold higher than that of transgenic lines. In the inoculated leaves of transgenic line 9, the rate of bacterial growth was much lower than that in transgenic lines 7 and 8.
Enhanced disease resistance in Arabidopsis is often accompanied by elevated transcript levels of PR genes (AtPR1 and AtPR5) associated with the SA-dependent pathway, whereas the induction of AtPDF1.2 is associated with the ethylene/JA-dependent pathway (Uknes et al., 1992; Rogers and Ausubel, 1997). To examine the possible involvement of stress genes in the enhanced pathogen resistance of CaAMP1-OX transgenic plants, the expression of AtPR1, AtPR5, and AtPDF1.2 was analyzed using RT-PCR (Fig. 7D). The expression of AtPR1 and AtPR5 was more rapidly and strongly induced by Pst DC3000 infection in transgenic plants than in wild-type plants. In particular, overexpression of CaAMP1 in transgenic line 9 was effective at inducing AtPR1 expression at an earlier time during Pst DC3000 (avrRpt2) infection. However, infection by this pathogen did not induce AtPDF1.2 expression in either wild-type or CaAMP1-OX transgenic plants. These data indicate that CaAMP1-OX plants effectively activate SA-dependent gene expression.
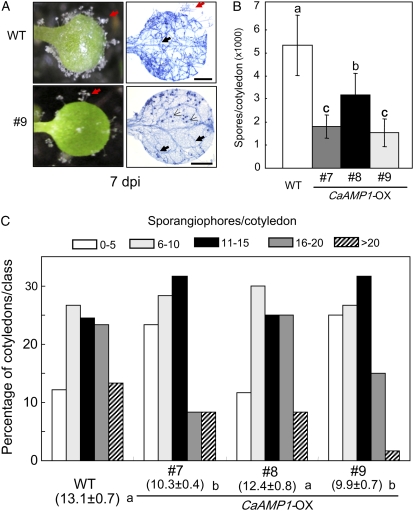
To determine whether the ectopic expression of CaAMP1 in Arabidopsis plants alters their resistance to biotrophic pathogens, we tested the response of CaAMP1-OX transgenic plants to the biotrophic oomycete Hyaloperonospora parasitica isolate Noco2 (Fig. 8). Wild-type (Col-0) and transgenic seedlings were inoculated with a suspension of an asexual inoculum of H. parasitica isolate Noco2 (5 × 104 conidiospores mL−1), which is virulent on Arabidopsis Col-0. Over 200 Arabidopsis seedlings were inoculated and the number of sporangiophores produced on cotyledons was scored at 5 d after inoculation. As shown in Figure 8A, the cotyledons of wild-type (Col-0) and transgenic seedlings responded to H. parasitica infection by stimulating parasite sporulation. The cotyledons of wild-type and transgenic plants supported different levels of mycelial growth, oospores, and sporangiophores, as judged by trypan blue staining (Fig. 8A). CaAMP1 overexpression resulted in decreased pathogen growth. Five and 6 d after inoculation, CaAMP1-OX transgenic lines showed enhanced resistance to H. parasitica infection (Fig. 8, B and C). Spore counts allow accurate quantification of pathogen reproduction, especially under conditions of high sporangiophore coverage of the cotyledons. As expected, wild-type plants permitted heavy sporulation (Fig. 8B), while spore production was significantly reduced in CaAMP1-OX transgenic lines. The production of spores on inoculated CaAMP1-OX transgenic lines was approximately 1.5- to 3-fold greater than that on wild-type plants. As shown in Figure 8C, quantitative disease ratings are expressed as the mean number of sporangiophores per cotyledon. Enhanced resistance resulted in less sporangiophore formation in cotyledons of CaAMP1-OX transgenic lines compared with wild-type plants. In transgenic plant lines, the level of susceptibility was somewhat less than in wild-type plants infected by H. parasitica isolate Noco2. The frequency of cotyledons with over 20 sporangiophores and the average number of sporangiophores were reduced in transgenic plants relative to wild-type plants. In particular, the frequency of cotyledons with over 20 sporangiophores (full susceptibility) was 13% in wild-type plants but only 8%, 8%, and 2% in transgenic lines 7, 8, and 9, respectively (Fig. 8C). Induction of reactive oxygen species (ROS) generation and the HR may play a critical role in the defense of plants against invading pathogens, including H. parasitica (Alvarez et al., 1998; McDowell et al., 2000; Knoth et al., 2007; Jung et al., 2008). To determine whether the enhanced resistance of CaAMP1-OX transgenic lines is associated with the HR or H2O2 accumulation, we also performed trypan blue and 3,3′-diaminobenzidine staining at 18 and 24 h after inoculation with H. parasitica. However, we could not find any significant differences between wild-type plants and CaAMP1-OX transgenic lines in the HR or in ROS formation (data not shown).
Figure 8.
Enhanced resistance of CaAMP1-OX transgenic Arabidopsis plants to H. parasitica isolate Noco2 infection. A, Disease symptoms and trypan blue-stained pathogen structures on 7-d-old cotyledons of wild-type (Col-0) and transgenic plants at 7 d after inoculation. Sporangiophores, hyphae, and oospores of H. parasitica are indicated with red arrows, bold black arrows, and thin black arrows, respectively. dpi, Days after inoculation. Bars=0.5 mm. B, Average number of spores per cotyledon at 6 d after H. parasitica infection. Each experiment contained the spore counts from 40 inoculated cotyledons of wild-type and transgenic lines. C, Number of sporangiophores per cotyledon of wild-type and transgenic plants at 5 d after inoculation with H. parasitica. Sporangiophores were counted on over 50 cotyledons per genotype. Average numbers of sporangiophores produced on the cotyledons of wild-type and transgenic lines are shown below each of the lines tested. Values with different letters show significant differences at P=0.05 according to the lsd test. The experiments were repeated three times with similar results. [See online article for color version of this figure.]
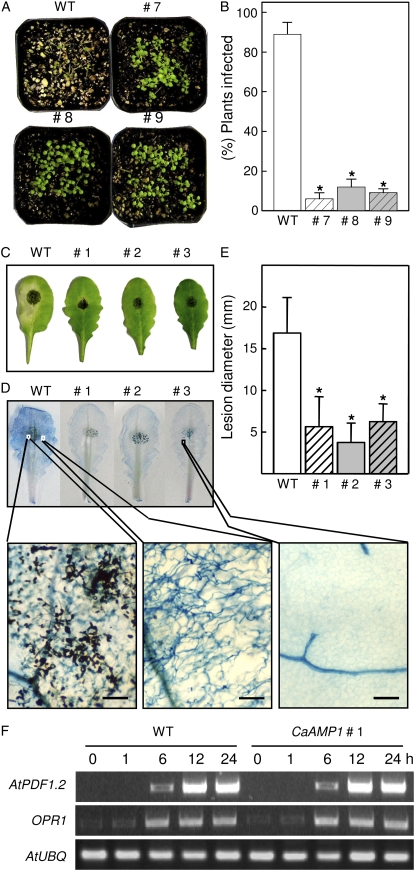
As shown in Figure 9, CaAMP1-OX transgenic lines were evaluated for their levels of resistance to F. oxysporum f. sp. matthiolae and A. brassicicola. Wild-type (Col-0) plants were severely damaged by F. oxysporum f. sp. matthiolae infection. However, typical disease symptoms did not appear in CaAMP1-OX lines at 7 d after inoculation (Fig. 9A). When inoculated with F. oxysporum f. sp. matthiolae, only 4% to 11% of inoculated transgenic plants showed disease symptoms at 7 d after inoculation, in contrast to 90% of wild-type plants (Fig. 9B).
Figure 9.
Inhibition of fungal pathogen infection in CaAMP1-OX transgenic Arabidopsis lines. A, Disease symptoms developed on wild-type (Col-0) and transgenic plants at 7 d after inoculation with F. oxysporum f. sp. matthiolae strain 247.61 (5 × 105 spores mL−1). B, Number of sporangiophores per cotyledon of wild-type and transgenic plants at 7 d after inoculation with H. parasitica. Sporangiophores were counted as percentages of wild-type and transgenic plants showing typical disease symptoms at 5 d after inoculation with F. oxysporum f. sp. matthiolae. Data points represent average frequencies of dead seedlings ± sd; a minimum of 50 plants were measured. Asterisks indicate significant differences as determined by Student's t test (P < 0.05). C, Leaves from wild-type (Ws-0) and CaAMP1 transgenic plants at 5 d after inoculation with a 5-μL droplet of A. brassicicola spores (105 spores mL−1). D, Leaves inoculated with A. brassicicola stained with lactophenol-trypan blue. Bars=25 μm. E, Lesion diameters were measured at 5 d after inoculation with A. brassicicola. Data points represent average lesion sizes ± sd; a minimum of 40 lesions were measured. The experiment was carried out three times with similar results. Asterisks indicate significant differences as determined by Student's t test (P < 0.05). WT, Ws-0 plants. F, RT-PCR analysis of the expression of the Arabidopsis AtPDF1.2 and OPR1 genes in wild-type (Ws-0) and transgenic plants inoculated with A. brassicicola. Ubiquitin (UBQ) expression level was analyzed as a quantification control. WT, Col-0 plants.
Arabidopsis Col-0 and Wassilewskija (Ws-0) were used as natural wild-type ecotypes that are resistant and susceptible to A. brassicicola, respectively (Kagan and Hammerschmidt, 2002). We generated Arabidopsis (Ws-0) transgenic plants overexpressing CaAMP1 to determine the role of CaAMP1 during A. brassicicola infection. CaAMP1 transgene transcription was confirmed in independent transgenic lines by northern-blot analysis (data not shown). Five days after inoculation with A. brassicicola, wild-type leaves showed significantly larger disease lesions than did transgenic leaves (Fig. 9E). As visualized by trypan blue staining, the spreading lesions were heavily colonized by fungal hyphae. Five days after inoculation, more than 50% of the area of the inoculated wild-type leaves was killed by the infection, with clearly visible concentric brown infection rings typical of A. brassicicola (Fig. 9, C and D). CaAMP1-OX transgenic plants were resistant to A. brassicicola, exhibiting only restricted lesions that affected less than 10% of the inoculated leaf areas.
It is well established that JA rather than SA is involved in a major signaling pathway mediating the resistance of Arabidopsis to A. brassicicola (Penninckx et al., 1998; van Wees et al., 2003). Two genes, PDF1.2 and OPR1, which were previously used as markers of the JA pathway, were selected to determine whether the JA signaling pathway affects the resistance of CaAMP1-OX transgenic plants to A. brassicicola. PDF1.2, which encodes a plant defensin (Penninckx et al., 1998), and OPR1, which encodes 12-oxophytodienoate reductase, are involved in JA biosynthesis (Biesgen and Weiler, 1999). Leaf tissues were harvested immediately and at 6, 12, and 24 h after inoculation, and expression levels were determined by RT-PCR analysis (Fig. 9F). The PDF1.2 and OPR1 genes were induced in wild-type plants and exhibited expression patterns similar to those seen for CaAMP1-OX transgenic plants (Fig. 9F). These results suggest that the resistance of CaAMP1-OX plants to A. brassicicola may be due to the antimicrobial activity of the CaAMP1 protein rather than to a JA-mediated defense response.
DISCUSSION
In this study, we have isolated the pathogen-induced CaAMP1 gene, which encodes a novel antimicrobial protein from pepper. Pepper leaves inoculated with the Xcv virulent strain Ds1 exhibited no visible symptoms at 18 h after inoculation. In contrast, the HR appeared in pepper leaves at 18 h after inoculation with the avirulent strain Bv5-4a. Many of the defense responses of plants to microbial infection are triggered by signals generated during the initial stages of the host-pathogen interaction (Lamb and Dixon, 1997). The CaAMP1 gene was induced by Xcv infection and by treatment with abiotic elicitors such as ethylene, MeJA, SA, BTH, and H2O2. In incompatible interactions, a marked increase in CaAMP1 expression was observed during the early stages of infection. Plant defense-related genes are known to be activated by the generation of endogenous plant signal molecules following the primary recognition of pathogen-encoded elicitors (Yang et al., 1997; Maffei et al., 2007). CaAMP1 transcripts were induced rapidly and strongly during incompatible interactions, which suggests that the CaAMP1 gene may be involved in defense responses. Furthermore, dead cells of virulent and avirulent Xcv and nonhost pathogen Pst strongly induced CaAMP1 expression in pepper leaves compared with the live cells. Recently, many pathogen effectors were proposed to suppress plant defense responses triggered either by PAMPs or by other effectors due to recognition by cognate resistance (R) proteins (Kjemtrup et al., 2000; Alfano and Collmer, 2004; Chisholm et al., 2006; Jones and Dangl, 2006). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility by suppressing PAMP-triggered callose deposition and ROS generation in Arabidopsis plants (Sohn et al., 2007). Thus, enhanced expression of CaAMP1 in pepper leaves infected with dead cells of Xcv and Pst supports the notion that the CaAMP1 gene may be induced by PAMPs of those bacterial pathogens but suppressed by unknown pathogenesis factors of living bacterial cells, including effector proteins. However, the knockout mutants of the type III secretion system of Xcv, which cannot deliver effector proteins into host cells, are required to resolve this issue clearly (Keshavarzi et al., 2004). Northern-blot analyses also revealed that the expression of CaAMP1 could be induced by JA and ethylene. Some abiotic elicitors, including SA, ethylene, and JA, are known to be significant in triggering disease resistance in plants (Robert-Seilaniantz et al., 2007).
Many of plant PR proteins are localized in extracellular regions, including PR-2 (1,3-β-glucanase), PR-3 (endochitinase), and PR-4 (endochitinase; Fritig et al., 1998). When plant pathogenic microorganisms invade plants, they initially multiply in the intercellular spaces of the plant cells. Therefore, the presence of antimicrobial proteins in the intercellular space is important for their antimicrobial ability to inhibit the invading pathogens. Analysis of the subcellular localization of smGFP-tagged CaAMP1 protein revealed direct secretion of CaAMP1 protein from onion epidermal cells to the intercellular space. Proteins in cells are often targeted to a specific cellular location by signal peptides found at their N or C termini. Our computational analyses with the predicted amino acid sequence of CaAMP1 did not reveal any signal peptide sequence motif in CaAMP1 protein. Therefore, unexpected extracellular localization of CaAMP1 remains to be verified in further studies. Strikingly, expression of CaAMP1 led to the lethal effects on onion epidermal cells. As observed with a confocal microscope, the CaAMP1-expressing cells exhibited abnormal cell morphologies, including the shrinkage and darkening of the cells. This suggests that the CaAMP1 protein exerts cytotoxic effects on onion epidermal cells as well as in vitro antimicrobial activities against various phytopathogens. Furthermore, nuclei of neighboring cells surrounding CaAMP1-expressing cells moved toward the CaAMP1-expressing cells. Cell polarization, rearrangements of cytoskeletons, and translocation of nuclei constitute a crucial process in development, morphogenesis, and defense (Schmelzer, 2002). Recognition of attacking pathogens triggers cell polarization, which thus allows the localized delivery of barrier materials and cargo proteins for defense execution around the penetration site (Gross et al., 1993; Škalamera and Heath, 1998). Therefore, we can propose a possible mechanism for CaAMP1-induced cell polarization of neighboring cells. To minimize potential damage, neighboring cells may undergo cell polarization and accumulate barrier materials or defense-related proteins to the cytotoxic CaAMP1-exposed region. However, we could not find any deleterious effect of the expressed CaAMP1 protein on the cellular structure of Arabidopsis, possibly due to the small amount expressed in the intercellular spaces of cells, when overexpressed in transgenic Arabidopsis plants (data not shown). These results also support the possibility that other host factors in Arabidopsis may prevent abnormal cell morphologies caused by CaAMP1.
The generation of stable transgenic mutants in pepper is time consuming and very difficult. The VIGS technique provided a fast and effective means to mimic loss-of-function phenotypes and allowed us to identify the role of defense-related genes in pepper plants (Choi et al., 2007; Hong et al., 2008). Since the CaAMP1 gene was distinctively induced by Xcv infection and abiotic elicitor treatments that trigger defense signaling, we tested whether the silencing of CaAMP1 alters the resistance of pepper plants to bacterial and fungal pathogen infection. Phenotypic differences were not observed between empty vector control (TRV:00) and CaAMP1-silenced (TRV:CaAMP1) pepper plants during growth and development. However, infection of empty vector control and CaAMP1-silenced pepper plants with the bacterial pathogen Xcv and the fungal pathogen C. coccodes showed that CaAMP1-silenced plants exhibited increased susceptibility to both bacterial spot and anthracnose diseases. Furthermore, CaBPR1 and CaPR10 expression was significantly compromised in CaAMP1-silenced pepper plants after inoculation with Xcv. Consistent with the observed in vitro antimicrobial activity of CaAMP1, these findings strongly support the notion that CaAMP1 may be required for the PAMP-triggered resistance of pepper plants to bacterial and fungal diseases. PAMP-triggered resistance is considered to be a weak response to virulent pathogens, and its molecular mechanism is not fully understood (Jones and Dangl, 2006).
Although CaAMP1 was strongly induced in pepper leaves by avirulent Xcv infection, silencing of CaAMP1 did not significantly alter the susceptibility of pepper plants to avirulent Xcv infection. These findings suggest a pivotal role of CaAMP1 in basal resistance rather than in R gene-mediated resistance (Nimchuk et al., 2003; Jones and Dangl, 2006). However, the lack of a role of CaAMP1 for HR-based immunity in pepper plants may be due to the inefficient silencing of CaAMP1 in pepper leaves inoculated with avirulent Xcv. VIGS of CaAMP1 did not completely eliminate all CaAMP1 transcripts from pepper leaves infected with avirulent Xcv. Residual CaAMP1 transcripts may suffice in the defense response to avirulent Xcv in pepper plants. Thus, a role for CaAMP1 in HR-based immunity cannot be excluded. The knockout mutants of CaAMP1 are required to resolve this issue. Together, these VIGS findings suggest that CaAMP1 expression positively contributes to the basal defense response of pepper plants to bacterial and fungal pathogens.
We revealed the involvement of CaAMP1 in plant defense against a variety of pathogens. These results clearly showed that CaAMP1-OX transgenic plants were more resistant to biotrophic and necrotrophic pathogens than were wild-type plants. The production of antimicrobial compounds including proteins may be involved in disease resistance (Ferrari et al., 2003; Denby et al., 2004). We next found that treatment with the CaAMP1 protein inhibited spore germination of plant pathogenic fungi, suggesting that CaAMP1 may directly interfere with an early process of fungal infection. Ectopic expression of CaAMP1 in Arabidopsis strongly enhanced the PAMP-triggered resistance of these plants to bacterial (Pst DC3000), oomycete (H. parasitica), and fungal (F. oxysporum and A. brassicicola) pathogens. Consistent with our results, the overexpression of THI2.1, which encodes the thionin protein in Arabidopsis, enhanced resistance to F. oxysporum, possibly due to a direct effect on the growth of fungal hyphae (Epple et al., 1997). Earlier studies showed that antimicrobial proteins derived from plants and animals are inhibitory to either fungi or bacteria but not to both (Hancock and Lehrer, 1998). However, the CaAMP1 protein exhibited a broad-spectrum antibiotic activity against fungi, oomycetes, and bacteria. Moreover, any abnormal growth phenotypes were not observed in CaAMP1-OX transgenic plants. These results suggest that enhanced resistance of CaAMP1-OX plants to fungal and bacterial diseases may not be due to metabolic perturbation.
The CaAMP1 protein may directly inhibit pathogen infection and/or trigger the production of SA-dependent defense signaling molecules. CaAMP1-OX Arabidopsis plants were significantly resistant to infection by the virulent strain Pst DC3000, accompanied by rapid expression of the AtPR1 and AtPR5 genes. SA is not only associated with plant resistance to biotrophic pathogen infection, such as by P. syringae and H. parasitica, but also with cross talk with other defense signaling molecules, including ethylene and MeJA (Veronese et al., 2004; Liu et al., 2005). SA also is known to regulate the expression of different sets of PR genes, such as PR1 and PR5 (Uknes et al., 1992; Gu et al., 2002). Members of the PR1 and PR5 families, which include thaumatin, osmotin, and related proteins, have been shown to possess antimicrobial activity against several pathogens (Fritig et al., 1998; Coca et al., 2000; Shatters et al., 2006). However, CaAMP1-OX transgenic plants did not constitutively express Arabidopsis PR genes. Therefore, the enhanced resistance of CaAMP1-OX plants to bacterial infection most likely is not due to the constitutive activation of defense-related genes. In Arabidopsis, defense responses under SA control are critical for resistance to the bacterial pathogen Pst DC3000 (Glazebrook, 2001). The significant expression of the AtPR1 and AtPR5 genes may be due, in part, to an increased SA level. Induction of the AtPR1 and AtPR5 genes may also be triggered by indirect effects of the overexpressed transgene, which thus contributes to the induction of resistance to pathogen infection. Another possibility is that CaAMP1 overexpression in Arabidopsis primes the defense pathway to trigger a PAMP-triggered resistance response to pathogen attacks. Such a “priming” of the defense pathway has been observed in NPR1-OX transgenic plants, in which SA-dependent PR genes are activated more strongly and rapidly than in wild-type plants (Cao et al., 1998). These results suggest that CaAMP1 may play dual roles in the defense response to pathogenic bacterial infection. Namely, CaAMP1 may not only directly inhibit bacterial and fungal pathogens but also may influence SA-dependent defense signaling in plants. SA is necessary for systemic acquired resistance in plants (Nandi et al., 2004; Block et al., 2005; Mishina and Zeier, 2007), which provides protection against secondary challenge infection by biotrophic pathogens and is correlated with the expression of PR proteins, including some with antimicrobial activity (Cao et al., 1998; Dewdney et al., 2000).
JA-regulated defense signaling is an important component of plant resistance to necrotrophic fungi (van Wees et al., 2003; Veronese et al., 2004; Coego et al., 2005). CaAMP1 overexpression in Arabidopsis conferred enhanced resistance to infection by the necrotrophic fungi F. oxysporum f. sp. matthiolae or A. brassicicola. The PDF1.2 and OPR1 genes involved in JA biosynthesis were similarly induced in both wild-type and CaAMP1-OX transgenic plants, although these genes were not constitutively expressed in CaAMP1-OX transgenic plants. The hevein-like protein Ac-AMPs, isolated from amaranth (Amaranthus caudatus), has been reported to show potent antifungal activity in vitro (Broekaert et al., 1990). However, overexpression of the Ac-AMP gene in tobacco plants did not confer resistance to B. cinerea or Alternaria longipes (De Bolle et al., 1996). Our data support the notion that the enhanced resistance of CaAMP1-OX plants to necrotrophic fungal pathogens may be due to the antimicrobial activity of the CaAMP1 protein rather than to a JA-dependent defense mechanism.
Taking all of the available evidence together, we have identified a novel antimicrobial protein gene, CaAMP1, which was isolated from pepper leaves infected with the Xcv avirulent strain Bv5-4a. Silencing of CaAMP1 led to an enhanced susceptibility of pepper plants to bacterial and fungal pathogen infection. In contrast, CaAMP1 overexpression conferred enhanced broad-spectrum disease resistance in plants. The CaAMP1 gene, encoding a protein with a high antimicrobial activity, may represent a new prospective transgene for engineering disease resistance in crop plants. However, identification of the target pathway for the CaAMP1 gene is required to gain insights into the plant defense mechanisms associated with CaAMP1 expression.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pepper seeds (Capsicum annuum ‘Hanbyul’) were sown in a steam-sterilized compost soil mix (peat moss, perlite, and vermiculite, 5:3:2, v/v/v), sand, and loam soil (1:1:1, v/v/v). The pepper plants were raised in a growth room at 27°C ± 1°C with approximately 80 μmol photons m−2 s−1 (white fluorescent lamps) for 16 h per day.
Arabidopsis (Arabidopsis thaliana; ecotypes Col-0 and Ws-0) wild-type and mutant plants were grown in a 9:1:1 mix of peat moss, perlite, and vermiculite under controlled environmental conditions (130 μmol photons m−2 s−1, 24°C/21°C with 60% relative humidity, and a 12-h-light/12-h-dark cycle). All seeds were vernalized at 4°C for 2 d before transfer to growth conditions.
Pathogen Inoculation, Disease Rating, and Tissue Staining
The Xanthomonas campestris pv vesicatoria (Xcv) virulent strain Ds1 and avirulent strain Bv5-4a and the nonpathogenic strain Pseudomonas syringae pv tomato (Pst) DC3000 were used in this study. To prepare bacterial suspensions for inoculation of pepper leaves, bacteria were cultured overnight in yeast-nutrient broth (5 g of yeast extract, 8 g of nutrient broth, and 1 L of water) at 28°C. Bacterial suspensions were adjusted to 108 cfu mL−1 with sterile tap water prior to inoculation. Pepper plants at the six-leaf stage were inoculated by infiltrating bacterial suspensions into the abaxial side of fully expanded leaves (Chung et al., 2007; Kim et al., 2007). To prepare dead bacterial cells of Xcv and Pst, bacterial suspensions (108 cfu mL−1; optical density at 660 nm [OD660]=0.06) were autoclaved at 121°C for 15 min. The bacteria-inoculated pepper plants were incubated in a growth room as described previously (Lee and Hwang, 1996).
Colletotrichum coccodes isolate 2-25 grown in potato dextrose agar medium was sporulated on oatmeal agar medium at 28°C. After harvesting spores in sterile tap water, the concentrations of the conidia suspensions were adjusted to 105 conidia mL−1. CaAMP1-silenced (TRV:CaAMP1) and unsilenced (TRV:00) pepper plants were inoculated by spraying conidia suspensions.
Pst DC3000 virulent to Arabidopsis ecotype Col-0 were used in this study. To prepare bacterial suspensions for inoculation of Arabidopsis leaves, Pst DC3000 was grown as described previously (Lee and Hwang, 2006). The density of the bacterial population was determined by plating serial dilutions on King's B medium supplemented with rifampicin (50 μg mL−1) at 28°C. Infected leaves were sampled immediately and at 1, 3, and 5 d after inoculation. Data are means ± sd of the log (cfu cm−2) of six replicates.
Hyaloperonospora parasitica isolate Noco2 was maintained in Arabidopsis (Col-0) by subculturing weekly. Seven- to 10-d-old seedlings were inoculated to produce a large quantity of inoculum. Spores on leaves inoculated with H. parasitica isolate Noco2 were collected in water. Seven-day-old seedlings were challenge inoculated by spraying with a suspension of asexual inoculum (5 × 104 conidiospores mL−1). The inoculated seedlings were covered with a transparent dome to maintain high humidity (80%–100%) and grown for 7 d at 17°C. Six days after inoculation, the degree of infection was determined by harvesting 40 inoculated cotyledons in 1 mL of water. After vigorous vortex mixing, the numbers of spores in three 10-μL aliquots from each spore suspension were counted using a hemocytometer. Asexual sporulation of H. parasitica also was visually assessed by counting the number of sporangiophores on both sides of cotyledons at 5 d after inoculation. A visual disease rating consisted of five classes: 0 to 5, 6 to 10, 11 to 15, 16 to 20, and over 20 sporangiophores per cotyledon. The infection and development of H. parasitica isolate Noco2 were assessed by staining inoculated seedlings with lactophenol-trypan blue (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue, dissolved in 10 mL of distilled water). Seven days after inoculation with H. parasitica, infected cotyledons were boiled for 5 min in the staining solution and destained overnight in chloral hydrate (2.5 g of chloral hydrate dissolved in 1 mL of distilled water). The destained cotyledons were then mounted in 70% glycerol for observation with a microscope.
Fusarium oxysporum f. sp. matthiolae strain 247.61 (Centraalbureau voor Schimmelcultures) and Alternaria brassicicola strain KACC40631 were grown on potato dextrose agar at 26°C for 2 to 3 weeks. To inoculate plants, the fungal spore density was adjusted to 5 × 105 spores mL−1 in sterile water. Arabidopsis plants grown as described above were soil drenched with the F. oxysporum spore suspension. Inoculation with A. brassicicola was performed by applying a 10-μL drop of spore suspension (5 × 105 spores mL−1) to the leaves. The inoculated plants were kept under a transparent cover to maintain high humidity and transferred to a growth chamber with 24°C day and 21°C night temperatures.
Isolation and Sequence Analysis of CaAMP1 cDNA
A pathogen-induced cDNA library was constructed using poly(A)+ mRNA extracted from pepper leaves inoculated with the avirulent strain Bv5-4a of Xcv (Choi et al., 2007). The pathogen-induced CaAMP1 cDNA was isolated from the pepper cDNA library using the differential hybridization method (Jung and Hwang, 2000) and sequenced on the ABI 310 DNA sequencer (Applied Biosystems).
Application of Elicitors
Stoppered 500-mL glass bottles containing pepper plants at the four-leaf stage were injected with ethylene to yield a final concentration of 5 μL L−1. MeJa (100 μm), SA (5 mm), BTH CGA245704 (10 μm), and H2O2 (100 μm) were sprayed onto pepper plants at the six-leaf stage. Pepper plants treated with MeJA were incubated in a vinyl bag. Control plants were sprayed with water. For wounding stress, the leaves were pricked with a needle.
Particle Bombardment
The coding region of the CaAMP1 gene was cloned between the cauliflower mosaic virus 35S promoter and the smGFP region of the binary vector p326GFP to generate a C-terminal fusion of smGFP to CaAMP1. For particle bombardment, the plasmids were purified using Qiagen Plasmid Maxi Kits according to the manufacturer's instructions (Qiagen). Onion (Allium cepa) epidermis was bombarded with gold particles coated with plasmids using a Bio-Rad PDS-1000/He particle delivery system. Bombarded specimens were incubated for 24 h on 1× Murashige and Skoog (MS) agar medium at 24°C and observed using a MRC-1024 confocal laser scanning microscope (Bio-Rad).
VIGS
The TRV-based VIGS system was used to silence CaAMP1 in pepper plants, as described by Liu et al. (2002). CaAMP1 sequences were amplified by PCR with the primers 5′-GAATTCATGATGAATGCTAATGGATT-3′ (forward) and 5′-GAATTCAGTCTGTGATCCCCGC-3′ (reverse). Fragments of 338 bp corresponding to the N-terminal coding region of the pepper CaAMP1 gene were cloned into the vector pTRV2 to yield pTRV2:CaAMP1, which was transformed into Agrobacterium tumefaciens strain GV3101. Transformants carrying pTRV1 or pTRV2:CaAMP1 were coinfiltrated into the fully expanded cotyledons of pepper plants (OD600=0.2). Plants were placed in a greenhouse at 25°C with a 16-h-light/8-h-dark photoperiod cycle to allow growth and viral spread. Pepper plants silenced for 4 to 5 weeks were used for pathogen inoculation.
Vector Construction and Arabidopsis Transformation
The binary vector pBIN35S was used to generate a plasmid for Arabidopsis transformation. The full-length CaAMP1 cDNA sequence from pBluescript SK− was obtained by digestion with BamHI and KpnI and cloned into pBIN35S. The recombinant plasmids were verified by sequencing. The binary plasmids were electroporated into A. tumefaciens strain AH109. The resulting agrobacteria were used to transform Arabidopsis (Col-0 and Ws-0) plants using the floral dipping procedure (Clough and Bent, 1998). Transgenic plants containing the pBIN35S constructs were selected on MS agar plates containing 50 μg mL−1 kanamycin.
Prior to pathogen inoculation, surface-sterilized seeds of Arabidopsis transgenic plants were plated on MS agar medium containing 50 μg mL−1 kanamycin and vernalized at 4°C for 2 d. After selection for 10 d, the kanamycin-resistant transgenic seedlings were grown in soil.
RNA-Blot and RT-PCR Analyses
Total RNA was prepared from Arabidopsis leaves using Trizol RNA extraction buffer (Invitrogen). To analyze the level of gene expression by northern blotting, equal quantities of RNA were separated on 1.2% formaldehyde-agarose gels in the presence of ethidium bromide and transferred to nylon membranes (Hybond N+; Amersham). The CaAMP1 cDNA was 32P labeled using a random priming kit (Boehringer Mannheim). Prehybridization and hybridization were performed at 65°C in 5% (w/v) dextran sulfate, 0.25 m disodium phosphate, pH 7.2, 7% (w/v) SDS, and 1 mm EDTA. The membranes were washed twice with 2× SSC, 0.1% SDS for 10 min each at room temperature and three times with 0.1× SSC, 0.1% SDS for 5 min each at 65°C. The hybridized blots were exposed to x-ray films.
To analyze gene expression in transgenic Arabidopsis plants by RT-PCR, total RNA (2 μg) from wild-type and transgenic plants was reverse transcribed using RT-AMV transcriptase (Roche) with oligo(dT) for 1 h at 42°C. PCR was carried out using ExTaq DNA polymerase (TaKaRa). The primers used to amplify AtPR1 were 5′-ATGAATTTTACTGGCTTCCAT-3′ (forward) and 5′-AACCCACATGTTCACGGCGGA-3′ (reverse). The primers used to amplify AtPR5 were 5′-TTCACATTCTCTTCCTCGTGTTCA-3′ (forward) and 5′-TCGTAGTTAGCTCCGGTACAAGTG-3′ (reverse). The primers used to amplify PDF1.2 were 5′-ATGGCTAAGTTTGCTTCCAT-3′ (forward) and 5′-ACATGGGACGTAACAGATAC-3′ (reverse). The primers used to amplify OPR1 were 5′-GGTCGATGGTTTCTAGCCAA-3′ (forward) and 5′-GCATGATCACATCAAACAGA-3′ (reverse). To detect AtPR1 and AtPR5 expression, amplification was programmed for 27 cycles, with each cycle consisting of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. For PDF1.2 and OPR1, amplification was also programmed for 25 cycles. The ubiquitin mRNA expression level was used as a quantitative control. Aliquots of individual PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under UV light.
Expression and Purification of Recombinant CaAMP1
pET32a vectors were used for expression of the CaAMP1-thioredoxin fusion protein in Escherichia coli. For construction of the CaAMP1:pET32a plasmid, the CaAMP1 coding region was amplified using the primers 5′-GAATTCATGGTTTCCAAAAGTAGTATTTTT-3′ (forward) and 5′-CTCGAGTTAGGCACAACAATAGTCACAACG-3′ (reverse). The DNA fragment was excised by digestion with EcoRI and XhoI and ligated into the expression vector pET32a. The resulting plasmid was used to transform E. coli BL21 and was verified by sequencing. Bacteria were grown to an appropriate density (OD600=0.6) at 37°C and induced with 0.1, 0.2, 0.5, 1.0, or 2.0 mm isopropyl thiogalactoside (IPTG) for various time periods to find optimum expression conditions. Optimal induction of CaAMP1 protein expression was achieved with 1 mm IPTG, and the culture was incubated for 4 h. Bacteria were harvested by centrifugation at 5,000g for 5 min at 4°C and stored at −70°C. The CaAMP1 protein was purified using the nickel-nitrilotriacetic acid agarose purification system kit (Invitrogen). The bacteria were suspended in a native binding buffer (50 mm NaPO4 and 500 mm NaCl, pH 8.0) and 8 mg of lysozyme for 30 min on ice, lysed by sonication on ice, and centrifuged at 5,000g for 15 min. Protein concentrations were assayed and SDS-PAGE was done. Soluble recombinant proteins (8 mL) were added to a superflow resin column (polypropylene column) equilibrated with native binding buffer. The column was washed with a native wash buffer (50 mm NaPO4, 500 mm NaCl, and 20 mm imidazole, pH 8.0), and the recombinant protein was eluted with 5 mL of native elution buffer (50 mm NaPO4, 500 mm NaCl, and 250 mm imidazole, pH 8.0).
Evaluation of in Vitro Antimicrobial Activity of the CaAMP1 Protein
The in vitro antimicrobial activity of the CaAMP1 protein was measured against plant pathogenic fungi, oomycetes, yeast, and bacteria. Minimum inhibitory concentrations of the CaAMP1 protein against these microorganisms were determined in a 48-well microtiter dish (Corning Glass Works). Protein samples were dialyzed for 24 h against 10 mm sodium phosphate buffer, pH 7.8, at 4°C and subsequently filter sterilized through 0.2 μm filters. Protein concentrations were determined and adjusted appropriately. Protein samples (100 μL) were pipetted into each well to give a total volume of 150 μL. The inocula used in this test were a zoospore suspension (104 zoospores mL−1) of Phytophthora capsici, a mycelial suspension of Rhizoctonia solani, spore suspensions (104 spores mL−1) of other plant pathogenic fungi (Alternaria brassicicola, Alternaria mali, Botrytis cinerea, Cladosporium cucumerinum, Colletotrichum coccodes, F. oxysporum f. sp. matthiolae, Magnaporthe grisea, and Sclerotinia sclerotiorum), yeast (104 cells mL−1; Candida albicans and Saccharomyces cerevisiae), and bacteria (104 cells mL−1; Bacillus subtilis, Micrococcus luteus, Pseudomonas syringae pv tomato, Ralstonia solanacearum, and Xcv). A volume of 10 μL of germ suspensions was added per well. The inoculated well plates were incubated at 28°C on a rotary shaker. Growth of the test microorganisms was evaluated after incubation for 2 to 4 d.
Bioassay of the Inhibition of Conidial Germination and Hyphal Growth by the CaAMP1 Protein
Conidial suspensions of A. brassicicola and F. oxysporum f. sp. matthiolae were shaken vigorously for 2 min. A 0.1-mL aliquot of the suspensions was dispensed into a microtube containing potato dextrose broth (Difco; 0.4 mL, 0.5%), and the CaAMP1 protein was added. After incubating F. oxysporum f. sp. matthiolae and A. brassicicola conidia for 6 h at 28°C, germinated conidia were counted on a hemocytometer using a light microscope.
Conidial suspensions of F. oxysporum f. sp. matthiolae and A. brassicicola in potato dextrose broth were incubated in microtubes until the hyphae had an average length of 30 μm. A 0.1-mL sample of the CaAMP1 protein at various concentrations was then added to the conidial suspensions. The mixtures were incubated until the control germlings attained an average length of approximately 400 μm. The lengths of 50 individual hyphae were determined using a light microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY548741 (CaAMP1), At2g14610 (PR1), At1g75040 (PR5), At5g44420 (PDF1.2), and At4g05320 (UBQ).
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors and Dr. U. Bonas (Martin-Luther-Universitaet) for A. tumefaciens strain GV3101.
This work was supported by the Crop Functional Genomics Center of the 21st Century (grant no. CG1133), Frontier Research Program, funded by the Ministry of Science and Technology, Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Byung Kook Hwang (bkhwang@korea.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42 385–414 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 [DOI] [PubMed] [Google Scholar]
- Bieri S, Potrykus I, Futterer J (2003) Effects of combined expression of antifungal barley seed proteins in transgenic wheat on powdery mildew infection. Mol Breed 11 37–48 [Google Scholar]
- Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208 155–165 [DOI] [PubMed] [Google Scholar]
- Block A, Schmelz E, O'Donnell PJ, Jones JB, Klee HJ (2005) Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiol 138 1481–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert I, Lee HI, Kush A, Chua NH, Raikhel N (1990) Wound-induced accumulation of mRNA containing a hevein sequence in laticifers of rubber tree (Hevea brasiliensis). Proc Natl Acad Sci USA 87 7633–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais CJ, Broglie R (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254 1194–1197 [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X (1998) Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA 95 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Oh SK, Park JM, Choi D (2007) Expression and promoter analyses of pepper CaCDPK4 (Capsicum annuum calcium dependent protein kinase 4) during plant defense response to incompatible pathogen. Plant Pathol J 23 76–89 [Google Scholar]
- Chung E, Seong E, Kim YC, Chung EJ, Oh SK, Lee S, Park JM, Joung YH, Choi D (2004) A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol Cells 17 377–380 [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coca MA, Damsz B, Yun DJ, Hasegawa PM, Bressan RA, Narasimhan ML (2000) Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J 22 61–69 [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P (2005) An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bolle MF, Osborn RW, Goderis IJ, Noe L, Acland D, Hart CA, Torrekens S, Van Leuven F, Broekaert WF (1996) Antimicrobial peptides from Mirabilis jalapa and Amaranthus caudatus: expression, processing, localization and biological activity in transgenic tobacco. Plant Mol Biol 31 993–1008 [DOI] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H (2001) Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol 127 852–862 [PMC free article] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38 473–486 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24 205–218 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y (2003) Virus-induced gene silencing. Methods Mol Biol 236 287–294 [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1997) Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritig B, Heitz T, Legrand M (1998) Antimicrobial proteins in induced plant defense. Curr Opin Immunol 10 16–22 [DOI] [PubMed] [Google Scholar]
- Gao AG, Hakimi SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CM (2000) Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat Biotechnol 18 1307–1310 [DOI] [PubMed] [Google Scholar]
- Garcia-Olmedo F, Molina A, Alamillo JM, Rodriguez-Palenzuela P (1998) Plant defense peptides. Biopolymers 47 479–491 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4 301–308 [DOI] [PubMed] [Google Scholar]
- Gonorazky AG, Regente MC, de la Canal L (2005) Stress induction and antimicrobial properties of a lipid transfer protein in germinating sunflower seeds. J Plant Physiol 162 618–624 [DOI] [PubMed] [Google Scholar]
- Gross P, Julius C, Schmelzer E, Hahlbrock K (1993) Translocation of cytoplasm and nucleus to fungal penetration site is associated with depolymerization of microtubules and defense gene activation in infected, cultured parsley cells. EMBO J 12 1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, Götz F, Glawischnig E, Lee J, Felix G, et al (2007) Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem 282 32338–32348 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14 177–193 [DOI] [PubMed] [Google Scholar]
- Hancock RE, Lehrer R (1998) Cationic peptides: a new source of antibiotics. Trends Biotechnol 16 82–88 [DOI] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi DS, Kim YJ, Hwang BK (2008) Novel GDSL-type pepper lipase gene, CaGLIP1, involved in bacterial disease susceptibility and abiotic stress tolerance. Planta 227 539–558 [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J (2003) Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Jung EH, Jung HW, Lee SC, Han SW, Heu S, Hwang BK (2004) Identification of a novel pathogen-induced gene encoding a leucine-rich repeat protein expressed in phloem cells of Capsicum annuum. Biochim Biophys Acta 1676 211–222 [DOI] [PubMed] [Google Scholar]
- Jung HW, Hwang BK (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13 136–142 [DOI] [PubMed] [Google Scholar]
- Jung HW, Lim CW, Lee SC, Choi HW, Hwang CH, Hwang BK (2008) Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227 409–425 [DOI] [PubMed] [Google Scholar]
- Kagan IA, Hammerschmidt R (2002) Arabidopsis ecotype variability in camalexin production and reaction to infection by Alternaria brassicicola. J Chem Ecol 28 2121–2140 [DOI] [PubMed] [Google Scholar]
- Keshavarzi M, Soylu S, Brown I, Bonas U, Nicole M, Rossiter J, Mansfield J (2004) Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17 805–815 [DOI] [PubMed] [Google Scholar]
- Kim BS, Kim YC, Shin KS, Kim JH (2007) Near-isogenic lines for genes conferring hypersensitive resistance to bacterial spot in chili pepper. Plant Pathol J 23 155–160 [Google Scholar]
- Kim YJ, Martin GB (2004) Molecular mechanisms involved in bacterial speck disease resistance of tomato. Plant Pathol J 20 7–12 [Google Scholar]
- Kjemtrup S, Nimchuk Z, Dangl JL (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr Opin Microbiol 3 73–78 [DOI] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T (2007) Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact 20 120–128 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48 251–275 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang BK (2005) Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta 221 790–800 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang BK (2006) CASAR82A, a pathogen-induced pepper SAR8.2, exhibits an antifungal activity and its overexpression enhances disease resistance and stress tolerance. Plant Mol Biol 61 95–109 [DOI] [PubMed] [Google Scholar]
- Lee SC, Kim YJ, Hong JK, Hwang BK (2000) Pepper gene encoding thionin is differentially induced by pathogens, ethylene and methyl jasmonate. Physiol Mol Plant Pathol 56 207–216 [Google Scholar]
- Lee YK, Hwang BK (1996) Differential induction and accumulation of β-1,3-glucanase and chitinase isoforms in the intercellular space and leaf tissues of pepper by Xanthomonas campestris pv. vesicatoria infection. J Phytopathol 144 79–87 [Google Scholar]
- Li Q, Lawrence CB, Xing HY, Babbitt RA, Bass WT, Maiti IB, Everett NP (2001) Enhanced disease resistance conferred by expression of an antimicrobial magainin analog in transgenic tobacco. Planta 212 635–639 [DOI] [PubMed] [Google Scholar]
- Liu G, Holub EB, Alonso JM, Ecker JR, Fobert PR (2005) An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J 41 304–318 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30 415–429 [DOI] [PubMed] [Google Scholar]
- Lorito M, Woo SL, Garcia I, Colucci G, Harman GE, Pintor-Toro JA, Filippone E, Muccifora S, Lawrence CB, Zoina A, et al (1998) Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci USA 95 7860–7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei ME, Mithofer A, Boland W (2007) Before gene expression: early events in plant-insect interaction. Trends Plant Sci 12 310–316 [DOI] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact 19 123–129 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22 523–529 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50 500–513 [DOI] [PubMed] [Google Scholar]
- Nandi A, Welti R, Shah J (2004) The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 18 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37 579–609 [DOI] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S (2000) Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat Biotechnol 18 1162–1166 [DOI] [PubMed] [Google Scholar]
- Park JM (2005) The hypersensitive response: a cell death during disease resistance. Plant Pathol J 21 99–101 [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC (2002) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29 569–579 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerré-Tugayé MT, Rosahl S, et al (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10 372–379 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55 495–519 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM (1997) Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareena S, Poovannan K, Kumar KK, Raja JAJ, Samiyappan R, Sudhakar D, Balasubramanian P (2006) Biochemical responses in transgenic rice plants expressing a defence gene deployed against the sheath blight pathogen, Rhizoctonia solani. Curr Sci 91 1529–1532 [Google Scholar]
- Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DJ (2003) Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol 132 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E (2002) Cell polarization, a crucial process in fungal defense. Trends Plant Sci 7 411–415 [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitrennikoff CP (2001) Antifungal proteins. Appl Environ Microbiol 67 2883–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Hema R, Anand A, Kang L, Udayakumar M, Mysore KS (2007) A systemic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol 176 782–791 [DOI] [PubMed] [Google Scholar]
- Shatters RG Jr, Boykin LM, Lapointe SL, Hunter WB, Weathersbee AA III (2006) Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. J Mol Evol 63 12–29 [DOI] [PubMed] [Google Scholar]
- Škalamera D, Heath MC (1998) Changes in the cytoskeleton accompanying infection-induced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. Plant J 16 191–200 [DOI] [PubMed] [Google Scholar]
- Soares-Costa A, Beltramini LM, Thiemann OH, Henrique-Silva F (2002) A sugarcane cystatin: recombinant expression, purification, and antifungal activity. Biochem Biophys Res Commun 296 1194–1199 [DOI] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK (2006) Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61 897–915 [DOI] [PubMed] [Google Scholar]
- Sohn KH, Lei R, Nemri A, Jones JD (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19 4077–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink RG, Dilmac N, Allen A, Smith TJ, Shah DM, Hockerman GH (2004) Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol 135 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Metraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35 235–270 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Chang HS, Zhu T, Glazebrook J (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Chen X, Bluhm B, Salmeron J, Dietrich R, Mengiste T (2004) The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J 40 558–574 [DOI] [PubMed] [Google Scholar]
- Wang H, Ye XY, Ng TB (2001) Purification of chrysancorin, a novel antifungal protein with mitogenic activity from garland chrysanthemum seeds. Biol Chem 382 947–951 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Genes Dev 11 1621–1639 [DOI] [PubMed] [Google Scholar]
- Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415 389–395 [DOI] [PubMed] [Google Scholar]
- Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, Dong X, Mou Z (2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J 52 1066–1079 [DOI] [PubMed] [Google Scholar]