Abstract
Plant cells contain several thioredoxin isoforms that are characterized by subcellular localization and substrate specificity. Here, we describe the functional characterization of a rice (Oryza sativa) thioredoxin m isoform (Ostrxm) using a reverse genetics technique. Ostrxm showed green tissue-specific and light-responsive mRNA expression. Ostrxm was localized in chloroplasts of rice mesophyll cells, and the recombinant protein showed dithiothreitol-dependent insulin β-chain reduction activity in vitro. RNA interference (RNAi) of Ostrxm resulted in rice plants with developmental defects, including semidwarfism, pale-green leaves, abnormal chloroplast structure, and reduced carotenoid and chlorophyll content. Ostrxm RNAi plants showed remarkably decreased Fv/Fm values under high irradiance conditions (1,000 μmol m−2 s−1) with delayed recovery. Two-dimensional electrophoresis and matrix-assisted laser-desorption/ionization time-of-flight analysis showed that the levels of several chloroplast proteins critical for photosynthesis and biogenesis were significantly decreased in Ostrxm RNAi plants. Furthermore, 2-Cys peroxiredoxin, a known target of thioredoxin, was present in oxidized forms, and hydrogen peroxide levels were increased in Ostrxm RNAi plants. The pleiotropic effects of Ostrxm RNAi suggest that Ostrxm plays an important role in the redox regulation of chloroplast target proteins involved in diverse physiological functions.
Thioredoxins are ubiquitously distributed proteins that regulate numerous cellular processes. All thioredoxins have a highly conserved active-site sequence that contains two redox-active Cys residues in a conserved motif, Trp-Cys-Gly-Pro-Cys (Holmgren, 1989). In the reduced form, thioredoxins are excellent catalysts for the reduction of intramolecular disulfide bonds of target proteins (Jacquot et al., 2002). In contrast to other eukaryotic cells, plants contain several thioredoxin variants (Balmer and Buchanan, 2002; Collin et al., 2004; Gelhaye et al., 2004). The cytosolic or type h thioredoxins are reduced by NADPH with NADPH-thioredoxin reductase, whereas a set of chloroplast thioredoxins, types f and m, is reduced by a ferredoxin-dependent system (Ruelland and Miginiac-Maslow, 1999). The Arabidopsis (Arabidopsis thaliana) genome contains at least eight genes for thioredoxin h, two for thioredoxin f, and four for thioredoxin m (Mestres-Ortega and Meyer, 1999; Collin et al., 2003, 2004).
Chloroplast thioredoxins are implicated in the light-dependent enzyme regulation of photosynthetic organisms from cyanobacteria to higher plants (Ruelland and Miginiac-Maslow, 1999). In the light, chloroplast thioredoxins reduce specific disulfide bridges in target proteins to activate them (Clancey and Gilbert, 1987; Droux et al., 1987a, 1987b). In the dark, thioredoxins and their substrates are present in the oxidized state and are inactive (Scheibe and Anderson, 1981). Originally, thioredoxins f and m were distinguished by their ability to activate Fru-1,6-bisphosphatase (FBPase) and NADP-dependent malate dehydrogenase (NADP-MDH), respectively (Maeda et al., 1986; Kamo et al., 1989). More recently, thioredoxin f has been shown to activate several chloroplast enzymes, including those involved in the Calvin cycle (FBPase, sedoheptulose-1,7-bisphosphatase, phosphoribulokinase, glyceraldehyde-3-P dehydrogenase, Rubisco [via Rubisco activase], ATP synthesis [CF1ATP synthase], and fatty acid synthesis [acetyl-CoA carboxylase]; Nishizawa and Buchanan, 1981; Wolosiuk et al., 1993; Sasaki et al., 1997; Schwarz et al., 1997; Zhang and Portis, 1999). Thioredoxin m has been shown to be involved in the inactivation of Glc-6-P dehydrogenase, the first enzyme of the oxidative pentose phosphate cycle (Wenderoth et al., 1997). Both thioredoxins f and m can activate FBPase and NADP-MDH in vitro, suggesting that there is some overlap in the substrate specificity of the isoforms (Hodges et al., 1994; Geck et al., 1996). However, disruption of the thioredoxin m gene in cyanobacteria (Aspergillus nidulans) is lethal, suggesting that the protein may have a distinctive in vivo function. This result also highlights the importance of using genetic tools to define the role of thioredoxins in intact organisms (Muller and Buchanan, 1989).
Although the molecular characteristics and biochemical properties of chloroplast thioredoxins have been well studied in vitro (Scheibe and Anderson, 1981; Geck et al., 1996; Mestres-Ortega and Meyer, 1999; Duek and Wolosiuk, 2001; Issakidis-Bourguet et al., 2001; Collin et al., 2003, 2004), less is known about their physiological role in intact plants. In this study, we have cloned a rice (Oryza sativa) gene encoding a thioredoxin m and used RNA interference (RNAi) to investigate the function of Ostrxm in rice. The results indicate that Ostrxm has an essential role in chloroplasts for plant growth and development.
RESULTS
Isolation and Expression Pattern of Ostrxm
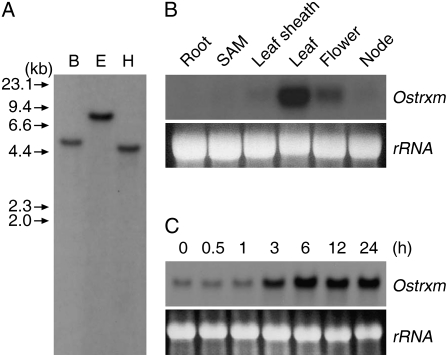
A search of The Institute for Genomic Research rice genome database (http://www.tigr.org/tigr-scripts/e2k1/osa1/putative_function_search.pl) with the search term “thioredoxin m-type” revealed matches with three genes (Os02g42700, Os04g44830, and Os12g08730). Alignment and phylogenetic analysis of the corresponding cDNA sequences of the three genes revealed that AK099517 (Os02g42700) and AK069195 (Os04g44830) share a sequence identity of 52.8%, but that they significantly diverge from AJ005841 (Os12g08730), sharing only 24.2% and 23.9% sequence identity with this gene, respectively (Supplemental Fig. S1). A cDNA corresponding to Os12g08730 designated as Ostrxm was isolated from a rice leaf cDNA library with the goal of investigating the in vivo function of the protein. A rice functional genomics database (http://signal.salk.edu/cgi-bin/RiceGE) search using the Ostrxm cDNA sequence as a query revealed that the gene comprises two exons interrupted by an intron and that it spans about 1 kb. The high degree of cDNA sequence divergences of Os02g42700 and Os04g44830 from Ostrxm are consistent with a Southern-blot analysis of rice genomic DNA using the Ostrxm cDNA as a probe, which revealed a single hybridizing band (Fig. 1A) and with a BLAST search of a rice genomic DNA database using the cDNA sequence as a query, which hit a single highly homologous gene. When expression of the three genes was evaluated by reverse transcription (RT)-PCR analysis, Ostrxm showed relatively higher expression and Os02g42700 and Os04g44830 were expressed at very low levels in 12-d-old seedlings. Northern-blot analysis was then performed to examine the expression pattern of the Ostrxm gene. As seen in Figure 1B, Ostrxm is predominantly expressed in leaf and floral tissue. To examine the light-dependent expression of Ostrxm, 12-d-old rice seedlings were allowed to adapt to the dark for 24 h and then exposed to light for specified times. Expression of Ostrxm transcripts in the seedlings began to increase after 3 h of light treatment and it was maintained continuously up to 24 h (Fig. 1C).
Figure 1.
Genomic complexity and mRNA expression of Ostrxm. A, Southern-blot analysis of the Ostrxm gene. Genomic DNA was digested with BamHI (B), EcoRI (E), or HindIII (H). B, Northern-blot analysis of Ostrxm gene expression. Total RNA was prepared from 21-d-old root, shoot apical meristem (SAM), leaf sheath and leaf, late boot to heading stage flower and reproductive stage node. C, Induction of the Ostrxm mRNA transcript by de-etiolation. Twelve-day-old rice seedlings were etiolated for 24 h and exposed to light for the indicated times under a photosynthetic flux of 200 μmol m−2 s−1 at 30°C. RNA was prepared from leaf tissues.
Subcellular Localization and Disulfide Oxidoreductase Activity of Recombinant Ostrxm
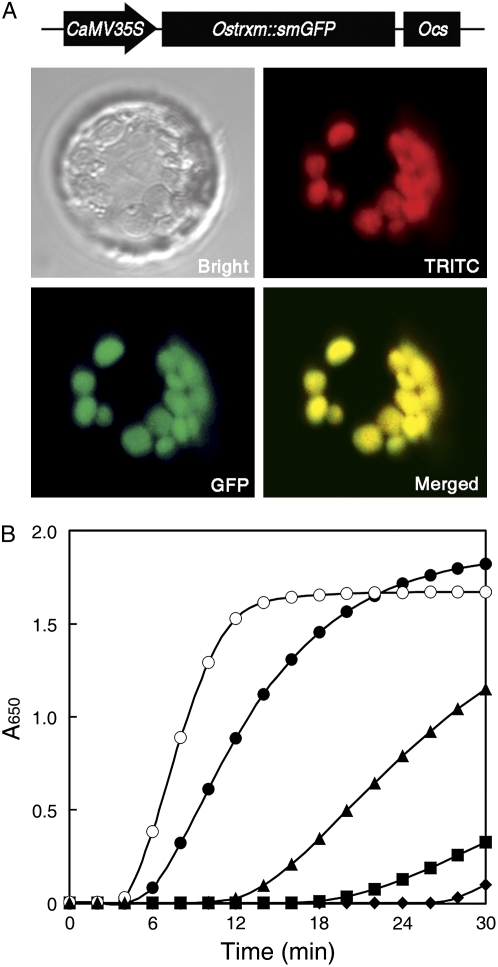
To determine the subcellular localization of Ostrxm in rice, we generated transgenic rice plants expressing Ostrxm-smGFP or ΔNOstrxm-smGFP (Ostrxm without the N-terminal transit peptide) and examined their localization in protoplasts by confocal laser-scanning microscopy. Ostrxm-smGFP appeared to colocalize with the red autofluorescence of chloroplasts, whereas ΔNOstrxm-smGFP localized to the cytosol (data not shown), indicating that Ostrxm synthesized in the cytosol is translocated into chloroplasts by virtue of its N-terminal transit peptide (Fig. 2A).
Figure 2.
Subcellular localization of Ostrxm and insulin reduction activity of the recombinant protein. A, Subcellular localization of Ostrxm. Ostrxm∷smGFP is indicated by green fluorescence (GFP) and the auto fluorescence of chloroplasts is indicated by red fluorescence (TRITC). The bright-field image (Bright) was obtained using the bright-field filter. Colocalized GFP and chloroplasts (Merged) appear as yellow. B, DTT-dependent insulin reduction activity of Ostrxm. Reaction mixtures contained 10 μg of bovine serum albumin as a control (♦), 0.5 μg of E. coli thioredoxin (○), 1 μg of Ostrxm (▪), 5 μg of Ostrxm (▴), or 10 μg of Ostrxm (•). [See online article for color version of this figure.]
In an experiment to investigate the disulfide reductase activity of the protein, recombinant Ostrxm was tested for its ability to reduce the insulin β-chain, an artificial substrate of thioredoxins. Recombinant Ostrxm reduced the insulin β-chain only in the presence of dithiothreitol (DTT; Fig. 2B). In the absence of DTT or when DTT was replaced with ascorbate, the protein was completely unable to reduce insulin (data not shown), indicating that Ostrxm possesses thiol-dependent disulfide reductase activity. However, the disulfide reductase activity of Ostrxm was lower than that of Escherichia coli thioredoxin. These results indicate that Ostrxm is a bioactive disulfide oxidoreductase in chloroplasts.
Phenotypic Analysis of Ostrxm RNAi Transgenic Rice
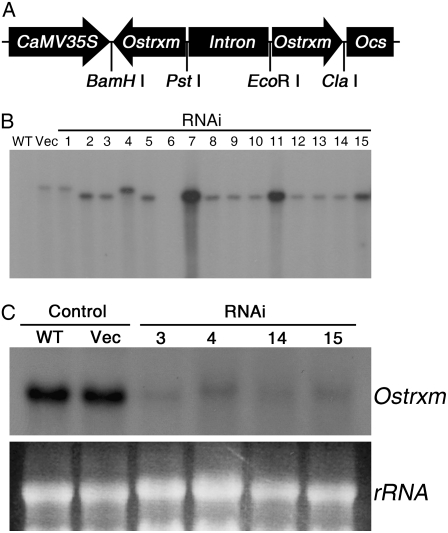
To study the physiological role of Ostrxm in rice, we generated Ostrxm RNAi transgenic plants that showed a significantly reduced expression pattern of Ostrxm (Fig. 3). Fifteen hygromycin-resistant T0 transformants were transferred into soil and grown in a greenhouse and self-pollinated to obtain segregating T1 progeny for genetic analysis. Integration and copy number of the T-DNA were confirmed by Southern-blot analysis (Fig. 3B). Among the 15 hygromycin-resistant T0 transformants, only four lines survived and produced T1 seeds. The four lines showed a segregation pattern of 3:1 for hygromycin resistance (Table I). Northern-blot analysis of the four surviving transgenic lines showed that Ostrxm expression was efficiently suppressed in all the RNAi transgenic lines (Fig. 3C). Two lines, 4 and 14, were selected for further analysis. T2 and T3 Ostrxm RNAi transgenic lines used for experiments were easily distinguished from segregating wild-type plants by their pale-green and semidwarf phenotypes. When gene-specific RT-PCR was performed to check the specificity and efficiency of the targeted knock-down of the Ostrxm gene, about 90% of Ostrxm transcripts was found to be specifically decreased, whereas transcripts of Os0242700 and Os04g44830 were not affected by Ostrxm RNAi (Supplemental Fig. S1C).
Figure 3.
The Ostrxm RNAi construct and analysis of Ostrxm expression in transgenic rice. A, Schematic representation of the Ostrxm RNAi construct. B, Southern-blot analysis of the T0 progeny of Ostrxm RNAi transgenic and wild-type plants using an [α-32P]ATP-labeled HPT (hygromycin phosphotransferase) cDNA as a probe. C, Northern-blot analysis of the T1 progeny of Ostrxm RNAi transgenic and wild-type plants.
Table I.
Segregation analysis of the T1 generation of Ostrxm RNAi plants
| Line | Hygromycinr | Ratio | Pale-Green Seedlings |
|---|---|---|---|
| T1-3 | 22+/8− | 2.93:1.07 | 22 |
| T1-4 | 24+/6− | 3.20:0.80 | 24 |
| T1-14 | 23+/7− | 3.07:0.93 | 23 |
| T1-15 | 22+/8− | 2.93:1.07 | 22 |
Germination was scored following seed imbibition and the subsequent development of roots and shoots was monitored. Both wild-type and Ostrxm RNAi seeds had germinated, with slight differences in germination time (Fig. 4A). When roots and shoots were examined, the RNAi transgenic rice seedlings exhibited marked dwarf phenotype compared to the wild-type seedlings (Fig. 4B). The Ostrxm RNAi transgenic lines also had pale-green leaves with light-yellow stripes, and exhibited reduced plant mass, tiller number (about 70%), and semidwarfism (Fig. 4, C and D). However, there was no significant difference in the leaf number. Ostrxm RNAi transgenic rice produced immature panicles and empty spikelets and had significantly reduced grain yields compared to wild-type plants (Fig. 4E). The average spikelet number per panicle in Ostrxm RNAi transgenic rice was 46.7 ± 3.6 compared with 67.0 ± 4.1 in the wild type.
Figure 4.
Phenotypic analysis of Ostrxm RNAi rice plants. A and B, Wild-type and transgenic rice plants are shown at 3 (A) or 10 (B) d postgermination. C, Wild-type and transgenic rice plants grown for 70 d. D, Leaf blade phenotypes of wild-type and transgenic rice plants grown for 70 d. Secondary leaves from primary tillers are shown. E, Main panicle of wild-type and Ostrxm RNAi rice plants grown for 115 d. Wild-type and transgenic rice plants were grown in a growth chamber (A and B) conditioned with 8 h dark/16 h light at 200 μmol m−2 s−1 and a day/night temperature regime of 30°C/25°C, or under sunlight conditions in a greenhouse (C–E). In each image, wild-type, Vec, and Ostrxm RNAi 4 and 14 plants are arranged from left to right (A–E).
Structural Defect of the Photosynthetic Apparatus in Ostrxm RNAi Plants
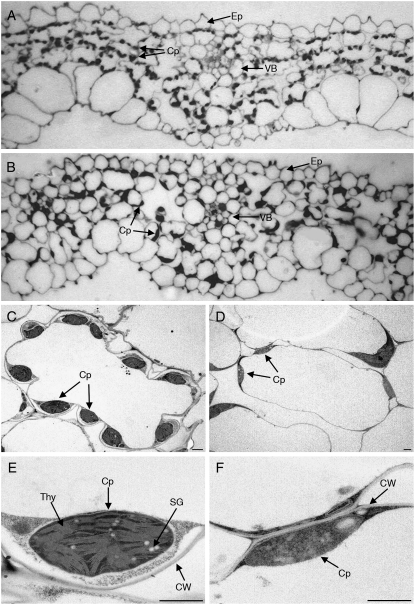
The pale-green and semidwarf phenotypes urged us to investigate chloroplast structure and the photosynthetic apparatus of Ostrxm RNAi plants. When leaves were examined under a light microscope, we observed decreased numbers of chloroplasts per mesophyll cell in Ostrxm RNAi plants compared to wild-type plants, and the shape and size of chloroplasts were irregular in Ostrxm RNAi plants compared to those of wild-type plants (Fig. 5, A and B). When chloroplasts of wild-type leaves were investigated by transmission electron microscopy, well-defined grana and stroma-thylakoid boundaries and regularly shaped starch grains were observed (Fig. 5, C and E). In contrast, chloroplasts from Ostrxm RNAi plants were of abnormal shape, their thylakoid boundaries were vague, and they lacked regularly shaped starch grains (Fig. 5, D and F). Overall, the ultrastructural defects observed in the leaves and chloroplasts of Ostrxm RNAi transgenic plants suggested that Ostrxm is required for chloroplast biogenesis and differentiation.
Figure 5.
Ultrastructural examination of Ostrxm RNAi transgenic rice. A and B, Light microscope images of leaves from wild-type and Ostrxm RNAi rice plants, respectively (magnification = 1,000×). C to F, Transmission electron micrographs of chloroplasts from wild-type (C and E) and Ostrxm RNAi rice plants (D and F). Scale bar = 1 μm (C–F). Cp, Chloroplast; VB, vascular bundle cell; Ep, epidermis; SG, starch grain; CW, cell wall; Thy, thylakoid. Plant tissues were harvested from 10-d-old seedlings grown in a growth chamber with supplemental lighting (8 h dark/16 h light at 200 μmol m−2 s−1 and a day/night temperature regime of 30°C/25°C).
Pigment Content and Photosynthetic Efficiency of Ostrxm RNAi Plants
In Ostrxm RNAi plants, chlorophyll (Chl) content per fresh weight was significantly decreased to 76% in the case of RNAi 4 or 53% in the case of RNAi 14. The amount of carotenoids and xanthophyll cycle pigments was also decreased; however, their relative levels with respect to total Chl were different. Thus, the relative amounts of neoxanthin and lutein were decreased in Ostrxm RNAi plants compared to wild-type and vector plants, but those of β-carotene and the pool size of the xanthophyll cycle pigments were increased in Ostrxm RNAi plants (Table II). In addition, the Chl a/b ratio was slightly higher in the leaves of Ostrxm RNAi plants compared to wild-type plants.
Table II.
Pigment content in the leaves of wild-type, Vec, and Ostrxm RNAi transgenic rice plants (mmol/mol Chl)
| Pigmenta | Wild Type | Vec | RNAi 4 | RNAi 14 |
|---|---|---|---|---|
| Neoxanthin | 53.4 ± 0.8 | 55.6 ± 1.3 | 42.2 ± 0.4 | 41.7 ± 0.3 |
| Violaxanthin | 134.2 ± 1.6 | 165.6 ± 1.0 | 165.1 ± 3.9 | 167.5 ± 0.5 |
| Antheraxanthin | 14.6 ± 1.7 | 20.1 ± 1.8 | 50.3 ± 0.4 | 40.6 ± 1.2 |
| Lutein | 213.0 ± 2.5 | 213.5 ± 0.8 | 178.9 ± 2.7 | 160.5 ± 2.4 |
| Zeaxanthin | 9.4 ± 0.1 | 12.2 ± 0.4 | 29.6 ± 1.2 | 18.8 ± 0.4 |
| β-Carotene | 135.7 ± 1.8 | 151.6 ± 2.0 | 195.7 ± 4.7 | 179.7 ± 3.5 |
| V + A + Zb | 158.1 ± 3.4 | 198.0 ± 3.2 | 245.0 ± 5.5 | 226.8 ± 2.1 |
| Chl a/b | 3.6 ± 0.1 | 3.6 ± 0.1 | 3.9 ± 0.2 | 3.9 ± 0.1 |
Pigments were separated and quantified by HPLC as described in “Materials and Methods”. All values are the mean ± sd of at least three independent experiments. Secondary leaves from primary tillers of 56-d-old plants are used for analysis.
Total xanthophyll cycle pigments including violaxanthin (V), antheraxanthin (A), and zeaxanthin (Z).
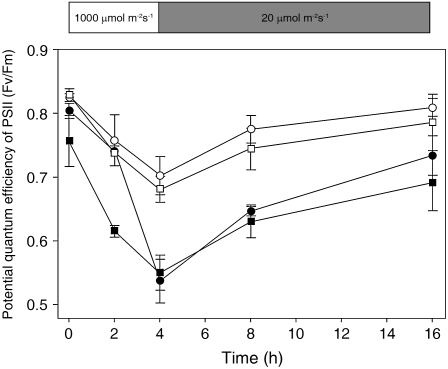
Because Ostrxm RNAi plants had reduced pigment content and structurally defective chloroplasts, we measured the potential quantum yield of PSII (Fv/Fm). Under high light irradiance of 1,000 μmol m−2 s−1 for 4 h, Ostrxm RNAi plants showed significant decreases in Fv/Fm by about approximately 30%, whereas wild-type plants showed about approximately 15% decrease (Fig. 6). After 4 h, the Fv/Fm of wild-type plants recovered to about 95%, whereas that of Ostrxm RNAi plants recovered to about 75%, and it recovered thereafter almost fully, but relatively slowly.
Figure 6.
Potential quantum efficiency of PSII in wild-type and Ostrxm RNAi plants. PSII photoinhibition was monitored by a decrease in the maximum photochemical efficiency of PSII (Fv/Fm) during exposure to 1,000 μmol m−2 s−1 irradiance for 4 h at 28°C and recovery at 20 μmol m−2 s−1 for the indicated time periods. Leaf segments of wild-type (○), Vec (□), and Ostrxm RNAi 4 (▪) and 14 plants (•) grown for 56 d in a greenhouse were used. Prior to a 4-h light treatment, plants were adapted for 2 d in a growth chamber with a light condition of 8 h dark and 16 h light at 200 μmol m−2 s−1 and a day/night temperature regime of 30°C/25°C. Data points represent the mean ± sd of three independent experiments.
Redox State of Ostrxm RNAi Plants
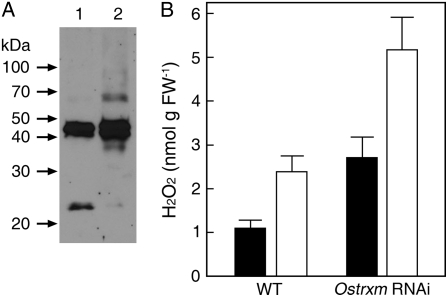
Thioredoxin m, acting as an electron donor for Cys-containing proteins, may regulate the activities of target proteins by reducing their disulfide bonds. To explore this hypothesis, the redox state of endogenous 2-Cys peroxiredoxin (2-Cys Prx; Motohashi et al., 2001; König et al., 2002) was monitored by nonreducing electrophoresis and western blotting using an antibody against 2-Cys Prx. In wild-type plants, both the 24-kD reduced monomeric form and the 48-kD oxidized dimeric form of 2-Cys Prx were detected, whereas only the oxidized dimeric form was detected in Ostrxm RNAi plants (Fig. 7A). This result suggests that Ostrxm regulates the catalytic activity of 2-Cys Prx by reducing redox-active Cys residues. Because the amount of reduced 2-Cys Prx is related to hydrogen peroxide (H2O2) catabolism, we wondered whether the H2O2 level in Ostrxm RNAi plants was increased. The level of H2O2 in leaves of Ostrxm RNAi plants in the dark or sunlight conditions was about 2-fold higher than that of wild-type plants (Fig. 7B). The increased H2O2 level in Ostrxm RNAi plants suggests that Ostrxm acting as a disulfide reductase plays a crucial role in the 2-Cys Prx-coupled detoxification of plastid-derived peroxides. Therefore, the defective flow of electrons to Ostrxm substrates in Ostrxm RNAi plants might be considerably implicated in their severe phenotypes.
Figure 7.
Redox state and oxidative stress levels of Ostrxm RNAi plants. A, Redox state of endogenous 2-Cys Prx in wild-type and Ostrxm RNAi plants. Soluble proteins from leaves of wild-type (lane 1) and Ostrxm RNAi (lane 2) plants were separated by SDS-PAGE under nonreducing conditions. Western blotting was performed with an antibody against 2-Cys Prx. B, H2O2 content in leaves of wild-type and Ostrxm RNAi plants in the dark (black bar) or sunlight (white bar) conditions. Data are the mean ± sd of at least three independent experiments. Sixty-day-old rice plants grown in a greenhouse (sunlight) were used.
DISCUSSION
Despite extensive in vitro characterization of thioredoxin m in plants, its in vivo function remains to be determined. Thus, we generated Ostrxm RNAi transgenic rice plants to investigate the in vivo function of Ostrxm in planta. Ostrxm RNAi plants showed phenotypes such as pale-green leaves, growth inhibition, reduced Chl and pigment content, decreased photosynthetic efficiency, defective chloroplast development, and inactivation of 2-Cys Prx. These results raise the possibility that some of the important biosynthetic pathways in chloroplasts are linked to the light-dependent thioredoxin electron transfer chain.
In higher plants, carotenoids mainly accumulate in the thylakoid membrane of chloroplasts, where they function to harvest light and protect the photosynthetic apparatus from oxidative damage by quenching the triplet excited state of Chl (3Chl) and reactive singlet oxygen (1O2) and dissipating excess energy (Demmig-Adams et al., 1996; Tracewell et al., 2001). Thus, the reduced photosynthetic pigments of Ostrxm RNAi plants might cause structural defects of chloroplasts and reduced photosynthetic efficiency under high irradiance conditions, finally producing the observed pale-green leaf and growth defect phenotypes. It has been experimentally demonstrated that the xanthophyll cycle is closely associated with nonphotochemical quenching (NPQ), which is an essential protective mechanism against high light stress (Gilmore and Yamamoto, 1993; Müller et al., 2001; Külheim et al., 2002). Formation and relaxation of NPQ is controlled not by the concentration of zeaxanthin, but by the extent of de-epoxidation of the xanthophyll pool (Johnson et al., 2008). The relative xanthophyll pool size (V + An + Z)/Chl, where V is violaxanthin, A is antheraxanthin, and Z is zeaxanthin, and the de-epoxidation state of xanthophyll was significantly higher in Ostrxm RNAi transgenic plants than in wild-type plants and empty-vector plants (Table II), indicating a higher capacity for xanthophyll cycle-dependent thermal energy dissipation by NPQ in Ostrxm RNAi plants. Xanthophyll pool size and the de-epoxidation state seem to be regulated to protect against the damage caused by suppression of Ostrxm in RNAi transgenic rice plants. Thus, a limitation in the acceptor side of PSI in Ostrxm RNAi plants results in a reduced antenna size of PSII, finally making Ostrxm RNAi plants more susceptible to high irradiance.
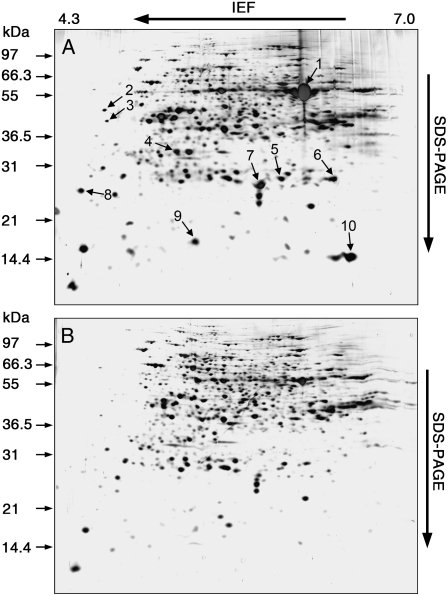
Recent approaches for identifying targets of chloroplast thioredoxins have provided new insights into the functions of plant thioredoxins (Motohashi et al., 2001; Balmer et al., 2003). An approach using immobilized thioredoxins m and f as baits, 26 identified targets were related to established (Calvin cycle, nitrogen and sulfur metabolism, translation, and pentose phosphate cycle) or unrecognized (isoprenoid, tetrapyrrole and vitamin biosynthesis, protein assembly/folding, protein and starch degradation, glycolysis, HCO3−/CO2 equilibration, plastid division, and DNA replication/transcription) cellular reactions (Balmer et al., 2003). Interestingly, two-thirds of the identified targets contained conserved Cys residues, increasing the probability that chloroplast thioredoxin regulates the catalytic activities of targets through the reduction of specific disulfide groups. To determine whether Ostrxm affects the expression of proteins that are involved in photosynthesis and chloroplast biogenesis, we examined the expression levels of both nuclear-encoded and chloroplast-encoded chloroplast genes by northern-blot analysis. However, we could not detect any significant differences in the transcript expression levels of these genes in Ostrxm RNAi transgenic plants as compared to wild-type plants (Supplemental Fig. S2). Thus, we performed two-dimensional electrophoresis (2-DE) and matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) analyses to identify differentially expressed proteins in Ostrxm RNAi plants. Ten protein spots resolved on 2-DE gels, which showed decreased levels in Ostrxm RNAi plants were subjected to MALDI-TOF analysis (Fig. 8), which led to the identification of eight proteins (Table III). Interestingly, the localization of seven identified proteins coincided with that of Ostrxm, and Rubisco small subunit and peptidyl-prolyl cis-trans isomerase, which is designated TLP40, were previously identified as target proteins of thioredoxin (Motohashi et al., 2001). Their possible implications in photosynthetic metabolisms help explain the observed phenotypic changes of Ostrxm RNAi plants although the exact underlying mechanisms remain to be elucidated.
Figure 8.
2-DE analysis of differentially expressed proteins in Ostrxm RNAi plants. Protein samples were separated on 2-DE gels (pI 4–7) and silver-stained. Differentially expressed proteins (decreased levels) in Ostrxm RNAi plants are indicated by arrows. A, Wild type; B, Ostrxm RNAi plants. Relative molecular masses are indicated on the left side (kD). The number of each protein spot (1–10) corresponds to its listing in Table III. Thirty-day-old rice plants grown in a greenhouse (sunlight) were used.
Table III.
Identification of differentially expressed proteins in Ostrxm RNAi plants
ND, Not determined; Est. Z, estimated Z score as a measure of confidence for profound-based identification. MM, Molecular mass.
| Spot | Identified Protein | Accession No. | Est. Z | pI/MM |
|---|---|---|---|---|
| 1 | Ribulose bisphosphate carboxylase large chain | CAG34174 | 1.15 | 6.2/53.35 |
| 2 | ND | – | – | – |
| 3 | Peptidyl-prolyl cis-trans isomerase | BAD05657 | 2.31 | 4.8/46.81 |
| 4 | 33-kD oxygen evolving protein of PSII | BAB64069 | 2.39 | 5.2/30.10 |
| 5 | Ribulose-5-P 3-epimerase | AAL84303 | 0.99 | 5.7/25.02 |
| 6 | Carbonic anhydrase 3 | AAD56038 | 2.24 | 6.3/22.30 |
| 7 | 23-kD polypeptide of PSII | AAC98778 | 2.13 | 6.0/23.33 |
| 8 | Putative benzothiadiazole-induced somatic embryogenesis receptor kinase 1 | BAD53863 | 1.15 | 5.6/44.21 |
| 9 | ND | – | – | – |
| 10 | Rubisco small subunit | AAR19268 | 1.91 | 6.8/15.05 |
Photoreduction of oxygen in PSI of chloroplasts generates superoxide anion (O2−) as the primary product of reduced oxygen species. O2− is converted to H2O2 and O2 by superoxide dismutase. To avoid photoinhibition and protect target molecules in chloroplasts, the immediate removal of H2O2 at the site of production is essential. H2O2 in chloroplasts is rapidly reduced to water by ascorbate catalyzed with ascorbate peroxidase in PSI (Asada, 2006). Peroxiredoxin, which is activated by thioredoxin, may also participate in the scavenging of H2O2 in chloroplasts. The increase of oxidized 2-Cys Prx and H2O2 levels in Ostrxm RNAi plants and the slowing down of the Calvin cycle under oxidative conditions (Balmer et al., 2003) suggest that Ostrxm is an important regulator of redox homeostasis in chloroplasts and that it is important for photosynthetic CO2 assimilation. Our study using Ostrxm RNAi plants indicates that Ostrxm, an active disulfide oxidoreductase in chloroplasts, is important for photosynthesis and chloroplast development. The observed severe phenotypes and physiological changes in Ostrxm RNAi plants indicate that Ostrxm may regulate target proteins through the reduction of Cys disulfides of targets involved in various chloroplast processes such as isoprenoid biosynthesis, Calvin cycle, and pentose phosphate cycle, etc. However, further genetic investigations are required to expand our understanding of the pleiotropic effects observed in Ostrxm RNAi plants.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa ‘Dong-jin’) was used for all experiments. Ostrxm RNAi transgenic rice was obtained by Agrobacterium-mediated transformation of embryonic callus using the RNAi technique. Hygromycin selection and plant regeneration were carried out as described (Dai et al., 2001). Surface-sterilized wild-type, vector-transformed (Vec), and transgenic seeds were germinated in one-half-strength Murashige and Skoog solid medium with 1% Suc. Rice plants were grown in a growth chamber conditioned with supplemental lighting (8 h dark/16 h light; 200 μmol m−2 s−1) and a day/night temperature regime of 30°C/25°C, or in a greenhouse (sunlight).
RT-PCR Analysis
RT-PCR was performed to measure the mRNA expression of the Os02g42700, Os04g44830, and Os12g08730 genes and to clone the Ostrxm cDNA (AJ005841). PCRs were performed on a PTC-0220 PCR machine (MJ Research) using 100 ng cDNA, 5 pmol each oligonucleotide primer, 200 μm each dNTP, 1 unit Ex Taq polymerase (TaKaRa Bio) and 1× Ex Taq polymerase buffer in a 20-μL volume. The PCR program initially started with a 95°C denaturation for 5 min, followed by 25 to 35 cycles of 95°C/1 min, 60°C/30 s, 72°C/1 min. The less expressed transcripts of Os02g42700 and Os04g44830 required more than 30 cycles of PCR for detection, whereas 25 cycles were enough for Os12g08730. Specific PCR primers for genes encoding Ostrxm isoforms were used (Os02g42700F: 5′-TCGCCAAGCCAGCCGCGTCTC-3′, Os02g42700F: 5′-CAGGGCTCAGCCTTGAATTGCATTTAT-3′, Os04g44830F: 5′-CCGTCTGCTCCTCTCCCCTCG-3′, Os04g44830F: 5′-CAGGCTTCCTTGCGACTTTCATGATTT-3′, Os12g08730F: 5′-ATGGCGTTGGAGACGTGCTTC-3′, and Os12g08730R: 5′-GCCACAGTATTGATTCCATGGAGTAGTAGGAT-3′).
Southern and Northern-Blot Analysis
For Southern-blot analysis, 10 μg of rice genomic DNA were digested with BamHI, EcoRI, and HindIII, separated on a 0.8% agarose gel, and transferred to a nylon membrane (Amersham). The membrane was hybridized and washed as described previously (Cheong et al., 1999). Total RNA (15 μg) was separated on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane. The rRNA bands were used as loading controls. The blot was hybridized with the same probe used for Southern-blot analysis.
Expression of Recombinant Ostrxm and Insulin Reduction Assay
cDNA encoding mature Ostrxm was subcloned into pGEX and expressed in Escherichia coli strain BL21 (DE3), and recombinant protein was purified as described previously (Jang et al., 2004). The disulfide oxidoreductase activity of the recombinant Ostrxm protein was assayed at 25°C in the presence of 0.5 mm DTT (Holmgren, 1989). Reactions were initiated by the addition of DTT to 1 mL reaction mixture, which contained 100 mm potassium phosphate buffer, pH 7.0, 0.1 mm EDTA, and 0.5 mg/mL insulin and various amounts of Ostrxm. Oxidoreductase activity was expressed as an increase in A650 using a spectrophotometer (Beckman DU 800).
Subcellular Localization of the Recombinant smGFP-Ostrxm
PCR was used to generate cDNA fragments encoding the full-length Ostrxm protein (amino acids 1–172) and the mature form of Ostrxm (amino acids 65–172). The cDNA fragments were ligated in frame with smGFP to create Ostrxm∷smGFP and ΔNOstrxm∷smGFP fusion constructs. The Ostrxm-smGFP and ΔNOstrxm∷smGFP cassettes were then ligated into the expression vector pCAMBIA1300. Transgenic rice plants expressing smGFP fusion constructs were generated by Agrobacterium-mediated transformation as described (Dai et al., 2001). Protoplasts were isolated from the leaves of 10-d-old transgenic rice plants grown on Murashige and Skoog plates in a growth chamber as described previously (Gupta and Pattanayak, 1993). Isolated protoplasts from transgenic plants were examined by laser-scanning confocal microscopy using an Olympus system (FV500 with IX71 microscope). The U-MGFPHQ filter set (excitation 460–480HQ; dichroic, 485; and emitter, 495–540HQ) and U-MWIG2 filter set (excitation 520–550; dichroic, 565; and emitter, 580IF) were used to image smGFP and chloroplasts, respectively.
Generation of Ostrxm RNAi Transgenic Rice
A 1-kb PCR fragment derived from the first intron of OsCDPK (accession no. AF 194414) was ligated into the PstI/EcoRI restriction sites of pBluescript (Stratagene). An open reading frame (519 bp) of the Ostrxm cDNA with EcoRI/ClaI restriction sites was inserted downstream of the intron, in the sense orientation. An open reading frame of the Ostrxm cDNA with BamHI/PstI restriction sites was inserted upstream of the intron in the antisense orientation. The complete Ostrxm RNAi construct contained an antisense fragment of the Ostrxm cDNA, followed by the OsCDPK intron, and then by the sense fragment of the Ostrxm cDNA, which included a stop codon. The Ostrxm RNAi construct was inserted into the BamHI/ClaI sites of pCAMBIA1300 between the cauliflower mosaic virus 35S promoter and the Ocs terminator to generate pCAMBIA1300∷Ostrxm. The Ostrxm RNAi plasmid was then transformed into rice using Agrobacterium strain LBA4404, as described previously (Dai et al., 2001).
Pigment Analysis
Photosynthetic pigments were analyzed according to Gilmore and Yamamoto (1991a). Briefly, leaf segments from 56-d-old plants grown in a greenhouse were collected after a 3-h dark adaptation period. Three individual plants were used for an experiment and the segments were immediately frozen in liquid nitrogen and ground with a Mixer-Mill (Qiagen). Pigments were extracted from the leaf powder by gentle agitation in ice-cold acetone for 1 h. To minimize pigment degradation, extraction was performed in darkness at 4°C. Cell debris was removed by centrifugation two times at 19,000g for 10 min at 4°C. Extracts were filtered through a 0.2-μm syringe filter and pigments were separated using an HPLC system (HP 1100 series; Hewlett-Packard) equipped with Spherisorb ODS-1 columns (Altech). The pigment concentration was estimated by using the conversion factors for peak areas (in nmol) that were previously calculated for this solvent mixture (Gilmore and Yamamoto, 1991b).
Microscopy
Plant tissues were fixed in a solution containing 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4) at 4°C for 4 h. They were then rinsed in 0.1 m phosphate buffer (pH 7.4) and treated with 1% (w/v) osmium tetraoxide for 4 h at 4°C. After rinsing with the phosphate buffer, the samples were dehydrated and embedded in LR White resin (London Resin Company). Thin sections (40- to 50-nm thickness) were prepared with an ultramicrotome (RMC MTXL) and collected on nickel grids (1-GN, 150 mesh). The sections were stained with uranyl acetate and lead citrate and examined using a JEM 100CXII transmission electron microscope (Gothandam et al., 2005).
Photoinhibitory Treatment and Chl Fluorescence Measurement
Leaf segments from the same plants described in the pigment analysis section were floated on tap water to avoid water loss, and photoinhibitory treatments were performed using a metal-halide lamp at 28°C. The photon flux density reaching the leaf surfaces was about 1,000 μmol m−2 s−1, and a 15-cm-deep water bath was placed under the lamp to avoid overheating. Fv/Fm is a Chl fluorescence parameter used to evaluate the maximum or potential quantum efficiency of PSII and its decrease in plant leaves is reliable evidence that the leaves are subjected to stress. Therefore, photoinhibition was assessed by measuring Fv/Fm before and after high light treatments using a portable fluorometer. A saturating light pulse (3,000 μmol m−2 s−1; 800-ms pulse duration) was applied to measure Fm. Fv/Fm was measured using leaf blade after dark adaptation for 15 min. After photoinhibition, leaves were allowed to recover at 20 μmol m−2 s−1 for the indicated time periods. At least three leaf segments were used and three independent experiments were carried out, giving nine replications for each time point.
2-DE, In-Gel Digestion, and Protein Identification
Two grams of rice plant leaves ground in liquid nitrogen were homogenized in 10 mL ice-cold Mg/Nonidet P-40 (NP-40) extraction buffer containing 0.5 m Tris-HCl (pH 8.3), 2% (v/v) NP-40, 20 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 1% polyvinyl polypyrrolidone, and 0.7 m Suc. 2-DE, in-gel digestion, and protein identification were performed according to previously reported methods (Kim et al., 2004).
H2O2 Measurement
To measure H2O2 levels, plants grown in sunlight conditions were used. Leaves of 60-d-old rice harvested at the midnight (dark) or midday (sunlight, 1,750 μmol m−2 s−1) were analyzed. Leaves (0.2 g) were ground with mortar and pestle in 2 mL of 0.2 m HClO4. The slurry was centrifuged at 20,000g for 5 min. The supernatant was neutralized with 4 m KOH to pH 7.5 and centrifuged at 20,000g for 1 min. An aliquot (100 μL) of supernatant was applied to a 2-mL column of AG-1-X2 resin (Bio-Rad) and the column was eluted with 2.5 mL of distilled water. The eluate was used for the assay of H2O2. Assays were performed with an Amplex red H2O2/peroxidase assay kit (Molecular Probes) according to the manufacturer's instructions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of rice thioredoxin m isoforms and their expression in Ostrxm RNAi transgenic rice.
Supplemental Figure S2. Northern-blot analysis of nuclear- and chloroplast-encoded chloroplast genes.
Supplementary Material
This work was supported by grants from the KOSEF/MOST for the EB-NCRC (R15–2003–012–01001–0), CFGC (CG3313–1), NRL (M10600000205–06J0000–20510), Health 21 R&D Project (A060549), the Korea Research Foundation (KRF–2003–908–F00003 to Y.H.C.), and the BK21 Program, the Ministry of Education and Human Resources Development, Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kyun Oh Lee (leeko@gnu.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Buchanan BB (2002) Yet another plant thioredoxin. Trends Plant Sci 7 191–193 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong NE, Choi YO, Lee KO, Kim WY, Jung BG, Chi YH, Jeong JS, Kim K, Cho MJ, Lee SY (1999) Molecular cloning, expression, and functional characterization of a 2Cys-peroxiredoxin in Chinese cabbage. Plant Mol Biol 40 825–834 [DOI] [PubMed] [Google Scholar]
- Clancey CJ, Gilbert HF (1987) Thiol/disulfide exchange in the thioredoxin-catalyzed reductive activation of spinach chloroplast fructose-1,6-bisphosphatase. Kinetics and thermodynamics. J Biol Chem 262 13545–13549 [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol 136 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Zheng P, Marmey P, Zhang S, Tian W, Chen S, Beachy RN, Fauquet C (2001) Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breed 7 25–33 [Google Scholar]
- Demmig-Adams B, Gilmore AM, Adams WWIII (1996) In vivo functions of carotenoids in higher plants. FASEB J 10 403–412 [DOI] [PubMed] [Google Scholar]
- Droux M, Jacquot JP, Miginac-Maslow M, Gadal P, Huet JC, Crawford NA, Yee BC, Buchanan BB (1987. a) Ferredoxin-thioredoxin reductase, an iron-sulfur enzyme linking light to enzyme regulation in oxygenic photosynthesis: purification and properties of the enzyme from C3, C4, and cyanobacterial species. Arch Biochem Biophys 252 426–439 [DOI] [PubMed] [Google Scholar]
- Droux M, Miginiac-Maslow M, Jacquot JP, Gadal P, Crawford NA, Kosower NS, Buchanan BB (1987. b) Ferredoxin-thioredoxin reductase: a catalytically active dithiol group links photoreduced ferredoxin to thioredoxin functional in photosynthetic enzyme regulation. Arch Biochem Biophys 256 372–380 [DOI] [PubMed] [Google Scholar]
- Duek PD, Wolosiuk RA (2001) Rapeseed chloroplast thioredoxin-m. Modulation of the affinity for target proteins. Biochim Biophys Acta 1546 299–311 [DOI] [PubMed] [Google Scholar]
- Geck MK, Larimer FW, Hartman FC (1996) Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. J Biol Chem 271 24736–24740 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42 265–271 [DOI] [PubMed] [Google Scholar]
- Gilmore AM, Yamamoto HY (1991. a) Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol 96 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AM, Yamamoto HY (1991. b) Resolution of lutein and zeaxanthin using a non-endcapped, lightly carbon-loaded C18 high-performance liquid chromatographic column. J Chromatogr 543 137–145 [Google Scholar]
- Gilmore AM, Yamamoto HY (1993) Linear models relating xanthophylls and lumen activity to non-photochemical fluorescence quenching: evidence that antheraxanthin explains zeaxanthin-independent quenching. Photosynth Res 35 67–68 [DOI] [PubMed] [Google Scholar]
- Gothandam KM, Kim ES, Cho H, Chung YY (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58 421–433 [DOI] [PubMed] [Google Scholar]
- Gupta HS, Pattanayak A (1993) Plant regeneration from mesophyll protoplasts of rice (Oryza sativa L.). Bio/Technology 11 90–94 [Google Scholar]
- Hodges M, Miginiac-Maslow M, Decottignies P, Jacquot JP, Stein M, Lepiniec L, Cretin C, Gadal P (1994) Purification and characterization of pea thioredoxin f expressed in Escherichia coli. Plant Mol Biol 26 225–234 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264 13963–13966 [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Mouaheb N, Meyer Y, Miginiac-Maslow M (2001) Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J 25 127–135 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Gelhaye E, Rouhier N, Corbier C, Didierjean C, Aubry A (2002) Thioredoxins and related proteins in photosynthetic organisms: molecular basis for thiol dependent regulation. Biochem Pharmacol 64 1065–1069 [DOI] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, et al (2004) Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117 625–635 [DOI] [PubMed] [Google Scholar]
- Johnson MP, Davison PA, Ruban AV, Horton P (2008) The xanthophyll cycle pool size controls the kinetics of non-photochemical quenching in Arabidopsis thaliana. FEBS Lett 582 262–266 [DOI] [PubMed] [Google Scholar]
- Kamo M, Tsugita A, Wiessner C, Wedel N, Bartling D, Herrmann RG, Aguilar F, Gardet-Salvi L, Schurmann P (1989) Primary structure of spinach-chloroplast thioredoxin f. Protein sequencing and analysis of complete cDNA clones for spinach-chloroplast thioredoxin f. Eur J Biochem 182 315–322 [DOI] [PubMed] [Google Scholar]
- Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH, Lee JJ, Kang KY (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4 3569–3578 [DOI] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz KJ (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 99 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külheim C, Ågren J, Jansson S (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297 91–93 [DOI] [PubMed] [Google Scholar]
- Maeda K, Tsugita A, Dalzoppo D, Vilbois F, Schurmann P (1986) Further characterization and amino acid sequence of m-type thioredoxins from spinach chloroplasts. Eur J Biochem 154 197–203 [DOI] [PubMed] [Google Scholar]
- Mestres-Ortega D, Meyer Y (1999) The Arabidopsis thaliana genome encodes at least four thioredoxins m and a new prokaryotic-like thioredoxin. Gene 240 307–316 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG, Buchanan BB (1989) Thioredoxin is essential for photosynthetic growth. The thioredoxin m gene of Anacystis nidulans. J Biol Chem 264 4008–4014 [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol 125 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa AN, Buchanan BB (1981) Enzyme regulation in C4 photosynthesis. Purification and properties of thioredoxin-linked fructose bisphosphatase and sedoheptulose bisphosphatase from corn leaves. J Biol Chem 256 6119–6126 [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M (1999) Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci 4 136–141 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M (1997) Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA 94 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R, Anderson LE (1981) Dark modulation of NADP-dependent malate dehydrogenase and glucose-6-phosphate dehydrogenase in the chloroplast. Biochim Biophys Acta 636 58–64 [DOI] [PubMed] [Google Scholar]
- Schwarz O, Schurmann P, Strotmann H (1997) Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem 272 16924–16927 [DOI] [PubMed] [Google Scholar]
- Tracewell CA, Vrettos JS, Bautista JA, Frank HA, Brudvig GW (2001) Carotenoid photooxidation in photosystem II. Arch Biochem Biophys 385 61–69 [DOI] [PubMed] [Google Scholar]
- Wenderoth I, Scheibe R, von Schaewen A (1997) Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. J Biol Chem 272 26985–26990 [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Ballicora MA, Hagelin K (1993) The reductive pentose phosphate cycle for photosynthetic CO2 assimilation: enzyme modulation. FASEB J 7 622–637 [DOI] [PubMed] [Google Scholar]
- Zhang N, Portis AR, Jr (1999) Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci USA 96 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.