Abstract
Pyrrolizidine alkaloids (PAs) are typical compounds of plant secondary metabolism and are believed to be part of the plant's chemical defense. Within the monocotyledonous plants, PAs have been described in only a few genera, mainly orchids, including Phalaenopsis. Because phylogenetic analyses suggest an independent origin of PA biosynthesis within the monocot lineage, we have analyzed the developmentally regulated expression of homospermidine synthase (HSS), the first pathway-specific enzyme of PA biosynthesis, at the cell level. HSS is expressed in the tips of aerial roots exclusively in mitotically active cells. Raphide crystal idioblasts present within the root apical meristem do not show HSS expression. In addition, young flower buds, but not mature flowers, express HSS and have been shown by tracer feeding experiments to be able to catalyze PAs. This second site of PA biosynthesis ensures high concentrations of PAs in the reproductive structures of the Phalaenopsis flower, even after the flower opens. Thus, in spite of its identical function in PA biosynthesis, HSS shows in Phalaenopsis a completely different spatial and developmental expression pattern in comparison to other PA-producing species. These results show that the proverbial diversity of plant secondary metabolism is not just a matter of structural diversity, but is also multifaceted in terms of pathway regulation and expression.
Typical features of plant secondary metabolism are its diversity and variability (Pichersky and Gang, 2000). About 200,000 chemicals that are synthesized by various biosynthetic pathways have been identified in plants (Ober, 2005). In the case of the pyrrolizidine alkaloids (PAs), we have found evidence that the ability to produce these compounds has been recruited several times independently during angiosperm evolution (Reimann et al., 2004). PAs are constitutively produced by certain plants as a defense against herbivores. The toxicity of PAs is responsible for the recurrent poisoning of livestock, wild life, and humans by widespread PA-containing plants (Mattocks, 1986; Stegelmeier et al., 1999). More than 400 structures have been elucidated from species belonging to some unrelated families of the angiosperms. Major occurrences are found within the Asteraceae (the tribes Senecioneae and Eupatorieae), the Boraginaceae and Heliotropiaceae (both Boraginales), the Apocynaceae, some genera of the Orchidaceae, and the subtropical genus Crotalaria of the Fabaceae (Hartmann and Witte, 1995; Hartmann and Ober, 2000). PAs with certain structural features, among others, a 1,2-double bond, are bioactivated by P-450 enzymes that convert the protoxic alkaloids into pyrrolic intermediates that easily react with biological nucleophiles (i.e. proteins and nucleic acids, causing severe cell toxicity and liver cancer; Fu et al., 2004). P-450 enzymes involved in bioactivation are found not only in the liver of vertebrates, but also in insects (Brattsten, 1992), for which PAs are also cytotoxic and genotoxic (Frei et al., 1992; Narberhaus et al., 2005). Orchids contain 1,2-saturated PAs that are regarded as nontoxic, at least as they cannot be activated by P-450 enzymes. Nevertheless, tissue distribution of PAs in Phalaenopsis suggests that PA accumulation represents a strategy one would expect for a plant chemical defense system that evolved under the selection pressure of herbivory (Frölich et al., 2006).
Our analyses of the evolution of PA biosynthesis within angiosperm plants have enabled us to show that the gene encoding homospermidine synthase (HSS) has been recruited at least four times independently from a gene encoding deoxyhypusine synthase (DHS), an enzyme of primary metabolism that is involved in the posttranslational activation of the eukaryotic initiation factor 5A (eIF5A; Ober and Hartmann, 1999b; Reimann et al., 2004). HSS and DHS share not only a high degree of sequence similarity and an almost identical genomic structure (Reimann et al., 2004), but also properties such as protein architecture, pH optimum, reaction mechanism, and even substrate specificity. The HSS can be interpreted as a DHS that has lost its intrinsic ability to bind the eIF5A precursor protein, but has retained its side activity, namely, the ability to synthesize homospermidine (Ober et al., 2003). A comparative analysis of the expression patterns of HSS and DHS within Senecio vernalis (Asteraceae, tribe Senecioneae) has shown that, during gene recruitment of the HSS-encoding gene, the regulatory elements were adapted to the specific needs of alkaloid biosynthesis, resulting in completely different expression patterns of these two related enzymes (Moll et al., 2002). Whereas DHS is expressed at low levels in all analyzed tissues, HSS is expressed exclusively in groups of specific cells within the roots of S. vernalis in the direct vicinity of the phloem tissue (Moll et al., 2002). Within Senecio, PAs are transported via the phloem from the roots, as the site of synthesis, to the shoots and the flower heads, as the site of accumulation (Hartmann et al., 1989). Comparing the expression patterns of HSS within the closely related species S. vernalis and Eupatorium cannabinum (Asteraceae, tribe Eupatorieae), we have been able to demonstrate that HSS is individually regulated despite its identical function in both plant species (i.e. catalysis of the first specific step within PA biosynthesis). In contrast to Senecio, HSS within E. cannabinum is expressed in all cells of the cortex parenchyma, excluding the endodermis (Moll et al., 2002; Anke et al., 2004). We have interpreted this observation as providing further support for the independent origin of PA biosynthesis in these two lineages of the Asteraceae (Anke et al., 2004; Reimann et al., 2004).
Here, we describe the tissue-specific expression of HSS within the orchid Phalaenopsis. As mentioned above, we have recently shown that HSS was recruited independently within the monocot lineage (Reimann et al., 2004). Despite the putative simple structure of phalaenopsine-type PAs, recent data suggest that the ability to produce PAs is an old feature within monocot plants with an origin close to the base of the monocot lineage (N. Nurhayati and D. Ober, unpublished data). Immunolabeling techniques with polyclonal antibodies raised against the HSS have revealed that, within Phalaenopsis, HSS is expressed in the mitotically active cells of the root apical meristem supplying the vegetative parts of the plants with PAs. Furthermore, the young flower buds have been identified by tracer feeding experiments and by immunolabeling experiments as additional sites of PA biosynthesis, boosting the PA content of this reproductive tissue just before the flowers open to attract pollinators. Thus, in comparison with other previously analyzed PA-producing plants, Phalaenopsis HSS shows a completely different spatial and developmental expression pattern, in spite of its identical function in the biosynthesis of PAs. Obviously, the proverbial diversity of plant secondary metabolism comprises not only structural diversity, but also diversity in pathway regulation.
RESULTS
Tissue-Specific Expression of HSS
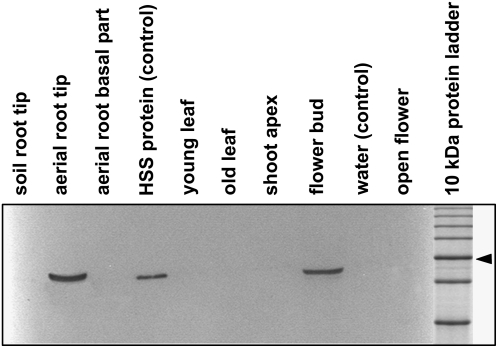
Recombinant HSS was purified and used to raise polyclonal antibodies in rabbits and to prepare an agarose gel matrix with covalently coupled HSS for affinity purification of the antibodies. These were used for western-blot analyses of various tissues of Phalaenopsis plants. HSS protein was detected only in the tips of aerial roots and in the flower bud. In contrast, no signal with the HSS-specific antibody was seen in the basal part of the aerial roots, in roots entering into the substrate, or in open flowers, suggesting the specific expression of HSS in space and time (Fig. 1). Moreover, young and old leaves and the shoot tip of an inflorescence showed no label. These are tissues from which several pseudogenes have been identified by approaches involving reverse transcription-PCR (N. Nurhayati and D. Ober, unpublished data), suggesting that these pseudogenes, of which some appear to be transcribed, are not expressed at the protein level. To determine whether HSS was expressed throughout the flower bud and the aerial root tip, longitudinal sections of these tissues were printed onto a nitrocellulose membrane. These tissue prints are presented in Figure 2, which indicates that HSS expression is restricted to the very end of the root tip and to specific tissues within the flower bud.
Figure 1.
Expression analysis of HSS in various tissues of Phalaenopsis. Soluble protein (20 μg) extracted from the tissues was separated by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. As a positive control, 10 ng of purified HSS protein was applied. Detection was performed with affinity-purified antibody against HSS of Phalaenopsis. Soil roots were roots that penetrated the substrate, whereas aerial roots did not. Young and old leaves were approximately 3 and 10 cm long, respectively. The 50-kD band of the marker protein is labeled with an arrowhead.
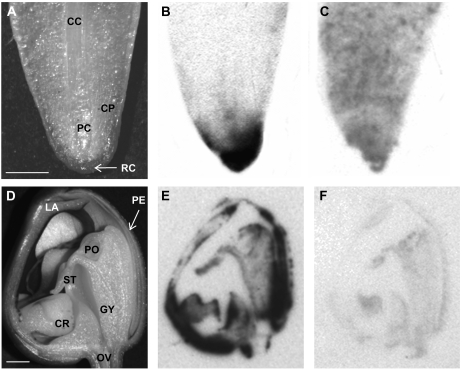
Figure 2.
Longitudinal sections of an aerial root tip (A–C) and of a flower bud (D–F). A and D, Sections documented by stereomicroscopy. B and E, Tissue prints of the sections in A and D, respectively, labeled with HSS-specific antibody. C and F, Tissue prints of the sections in A and D, respectively, labeled with preimmune serum. CC, Central cylinder; CR, callus on the labellum; GY, gynostemium (column); CP, cortex parenchyma; LA, labellum; OV, ovary; PC, procambium; PE, perigone; PO, pollinium; RC, root cap; ST, stigma. Size bars = 2 mm.
Immunolocalization of HSS in Aerial Root Tips and Flower Buds of Phalaenopsis
To identify the cells that expressed HSS, longitudinal sections of resin-embedded tissues were labeled with the HSS-specific antibody. Figure 3A shows a longitudinal section of the meristem of an aerial root as visualized by UV microscopy. Bound primary antibody was detected with secondary antibodies labeled with fluorescein isothiocyanate (FITC). The root cap as the most distal tissue of the root protecting the meristem was not as well labeled as the procambium. Intense labeling was found in the cortical tissues and the epidermis close to the root apex. Higher magnifications of the root apex (Fig. 3C) showed the closed organization of the root tip. The labeling intensity decreased rapidly behind the apex, being undetectable about 2 mm behind this structure.
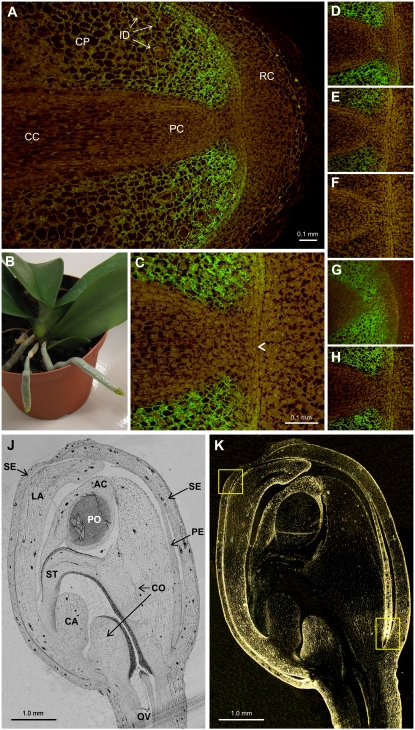
Figure 3.
Immunolabeling of HSS in aerial root tips and in a flower bud of Phalaenopsis. A, Longitudinal section of a root tip incubated with HSS-specific antibody before detection with a FITC-conjugated secondary antibody. B, Phalaenopsis plant with two aerial roots that were found to express HSS. C, Detail of A indicating the initials (arrowhead). D to H, Detail of A after incubation with HSS-specific antibody without additives (D) or in the presence of purified HSS (E and F), DHS (G), or BSA (H) in a molar ratio of antibody to added protein of 10:1 (E) and 1:3 (F–H). J, Unlabeled longitudinal section of a flower bud visualized by bright-field microscopy. K, Section as J, labeled with HSS-specific antibody and immunogold detection after silver enhancement visualized by dark-field microscopy. Yellow boxes denote the position of the details shown in Figure 4, E and F. AC, Anther cap; CA, callus; CC, central cylinder; CO, column; CP, cortex parenchyma; ID, idioblasts; LA, labellum; OV, ovary; PC, procambium; PE, petal; PO, pollinium; RC, root cap; SE, sepal; ST, stigma.
A better resolution of the label at the single-cell level was achieved by silver enhancement after use of gold-labeled secondary antibodies (Fig. 4). Figure 4A shows a cross section of the root tip, confirming the restriction of the label to the cortical cells. The cells of the central cylinder were devoid of label. We assumed that the endodermis was the innermost cell layer expressing HSS (Fig. 4A). Because of the early stage of differentiation of these cells, the Casparian strip that would have allowed the unequivocal identification of the endodermis was not detectable. Longitudinal sections of the root tip exhibited, within the labeled cortical cells, unlabeled idioblasts containing bundles of raphides (Fig. 4B). HSS expression, which was found in the young cells of the epidermal cell layer, diminished much faster with increasing distance to the root apex than that observed in the cortical cell layer (Fig. 4C). Figure 4D is a higher magnification view of those labeled cells in Figure 4C that lie in the direct vicinity of the initials (stem cells; see below). HSS protein is detectable in the young cortex cells as soon as the vascular and the cortical cell layers are distinguishable.
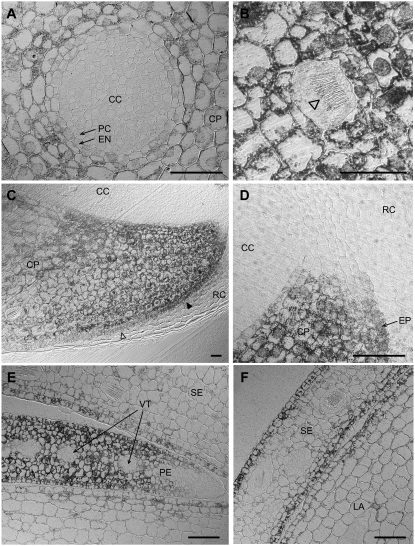
Figure 4.
Details of aerial root (A–D) and flower bud (E and F) sections after immunogold labeling and silver enhancement. A, Cross section of an aerial root approximately 500 μm behind the root tip. B to D, Longitudinal section of an aerial root tip. B, Unlabeled idioblast with a bundle of raphides (arrowhead) within labeled cortex parenchyma cells. C, Lower half of a section similar to that in Figure 3A. The labeling intensity decreases rapidly with increasing distance from the root tip, especially in the epidermal cell layer (filled arrowhead, labeled epidermis; open arrowhead, unlabeled epidermis). D, Detail of Figure 4C showing HSS expression directly behind the initials at the root apex. E and F, Details of the section in Figure 3K (yellow boxes, bottom right and top left, respectively). Size bar = 50 μm. CC, Central cylinder; CP, cortex parenchyma; EN, endodermis; EP, epidermis; LA, labellum; PC, pericycle; PE, petal; RC, root cap; SE, sepal; VT, vascular tissue.
To localize HSS in Phalaenopsis flower buds, longitudinal sections were incubated with HSS-specific antibody, which was detected with silver enhancement by dark-field microscopy (Fig. 3K). The images so obtained were then compared with those of the same unlabeled section documented by bright-field microscopy (Fig. 3J). HSS was detected in the epidermal cell layers of the sepals and the petals, including the labellum (Figs. 3K and 4F). Furthermore, HSS expression was found in the anther cap, the ventral side of the column, and within the ovary (Fig. 3K). In the tip of a petal, the epidermis and all the mesophyll were intensely labeled, whereas the vascular tissue and the end of the petal were unlabeled (Figs. 3K and 4E).
The specificity of the label within the root apex and the flower bud was confirmed by the incubation of several successive sections, under identical conditions as above, with the HSS-specific antibody to which increasing amounts of soluble HSS were added. The decreasing intensity of the label after preincubation of the antibody with increasing amounts of soluble HSS established the specificity of the HSS antibody (Fig. 3, D–H). Preincubation of the antibody with soluble DHS and with bovine serum albumin (BSA) had no effect on labeling intensity (Fig. 4, G and H, respectively), excluding any cross-detection of DHS and any nonspecific interactions of the antibody with proteins.
Capacity of Phalaenopsis Flower Buds to Synthesize PAs
The restricted expression of HSS in the tips of aerial roots nicely confirms previous results obtained by tracer feeding experiments (Frölich et al., 2006). By contrast, the detection of HSS protein in flower buds of Phalaenopsis was an unexpected result because this tissue was not thought to be able to catalyze the formation of PAs. Therefore, we tested Phalaenopsis flower buds to determine whether they expressed only HSS or the whole PA biosynthetic pathway. In tracer feeding experiments, we applied 14C-labeled putrescine as the tracer to isolated flower buds of two inflorescences (Fig. 5; Table I). These experiments clearly showed that the youngest flower buds had the highest capacity to produce PAs. With increasing size of the flower buds, the efficiency of the incorporation of the tracer into PAs decreased until the ability of the buds to produce PAs was lost (flower bud 3 and flower bud 6 of inflorescences A and B, respectively, in Fig. 5). The radiolabeled PAs detected by these tracer feeding experiments were 1R-phalaenopsine and 1S-phalaenopsine, two isomers possessing trachelanthamidine and isoretronecanol as the necine base moiety, respectively (Table I).
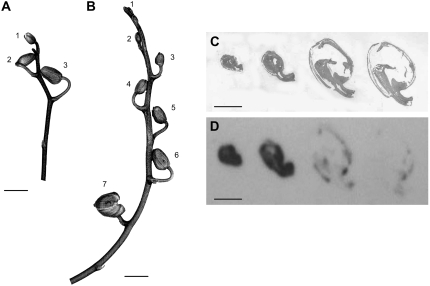
Figure 5.
Inflorescences of Phalaenopsis plants. A and B, The numbers labeling the buds refer to the tracer feeding experiments shown in Table I. C and D, Tissue prints of longitudinal sections of Phalaenopsis buds on nitrocellulose membrane developed with affinity-purified antibody against HSS of Phalaenopsis (D) and stained for total protein by Indian ink (C). Size bars in A to D = 1 cm.
Table I.
Feeding of 14C-labeled putrescine as a precursor of PA biosynthesis to Phalaenopsis flower buds at various developmental stages
The numbers of the flower buds of the inflorescences A and B refer to Figure 5.
| Analyzed Plant Organ | Recovery of Applied Radioactivity (%)a | % of Recovered Radioactivityb
|
Total Incorporation into PAs (%)c | ||
|---|---|---|---|---|---|
| 1R-Phalaenopsine | 1S-Phalaenopsine | Total PA | |||
| Inflorescence A | |||||
| Flower bud 1 | 10 | 8 | 14 | 22 | 2 |
| Flower bud 2 | 21 | 5 | 3 | 8 | <2 |
| Flower bud 3 | 17 | 0 | 0 | 0 | 0 |
| Inflorescence B | |||||
| Flower bud 1 | 13 | 28 | 28 | 56 | 7 |
| Flower bud 2 | 17 | 22 | 7 | 29 | 5 |
| Flower bud 3 | 17 | 20 | 11 | 31 | 5 |
| Flower bud 4 | 11 | 11 | 0 | 11 | 1 |
| Flower bud 5 | 14 | 9 | 0 | 9 | 1 |
| Flower bud 6 | 13 | 0 | 0 | 0 | 0 |
| Flower bud 7 | 3 | 0 | 0 | 0 | 0 |
In the methanol extract (incorporated radioactivity = 100%).
Radioactivity in methanol extract = 100%.
Incorporated radioactivity.
DISCUSSION
Phalaenopsis orchids and some related species have, for a long time, been considered the only monocotyledonous plants known to produce PAs, suggesting restricted occurrence of these alkaloids within the monocot lineage. However, phylogenetic analyses suggest that HSS, as the first specific enzyme of PA biosynthesis, was recruited early in the evolution of the monocots (N. Nurhayati and D. Ober, unpublished data). Using affinity-purified polyclonal antibodies raised against recombinant HSS of Phalaenopsis and by tracer feeding experiments, we have been able to show that HSS and the other enzymes of PA biosynthesis are expressed in two completely different tissues tightly regulated in space and time. Thus, the expression pattern of PA biosynthesis within Phalaenopsis is completely different from that found in other PA-producing species (i.e. S. vernalis and E. cannabinum of the Senecioneae and Eupatorieae tribe within the Asteraceae, respectively (Moll et al., 2002; Anke et al., 2004). These results add a further facet to the diversity of plant secondary metabolism and indicate the complexity of processes that were necessary in evolution to integrate new pathways within plant metabolism.
Root-Specific Expression of HSS in Phalaenopsis
Enzymes synthesizing alkaloids are expressed in many different tissues of the plant that, in most cases, are not identical with the preferential site of alkaloid accumulation. The roots are recognized as a site with the ability to synthesize a remarkable diversity of secondary metabolites, thereby adjusting their metabolic activities in response to biotic and abiotic stresses (Flores et al., 1999). These below-ground processes are often essential components of ecosystem productivity and stability. Nicotine and tropane alkaloids are synthesized in the roots of the Solanaceae and are transported via the xylem to the shoots as the preferential site of accumulation (for review, see De Luca and St-Pierre, 2000). Furthermore, in Senecio spp., PAs are exclusively synthesized in the roots (Toppel et al., 1987) and are transported via the phloem to the inflorescences, which accumulate about 90% of all the PAs found in the plant (Hartmann and Zimmer, 1986; Hartmann et al., 1989). Recently, we have been able to show that the expression pattern of HSS is highly specific, but completely different between the two Asteraceae species S. vernalis (Senecioneae) and E. cannabinum (Eupatorieae). We have interpreted this difference as additional support for the independent origin of the HSS-coding gene within the two asteraceous tribes (Moll et al., 2002; Anke et al., 2004). According to tracer feeding experiments performed by Frölich et al. (2006), the site of PA biosynthesis in Phalaenopsis is probably the tip of the aerial roots from which the alkaloids are transported to the basal parts of the roots and to young leaves. These data are in accordance with the immunolocalization of HSS in the apical meristem of the roots tips. Figure 3C suggests that Phalaenopsis has a closed meristem type. Roots of Arabidopsis (Arabidopsis thaliana) also have a closed apical meristem type and have been studied intensively over the last decade (Scheres et al., 1996, 1997, 2004; Benfey and Scheres, 2000; Doerner, 2003). This type of meristem is characterized by cell files converging on an apparent pole at the center of the meristem (Jiang and Feldman, 2005). The cells located at the apical pole of the files are the initials or stem cells. Initials abut onto cells of the quiescent center, cells that have greatly reduced rates of mitosis and that are responsible for maintaining the “stemness” of the initials, as has been shown by laser ablation experiments (van den Berg et al., 1995; Doerner, 1998). After each initial cell division, one daughter is disconnected from the quiescent center and is allowed to differentiate, adding one cell to the plant body, whereas the initial cell retains its position within the meristem. Within Arabidopsis, the cell files originating at four sets of initials differentiate to the epidermis and the lateral root cap, to the cortical tissues of endodermis and cortex parenchyma, to the vascular tissue, and to the columella (the central root cap), respectively. HSS is expressed in Phalaenopsis root tips in the epidermis and in the cortex, most probably including the endodermis (Figs. 3A, and 4, C and D). The presence of the label, even in cells close to the apex, suggests that the cortex and epidermis cells express HSS as soon as their fate is determined, but only as long as these cells are meristematic because the signal fades away a few millimeters behind the apex. Further support that only those cells that are mitotically active express HSS is provided by the raphide idioblasts, which are devoid of HSS immunosignals.
Raphide Crystal Idioblasts Do Not Express HSS
The raphide crystal idioblasts are the only cells within the root apical meristem that are not labeled for HSS. Cells that form intravacuolar calcium oxalate crystals have been reported in diverse tissues and organs of a large number of plant species (Arnott and Pautard, 1970; Franceschi and Horner, 1980), including the meristematic region in the root apices of grapevines (Storey et al., 2003) and orchids (Kausch and Horner, 1983b, 1984). Raphide crystals were originally regarded as excretory products or as providing protection against foraging animals (Franceschi and Horner, 1980). However, recent data suggest that they act as a calcium sink involved in the regulation of cytoplasmic calcium levels in the meristematic cells of root apices (Franceschi, 1989; Storey et al., 2003). Crystal idioblast initials are usually formed in meristematic tissues and differentiate precociously in comparison with the surrounding cells (Arnott and Pautard, 1970; Franceschi and Horner, 1980; Kausch and Horner, 1983a). Once initiated, an idioblast differentiates and does not undergo mitosis or nuclear fusion, as has been shown for crystal idioblasts in the cortex of roots of Vanilla planifolia (Orchidaceae; Kausch and Horner, 1983b, 1984). These crystal idioblasts are also easy to distinguish in the meristem of Phalaenopsis, as they are larger than the surrounding cells, a typical feature of this type of idioblast. The enlargement of the nucleus and the elongation of the whole cell to a final size that is larger than that of the surrounding cells that are still dividing are the first observed changes in developing crystal idioblasts (Franceschi and Horner, 1980; Kausch and Horner, 1983a). For raphide crystal idioblasts of V. planifolia (Orchidaceae) and of Vanda, a close relative of Phalaenopsis, this enlargement is coupled with endopolyploidization (Kausch and Horner, 1984; Lim and Loh, 2003), which is caused by a switch of their cell cycle program from growth to endopolyploidization and cell enlargement (Sugimoto-Shirasu and Roberts, 2003; Jakoby and Schnittger, 2004). The observation that HSS is not expressed in these specialized cells suggests that HSS expression and most probably PA biosynthesis are restricted to meristematic (i.e. mitotically active, cells within the apical meristem of aerial roots of Phalaenopsis).
In the epidermal cell layer, HSS expression fades away at a shorter distance to the apex than in the cortex (Fig. 4C). Probably the cells of the epidermis differentiate earlier than the surrounding cells; this might result in a loss of mitotic activity and of HSS expression. The distribution of meristematic activity is well known to differ in the various root regions (Esau, 1965). A global gene expression analysis of three developmental stages in Arabidopsis has revealed that epidermis cells achieve their tissue-specific gene expression pattern earlier than the cells of the cortex or the vascular cells (Birnbaum et al., 2003; Scheres et al., 2004). This suggests that, within the epidermis, the switch from mitosis to cell differentiation occurs earlier than in the adjacent tissues.
Flower Buds of Phalaenopsis Are a Second Site for PA Biosynthesis
Recently, we have characterized the distribution and the concentration of PAs within Phalaenopsis plants (Frölich et al., 2006). The highest concentrations are found in young flower buds. Within the open flower, the highest concentrations occur in the column and the pollinia, the reproductive organs of the orchids. Lower concentrations of PAs have been detected in the vegetative tissues, with the highest concentrations in the tips of aerial roots and in young leaves. Tracer feeding experiments have identified the tips of the aerial roots as a site of PA biosynthesis. Because the PAs of no other tissues could be labeled, their presence in these tissues can only be accounted for by transport from the sites of biosynthesis in the root tips to sites of accumulation. The localization of HSS in flower buds has therefore been an unexpected result, prompting further tracer feeding experiments with flower buds at different developmental stages. The results clearly show the youngest flower buds as being the most effective in PA biosynthesis, an ability that disappears with the increasing age of the flower buds, and is lost well before the buds open. Thus, in addition to the PA biosynthesis in the tips of aerial roots that has been suggested to be responsible for PAs found in the vegetative tissues (Frölich et al., 2006), flower buds boost their PA biosynthetic capacity during flower development. The resulting high concentrations of PAs found in young buds (up to 6.3 mg/g fresh weight) are increasingly diluted by the expanding tissue mass of the developing flower, but remain at a high level (about 1 mg/g fresh weight) when the flower opens. The conflict with the data of Frölich et al. (2006) in which flower buds have not been identified as the site of PA biosynthesis is most likely attributable to the correlation of HSS expression and developmental stage of the flower bud, suggesting that, in the flower buds used for the former tracer feeding experiments, the PA biosynthesis had already been switched off.
Diverse Expression of PA Biosynthesis within Angiosperms
HSS expression in Phalaenopsis adds a further facet to the diversity of the expression patterns that we have identified so far for a key enzyme in the pathway of PA biosynthesis: Two organs of a plant express all genes involved in the same biosynthesis depending on developmental cues. The developmental regulation of HSS expression has also been found within E. cannabinum. HSS is expressed in the root cortex only in the young growing plant. The moment that the flowers open, HSS expression is switched off (Anke et al., 2004). In S. vernalis, in which HSS is expressed in groups of specialized cells of the endodermis and the adjacent cortex parenchyma directly opposite the phloem (Moll et al., 2002), no link between plant development and HSS expression has been observed. The findings that PAs are accumulated within Phalaenopsis preferentially in young roots and young leaves, and that an accessory biosynthesis in young flower buds ensures high concentrations of PAs within the reproductive tissues of the orchid flower, suggest an ecological role for the 1,2-saturated PAs within Phalaenopsis; this role might have been one of the factors shaping this thoroughly regulated biosynthesis during evolution. Further support for an ecological role of PAs is provided by the data of specialized insects that developed highly specific adaptations enabling them to accumulate und use the plant-derived PAs for their own benefit, particularly as defense against their predators (Hartmann and Ober, 2000). Among these insects are species belonging to the tiger moths (Arctiidae) that sequester not only 1,2-unsaturated, but also 1,2-saturated, PAs like phalaenopsine (Hartmann et al., 2005) that are converted by specific enzymes to insect alkaloids to be transmitted to the adult stage during metamorphosis. Favorable effects of 1,2-saturated PAs on plant performance have recently been suggested by Koulman et al. (2008), who detected conjugates of this kind of PAs in a variety of commercial cultivars of grasses, in which they might have been inadvertently selected by breeders. A challenge for the future will be not only a better understanding of the biological activity of 1,2-saturated PAs, but also a better knowledge of the origin of the elements that control the complex expression patterns of PA biosynthesis in the various PA-producing species for which it was shown that the genes encoding HSS are of independent origin (Reimann et al., 2004).
MATERIALS AND METHODS
Plant Material
An interspecific hybrid of Phalaenopsis equestris × (Phalaenopsis aphrodite × Phalaenopsis mannii) was obtained as mature plants from Wichmann Orchideen. Plants were grown and maintained at 23°C and approximately 50% humidity in a greenhouse.
Tracer Feeding Experiments
Flower buds cut from inflorescences A and B (Fig. 5) were allowed to take up a [1,4-14C]putrescine (GE Healthcare) tracer (185 kBq each) with a final concentration of 100 μm from 400 μL of tap water. After complete uptake of the tracer (approximately 24 h), the flower buds were incubated with tap water devoid of tracer for 7 and 5 d in the case of flower buds of inflorescences A and B, respectively. The organs were washed with water, dabbed dry, weighed, and ground in a mortar before they were extracted twice for 30 min with methanol. The supernatant of the combined methanol extracts was analyzed for total radioactivity by scintillation counting. Aliquots were subjected to thin-layer chromatography, HPLC, or gas chromatography/mass spectrometry analysis for the identification and quantification of the labeled products according to Hartmann and Dierich (1998).
Polyclonal Antibody Preparation and Affinity Purification
To generate polyclonal antibodies, recombinant untagged HSS was purified by a three-step purification procedure similar to that previously described for tobacco (Nicotiana tabacum; DHS; Ober and Hartmann, 1999a). Cells of a 100-mL Escherichia coli BL21(DE3) culture expressing HSS of Phalaenopsis were harvested and sonicated and the supernatant applied to a 2.5 × 5.0 cm DEAE-Fractogel column (Merck) in buffer A (50 mm potassium phosphate, pH 8.7, 2 mm dithioerythritol, 0.1 mm EDTA). Under these conditions, HSS did not bind to the matrix. The flow-through fraction was applied directly onto a phenyl Sepharose CL-4B column (Amersham Biosciences) equilibrated with buffer B (5 mm potassium phosphate, pH 8.7, 2 mm dithioerythritol, 0.1 mm EDTA, 1 m sodium chloride). Elution of HSS was achieved with buffer B without sodium chloride. Peak fractions were loaded onto a MonoQ column (Amersham Biosciences) equilibrated with buffer A and eluted with a gradient from 0 to 1.0 m sodium chloride. Active fractions were pooled and concentrated by using an Amicon Stirred Cell (Millipore). The purified protein was rebuffered to 5 mm potassium phosphate, pH 8.0, by using an Amicon Stirred Cell (Millipore) and freeze dried. One aliquot of 500 μg and two aliquots of 250 μg of pure HSS were used to raise polyclonal antibodies in rabbits by repeated subcutaneous injections (performed by Bioscience). Purified HSS (1 mg) was coupled to activated Sepharose 4B (GE Healthcare) according to the manufacturer's instructions. The resulting affinity matrix was incubated overnight at room temperature with the serum, washed with 0.1 m sodium acetate (pH 4.5) containing 0.5 m NaCl, and eluted with 0.2 m sodium acetate (pH 2.7) containing 0.5 m NaCl. The eluting antibodies were rebuffered to phosphate-buffered saline (PBS), concentrated, and stored at −20°C until further use.
Protein Gel-Blot Analysis and Tissue Print
Tissue samples were pulverized in a mortar in the presence of liquid nitrogen and the powder was extracted in PBS supplemented with 2.5% (w/v) polyvinylpyrrolidone and 2.5% (w/v) sodium ascorbate. The extract was centrifuged, and the protein in the supernatant was quantified by the method of Bradford (1976). By using a discontinuous buffer system, proteins were separated on 12% (w/v) SDS-PAGE gels at 200 V constant voltage. Protein gels were electroblotted onto polyvinylidene fluoride membrane (Immobilon P; Millipore) with a current density of 2.5 mA cm−2. The lane with the molecular weight marker was cut out and stained with Indian ink (Hancock and Tsang, 1983). The remainder of the membrane was used for immunodetection at room temperature. First, the membrane was blocked for 1 h with Tris-buffered saline (TBS) supplemented with 0.1% Tween 20 (TBS-T) and 5% (m/v) milk powder. Afterward, this solution was exchanged and the membrane was incubated with the affinity-purified polyclonal antibody (OD280 = 1.1 × 10−4) for 1 h. Following several washing steps (3 × 7 min with TBS-T), the membrane was incubated with a goat-anti-rabbit secondary antibody conjugated to horseradish peroxidase (diluted 1:3,300 [v/v]; Dianova) for 1 h. The washing steps were repeated before chemiluminescence detection was performed with the ECL western-blotting system (GE Healthcare) and documented on XAR5 x-ray film (Eastman-Kodak).
Tissue prints were prepared according to the method of Cassab and Varner (1987). Longitudinal sections of Phalaenopsis flower buds and aerial root tips were pressed for 30 s onto a nitrocellulose paper (Hybond-C extra; Amersham Biosciences) that had been equilibrated beforehand in 0.2 m calcium chloride solution for 30 min. The membrane was dried at room temperature and the detection of the tissue print with primary and secondary antibodies was carried out as described above. Proteins bound to the membrane were stained with India ink according to Hancock and Tsang (1983).
Immunocytochemical Localization
The fixation and embedding of tissues were performed as described previously (Anke et al., 2004). Briefly, segments of plant organs (maximum 1 cm in diameter) were fixed under reduced pressure for 2 h in ice-cold buffered fixative (2% [w/v] glutaraldehyde, 0.1% Triton X-100 [w/v], and 2% soluble polyvinylpyrrolidone [Kollidon 30; BASF] in 0.05 m potassium phosphate buffer, pH 7.2). After dehydration in a graded ethanol series, the sections were embedded in Technovit 7100 resin (Heraeus-Kulzer) for light microscopy analyses and in Unicryl resin (Plano) for analyses by transmission electron microscopy. Unicryl was polymerized under UV light at 4°C within 3 d. For UV and light microscopy, sections of 3 to 4 μm were cut on a microtome (HM355S; Microm) and mounted on glass slides coated with Teflon (Roth). Before being blocked with 10% (w/v) BSA and 0.1% fish gelatin, sections were washed successively at 37°C with 50 mm ammonia chloride and 50 mm Gly (20 min each). Afterward, sections were washed again (3 × 7 min PBS) and incubated with affinity-purified primary antibody (OD280 = 4.9 × 10−2) at 37°C for 3 h in a humid chamber. Washing steps with PBS were repeated. Incubation with a secondary goat anti-rabbit antibody labeled with FITC for fluorescence detection (1:100 [v/v] diluted in PBS; Sigma) or labeled with 18-nm gold particles for immunogold labeling (1:75 [v/v] diluted in PBS; Dianova) was performed for 1 h in the dark. FITC-labeled sections were protected with mounting medium (Citifluor; Agar Scientific) against bleaching and excited by UV light of 450 to 490 nm on an Axioskop 2 epifluorescence microscope (Zeiss). Sections labeled with gold particles were enhanced with silver-enhancing reagent (Amersham Biosciences) before documentation. Images were recorded with an AxioCam HRc digital camera (Zeiss).
Acknowledgments
We thank Thomas Hartmann for helpful discussions of the data and Ben Scheres (Utrecht, The Netherlands) for his support in interpreting our first longitudinal sections of the root tips. We are also grateful to B. Schemmerling for her excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (grant to D.O.) and the Austrian Academy of Sciences (scholarship to E.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dietrich Ober (dober@bot.uni-kiel.de).
Open Access articles can be viewed online without a subscription.
References
- Anke S, Niemüller D, Moll S, Hänsch R, Ober D (2004) Polyphyletic origin of pyrrolizidine alkaloids within the Asteraceae. Evidence from differential tissue expression of homospermidine synthase. Plant Physiol 136 4037–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott HJ, Pautard FG (1970) Calcification in plants. In H Schraer, ed, Biological Calcification: Cellular and Molecular Aspects. Appeton Century Crofts, New York, pp 375–446
- Benfey PN, Scheres B (2000) Root development. Curr Biol 10 R813–R815 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brattsten LB (1992) Metabolic defenses against plant allelochemicals. In GA Rosenthal, MR Berenbaum, eds, Herbivores—Their Interactions with Secondary Plant Metabolites, Vol 2. Academic Press, San Diego, pp 175–242
- Cassab GI, Varner JE (1987) Immunocytolocalization of extensin in developing soybean seed coats by immunogold-silver staining and by tissue printing on nitrocellulose paper. J Cell Biol 105 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, St-Pierre B (2000) The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci 5 168–173 [DOI] [PubMed] [Google Scholar]
- Doerner P (1998) Root development: quiescent center not so mute after all. Curr Biol 8 R42–R44 [DOI] [PubMed] [Google Scholar]
- Doerner P (2003) Plant meristems: a merry-go-round of signals. Curr Biol 13 R368–R374 [DOI] [PubMed] [Google Scholar]
- Esau K (1965) Plant Anatomy, Ed 2. John Wiley & Sons, New York
- Flores HE, Vivanco JM, Loyola-Vargas VM (1999) ‘Radicle’ biochemistry: the biology of root-specific metabolism. Trends Plant Sci 4 220–226 [DOI] [PubMed] [Google Scholar]
- Franceschi VR (1989) Calcium oxalate formation is a rapid and reversible process in Lemna minor L. Protoplasma 148 130–137 [Google Scholar]
- Franceschi VR, Horner HT (1980) Calcium oxalate crystals in plants. Bot Rev 46 361–427 [Google Scholar]
- Frei H, Luthy J, Brauchli J, Zweifel U, Wurgler FE, Schlatter C (1992) Structure/activity relationships of the genotoxic potencies of sixteen pyrrolizidine alkaloids assayed for the induction of somatic mutation and recombination in wing cells of Drosophila melanogaster. Chem Biol Interact 83 1–22 [DOI] [PubMed] [Google Scholar]
- Frölich C, Hartmann T, Ober D (2006) Tissue distribution and biosynthesis of 1,2-saturated pyrrolizidine alkaloids in Phalaenopsis hybrides (Orchidaceae). Phytochemistry 67 1493–1502 [DOI] [PubMed] [Google Scholar]
- Fu PP, Xia Q, Lin G, Chou MW (2004) Pyrrolizidine alkaloids—genotoxicity, metabolism enzymes, metabolic activation, and mechanisms. Drug Metab Rev 36 1–55 [DOI] [PubMed] [Google Scholar]
- Hancock K, Tsang VC (1983) India ink staining of proteins on nitrocellulose paper. Anal Biochem 133 157–162 [DOI] [PubMed] [Google Scholar]
- Hartmann T, Dierich B (1998) Chemical diversity and variation of pyrrolizidine alkaloids of the senecionine type: Biological need or coincidence? Planta 206 443–451 [Google Scholar]
- Hartmann T, Ehmke A, Eilert U, von Borstel K, Theuring C (1989) Sites of synthesis, translocation and accumulation of pyrrolizidine alkaloid N-oxides in Senecio vulgaris L. Planta 177 98–107 [DOI] [PubMed] [Google Scholar]
- Hartmann T, Ober D (2000) Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. In FJ Leeper, JC Vederas, eds, Topics in Current Chemistry, Vol 209. Springer, Berlin, pp 207–244
- Hartmann T, Theuring C, Beuerle T, Klewer N, Schulz S, Singer MS, Bernays EA (2005) Specific recognition, detoxification and metabolism of pyrrolizidine alkaloids by the polyphagous arctiid Estigmene acrea. Insect Biochem Mol Biol 35 391–411 [DOI] [PubMed] [Google Scholar]
- Hartmann T, Witte L (1995) Chemistry, biology and chemoecology of the pyrrolizidine alkaloids. In SW Pelletier, ed, Alkaloids: Chemical and Biological Perspectives, Vol 9. Pergamon Press, Oxford, pp 155–233
- Hartmann T, Zimmer M (1986) Organ-specific distribution and accumulation of pyrrolizidine alkaloids during the life history of two annual Senecio species. J Plant Physiol 122 67–80 [Google Scholar]
- Jakoby M, Schnittger A (2004) Cell cycle and differentiation. Curr Opin Plant Biol 7 661–669 [DOI] [PubMed] [Google Scholar]
- Jiang K, Feldman LJ (2005) Regulation of root apical meristem development. Annu Rev Cell Dev Biol 21 485–509 [DOI] [PubMed] [Google Scholar]
- Kausch AP, Horner HT (1983. a) The development of mucilaginous raphide crystal idioblasts in young leaves of Typhia angustifolia L. (Typhaceae). Am J Bot 70 691–705 [Google Scholar]
- Kausch AP, Horner HT (1983. b) Development of syncytial raphide crystal idioblasts in the cortex of adventious root s of Vanilla planifolia L. (Orchidaceae). Scan Electron Microsc II 893–903 [Google Scholar]
- Kausch AP, Horner HT (1984) Increased nuclear DNA content in raphide crystal idioblasts during development in Vanilla planifolia L. (Orchidaceae). Eur J Cell Biol 33 7–12 [PubMed] [Google Scholar]
- Koulman A, Seeliger C, Edwards PJB, Fraser K, Simpson W, Johnson L, Cao M, Rasmussen S, Lane GA (2008) E/Z-Thesinine-O-4′-α-rhamnoside, pyrrolizidine conjugates produced by grasses (Poaceae). Phytochemistry 69 1927–1932 [DOI] [PubMed] [Google Scholar]
- Lim WL, Loh CS (2003) Endopolyploidy in Vanda Miss Joaquim (Orchidaceae). New Phytol 159 279–287 [DOI] [PubMed] [Google Scholar]
- Mattocks AR (1986) Chemistry and Toxicology of Pyrrolizidine Alkaloids. Academic Press, London
- Moll S, Anke S, Kahmann U, Hänsch R, Hartmann T, Ober D (2002) Cell-specific expression of homospermidine synthase, the entry enzyme of the pyrrolizidine alkaloid pathway in Senecio vernalis, in comparison with its ancestor, deoxyhypusine synthase. Plant Physiol 130 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus I, Zintgraf V, Dobler S (2005) Pyrrolizidine alkaloids on three trophic levels—evidence for toxic and deterrent effects on phytophages and predators. Chemoecology 15 121–125 [Google Scholar]
- Ober D (2005) Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci 10 444–449 [DOI] [PubMed] [Google Scholar]
- Ober D, Harms R, Witte L, Hartmann T (2003) Molecular evolution by change of function: Alkaloid-specific homospermidine synthase retained all properties of deoxyhypusine synthase except binding the eIF5A precursor protein. J Biol Chem 278 12805–12812 [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T (1999. a) Deoxyhypusine synthase from tobacco: cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J Biol Chem 274 32040–32047 [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T (1999. b) Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc Natl Acad Sci USA 96 14777–14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5 439–445 [DOI] [PubMed] [Google Scholar]
- Reimann A, Nurhayati N, Backenköhler A, Ober D (2004) Repeated evolution of the pyrrolizidine alkaloid-mediated defense system in separate angiosperm lineages. Plant Cell 16 2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B (1997) Cell signaling in root development. Curr Opin Genet Dev 7 501–506 [DOI] [PubMed] [Google Scholar]
- Scheres B, McKhann HI, van den Berg C (1996) Roots redefined: anatomical and genetic analysis of root development. Plant Physiol 111 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, van den Toorn H, Heidstra R (2004) Root genomics: towards digital in situ hybridization. Genome Biol 5 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegelmeier BL, Edgar JA, Colegate SM, Gardner DR, Schoch TK, Coulombe RA, Molyneux RJ (1999) Pyrrolizidine alkaloid plants, metabolism and toxicity. J Nat Toxins 8 95–116 [PubMed] [Google Scholar]
- Storey R, Jones RGW, Schachtman DP, Treeby MT (2003) Calcium-accumulating cells in the meristematic region of grapevine root apices. Funct Plant Biol 30 719–727 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6 544–553 [DOI] [PubMed] [Google Scholar]
- Toppel G, Witte L, Riebesehl B, von Borstel K, Hartmann T (1987) Alkaloid patterns and biosynthetic capacity of root cultures from some pyrrolizidine alkaloid producing Senecio spp. Plant Cell Rep 6 466–469 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378 62–65 [DOI] [PubMed] [Google Scholar]