Abstract
Phytochrome A is the primary photoreceptor for mediating various far-red light-induced responses in higher plants. We recently showed that Arabidopsis (Arabidopsis thaliana) FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1), a pair of homologous proteins sharing significant sequence homology to Mutator-like transposases, act as novel transcription factors essential for activating the expression of FHY1 and FHL (for FHY1-like), whose products are required for light-induced phytochrome A nuclear accumulation and subsequent light responses. FHY3, FAR1, and Mutator-like transposases also share a similar domain structure, including an N-terminal C2H2 zinc finger domain, a central putative core transposase domain, and a C-terminal SWIM motif (named after SWI2/SNF and MuDR transposases). In this study, we performed a promoter-swapping analysis of FHY3 and FAR1. Our results suggest that the partially overlapping functions of FHY3 and FAR1 entail divergence of their promoter activities and protein subfunctionalization. To gain a better understanding of the molecular mode of FHY3 function, we performed a structure-function analysis, using site-directed mutagenesis and transgenic approaches. We show that the conserved N-terminal C2H2 zinc finger domain is essential for direct DNA binding and biological function of FHY3 in mediating light signaling, whereas the central core transposase domain and C-terminal SWIM domain are essential for the transcriptional regulatory activity of FHY3 and its homodimerization or heterodimerization with FAR1. Furthermore, the ability to form homodimers or heterodimers largely correlates with the transcriptional regulatory activity of FHY3 in plant cells. Together, our results reveal discrete roles of the multiple domains of FHY3 and provide functional support for the proposition that FHY3 and FAR1 represent transcription factors derived from a Mutator-like transposase(s).
As sessile organisms, higher plants have evolved a network of photoreceptors to sense changes in the ambient light environment and undergo adaptive growth and development. Among these photoreceptors, phytochromes are the best characterized and exist in two distinct forms, Pr and Pfr. Phytochromes are activated when their red light-absorbing Pr form is photoconverted into the biologically active, far-red (FR) light-absorbing Pfr form. The Pfr form is converted back to the biologically inactive Pr form on absorption of FR light to desensitize light signaling (Deng and Quail, 1999; Neff et al., 2000). In Arabidopsis (Arabidopsis thaliana), phytochromes are encoded by a five-member gene family, PHYTOCHROME A (PHYA) to PHYE. Among all phytochromes, phyA is unique in several respects. First, phyA is a type I (light-labile) phytochrome, whereas phyB to phyE are type II (light-stable) phytochromes (Hirschfeld et al., 1998). Second, phyA is not only responsible for the photoirreversible very low fluence response (such as promotion of seed germination) triggered by a broad spectrum of irradiations (UV, visible, and FR light), but also is the primary photoreceptor mediating the high irradiance response to continuous FR light (FRc), including inhibition of hypocotyl elongation, opening of the apical hook, expansion of cotyledons, accumulation of anthocyanin, and FRc-preconditioned blocking of greening (Whitelam et al., 1993; Botto et al., 1996).
Intensive molecular genetic studies with Arabidopsis have helped to establish a framework of the molecular events linking phyA activation to regulated nuclear gene expression. It is believed now that upon photoactivation, phyA converts into the active Pfr form and is translocated from cytoplasm, where it resides in darkness, into the nucleus (Kircher et al., 1999; Kim et al., 2000). In the nucleus, phyA could interact directly with a set of transcription factors, such as the basic helix-loop-helix proteins PIF3 (for PHYTOCHROME INTERACTING FACTOR3) and PIF1 (Ni et al., 1999; Huq et al., 2004; Duek and Fankhauser, 2005), to induce a transcriptional cascade necessary to implement FR light-mediated photomorphogenic responses (Tepperman et al., 2001; Khanna et al., 2006). On the other hand, genetic studies have identified over 20 signaling intermediates required for the full range of phyA-mediated FR light responses (for review, see Quail, 2002; Wang and Deng, 2003). Among all phyA signaling mutants identified so far, the phenotypes of fhy3 (for far-red elongated hypocotyl3) and fhy1 mutants most closely resemble that of the phyA photoreceptor mutant, and it has been suggested that FHY3 and FHY1 act early and close to the photoreceptor itself (Desnos et al., 2001; Wang and Deng, 2002; Wang et al., 2002). The Arabidopsis genome contains one close homolog for FHY1, named FHL (for FHY1-like). The gene products of FHY1 and FHL are localized in both the cytoplasm and nucleus (Desnos et al., 2001; Zhou et al., 2005). It was shown recently that FHY1 and FHL act together to mediate the light-dependent nuclear accumulation of phyA, possibly through direct physical interaction with the Pfr form of phyA (Hiltbrunner et al., 2005, 2006).
FHY3 and FAR1 (for FAR-RED-IMPAIRED RESPONSE1) were identified as two essential positive regulators of phyA signaling in forward genetic screens (Whitelam et al., 1993; Hudson et al., 1999). Molecular cloning revealed that FHY3 and FAR1 encode two homologous proteins sharing 47.3% amino acid identity and 79.4% amino acid similarity (Hudson et al., 1999; Wang and Deng, 2002). Mutations in FAR1 cause similar but weaker defects in hypocotyl elongation and anthocyanin accumulation, compared with fhy3 mutants. However, FRc-preconditioned greening block, cotyledon opening, and expansion are largely not affected in far1 mutants (Hudson et al., 1999; Wang and Deng, 2002). Strikingly, FHY3 and FAR1 share extensive sequence similarities with MURA, the transposase encoded by the Mutator element of maize (Zea mays; Hudson et al., 2003), and the predicted transposase of the maize mobile element Jittery (Xu et al., 2004). Both of these transposons are members of the superfamily of Mutator-like elements (MULEs; Lisch, 2002; Walbot and Rudenko, 2002). FHY3, FAR1, and Mutator-like transposases also share a similar domain structure, including an N-terminal C2H2 zinc finger domain, a central putative core transposase domain, and a C-terminal SWIM motif (named after SWI2/SNF and MuDR transposases; Makarova et al., 2002). It has been proposed that FHY3/FAR1 may control the expression of their target genes by a mechanism that has evolved from an ancestral, MURA-like transposase (Hudson et al., 2003; Lin and Wang, 2004; Babu et al., 2006). In a recent study, we demonstrated that FHY3 and FAR1 act together to directly activate the expression of FHY1 and FHL, whose products are essential for light-induced phyA nuclear accumulation and subsequent light responses (Lin et al., 2007).
To gain a better understanding of the molecular basis underpinning the observed partially overlapping function of FHY3 and FAR1, we performed a promoter-swap analysis of FHY3 and FAR1. Our results suggest that the promoter activities of this pair of homologous genes have diverged and protein subfunctionalization has occurred. We also performed a detailed structure-function analysis of Arabidopsis FHY3 using site-directed mutagenesis. Our results suggest that the multiple domains of FHY3 perform discrete but essential functions in mediating phyA signal transduction. Additionally, our results provide functional support for the proposition that FHY3 and FAR1 define a novel class of transcription factors derived from an ancient Mutator-like transposase(s).
RESULTS
FHY3 and FAR1 Play Partially Overlapping Roles in phyA Signaling
Phylogenetic analysis suggested that FAR1 and FHY3 most likely arose from a gene duplication event that predates the divergence of core eudicots, at least approximately 110 million years ago (Lin et al., 2007). However, despite their relatively ancient divergence in evolution, the similar yet distinct phenotypes displayed by loss-of-function fhy3 and far1 mutants suggest that FHY3 and FAR1 likely play partially overlapping functions in phyA signaling. Consistent with this, we previously reported that the fhy3-1 far1-2 double mutants display an additive effect. In addition, we previously showed that overexpression of FAR1 or FHY3 (driven by the 35S promoter) can largely suppress each other's loss-of-function mutant phenotype (Wang and Deng, 2002). Furthermore, we showed that overexpression of FHY3 and FAR1 can essentially restore the genome expression profiles abolished by the far1-2 and fhy3-1 mutations, respectively (Wang et al., 2002). While these data clearly suggest a significant functional overlap between FHY3 and FAR1, such overexpression data should be interpreted with caution. We cannot formally eliminate the possibility that ectopic expression of these genes causes them to assume a novel role. Thus, the extent of functional overlap and the molecular basis for their functional divergence remain largely unknown.
To explore the molecular basis of partially overlapping function of FHY3 and FAR1, we generated FHY3 and FAR1 transgenes containing their endogenous promoter (approximately 2 kb from the ATG start codon, including the 5′-untranslated region) to drive the full-length genes plus 3′-untranslated region and introduced them into their respective mutant background. As expected, the FHY3p∷FHY3 and FAR1p∷FAR1 transgenes completely rescued the fhy3-4 (a putative null allele of fhy3; Wang and Deng, 2002) and far1-2 (a putative null allele of far1; Hudson et al., 1999) mutant phenotypes, respectively (Fig. 1, A and B). This observation suggests that the FHY3 and FAR1 promoters and their protein-coding regions are fully functional in rescuing their respective mutant phenotypes. We next tested the biological function of the FHY3 and FAR1 genes driven by each other's promoter. FAR1p∷FHY3 completely rescued the far1-2 mutant phenotype, but it only partially rescued the fhy3-4 mutant phenotype (Fig. 1, C and D), although the expression levels of the FAR1p∷FHY3 transgene in these lines were comparable (Supplemental Fig. S1, C and D). This observation suggests that FHY3 protein can substitute the function of FAR1 protein (at least in our assay conditions). The same observation also suggests that the FAR1 promoter cannot fully substitute the activity of FHY3 promoter. Similarly, FHY3p∷FAR1 can completely restore the far1-2 mutant phenotype, but it failed to rescue the fhy3-4 mutant phenotype (Fig. 1, E and F). The FAR1 transcript level in the FHY3p∷FAR1/fhy3-4 background is comparable to that in the FHY3p∷FAR1/far1-2 background (Supplemental Fig. S1, E and F). These observations suggest that FAR1 protein cannot fully substitute the function of FHY3 protein. On the other hand, the complete rescue of the far1-2 mutant phenotype by FHY3p∷FAR1 suggests that the FHY3 promoter can replace the function of FAR1 promoter. Together, our results suggest that FHY3 promoter and FHY3 protein can recapitulate the activity of FAR1 promoter and FAR1 protein, respectively; however, the FAR1 promoter and FAR1 protein cannot substitute the function of FHY3 promoter and FHY3 protein, respectively.
Figure 1.
Partially overlapping function of FHY3 and FAR1 in regulating phyA signaling. A, FHY3p∷FHY3 completely rescues the fhy3-4 mutant phenotype. B, FAR1p∷FAR1 completely rescues the far1-2 mutant phenotype. C, FAR1p∷FHY3 completely rescues the far1-2 mutant phenotype. D, FAR1p∷FHY3 partially rescues the fhy3-4 mutant phenotype. E, FHY3p∷FAR1 completely rescues the far1-2 mutant phenotype. F, FHY3p∷FAR1 fails to rescue the fhy3-4 mutant phenotype. Four-day-old FR light-grown seedlings are shown. Bars = 2 mm. [See online article for color version of this figure.]
The N-Terminal C2H2 Zinc Finger Domain of FHY3 Is Required for Its DNA-Binding Activity and Its Biological Function
To further define the structure-function relationship of FHY3, we performed mutagenesis studies of a group of highly conserved amino acids in the predicted N-terminal C2H2 zinc finger domain, the central core transposase domain, and the C-terminal SWIM motif (Fig. 2A; Supplemental Fig. S2). Our previous domain-deletion analysis suggested that the DNA-binding activity of FHY3 and FAR1 resides in the N-terminal domain containing a predicted C2H2-type zinc finger motif (Lin et al., 2007). This zinc-chelating finger motif is shared by the putative DNA-binding domain of MULE transposases and belongs to the WRKY-GCM1 family of zinc fingers (Babu et al., 2006; Lin et al., 2007). To provide direct evidence for the notion that this conserved C2H2 zinc finger motif is responsible for direct DNA binding, we generated a series of mutants in which the conserved Cys (C) and His (H) residues (C118, C157, H180, and H182), which are presumably involved in chelating the zinc atom, were changed into Ala (A): Bm1 for C157A single mutation, Bm11 for C118A single mutation, Bm14 for C157A H180A H182A triple mutation, and Bm13 for C118A C157A H180A H182A quadruple mutation. First, we tested whether these mutations affect their ability to bind and activate FHY1p∷LacZ and FHLp∷LacZ reporter gene expression in a yeast one-hybrid assay. As reported previously (Lin et al., 2007), FHY3 wild-type protein, in fusion with the GAL4 activation domain (GAD), could bind to the FHY1 and FHL promoters and activate both reporter genes, whereas GAD itself and all zinc finger mutant fusion proteins (GAD-Bm1, GAD-Bm11, GAD-Bm14, and GAD-Bm13) were not able to activate these reporter genes (Fig. 2B), even though these fusion proteins were expressed at similar levels (Fig. 2C). This result indicates that mutations in these conserved Cys and His residues of FHY3 abolish its DNA-binding activity.
Figure 2.
The C2H2 zinc finger motif is essential for direct DNA binding. A, A molecular diagram of FHY3 protein showing its three structural domains. The sites selected for mutagenesis studies are indicated. aa, Amino acids. B, Yeast one-hybrid assay showing that GAD-Bm1, GAD-Bm11, GAD-Bm13, and GAD-Bm14 lose the ability to activate FHY1p∷LacZ and FHLp∷LacZ reporter gene expression in yeast. C, Immunoblot analysis showing the similar levels of expression for the GAD-FHY3, GAD-Bm1, GAD-Bm11, GAD-Bm13, and GAD-Bm14 fusion proteins in yeast. D, EMSA showing that GST-FHY3N protein binds to the FHY1 promoter oligonucleotide probe and causes the formation of a shifted band (arrowhead), whereas GST-Bm1N, GST-Bm11N, and GST-Bm13N are unable to bind to the FHY1 oligonucleotide probe. FP, Free probe.
We next used an electrophoresis mobility shift assay (EMSA) to further test their effects on direct DNA binding. The N-terminal 200-amino acid fragment of FHY3 containing the C2H2 zinc finger domain was expressed as a glutathione S-transferase (GST) fusion protein. The wild-type FHY3 fusion protein (GST-FHY3N) can bind to the 32P-labeled FHY1 promoter fragment and caused a mobility shift, and the various mutant fusion proteins (GST-Bm1N, GST-Bm11N, and GST-Bm13N) all failed to bind to the FHY1 probe (Fig. 2D). This observation further supports the notion that the C2H2 zinc finger motif is directly involved in DNA binding.
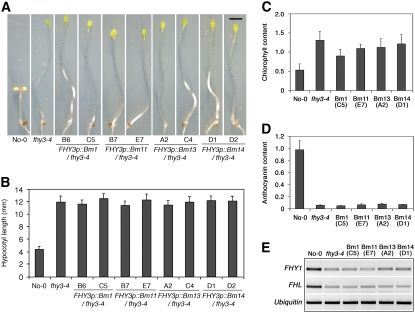
To test the function of the C2H2 zinc finger motif in vivo, we generated transgenic plants expressing the various FHY3 mutant forms (driven by the FHY3 endogenous promoter) in the fhy3-4 mutant background. All transgenes of zinc finger mutant forms failed to complement the hypocotyl growth inhibition and cotyledon-unfolding phenotype of the fhy3-4 mutants (over 40 independent transgenic lines were examined for each transgene, and two representative lines are shown; Fig. 3, A and B), although their transcription levels were higher than that of the endogenous FHY3 gene in a wild-type background (Supplemental Fig. S3A). As prolonged FR light treatment impairs subsequent greening upon transferring to white light conditions (Barnes et al., 1996), we also examined whether the various zinc finger mutant forms can rescue the FR light-preconditioned blocking of greening defect observed with fhy3-4 mutants (Wang and Deng, 2002). Wild-type and various transgenic seedlings were first grown in FRc for 3 d and then transferred to white light for 2 d to promote chloroplast maturation. Similar to the fhy3-4 mutants, all transgenic plants expressing these mutant forms of FHY3 accumulated much higher levels of chlorophyll than wild-type plants (Fig. 3C). In addition, transgenic plants harboring various zinc finger mutant forms of FHY3 displayed similar defects in FR light-induced anthocyanin accumulation (Fig. 3D). Furthermore, these transgenes failed to restore the expression of FHY1 and FHL to wild-type levels (Fig. 3E). Together, these results convincingly support the notion that the C2H2 zinc finger motif is involved in direct DNA binding and that this activity is essential for FHY3 function in planta.
Figure 3.
The zinc finger motif is essential for the biological function of FHY3. A, Representative images showing that FHY3p∷Bm1, FHY3p∷Bm11, FHY3p∷Bm13, and FHY3p∷Bm14 fail to rescue the fhy3-4 mutant phenotype. Bar = 2 mm. B, Hypocotyl lengths of the seedlings in A. Error bars represent sd from 20 seedlings. C, Quantification of chlorophyll accumulation in seedlings of various genotypes. Error bars represent sd of triplicate experiments. D, Quantification of anthocyanin accumulation in seedlings of various genotypes. Error bars represent sd of triplicate experiments. For A to D, seedlings were grown under FRc for 4 d. E, Semiquantitative RT-PCR of FHY1 and FHL genes in various genotypes. Seedlings were grown in darkness for 4 d and then transferred to FR light for 6 h before RNA extraction. RT-PCR of a Ubiquitin gene is shown at bottom as a positive control. [See online article for color version of this figure.]
The Transposase Catalytic Domain and SWIM Motif of FHY3 Are Required for Its Transcriptional Activation Activity
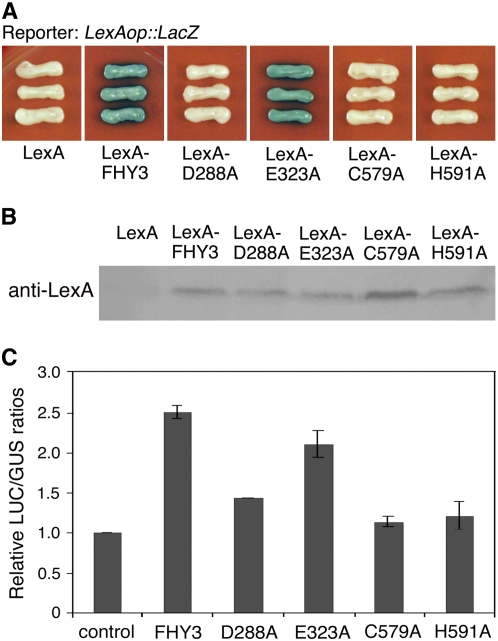
Our previous study suggested that the entire C-terminal region of FHY3 and FAR1 containing the central putative transposase catalytic domain and the SWIM zinc finger motif is required for their transcriptional activation activity (Lin et al., 2007). Notably, mutations in two conserved amino acids of FHY3, D283N and G305R, corresponding to the molecular lesions of the fhy3-10 and fhy3-9 mutant alleles (Wang and Deng, 2002), abolished the transcriptional activation activity of FHY3 (Lin et al., 2007). To further assess the role of the core catalytic transposase domain and the SWIM motif in transcriptional regulation of FHY3, we mutated two other conserved amino acids in the transposase catalytic domain (D288A and E323A) and two amino acids in the SWIM zinc finger domain (C579A and H591A; Fig. 2A). Both D288 and E323 are highly conserved in FHY3, FAR1, other FHY3/FAR1-related proteins (named FRS proteins; Lin and Wang, 2004) and Mutator-like transposases, and D288 is one of the three amino acids predicted to form a putative DDE catalytic triad motif shared by FHY3/FAR1 and Mutator-like transposases (Lin et al., 2007; Supplemental Fig. S2). These mutant forms of FHY3 were fused with the LexA DNA-binding domain and cotransformed with the LexAop∷LacZ reporter gene into the yeast strain EGY48 to test their effects on transcriptional activation of the reporter gene. Consistent with our previous study, LexA-FHY3 strongly activated the LexAop∷LacZ reporter gene expression, compared with LexA itself (Lin et al., 2007). Strikingly, LexA-D288A, LexA-C579A, and LexA-H591A failed to activate the reporter gene expression (similar to previously reported LexA-D283N and LexA-G305R; Lin et al., 2007), but LexA-E323A retained a similar activation activity as LexA-FHY3 (Fig. 4A). Immunoblot analysis confirmed that these fusion proteins were expressed at similar levels (Fig. 4B). These results suggest that D288, C579, and H591, but not E323, are essential for the transcriptional activation activity of FHY3, at least in the yeast cells.
Figure 4.
D288, C579, and H591 are required for the transcriptional activation activity of FHY3 in yeast and Arabidopsis protoplasts. A, Yeast one-hybrid assay showing that the LexA-FHY3 and LexA-E323A fusion proteins activate LexA operator-driven LacZ reporter gene (LexAop∷LacZ) expression in yeast to similar levels, whereas LexA by itself, LexA-D288A, LexA-C579A, and LexA-H591A fail to activate reporter gene expression. B, Immunoblot analysis showing similar expression levels of LexA-FHY3, LexA-D288A, LexA-E323A, LexA-C579A, and LexA-H591A fusion proteins in yeast. C, Wild-type FHY3 activates FHY1p∷LUC reporter gene expression in Arabidopsis protoplasts. E323A slightly reduces the reporter gene expression, D288A causes a modest reduction in the reporter gene expression, and C579A and H591A fail to activate the reporter gene expression. Error bars represent sd of triplicate experiments.
Next, we examined the effects of these mutations on transcriptional activation activity of FHY3 in plant cells by coexpressing the various mutant forms of FHY3 with the FHY1p∷LUC (for LUCIFERASE) reporter gene and the 35S∷GUS internal control plasmid in Arabidopsis protoplasts. The relative activation activity was measured as the ratio of LUC to GUS. Consistent with the observation made in yeast one-hybrid assay (Fig. 4A), E323A activated the reporter gene expression similar to a wild-type level, but the C579A and H591A mutations essentially abolished the ability to activate FHY1p∷LUC reporter gene expression in Arabidopsis protoplasts. Intriguingly, in contrast to the loss of activation activity observed in yeast cells, the D288A mutant form of FHY3 partially activated the FHY1p∷LUC reporter gene expression in Arabidopsis protoplasts (Fig. 4C). The discrepancy observed between the yeast one-hybrid assay and the Arabidopsis protoplast assay for the D288A mutant is likely due to the different experimental systems. In plant cells, there might be additional FHY3 functional partners to help restore the partial activity of D288A, whereas such factors might be missing in the yeast cells. Regardless, our results suggest that the SWIM zinc finger domain is essential for the transcriptional activation activity of FHY3. D288 might be required for its optimal activity, but E323 is not essential for transcriptional activation activity of FHY3.
To further test the in vivo functional significance of these conserved amino acids, we generated transgenic plants expressing these mutant versions of FHY3 (driven by the FHY3 endogenous promoter) in the fhy3-4 mutant background. In order to test a possible effect of these mutations on the subcellular localization of FHY3 protein (see below), these mutant proteins were tagged with yellow fluorescent protein (YFP) at their C termini. The wild-type FHY3p∷FHY3-YFP transgene successfully rescued the fhy3-4 mutant phenotype (Supplemental Fig. S4), suggesting that the YFP tag does not affect the normal function of FHY3 protein. Consistent with their transcriptional activation activities observed in Arabidopsis protoplasts, FHY3p∷E323A-YFP fully rescued the fhy3-4 mutant phenotype, including hypocotyl elongation, FRc-preconditioned block of greening, anthocyanin accumulation, and induction of FHY1/FHL expression (Fig. 5). Also consistent with its transcriptional activation activity observed in Arabidopsis protoplasts, FHY3p∷D288A-YFP partially rescued the fhy3-4 mutant hypocotyl elongation and FRc-preconditioned block of greening phenotype, but intriguingly, it nearly restored anthocyanin accumulation and FHY1/FHL expression to wild-type levels (Fig. 5). We repeated this experiment three times, and the results are reproducible. The reasons for the observed discrepancies of FHY3p∷D288A-YFP in rescuing various aspects of seedling photomorphogenesis are not clear. It is possible that FHY3 may regulate hypocotyl elongation and chlorophyll and anthocyanin accumulation through targeting different sets of downstream genes. Anthocyanin accumulation and FHY1/FHL gene expression might be more sensitive to the functional status or threshold level of FHY3 protein, whereas hypocotyl elongation inhibition and FRc-preconditioned blocking of greening may require full activity of FHY3.
Figure 5.
Phenotypic analysis of FHY3p∷D288A-YFP and FHY3p∷E323A-YFP transgenic plants. A, Representative images showing that FHY3p∷D288A-YFP partially rescues the fhy3-4 mutant phenotype, while FHY3p∷E323A-YFP completely rescues the fhy3-4 mutant phenotype. Bar = 2 mm. B, Hypocotyl lengths of the seedlings in A. Error bars represent sd from 20 seedlings. C, Quantification of chlorophyll accumulation in seedlings of various genotypes. Bars represent sd of triplicate experiments. D, Quantification of anthocyanin accumulation in seedlings of various genotypes. Error bars represent sd of triplicate experiments. For A to D, seedlings were grown under FRc for 4 d. E, Semiquantitative RT-PCR of FHY1 and FHL genes in various genotypes. Seedlings were grown in darkness for 4 d and then transferred to FR for 6 h before RNA extraction. RT-PCR of a Ubiquitin gene is shown at bottom as a positive control. [See online article for color version of this figure.]
In agreement with the observed loss of transcriptional activation activity in yeast and in Arabidopsis protoplasts, FHY3p∷C579A-YFP and FHY3p∷H591A-YFP were not effective in rescuing the fhy3-4 mutant phenotype (Fig. 6), even though the transcript levels of these transgenes were comparable and higher than the endogenous FHY3 gene in wild-type plants (Supplemental Fig. S3, B and C). These results indicate that C579 and H591 in the SWIM domain are essential for FHY3 function.
Figure 6.
The C-terminal SWIM domain is critical for FHY3 biological function. A, Representative images showing that FHY3p∷C579A-YFP and FHY3p∷H591A-YFP fail to rescue the fhy3-4 mutant phenotype. Bar = 2 mm. B, Hypocotyl lengths of the seedlings in A. Error bars represent sd from 20 seedlings. C, Quantification of chlorophyll accumulation in seedlings of various genotypes. Error bars represent sd of triplicate experiments. D, Quantification of anthocyanin accumulation in seedlings of various genotypes. Error bars represent sd of triplicate experiments. For A to D, seedlings were grown under FRc for 4 d. E, Semiquantitative RT-PCR of FHY1 and FHL genes in various genotypes. Seedlings were grown in darkness for 4 d and then transferred to FR light for 6 h before RNA extraction. RT-PCR of a Ubiquitin gene is shown at bottom as a positive control. [See online article for color version of this figure.]
Transcriptional Activation Activity of FHY3 Correlates with Its Dimerization Ability
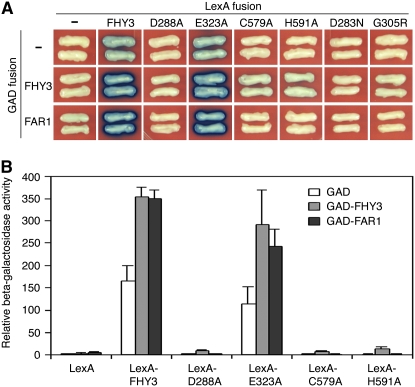
FHY3 and FAR1 proteins were previously shown to be able to form homodimers by self-association or heterodimers with each other (Wang and Deng, 2002, Hudson et al., 2003). To determine which of the structural domains of FHY3 is involved in dimerization, we performed a yeast two-hybrid assay. As previously reported, although both LexA-FHY3 and LexA-FAR1 can autoactivate the LexAop∷LacZ reporter gene to certain extents, when combined with GAD-FHY3 or GAD-FAR1, the activities of the reporter gene are significantly increased. Notably, LexA-D288A, LexA-C579A, LexA-H591A, LexA-D283N, and LexA-G305R failed to activate the LexAop∷LacZ reporter gene in combination with GAD-FHY3 or GAD-FAR1, but LexA-E323A retained the ability to activate LacZ reporter gene expression (Fig. 7). This observation suggests that both the central core transposase domain and the C-terminal SWIM motif are essential for FHY3 homodimerization or its heterodimerization with FAR1 and that this ability largely correlates with FHY3 transcriptional activation activity in yeast cells.
Figure 7.
The central transposase catalytic domain and the C-terminal SWIM domain are required for dimerization in yeast. A, Yeast two-hybrid assay showing that LexA-FHY3 and LexA-E323A enhance the activation of LexAop∷LacZ reporter gene expression in combination with GAD-FHY3 or GAD-FAR1, whereas LexA-D288A, LexA-C579A, LexA-H591A, LexA-D283N, and LexA-G305R fail to activate the reporter gene in combination with GAD-FHY3 or GAD-FAR1. B, Quantification of the relative β-galactosidase activities for various combinations shown in A. Error bars represent sd of six replicates.
FHY3 and FAR1 proteins are localized in the nucleus in plant cells (Hudson et al., 1999; Wang and Deng, 2002; Lin et al., 2007). To examine a possible effect of D288A, E323A, and H591A amino acid substitutions on FHY3 nuclear targeting, we examined the subcellular localization of FHY3-YFP, D288A-YFP, E323A-YFP, and H591A-YFP. Their homozygous transgenic plants were grown in darkness for 4 d, and the fluorescence signal was monitored using a confocal microscope. Like the wild-type FHY3-YFP fusion protein, the D288A-YFP, E323A-YFP, and H591A-YFP fusion proteins were all exclusively targeted into the nucleus (Fig. 8). This result indicates that the D288A, E323A, and H591A amino acid substitutions do not affect nuclear localization of the FHY3 protein.
Figure 8.
D288A, E323A, and H591A do not affect nuclear localization of FHY3. Confocal images of transgenic plants expressing FHY3p∷FHY3-YFP, FHY3p∷D288A-YFP, FHY3p∷E323A-YFP, and FHY3p∷H591A-YFP showing that the wild-type (WT) and mutant proteins (all fused with YFP) are exclusively localized in the root nucleus (left images). The middle images are bright-field images, and the right images are merged images of the left and middle images. Bars = 25 μm.
DISCUSSION
Partially Overlapping Function of FHY3 and FAR1 Entails Divergence of Their Promoter Activities and Subfunctionalization of the Proteins
Among the previously identified phyA signaling mutants, the phenotypes of fhy3 and fhy1 mutants most closely resemble that of the phyA photoreceptor mutant, including defects in hypocotyl growth, apical hook and cotyledon unfolding, anthocyanin accumulation, FRc-preconditioned block of greening, and induction of phyA-responsive gene expression. These observations suggest that FHY3 and FHY1 act early and close to the photoreceptor itself (Barnes et al., 1996; Desnos et al., 2001; Wang and Deng, 2002; Wang et al., 2002). Interestingly, mutations in FAR1 and FHL caused much weaker phenotypes than the fhy3 and fhy1 mutants, respectively (Hudson et al., 1999, 2003; Wang and Deng, 2002; Zhou et al., 2005). However, overexpression of FHY3 or FAR1 can suppress the mutant phenotypes of each other (Wang and Deng, 2002), and similarly, overexpression of FHY1 and FHL also can suppress each other's mutant phenotype (Zhou et al., 2005). In addition, the fhy3 far1 double mutant displays a stronger phenotype than their single mutant phenotypes (Wang and Deng, 2002) and the fhy1 fhl double mutant displays a stronger phenotype than the fhy1 and fhl single mutant phenotypes (Zhou et al., 2005). Moreover, FHY3 and FAR1 are capable of forming both homodimers and heterodimers with each other (Wang and Deng, 2002; Hudson et al., 2003). Similarly, FHY1 and FHL are also capable of self-association and heterodimeric interaction (Zhou et al., 2005). These results suggest that FHY3 and FAR1, as well as FHY1 and FHL, possess partially overlapping as well as distinct functions in mediating phyA signaling. Furthermore, it has been shown that FHY1 and FHL act together to regulate phyA nuclear accumulation in response to FR light, with FHY1 playing a more predominant role (Hiltbrunner et al., 2005, 2006). The functional divergence of FHY1 and FHL could be due, at least partially, to much lower levels of FHL expression (FHY1 transcript abundance is approximately 15-fold in excess of FHL transcript level in FR-grown seedlings; Zhou et al., 2005). The molecular basis for the observed partially overlapping function of FHY3 and FAR1 remains unknown. In a previous study, we showed that both FHY3 and FAR1 can directly bind to a CACGCGC motif present in the FHY1 and FHL promoters and that they act together to up-regulate FHY1 and FHL expression, and consequently phyA nuclear accumulation and light responses (Lin et al., 2007). In this study, we show that FAR1p∷FHY3 can completely rescue the far1-2 mutant phenotype but can only partially rescue the fhy3-4 mutant phenotype (Fig. 1, C and D). Notably, FHY3p∷FAR1 can completely restore the far1-2 mutant phenotype, but it fails to rescue the fhy3-4 mutant phenotype (Fig. 1, E and F). These results suggest that FHY3 promoter and FHY3 protein can recapitulate the activity of FAR1 promoter and FAR1 protein, respectively; however, the FAR1 promoter and FAR1 protein cannot substitute the function of FHY3 promoter and FHY3 protein, respectively. Thus, our results suggest that the partially overlapping function of FHY3 and FAR1 entails both divergence of promoter activities and protein subfunctionalization, with FHY3 playing a more predominant role. This notion is consistent with the much stronger morphological phenotype and more severe alteration in phyA-responsive gene expression observed with the fhy3 mutant compared with the far1 mutant (Hudson et al., 1999, 2003; Wang et al., 2002). Conceivably, divergence of the promoter activities and protein subfunctionalization between FHY3 and FAR1 may contribute to both fine-tuning of phyA signaling and ensuring greater developmental plasticity during seedling photomorphogenesis.
Discrete and Essential Roles of the Multiple Domains of FHY3
Based on phylogenetic and functional analyses, we and others have previously proposed that FHY3 and FAR1 likely define a novel class of transcription factors derived from an ancient Mutator-like transposase(s) (Hudson et al., 2003; Babu et al., 2006; Lin et al., 2007). Logically, we asked where the DNA-binding activity of FHY3 and FAR1 derived from. Notably, all eukaryotic transposases that have been biochemically characterized so far possess two functionally separable domains: an N-terminal region that binds to the terminal inverted repeats of the cognate transposons and a central or C-terminal region that catalyzes the cleavage and transfer reactions of the cut-and-paste transposition reaction (Haren et al., 1999; Lisch, 2002; Michel et al., 2003; Augé-Gouillou et al., 2005; Feschotte et al., 2005). FHY3 and FAR1 share an N-terminal C2H2 zinc finger domain of the WRKY-GCM1 family with MULE transposases (Babu et al., 2006; Lin et al., 2007). Our previous domain-deletion analysis suggested that this domain is required for the DNA-binding activity of FHY3 and FAR1 (Lin et al., 2007). In this study, using yeast one-hybrid assay and in vitro EMSA, we showed that the conserved C118, C157, H180, and H182 residues, presumably involved in forming the zinc finger motif, are essential for direct DNA binding (Fig. 2). Furthermore, transgenic analysis indicated that these amino acid residues are essential for proper FHY3 function in vivo (Fig. 3). These results provide further support that the N-terminal C2H2 zinc finger motif is responsible for direct DNA binding and that this domain is likely derived from the same region of MULE transposases. These observations also lead us to predict that this region of MULE transposases is likely involved in DNA binding to the terminal inverted repeats of the cognate transposons (Benito and Walbot, 1997). Consistent with this, recycling of the transposase DNA-binding domain to build transcription factors seems to be a recurrent theme. For example, it has been suggested that the DNA-binding domains of mammalian CENP-B and THAP proteins, Drosophila DREF protein, Arabidopsis DAYSLEEPER protein, and the primate SETMAR protein were derived from the DNA-binding domains of their respective transposase ancestors (Feschotte and Pritham, 2007, and refs. therein).
Next we asked where the transcriptional regulatory activity of FHY3 and FAR1 derived from. Interestingly, several different classes of transposases (such as MURA and TnpA, one of the two transposase proteins encoded by the maize Spm transposon) are known to function as transcriptional regulators that regulate their own expression (Raizada et al., 2001; Cui and Fedoroff, 2002). For example, Raizada et al. (2001) observed that active MuDR elements increase the expression of a LUC reporter gene driven by its terminal inverted repeat promoter by 2- to 10-fold, and they proposed that MURA may act as a weak transcriptional activator. Similarly, the transposase encoded by the maize Activator (Ac) transposable element has been shown to repress the Ac transposase gene promoter (a form of element “self-repression”) as well as nontransposable element promoters (Fridlender et al., 1996; MacRae, 2002). It is possible, therefore, that certain classes of transposases may possess a “second function” (i.e. a transcriptional regulatory activity; MacRae, 2002). In addition, previous studies indicated that dimerization or multimerization of transposase is essential for their transposition activity. This has been shown for several classes of transposable elements, such as Tn5, IS91, mariner, Ac, and Hermes (Essers et al., 2000; Michel et al., 2003, and refs. therein). Furthermore, it was shown that both Ac and Hermes transposases use their C-terminal domains for dimerization (Essers et al., 2000; Michel et al., 2003). We previously showed that the transcriptional activation activity of FHY3 is abolished by either one of two amino acid substitutions (G305R or D283N) located in the central domain of FHY3. This region shares the most extensive sequence similarity with the core catalytic transposase domain of MULE transposases and a group of bacterial transposases (Eisen et al., 1994; Babu et al., 2006; Lin et al., 2007). In this region, FHY3, FAR1, and Mutator-like transposases also carry a putative DDE catalytic triad motif (or closely related variants) that is critical for transposase/integrase function (Boeke, 2002; Lin et al., 2007). These conserved residues might be important for the function of FHY3 and FAR1 proteins and for catalytic activities of MULE transposases. In this study, we showed that mutation in D288, which corresponds to the first D residue in the predicted DDE catalytic triad of FHY3, caused a partial reduction in the transcriptional activation activity and abolished the ability to form homodimers or heterodimers with FAR1 in yeast cells (Figs. 4 and 7). Furthermore, the FHY3p∷D288A-YFP transgene conferred partial rescue of the fhy3-4 mutant phenotype (Fig. 5). Somewhat surprisingly, mutation in another conserved residue, E323, did not significantly affect its transcriptional activation activity in yeast and Arabidopsis protoplasts, and the FHY3p∷E323A-YFP transgene completely rescued the fhy3-4 mutant phenotype (Figs. 4 and 5). Together with the previously observed critical role of D283 and G305 (both in the central core transposase domain of FHY3), our results argue that both the central core transposase domain and the C-terminal SWIM motif are essential for the transcriptional activation activity and biological function of FHY3 in vivo.
In addition to the N-terminal DNA-binding domain and the central transcriptional activation domain, the C-terminal portions of both FHY3 and FAR1 also share a SWIM-type zinc finger motif of a CxCxnCXH pattern (where x stands for any amino acid residue) with plant MULE transposases (Makarova et al., 2002; Lin et al., 2007). This motif also exists in all known retroviruses (with the exception of spumaretroviruses), many nucleic acid-binding proteins, and some retrotransposons, such as copia-like retrotransposons from tobacco (Nicotiana tabacum) and Ty elements in yeast, and is predicted to involve DNA binding or protein-protein interaction (Yu et al., 2000; Makarova et al., 2002). The functional significance of the SWIM motif in MULE transposases and FHY3/FAR1 remains to be elucidated. In this study, we showed that amino acid substitution at the C579 (C579A) and H591 (H591A) of the SWIM motif completely abolished its transcriptional activation activity and its ability to form homodimers or heterodimers with FAR1 (Figs. 4 and 7). Furthermore, the FHY3p∷C579A-YFP and FHY3p∷H591A-YFP transgenes were not effective in rescuing the fhy3-4 mutant phenotype (Fig. 6). Based on these observations, we concluded that (1) both the central core transposase domain and the C-terminal SWIM motif of FHY3 are essential for mediating protein-protein interaction (both homodimerization and heterodimerization with FAR1); (2) both the central core transposase domain and the C-terminal SWIM motif of FHY3 are essential for its transcriptional regulatory activity; and (3) the ability of FHY3 to dimerize correlates with its transcriptional regulatory activity in plant cells. Collectively, these results support the proposition that the transcriptional regulatory activity of FHY3 and FAR1 is likely drawn from the ancient Mutator-like transposase(s). Similar to this scenario, MUSTANG possibly represents another class of transcription factors derived from Mutator-like transposases, although its biological function remains unknown (Cowan et al., 2005). Our results provide functional support and extend the observation that maize transposons could influence nearby gene expression in a heritable fashion, acting as “controlling elements” in the genome as proposed by McClintock over half a century ago (McClintock, 1956).
An intriguing issue for future studies is to understand the molecular and structural basis underlying the domestication process changing a Mutator-like transposase into a novel transcription factor, in this case FHY3 and FAR1, which are essential for phyA-mediated FR light signaling in Arabidopsis. It will also be interesting to determine the selection pressure that caused this domestication event and the adaptive advantage of having these domesticated host genes during evolution. Notably, FHY3/FAR1-like genes appear to be restricted to angiosperms (Lin et al., 2007). Coincident with this, innovation of phyA was proposed to occur prior to the origin of angiosperms, and this event has been hypothesized to confer adaptive advantage to the successful colonization of the first angiosperms (Mathews, 2005). It would be highly interesting to determine whether FHY3/FAR1-like genes exist and whether they play a similar role in regulating phyA signaling in early basal angiosperms, and how these domesticated genes might have conferred an increased fitness to the early angiosperms during their establishment on earth.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The fhy3-4 and far1-2 mutants of Arabidopsis (Arabidopsis thaliana) are of the Nossen-0 ecotype and were described previously (Wang and Deng, 2002). Seeds were sterilized by incubation in freshly prepared 30% bleach plus 0.01% (v/v) Triton X-100 for 15 min and then washed four times with sterile water. The surface-sterilized seeds were sown on Murashige and Skoog germination plates (1× Murashige and Skoog medium supplied with 1% Suc) and were cold treated for 3 d at 4°C. After exposure to white light for 24 h to stimulate germination, plates with seeds were transferred to FR light conditions (fluence rate of approximately 0.5 μmol m−2 s−1) for 4 d at 22°C before phenotypic analysis.
Site-Directed Mutagenesis and Plasmid Construction
To generate various mutations in the C2H2 zinc finger domain, the plasmid pTA-FHY3FL (Lin et al., 2007) was used as the template using the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instruction. The resulting plasmids are pTA-FHY3Bm1 containing C157A single mutation, pTA-FHY3Bm11 containing C118A single mutation, pTA-FHY3Bm13 containing C118A C157A H180A H182A quadruple mutation, and pTA-FHY3Bm14 containing C157A H180A H182A triple mutation. The primers used were as follows: C118A, 5′-GTTTATTGATGCGAAGTTTGCTGCCTCTAGGTATGGTACTAAAAGAG-03′; C157A, 5′-GAACATGTGCGAAAACCGATGCCAAAGCTAGTATGCATGTTAAGAG-3′; H180A-H182A, 5′-CACAGCTTTGTGAGGGAGGCTAATGCTGAGCTTTTACCTGCACAAGC-3′.
To generate TOPO-FHY3-D288A, TOPO-FHY3-E323A, TOPO-FHY3-C579A, and TOPO-FHY3-H591A constructs, the plasmid TOPO-FHY3 (Lin et al., 2007) was used as the template in the site-directed mutagenesis. The primers used were as follows: D288A, 5′-ATGTTGTTTCTCTAGCCACCACCTACGTAAG-3′; E323A, 5′-CCTTAATATCCGATGCGAGTGCAGCAACATATTC-3′; C579A, 5′-AGGCTGAAGTCTCTTGCATCGCTCGGTTATTTGAG-3′; H591A, 5′-AGGGTATCTCTGCAGAGCCACTCTTAATGTTCTTC-3′. All constructs were validated by sequencing.
To facilitate downstream cloning, the XhoI-HindIII fragments of FHY3-D288A, FHY3-E323A, FHY3-C579A, and FHY3-H591A were released from their corresponding TOPO vectors and then subcloned into pTA-FHY3FL plasmid at XhoI and HindIII sites, generating pGEM-FHY3-D288A, pGEM-FHY3-E323A, pGEM-FHY3-C579A, and pGEM-FHY3-H591A, respectively.
To make the FHY3p∷FHY3, FAR1p∷FAR1, FHY3p∷FAR1, and FAR1p∷FHY3 transgenes, the FHY3 and FAR1 genomic DNA (including the coding region and 3′-untranslated region) were amplified by PCR from Columbia wild-type DNA and then cloned into pGEM-T Easy vector (Promega), generating pGEM-FHY3G and pGEM-FAR1G, respectively. The primers used for the FHY3 gene are FHY3G1 (5′-TGGATCCATGGATATAGATCTTCGACTAC-3′) and FHY3G2 (5′-TGTCGACTGCCTAAAAGCTCTATTTGCC-3′). The primers used for the FAR1 gene are FAR1G1 (5′-TGGATCCATGGATTTGCAAGAGAATCTG-3′) and FAR1G2 (5′-TCTCGAGGCAAATGTTTCTTTTGACCAACTC-3′). A BamHI-SalI fragment containing FHY3 and a BamHI-XhoI fragment containing FAR1 were released from pGEM-FHY3G and pGEM-FAR1G and cloned into BamHI-SalI-digested pPZP-FHY3p intermediate vector (Lin et al., 2007) to produce FHY3p∷FHY3 and FHY3p∷FAR1, respectively. Similarly, the same FHY3 and FAR1 fragments were ligated into BamHI-SalI-digested pPZP-FAR1p intermediate vector (Lin et al., 2007) to produce FAR1p∷FHY3 and FAR1p∷FAR1, respectively.
To generate various FHY3 fusions with YFP, the coding region of YFP was first amplified from pCitrine-3 plasmid (Griesbeck et al., 2001) and then cloned into the pGEM-T Easy vector to produce pGEM-YFP. The YFP gene was released from pGEM-YFP by BamHI and SalI digestion and then subcloned into pPZP-FHY3p at BamHI and SalI sites, resulting in pPZP-FHY3p-YFP. FHY3 and its various mutant forms D288A, E323A, C579A, and H591A were released from their corresponding TOPO vectors by NheI and BamHI digestion and then subcloned into pPZP-FHY3p-YFP vector at XbaI and BamHI sites, giving rise to FHY3p∷FHY3-YFP, FHY3p∷D288A-YFP, FHY3p∷E323A-YFP, FHY3p∷C579A-YFP, and FHY3p∷H591A-YFP, respectively.
To construct binary vectors for C2H2 zinc finger domain mutations, FHY3Bm1, FHY3Bm11, FHY3Bm13, and FHY3Bm14 fragments were released from their corresponding pTA vectors digested by BamHI and SalI and then inserted into pPZP-FHY3p at BamHI and SalI sites, generating FHY3p∷Bm1, FHY3p∷Bm11, FHY3p∷Bm13, and FHY3p∷Bm14, respectively.
To construct vector for DNA binding in yeast one-hybrid assay, various FHY3 mutated genes were released from their pTA vectors cut by MfeI and SalI and then inserted into the JG4-5 (Clontech) digested by EcoRI and XhoI, resulting in GAD-Bm1, GAD-Bm11, GAD-Bm13, and GAD-Bm14, respectively. To generate plasmid for transcriptional activation assay, various FHY3 mutated genes were released from their pGEM vectors by MfeI and SalI and then cloned into EG202 (Clontech) at the EcoRI and SalI sites, producing LexA-D288A, LexA-E323A, LexA-C579A, and LexA-H591A, respectively.
To generate plasmids for protoplast transient expression assay, FHY3-D288A, FHY3-E323A, FHY3-C579A, and FHY3-H591A fragments were digested from their corresponding pGEM vectors by MfeI and SalI and then cloned into pUC-SPYCE (Walter et al., 2004) vector at the BamHI and SalI sites, resulting in pUC-D288A, pUC-E323A, pUC-C579A, and pUC-H591A, respectively.
To generate vectors for GST fusion protein expression, the N-terminal fragments containing the first 200 amino acids of FHY3Bm1, FHY3Bm11, and FHY3Bm13 were amplified by PCR using their corresponding pTA vector as the template with primers FHY3-BM (5′-GGATCCCAATTGATGGATATAGACCTTCGACTAC-3′) and FHY3-N200 (5′-CATGTCGACTCATGCAGCATATATCTTTCTGG-3′). The PCR fragments were digested with MfeI and SalI and then inserted into the pGEX 5X-1 vector (Amersham Biosciences) at the EcoRI and SalI sites, generating GST-Bm1N, GST-Bm11N, and GST-Bm13N, respectively.
The FHY1p∷LacZ, FHLp∷LacZ, FHY1p∷LUC, LexA-FHY3, LexA-D283N, LexA-G305R, GAD-FHY3, GAD-FAR1, pUC-FHY3, and GST-FHY3N plasmids were described previously (Lin et al., 2007).
Plant Transformation
The binary vectors were electroporated into the Agrobacterium tumefaciens strain GV3101 and transformed into the fhy3-4 or far1-2 mutant via the floral dip method (Clough and Bent, 1998). About forty kanamycin-resistant T1 transgenic lines for each transgene were selected. Phenotypic analyses were performed in the T2 generation and confirmed in the T3 generation. Homozygous plants with comparable expression levels were used in all experiments.
Yeast One-Hybrid and Two-Hybrid Assays
The yeast one-hybrid and two-hybrid assays were performed essentially as described by Lin et al. (2007) and Wang and Deng (2002), respectively.
The yeast strain EGY48 was used in these assays.
Semiquantitative Reverse Transcription-PCR
The procedures for plant total RNA extraction, reverse transcription (RT)-PCR, and primers were described previously (Lin et al., 2007).
Chlorophyll and Anthocyanin Measurement
Measurement of chlorophyll and anthocyanin content was performed as described previously (Lin and Wang, 2005). About 100 seedlings per genotype were used for each measurement.
Yeast Protein Extraction and Immunoblot Assay
Protein extraction from the yeast strain was performed according to the Yeast Protocols Handbook (Clontech). The LexA fusion protein was detected with anti-LexA antibody (Santa Cruz Biotechnology), and the GAD fusion protein was detected against anti-hemagglutinin-epitope antibody (Roche). The proteins were visualized by incubating with goat anti-mouse secondary antibody conjugated with alkaline phosphatase in the presence of 5-bromo-4-3-indolyl-phosphate and nitroblue tetrazolium as substrates.
Transient Expression in Arabidopsis Protoplast
The protoplast transient expression assay was carried out as described previously (Lin et al., 2007).
EMSA
Various GST-FHY3N mutant recombinant fusion proteins were expressed in the Escherichia coli BL21 (DE3) strain and purified using glutathione-Sepharose 4B beads (Amersham Bioscience). The recombinant proteins are of the same Mr based on SDS-PAGE analysis. The EMSA procedure was described previously (Lin et al., 2007). The sequence of the FHY1 oligonucleotide probe is 5′-AACACGTAGACTCTTTTCACGCGCCAAATCAAACACCAT-3′.
Confocal Microscopy Observation
Transgenic seedlings of various genotypes were grown in darkness for 4 d before examination. The whole seedlings were mounted on a slide, and YFP fluorescence was observed with a confocal microscope (SP5; Leica). Representative images were taken from the same region of roots with identical settings.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers (in parentheses): FHY3 (NP_188856), FAR1 (AAD51282), FHY1 (NP_001078018), FHL (NP_001078521), PHYA (NP_001117256).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR analysis of transgene expression.
Supplemental Figure S2. Amino acid alignment of Arabidopsis FHY3 and FAR1 with the maize MURA and Jittery transposases.
Supplemental Figure S3. RT-PCR analysis of FHY3 transgene expression.
Supplemental Figure S4. FHY3-YFP rescues fhy3-4 mutant phenotype.
Supplementary Material
Acknowledgments
We thank Elizabeth Estabrook and Mingqui Dai (Boyce Thompson Institute) for their critical reading of and comments on the manuscript.
This work was supported by the National Science Foundation (grant no. IOS–0641639 to H.W., Research Experiences for Undergraduates grant no. DBI–0453331 to C.B. [Reed College], and grant no. DBI–0618969 for the microscopy facilities at Boyce Thompson Institute) and by a China Scholarship Council award to Y.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Haiyang Wang (hw75@cornell.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Augé-Gouillou C, Brillet B, Germon S, Hamelin MH, Bigot Y (2005) Mariner Mos1 transposase dimerizes prior to ITR binding. J Mol Biol 351 117–130 [DOI] [PubMed] [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34 6505–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Quaggio RB, Whitelam GC, Chua NH (1996) fhy1 defines a branch point in phytochrome A signal transduction pathways for gene expression. Plant J 10 1155–1161 [DOI] [PubMed] [Google Scholar]
- Benito MI, Walbot V (1997) Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol Cell Biol 17 5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD (2002) Putting mobile DNA to work: the toolbox. In NL Craig, R Craigie, M Gellert, AM Lambowitz, eds, Mobile DNA II. American Society of Microbiology, Washington, DC, pp 24–37
- Botto JF, Sanchez RA, Whitelam GC, Casal JJ (1996) Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol 110 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cowan RK, Hoen DR, Schoen DJ, Bureau TE (2005) MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol 22 2084–2089 [DOI] [PubMed] [Google Scholar]
- Cui H, Fedoroff NV (2002) Inducible DNA demethylation mediated by the maize Suppressor-mutator transposon-encoded TnpA protein. Plant Cell 14 2883–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10 121–129 [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10 51–54 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Benito MI, Walbot V (1994) Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res 22 2634–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers L, Adolphs RH, Kunze R (2000) A highly conserved domain of the maize activator transposase is involved in dimerization. Plant Cell 12 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Osterlund M, Peeler R, Wessler SR (2005) DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res 33 2153–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomics. Annu Rev Genet 41 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender M, Harrison K, Jones JDG, Levy AA (1996) Repression of the Ac transposase gene promoter by Ac transposase. Plant J 9 911–917 [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J Biol Chem 276 29188–29194 [DOI] [PubMed] [Google Scholar]
- Haren L, Ton-Hoang B, Chandler M (1999) Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol 53 245–281 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schaefer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schaefer E (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34 453–471 [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941 [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH (2006) Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kircher S, Toth R, Adam E, Schafer E, Nagy F (2000) Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J 22 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schäfer E, Nagy F (1999) Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H (2004) Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol 136 4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H (2005) Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D (2002) Mutator transposons. Trends Plant Sci 7 498–504 [DOI] [PubMed] [Google Scholar]
- MacRae AF (2002) Experimental evidence that the maize Activator (Ac) transposase can act on a non-transposable element promoter to repress reporter gene expression in transient plant assays. Genetica 115 289–309 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Koonin EV (2002) SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci 27 384–386 [DOI] [PubMed] [Google Scholar]
- Mathews S (2005) Phytochrome evolution in green and nongreen plants. J Hered 96 197–204 [DOI] [PubMed] [Google Scholar]
- McClintock B (1956) Controlling elements and the genes. Cold Spring Harb Symp Quant Biol 21 197–216 [DOI] [PubMed] [Google Scholar]
- Michel K, O'Brochta DA, Atkinson PW (2003) The C-terminus of the Hermes transposase contains a protein multimerization domain. Insect Biochem Mol Biol 33 959–970 [DOI] [PubMed] [Google Scholar]
- Neff M, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14 257–271 [PubMed] [Google Scholar]
- Ni M, Halliday K, Tepperman J, Quail P (1999) Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly by light. Nature 400 781–784 [DOI] [PubMed] [Google Scholar]
- Quail P (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3 85–93 [DOI] [PubMed] [Google Scholar]
- Raizada MN, Brewer KV, Walbot V (2001) A maize MuDR transposon promoter shows limited autoregulation. Mol Genet Genomics 265 82–94 [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V, Rudenko GN (2002) MuDR/Mu transposable elements of maize. In NL Craig, R Craigie, M Gellert, AM Lambowitz, eds, Mobile DNA II. American Society of Microbiology, Washington, DC, pp 533–564
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Deng XW (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2003) Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci 8 172–178 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma L, Habashi J, Li J, Zhao H, Deng XW (2002) Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32 723–733 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Yan X, Maurais S, Fu H, O'Brien DG, Mottinger J, Dooner HK (2004) Jittery, a mutator distant relative with a paradoxical mobile behavior: excision without reinsertion. Plant Cell 16 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Wright SI, Bureau TE (2000) Mutator-like elements in Arabidopsis thaliana: structure, diversity and evolution. Genetics 156 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Hare PD, Yang SW, Zeidler M, Huang LF, Chua NH (2005) FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J 43 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.