Abstract
Telomeres, the essential structures at the ends of eukaryotic chromosomes, are composed of G-rich DNA and asociated proteins. These structures are crucial for the integrity of the genome, because they protect chromosome ends from degradation and distinguish natural ends from chromosomal breaks. The complete replication of telomeres requires a telomere-dedicated reverse transcriptase called telomerase. Paradoxically, proteins that promote the very activities against which telomeres protect, namely DNA repair, recombination and checkpoint activation, are integral to both telomeric chromatin and telomere elongation. This review focuses on recent findings that shed light on the roles of ATM-like kinases and other checkpoint and repair proteins in telomere maintenance, replication and checkpoint signaling.
Introduction
Checkpoint response to double-strand breaks
Genome integrity is dependent on the correct replication of chromosomes and their distribution to the two daughter cells in every cell division. However, normal cellular events (e.g. DNA replication, or exposure to exogenous agents such as radiation) can result in DNA damage. To ensure that cells do not proceed into subsequent cell cycles with damaged chromosomes, cells activate checkpoints that are capable of triggering cell cycle arrest, which provides time for repair of the damage before cell division. Although checkpoints can recognize different forms of DNA damage, the best-studied pathways are those that are activated in response to chromosomal double-strand breaks (DSBs) [1–3]. Eukaryotes are exquisitely sensitive to DSBs; in yeast and human cells, even a single chromosomal break is sufficient to elicit a checkpoint-mediated cell cycle arrest [4,5].
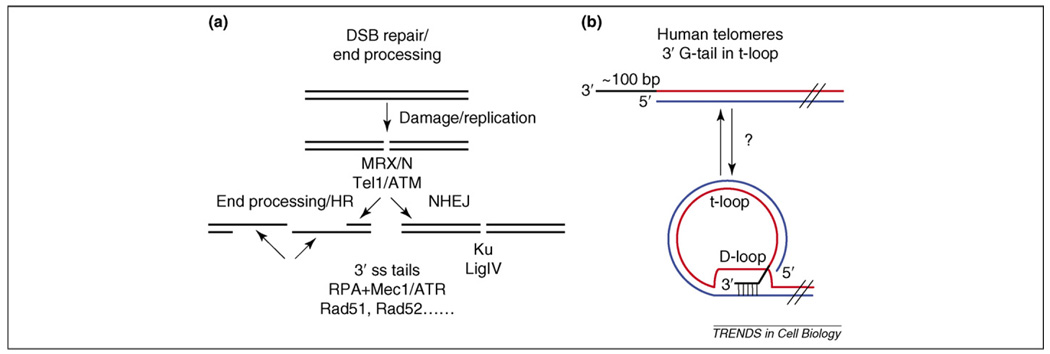
Many of the proteins involved in detecting and repairing DNA damage are conserved. The MRX or MRN complex comprises Mre11, Rad50 and either Xrs2 in the budding yeast Saccharomyces cerevisiae (for MRX) or NBS1 in mammals (for MRN). In all species, ranging from yeast to mammals, these proteins are the first to arrive at a DSB (see Table 1 for a list of the proteins discussed in this review) [2,6]. The DSB-bound MRX/N complex can recruit and activate the phosphoinositide 3-kinase (PI3K) family member ataxia telangiectasia mutated (ATM) in mammals, or its homolog Tel1 in S. cerevisiae. MRX/N promotes resection of the DSB, which generates 3′ single-stranded tails that are coated by the sequence non-specific single-stranded DNA-binding complex RPA, a heterotrimeric complex that has essential roles in DNA replication, recombination and repair [1–3,6–8]. RPA-coated single-stranded DNA then recruits and activates another ATM-related kinase called ATR in mammals, and Mec1 in S. cerevisiae. As a result of these events, ATM–ATR-like kinases initiate a Ser–Thr phosphorylation-mediated signal transduction cascade that leads to cell cycle arrest and repair of the DSB (Figure 1a) [1–3,9].
Table 1 .
Checkpoint/DNA damage and telomere proteins in eukaryotes
| Protein function | Baker’s Yeast | Human |
|---|---|---|
| Mre11 complex | Mre11 | MRE11 |
| Rad50 | RAD50 | |
| Xrs2 | NBS1 | |
| PI3K | Tel1 | ATM |
| Mec1 | ATR | |
| Telomerase, catalytic core | TLC1 | hTR |
| Est2 | TERT | |
| Telomerase, accessory proteins | Est1, Est3 | EST1A, EST1B |
| G-tail binding proteins | Cdc13 | POT1 |
S. cerevisiae genes: EST1, Ever Shorter Telomere 1; CDC13, Cell Division Cycle 13; TLC1, Telomerase Component 1; STN1, Suppressor of cdc Thirteen 1′; TEN1, Telomeric pathways in association with Stn1, number 1; RAP1, Repressor Activator Protein 1; RIF1, Rap1p Interacting Factor 1; RIF2, Rap1p Interacting Factor 2; CLB2, CycLin B 2; RPA, Replication Protein A.
Mammalian genes: ATM, Ataxia Telangiectasia Mutated; ATR, Ataxia Telangiectasia and Rad3-related; RPA, Replication Protein A; POT1, Protection Of Telomeres 1; hTR, human Telomerase RNA; TERT, Telomerase Reverse Transcriptase; NBS1, Nijmegen Breakage Syndrome 1.
Figure 1.
Telomere structure and DSB processing. (a) A simplified version of the events that occur after the formation of a DNA double-strand break (DSB). The ends are bound by the MRX/N complex, which recruits the Ser–Thr protein kinase Tel1 (also known as ATM). Subsequent processing and protein binding will convert the break into either long stretches of single-stranded DNA that are a substrate for binding of RPA and recruitment of Mec1 (also known as ATR) before homologous recombination (HR) (left) or shuttle it down the Ku-dependent non-homologous end joining (NHEJ) pathway (right). (b) Mammalian telomeres have long G-tails (~75–300 bases) at both chromosome ends throughout the cell cycle – whether or not the cells express telomerase. By contrast, S. cerevisiae telomeres have short (~15 base) G-tails throughout most of the cell cycle, except in late S–G2 phase, when G-tails are much longer [16,101]. Long G-tails in S. cerevisiae are also telomerase-independent [16]. In mammals and other organisms (but probably not in S. cerevisiae), the G-tail can invade the duplex region of the telomere, displacing an internal segment of single-stranded G-telomeric DNA and forming a terminal circle. In T-loops, the G-tail is base-paired to the C-strand within an otherwise duplex region of telomeric DNA, displacing the G-strand in the original duplex and generating an internal stretch of ~100 bases of single-stranded G-strand telomeric DNA called a D-loop. T-loops are thought to protect the end against degradation and other hazards.
Telomere structure
Telomeres are specialized nucleoprotein complexes at the ends of eukaryotic chromosomes, and they are essential for genome integrity [10–12]. In virtually all eukaryotes, telomeric DNA consists of tandem repeats of a short sequence that extends from several hundred base-pairs (~300 bp in S. cerevisiae) up to many thousands of base-pairs (in mammals). Although the exact sequence of the telomeric repeat varies from organism to organism, the 3′ strand is invariably G-rich and is extended to form a single-stranded tail known as the G-tail [10–12]. In mammals and other eukaryotes, but probably not in organisms such as S. cerevisiae that have relatively short telomeres, the G-tail can invade the duplex portion of the telomere to form a terminal circle called a T-loop (Figure 1b) [13]. G-tails are bound in vivo by sequence-specific DNA-binding proteins (i.e. Cdc13 in S. cerevisiae; POT1 in mammals), and the genes encoding these proteins are essential [10,11]. G-tail-binding proteins recruit additional proteins to the telomere. For example, S. cerevisiae Cdc13 is the DNA-binding subunit of a heterotrimeric complex that includes Stn1 and Ten1. Although the Cdc13 complex is specific for S. cerevisiae telomeric TG1–3 DNA, it shares structural features with the sequence non-specific RPA complex [14].
Telomere functions
Telomeric DNA, in concert with its complement of telomere-associated proteins, carries out multiple functions that are crucial for the maintenance of a stable genome. First, telomeres provide a substrate for a specialized mechanism of replication that counteracts the loss of DNA from chromosome ends that are replicated solely by semi-conservative DNA replication (Figure 2). In most eukaryotes, this ‘end replication problem’ is solved by telomerase, a specialized reverse transcriptase that uses its integral RNA subunit as a template to lengthen the telomeric G-tail. After telomerase acts, a conventional DNA polymerase fills in the complementary C-strand [10–12].
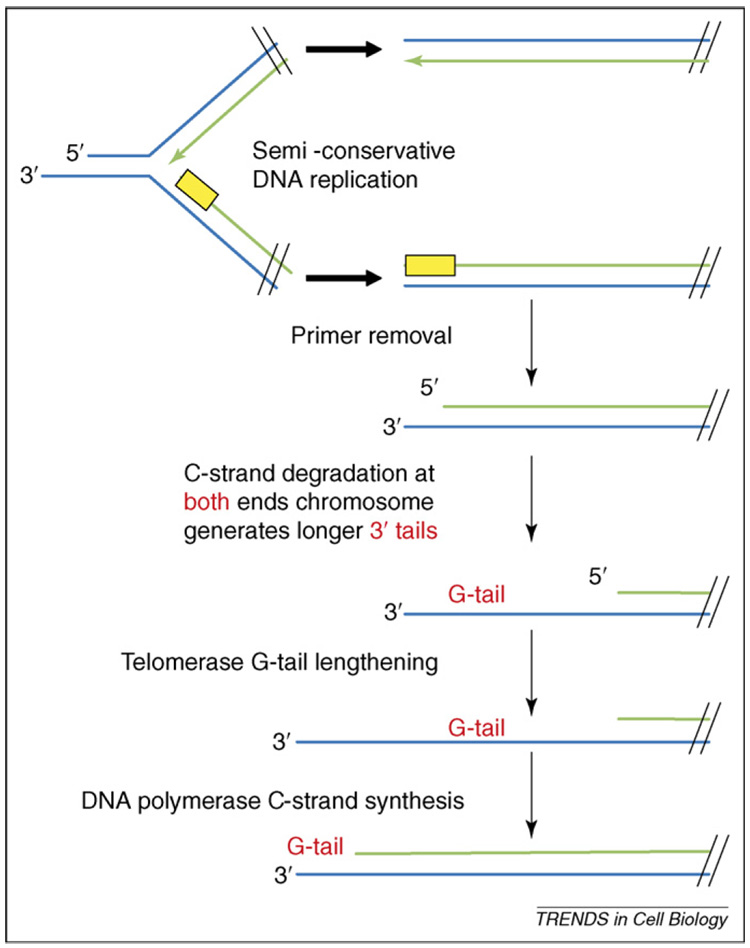
Figure 2.
End replication and C-strand degradation. The figure depicts an abbreviated view of what occurs at the very ends of a chromosomal DNA molecule during DNA replication. For convenience, only the left ends of the parent and progeny molecules are shown. Conventional DNA polymerases synthesize DNA only in the 5′ to 3′ direction, and in eukaryotes this synthesis is primed by 8–12 bases of RNA (yellow boxes). Removal of the terminal RNA primer leaves a gap at the 5′ end of newly replicated strands, and this gap cannot be repaired by a conventional DNA polymerase. Given the polarity of telomeric DNA, this gap exposes a short 3′ single-stranded G-tail on the complementary strand. Over time, incomplete replication would result in progressive loss of DNA and ultimately genetic information. After conventional replication is complete, the C-strand is degraded. This degradation occurs at both ends of the chromosome – although only one end is shown – and generates a long G-tail at both ends of the chromosome. The G-tail on both the leading and lagging strand of a subset of telomeres can be elongated by telomerase. Before cell division, the conventional replication machinery fills in the C-strand. After RNA-primed C-strand resynthesis, removal of the RNA primer regenerates a short G-tail at both ends of the chromosome. Although this view is based mainly on data from S. cerevisiae, mammalian chromosomes also have G-tails at both ends, even in cells that do not express telomerase [102].
In addition to serving as substrates for a specialized replication mechanism, telomeres also disguise chromosome ends from the checkpoint and repair machinery. Whereas even a single DSB can elicit a checkpoint-mediated cell cycle arrest [4,5], multiple natural ends (i.e. telomeres) are not perceived as DNA damage. This feature is especially surprising because single strand DNA is one of the main signals that trigger DNA damage checkpoints [1–3], yet telomeres end with a single-strand G-tail or contain an internal stretch of single-stranded DNA (Figure 1b, T-loop; 2A, G-tail). Telomeres also behave differently from DSBs. For example, telomeres fuse neither with each other nor with DSBs. Moreover, throughout most of the cell cycle, telomeres are not susceptible to degradation. However, the 5′ end or C-strand of telomeres is degraded during a narrow window in the cell cycle that is coupled temporally with semi-conservative replication of the telomere (Figure 2). This cell cycle-regulated degradation has been best demonstrated in S. cerevisiae, but it probably also occurs in higher eukaryotes [11,15]. C-strand degradation occurs on both ends of linear DNA molecules [16]. As a result, a G-tail is generated at both ends of the chromosome, not just at the end replicated by the lagging strand polymerase (Figure 2). This action creates a substrate at both chromosome ends for the G-tail-binding proteins Cdc13 (S. cerevisiae) and POT1 (mammals), and this substrate protects telomeres from further degradation [17–20]. In higher eukaryotes, sequestering the G-tail into a T-loop and/or into a G-quadruplex structure might contribute to the protective function of the telomere [13,21].
Telomerase expression and dysfunctional telomeres
In cells that do not express telomerase, telomeres slowly but progressively shorten, a phenomenon also known as the ever-shorter telomere (est) phenotype [10–12]. In S. cerevisiae, telomerase-deficient cells lose ~3 bp per telomere per cell cycle, whereas the rate of loss in mammals is higher, ~50–100 bp per cell division. Although S. cerevisiae cells express telomerase constitutively, in humans, most somatic cells do not have detectable telomerase activity [22,23]. However, telomerase is expressed in human stem cells and in the vast majority of human tumors, and this expression contributes to the growth potential of these cells [23–25]. In both S. cerevisiae and human cells in culture, telomerase is present in extremely low amounts; telomerase-proficient cells probably contain fewer active telomerase complexes than telomeres [26,27]. Thus, although human hematopoietic stem cells are telomerase-positive, their level of telomerase is not sufficient to prevent loss of telomeric DNA [28]. The impact of this low level of telomerase is highlighted by the recent discovery that heterozygous mutations in telomerase subunits cause several hereditary human diseases, such as dyskeratosis congenita and idiopathic pulmonary fibrosis. These mutations, which reduce telomerase activity as monitored by in vitro assays [29], have dire effects on the life span of an organism, probably owing to stem cell failure [30–33].
Because mammalian telomeres are many kilobases long, the slow loss of telomeric DNA that occurs in somatic tissues is tolerated for many cell divisions without apparent loss of telomere function. However, if telomeres become critically short in either S. cerevisiae or mammals, they lose their ability to protect chromosome ends and trigger a DNA damage response [12]. In S. cerevisiae, a Mec1-dependent checkpoint is activated in telomerase-deficient cells, but only when telomeres become extremely short or when Cdc13 inactivation leads to rampant C-strand degradation [34–36]. However, even wild type length mammalian telomeres can become dysfunctional, owing to loss of protein–telomeric DNA interactions, and these altered telomeres can trigger an ATM- or ATR-mediated checkpoint depending on which protein is removed [37]. Dysfunctional telomeres have many similarities to DSBs; in mammals, they form DNA damage-like foci called TIFs (telomere dysfunction-induced foci) [38]. TIFs are similar in appearance and protein content to the foci induced at DSBs, and they produce similar downstream effects (e.g. cell cycle arrest, senescence and/or cell death) [38–42].
To summarize this introduction, telomeric DNA and its associated proteins can be thought of as serving two crucial functions. First, they provide a substrate for non-canonical, RNA-templated replication. Second, their structure prevents telomeres from being seen as damaged DNA by checkpoint activities whose downstream effectors could promote their fusion or degradation, which can lead to cell cycle arrest and/or cell death. Given this view, it is paradoxical that checkpoint and repair proteins are implicated in telomerase-mediated telomere elongation. In the remainder of this article, we review recent work in S. cerevisiae and mammals regarding the role of ATM-like checkpoint kinases in telomerase-mediated telomere replication.
Checkpoint and repair proteins have telomere functions
The first mutations found to affect telomere length, tel1 and tel2, were identified when a collection of S. cerevisiae mutants was screened by Southern hybridization for strains with short telomeres [43]. When the yeast TEL1 gene was sequenced [44,45], its closest homolog in the database was the human gene for the checkpoint protein ATM. S. cerevisiae Tel1 has a key role in telomere length regulation, as tel1Δ cells have very short telomeres (~100 bp, approximately one-third of the length of wild type telomeres). Mutation of any one (or all three) of the proteins of the MRX complex has a similar effect on telomere length [10,11]. Deleting the major S. cerevisiae checkpoint kinase MEC1 on its own has very little effect on telomere length, whereas combining mec1Δ with either tel1Δ or mrxΔ leads to an est phenotype [46]. Furthermore, the kinase activities of Tel1 and Mec1 are required for their telomere effects [47]. Thus, S. cerevisiae telomerase fails to act without upstream events orchestrated by either the MRX complex and Tel1 or Mec1. As assessed by in vitro assays, telomerase activity is normal in mec1Δ tel1Δ cells, and their defect in telomerase replication can be bypassed by altering telomeric chromatin [48,49]. Thus, these kinases probably regulate the degree to which telomerase can access its substrates.
The mammalian homologs of the yeast checkpoint proteins also affect telomeres. Mutations in the genes encoding ATM, MRE11 and NBS1 in humans result in, respectively, ataxia telangiectasia (AT), AT-like syndrome, and Nijmegen breakage syndrome (NBS), neurodegenerative diseases that are associated with genome instability and a predisposition to cancer [50–52]. Cells from patients with either AT or NBS have short telomeres and elevated levels of aberrant telomeres [10,52,53]. The MRN complex probably affects telomeres directly, because the complex is telomere-associated [54]. Several mammalian recombination proteins, such as RAD54 [55] and RAD51D [56], also localize to and/or affect telomere length. To date, the effects that these proteins have on telomeres have not been linked directly to changes in the behavior of telomerase.
Both S. cerevisiae TEL2 and its mouse homolog, Tel2, are essential [57,58]. Yeast TEL2 was first identified because compromising its function results in short telomeres [43]. S. cerevisiae Tel2 affects Tel1 recruitment to DSBs [59], and it is possible that Tel2 has similar functions at telomeres. Although mouse Tel2 has not been linked directly to telomeres, its expression is required for the stability of all PI3K-related kinases [58]. The fission yeast Schizosaccharomyces pombe Tel2 probably behaves similarly to its mouse counterpart, because it also associates physically with all PI3K-like kinases [60].
S. cerevisiae telomerase is regulated during the cell cycle
The products of four S. cerevisiae genes, all of which are required for telomerase action in vivo, form the telomerase holoenzyme: Est2, the catalytic reverse transcriptase; TLC1, telomerase RNA; and two accessory proteins, Est1 and Est3 [10,11]. In addition, Sm proteins associate with TLC1 RNA in the active telomerase complex [61]. Although mutating TLC1 RNA so that it no longer binds to Sm proteins does not cause an est phenotype, it does reduce TLC1 RNA levels and telomere length.
Most of the telomere is replicated by semi-conservative DNA replication, which occurs at the very end of S phase and is followed by C-strand degradation [10–12]. Late S–G2 phase is also the time during the S. cerevisiae cell cycle when telomerase acts, yet in vitro assays reveal that protein extracts from G1 phase cells have as much telomerase activity as that found in extracts from cells arrested in mitosis [62,63]. Unexpectedly, Est2 and TLC1 are telomere-associated throughout most of the cell cycle, even in G1 and early S phase, when telomerase is inactive [64]. However, Est1 [64] and Est3 (K. Daumer and V.A. Zakian, unpublished) come to the telomere only in late S–G2 phase, and Cdc13 telomere-binding increases enormously at this time, concomitant with the appearance of long G-tails [64]. The cell cycle-regulated binding of both Est1 and Est3 is due at least in part to cell cycle-regulated abundance of Est1 [64,65].
Unlike replication of S. cerevisiae telomeres, which occurs at the very end of S phase, semi-conservative DNA replication of human telomeres occurs throughout S phase [66,67]. A small amount of telomeric DNA synthesis occurs in post-S phase human cells that do not express telomerase [68]. This late replication might be due to the restarting of forks that stall within telomeric DNA, or it might be caused by the re-synthesis of degraded C-strand DNA. Cytological studies in cultured cancer cells demonstrate that human telomerase localizes to a subset of telomeres during S phase, suggesting that the action of human telomerase is also cell cycle-regulated [69,70].
In S. cerevisiae and mammals, telomerase acts preferentially at short telomeres
Two types of assays demonstrate that short telomeres are preferential substrates for S. cerevisiae telomerase (Figure 3ab). In the first assay, recombination is used to generate a single, short telomere of ~100 bp in a cell with otherwise wild type length telomeres (Figure 3a) [71]. After ~50 cell divisions, telomerase restores the shortened telomere to a wild type length. However, its rate of lengthening is much faster in the first cell cycles after shortening than it is in subsequent cell cycles. Thus, telomere length exerts a negative effect on the rate of telomerase elongation.
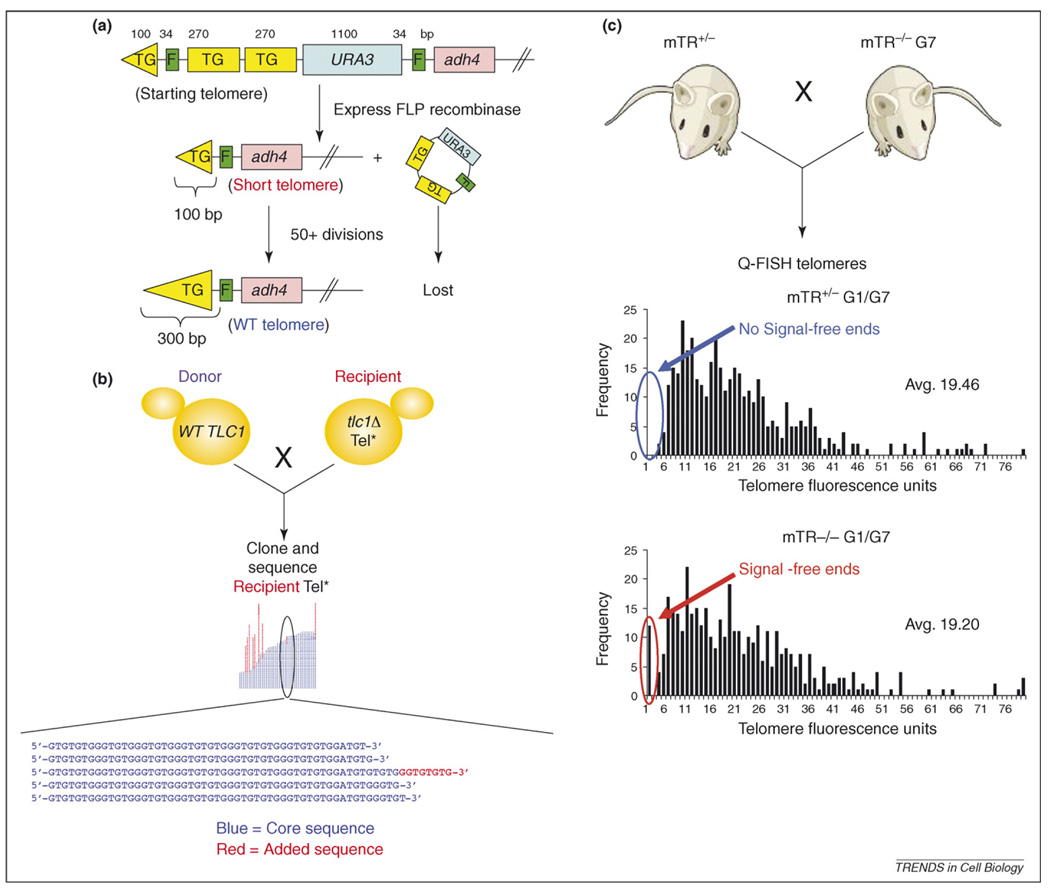
Figure 3.
Methods used to observe the preference of telomerase for short telomeres in S. cerevisiae and mice. (a) Preferential elongation of short telomeres in S. cerevisiae: Assay 1. As described by Marcand et al. [71], a single telomere is replaced with a construct containing telomeric repeats (TG, yellow boxes). These repeats are flanked by FRT sites (F, green boxes; top), which are recognition elements for the FLP recombinase. Because FRT sites are short (34 bp), the cell views the internal telomere repeats as part of the telomere. When the FLP recombinase is induced by adding galactose to the medium, the internal telomeric repeats are removed, leaving behind a ~100-bp tract of telomeric DNA (middle) in a cell with otherwise wild type length (~300 bp) telomeres. It takes multiple generations for telomerase to re-extend this short telomere to the wild type length of ~300 bp (bottom; large yellow triangle). (b) Preferential elongation of short telomeres in S. cerevisiae: Assay 2. As described in Teixeira et al. [72], telomerase-proficient wild type donor cells (TLC1, left) are mated to telomerase-deficient recipient cells (tlc1Δ, right) carrying one or more uniquely marked telomeres (Tel*). The resulting diploids are telomerase-proficient, which enables telomere-lengthening of the recipient telomeres. Polymerase chain reaction is used to clone and sequence Tel* from multiple diploids after the first S phase subsequent to mating. Because the sequence of S. cerevisiae telomeric DNA is not a precise repeat, and because a portion of the telomere is replicated by semi-conservative DNA replication, it is possible to distinguish the original telomere sequence at varying stages of attrition (blue) from DNA added by telomerase (red). (c) Preferential elongation of short telomeres in mice. As described by Hemann et al. [73], mice heterozygous for telomerase RNA deficiency (mTR+/−, top left) were mated with mice deficient for mTR for seven generations (mTR−/− G7, top right). The telomeres of the mTR−/− G7 mice have shortened to the point that many chromosomes are unstable, resulting in infertility and other defects. Quantitative fluorescence in situ hybridization (Q-FISH) was used to determine telomere length in the offspring. Despite the fact that both mTR+/− and mTR−/− offspring inherited similar average telomere lengths and distributions (graphs), a significant fraction of chromosomes with no detectable telomeric repeats at their ends (red arrow, bottom graph) is detected in mTR−/− offspring but not in mTR+/− offspring (blue arrow, top graph). Thus, very short telomeres are preferentially lengthened by mouse telomerase. (Figure 3c was kindly supplied by C. Greider).
The second assay, the single telomere extension assay (STEX) [72], involves mating a telomerase-deficient ‘recipient strain’ to a telomerase-proficient ‘donor’ strain to generate telomerase-proficient diploid cells. Individual telomeres originating from the recipient strain are cloned and sequenced after the newly formed telomerase-proficient diploids have completed their first S phase (Figure 3b). STEX reveals that, in a given S phase, telomerase acts on only a subset of telomeres, and its probability of acting on a given telomere is related to its length. Although fewer than 10% of wild type length telomeres (~300 bp) are extended in a given cell cycle, short telomeres (~100 bp) are extended much more frequently (~50% of cell cycles). Telomerase adds a variable amount of telomeric DNA, from 2 to 100 bases per telomere, but the amount does not correlate with telomere length unless telomeres are extremely short (<100 bp), in which case telomerase is more processive. Even the elimination of either Rif1 or Rif2, telomere-binding proteins that negatively regulate telomerase, affected only the frequency of elongation, not the amount of DNA added.
The preference of telomerase for short telomeres is also apparent in mammals [73]. Mice lacking telomerase RNA are viable for multiple generations, but their telomeres get progressively shorter with successive generations. Quantitative fluorescent in situ hybridization (Q-FISH) reveals that telomeres in splenocytes from seventh-generation telomerase-null mice are much shorter than wild type telomeres. Also the vast majority of these cells have several chromosome ends with no detectable telomeric DNA (Figure 3c). When these mice are mated to telomerase-proficient mice, their progeny have a modest increase in average telomere length, but chromosomes with no telomeric DNA are no longer detected. Thus, during mouse development, very short telomeres are preferential substrates for telomerase. Experiments in human cultured cells have also provided evidence that short telomeres undergo preferential lengthening [74–76].
S. cerevisiae telomerase preferentially binds to short telomeres
In S. cerevisiae, the regulation of telomerase by length is not an all-or-nothing phenomenon. In a given S phase, ~50% of short telomeres are not lengthened, and a small fraction (~10%) of wild type length telomeres are telomerase substrates [72]. It is possible that telomerase acts preferentially at short telomeres, because their structure promotes telomerase-binding. Alternatively, telomerase might bind equally well to short and wild type length telomeres, but it might be activated preferentially at short telomeres. The latter model is consistent with the finding that the catalytic core of telomerase, Est2 and the telomerase RNA, is telomere-associated throughout most of the cell cycle, even when telomerase is inactive [64]. Given that the accessory protein Est1, an essential telomerase subunit, binds to telomeres only when telomerase is active, its binding might activate Est2p protein that is already telomere-bound. However, the inducible short telomere assay reveals that both Est1 and Est2 – not just Est1 – bind to a much greater extent to short telomeres [77,78].
The inducible short telomere assay has been exploited to determine the protein content of short versus wild type length telomeres [77,78]. The heterodimeric Ku complex binds to S. cerevisiae telomeres throughout the cell cycle [79]. A specific interaction between Ku and telomerase RNA brings Est2 to the telomere in G1 phase. Wild type Cdc13, which binds to G-tails, is required to bring Est2 to the telomere in late S–G2 phase [64]. However, neither Ku [77] nor Cdc13 [77,78] is enriched at short telomeres, and telomere length has no effect on the cell cycle binding profiles of either of these proteins. This length-insensitive Cdc13-binding suggests that C-strand degradation occurs equally at short and wild type length telomeres.
Rap1 is a sequence-specific duplex telomere-binding protein that binds about once every 18 bp throughout the S. cerevisiae telomere [10,11]. As telomeres shorten, by definition they lose Rap1 sites. Rif1 and Rif2, two negative regulators of telomerase, bind to telomeres by associating with the carboxyl terminus of Rap1. The simplest hypothesis is that the levels all three proteins are reduced at short telomeres. Indeed, Rap1 and Rif2 are present at ~50% of wild type levels at short telomeres but, unexpectedly, Rif1 binds equally well to short and wild type length telomeres [77].
The S. cerevisiae ATM homologue Tel1 is needed to distinguish short from wild type length telomeres
In the absence of Tel1, telomeres are very short, and the binding of both Est1 and Est2 is low [80]. The inducible short telomere assay reveals that the ability to preferentially target Est1 and Est2 to short telomeres is lost in tel1Δ cells [77]. The STEX assay also finds that Tel1 is needed for preferential elongation of short telomeres, although this dependence varies among different telomeres depending on the identity of their subtelomeric DNA [81]. The increased processivity of telomerase at extremely short telomeres is also Tel1-dependent [82].
Tel1 binds to wild type length telomeres, but this binding is low and restricted to late S–G2 phase [77]. By contrast, Tel1 binds robustly to short telomeres. Preferential binding of Tel1p to short telomeres is seen both at inducibly short telomeres [77,78] and at telomeres made short by mutation [83]. Binding of Tel1 to short telomeres occurs in G1 and early S phase (i.e. before telomerase-binding), increases as cells move through the cell cycle, and stays high for at least two cell cycles [77].
Binding of Tel1 to telomeres is lost in cells expressing xrs2–664 [77], a mutation that generates a protein lacking the carboxyl terminus of Xrs2 and that eliminates binding of Tel1 to DSBs [84]. In addition, telomerase does not bind preferentially to short telomeres in xrs2–664 cells [77]. All three subunits of the MRX complex bind preferentially to short telomeres (M. Sabourin and V.A. Zakian, unpublished). Although binding of MRX and Tel1 are temporally indistinguishable, binding of Tel is Xrs2-dependent and thus it probably occurs after MRX association [77]. When Tel1 is overexpressed, it binds to wild type length as well as to short telomeres, and telomeres are hyperelongated by an MRX-dependent mechanism [77,83]. Taken together, these data make a strong argument in support of the notion that Tel1 is recruited to short telomeres by a specific interaction with Xrs2, and that this binding is required for the preferential binding and lengthening of short telomeres by telomerase. Given that Tel1 also binds at low levels to wild type length telomeres [77], it might also mark the subset of these telomeres that are extended by telomerase.
How does MRX recognize short telomeres? Before binding of MRX, low amounts of Rap1 and Rif2 are the only known features of telomeric chromatin that are different at short and wild type length telomeres [77]. Reduced binding of Rap1 and Rif2 might be the signal or part of the signal that directs MRX and ultimately telomerase to short telomeres. For example, perhaps telomeric DNA ends are more accessible to MRX when binding of Rap1 and Rif2 is low. Preferential binding of MRX to short telomeres is sufficient to explain preferential binding of Tel1 to these ends. Although the Tel1 kinase activity is not required for its telomeric association [83], its kinase activity is required for its effects on telomere length. Phosphorylation of telomere targets must require telomere-binding, because xrs2–664 cells, which have Tel1 that is catalytically active but unable to bind telomeres, have short tel1Δ length telomeres [77,84].
Identification of Tel1 targets is crucial for understanding how Tel1 promotes telomerase-binding. Although Tel1 phosphorylates Xrs2, this phosphorylation is not important for maintaining telomere length [85]. By contrast, Tel1 phosphorylates Cdc13, and the simultaneous mutation of two of its phosphorylation sites results in an est phenotype [86]. Phosphorylation of Cdc13 might promote the Cdc13–Est1 interaction, which is thought to recruit telomerase in late S–G2 phase [87,88]. The negative regulator Rif1 has 14 potential Tel1 targets, and Tel1 (and/or Mec1) phosphorylates at least one of these in response to DNA damage [89]. Binding of Tel1 to the inducible short telomere increases over at least two cell cycles [77], suggesting that Tel1-mediated phosphorylation of its targets increases the affinity of Tel1 for short telomeres.
In contrast to Tel1, Mec1 does not have a major role at S. cerevisiae telomeres. The absence of Mec1 has only modest effects on telomere length [44,46], telomerase-binding [80], and the frequency of telomere lengthening [81].
Checkpoint kinases at mammalian telomeres
Unlike S. cerevisiae Tel1, mouse ATM is not required for the preferential lengthening of short telomeres [90]. This function might be assumed by ATR, the mammalian homolog of S. cerevisiae Mec1. A precedent for this possibility is found in S. pombe, in which Rad3, the ATR–Mec1 homolog, has a much more crucial role in telomere maintenance than S. pombe Tel1 [91].
Just as it does in S. cerevisiae, the MRE11 complex binds to telomeres in telomerase-proficient human cells, as demonstrated by immunofluorescence in asynchronous cells [54]. In addition, in synchronized cells, MRE11 and RAD50 are telomere-associated throughout the cell cycle, whereas NBS1, the counterpart of S. cerevisiae Xrs2, is associated with the telomere only during S phase. In human fibroblasts that do not express telomerase, ATR, ATM and MRE11 are all telomere-associated in a cell cycle-dependent manner; binding of ATR occurs first, followed by MRE11 and then ATM [68,92]. Proteins involved in homologous recombination, such as RAD51 and RAD52, are also recruited to human telomeres during S phase. The association of these proteins is thought to promote T-loop formation after DNA replication [68]. However, these cell cycle experiments used the DNA replication inhibitor aphidicolin to synchronize cells [54,68,92]. Given that aphidicolin can cause telomere damage even at low doses (M. van Overbeek and T. de Lange, personal communication), the recruitment of checkpoint and repair proteins to human telomeres in synchronized cells might be a response to DNA damage rather than a step in normal telomere metabolism.
Are replicating telomeres seen as DSBs?
In the 1930s, pioneering work by Muller with flies and by McClintock with maize [103] led to the description of telomeres as structures that protect chromosomes from loss and end-to-end fusions. Telomere replication probably requires the transient loss of protective protein–DNA structure(s) to allow the replication machinery access to DNA. For telomeres organized in T-loops, passage of the replication fork would seem to demand a transient loss of the T-loop.
One possibility to explain the presence of checkpoint and repair proteins at telomeres is that, during their replication, telomeres are seen as DSBs. This view is supported by the fact that, soon after semi-conservative DNA replication, the C-strand of S. cerevisiae telomeres is specifically degraded [16], a clear loss of the normal role of the telomere in protecting ends from degradation. Telomeres in human fibroblasts that do not express telomerase also appear to lose their protective structure late in the cell cycle, because they become substrates for labeling by terminal transferase [92]. Given that this access to DNA-modifying activities occurs even in asynchronous cells, it cannot be attributed to aphidicolin-induced telomere damage. Nonetheless, if replicating telomeres are seen transiently as DSBs, their semi-conservative replication cannot elicit a classic checkpoint-mediated cell cycle arrest, because it happens at every telomere in every S phase.
Another possibility is that telomerase-mediated lengthening, which occurs at only a subset of telomeres, creates a structure that is seen as a DSB. Consistent with this view, the S. cerevisiae proteins that bind to DSBs are also crucial for telomerase-mediated lengthening and bind preferentially to the short telomeres that are the preferred substrates for telomerase. As discussed above, the order of appearance of these proteins at the telomere (MRX followed by Tel1), their genetic dependencies (binding of Tel1 requires the carboxyl end of Xrs2), and their consequences (MRX-promoted degradation of the 5′ strand) are identical to the early steps in DSB processing (Figure 1a).However, in the inducible short telomere system, the presence of a single short telomere does not affect cell cycle progression, as monitored by the timing of cell cycle landmarks, such as RNR1 RNA or cyclin Clb2 expression [77]. Likewise, STEX demonstrates that ~10% of wild type telomeres are lengthened by telomerase in a given cell cycle [72]. The most plausible scenario is that these lengthening telomeres are distributed among most cells in the population, rather than being concentrated in a small fraction of cells. If so, given that a haploid S. cerevisiae cell has 32 telomeres, cells must tolerate at least a few telomeres undergoing telomerase lengthening without triggering a checkpoint-mediated cell cycle arrest. However, the DNA damage checkpoint is activated if most telomeres in the cell are undergoing telomerase lengthening, as occurs as a result of sudden Tel1 overexpression in cells in which all telomeres are initially very short [93]. Even a single elongating S. cerevisiae telomere might provoke a subset of checkpoint-mediated downstream events (e.g. phosphorylation of the Rad53 checkpoint kinase [94]) and yet not trigger cell cycle arrest. In this respect, the response to a few short telomeres might be similar to what occurs in response to stalled replication forks during the S. cerevisiae S phase. A threshold number of stalled forks is needed to elicit an intra-S phase checkpoint, but a smaller number can cause a sub-threshold level of Rad53 phosphorylation [95–97]. Likewise, reducing POT1 levels in human tumor cells results in G-tail shortening and a transient DNA damage response but not a cell cycle arrest [98].
So how are telomeres distinguished from DSBs during their preparation for telomerase elongation? To date, the data providing an answer to this question come mainly from S. cerevisiae, in which the early events in telomerase lengthening are indistinguishable from what occurs at DSBs. MRX and Tel1 are recruited to both short telomeres and to DSBs (Figure 1a). However, the subsequent binding of Mec1p is readily detected at DSBs [6] but not at short telomeres (M. Sabourin and V.A. Zakian, unpublished). Moreover, when a DSB is generated near a tract of telomeric sequence that has been inserted at an internal site on a chromosome, both the binding of Mec1 to the break and the subsequent cell cycle arrest are abbreviated [99,100]. Given that the checkpoint response to DSBs is Mec1-dependent, the absence of Mec1 from elongating telomeres is probably sufficient to explain why telomerase action does not impede cell cycle progression.
MRX-promoted degradation of the 5′ strand at a DSB generates 3′ single-stranded DNA with a sequence that is determined by the site of the break. However, at telomeres, 5′ strand degradation always generates G-strand telomeric DNA. The Cdc13 and POT1 complexes bind specifically to these G-tails, whereas RPA binds non-specifically to single-stranded DNA at DSBs. RPA then recruits Mec1, but this does not occur at S. cerevisiae telomeres, presumably because the Cdc13 complex does not interact with Mec1p. Cdc13 also limits the extent of C-strand degradation, preventing it from reaching non-telomeric sequences. Reducing Cdc13 function by, for example, growing cdc13–1 cells at high temperatures, leads to unchecked C-strand degradation and activation of a checkpoint-mediated arrest [10,11,35]. Checkpoint activation in this context is due to the exposure of non-telomeric single-stranded DNA that can recruit RPA and Mec1. Thus, the binding of G-strand-specific binding proteins, such as Cdc13 and POT1, to the single-stranded DNA generated at telomeres – as opposed to binding of RPA to the single-stranded DNA exposed at a DSB – is probably the key event that distinguishes replicating telomeres from damaged DNA.
Concluding remarks
Given that their replication occurs in every S phase, by definition replicating telomeres cannot trigger a checkpoint-mediated cell cycle arrest. Nonetheless, in S. cerevisiae and mammalian cultured cells, many of the proteins that bind to DSBs and/or act in the checkpoint response also affect telomeres. We propose that the 5′ resection that occurs at telomeres as part of their normal replication does not trigger a checkpoint arrest, because telomere-specific single-strand binding proteins, such as Cdc13 (yeast) or POT1 (mammals), bind to the exposed telomeric G-tails. By contrast, the single-stranded DNA that is generated by resection of a DSB binds to the sequence non-specific RPA complex, and this binding of RPA is required for a checkpoint response. If this model is correct, it will be important to understand how RPA is excluded from telomeric G-tails, because RPA is probably involved in the replication of duplex telomeric DNA. In S. cerevisiae, preferential lengthening of short telomeres can be explained by their preferential binding of MRX, which recruits Tel1 and ultimately telomerase. What is not clear is how MRX distinguishes short from wild type length telomeres. Another important goal is to identify additional Tel1 targets that affect telomerase recruitment when phosphorylated. Finally, it is not known how much of the data generated in S. cerevisiae on the role of checkpoint proteins in telomerase recruitment applies to higher cells. Specifically, do the MRN complex and ATM–ATR kinases affect telomere length in mammals by channeling telomerase to short telomeres?
Acknowledgements
We thank C. Greider for providing the unpublished data in Figure 3c,j, J. Phillips for Figure 3b,c, C. Tuzon for his careful reading of the manuscript, T. de Lange and M. van Overbeek for discussions about the events at mammalian telomeres, and the National Institutes of Health for supporting the research in our laboratory.
References
- 1.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 2.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 3.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu. Rev. Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 4.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 5.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 6.Lisby M, Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87:579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Zou Y, et al. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning E, et al. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 10.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 11.Vega LR, et al. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 12.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 13.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 14.Gao H, et al. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 15.Baird DM. Telomere dynamics in human cells. Biochimie. 2008;90:116–121. doi: 10.1016/j.biochi.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Wellinger RJ, et al. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 17.Taggart AK, Zakian VA. Telomerase: what are the Est proteins doing? Curr. Opin. Cell Biol. 2003;15:275–280. doi: 10.1016/s0955-0674(03)00040-1. [DOI] [PubMed] [Google Scholar]
- 18.Lei M, et al. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, et al. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 20.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 21.Paeschke K, et al. Cell cycle dependent regulation of telomere tethering in the nucleus. Nat. Struct. Mol. Biol. doi: 10.1007/s10577-008-1222-x. (in press) [DOI] [PubMed] [Google Scholar]
- 22.Cong YS, et al. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsyth NR, et al. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 24.Sealey D, et al. Telomere DNA replication, telomerase and human disease. In: DePamphilis M, editor. DNA Replication and Human Disease. CSHL Press; 2006. pp. 561–592. [Google Scholar]
- 25.Shay J, Wright W. Telomerase and human Cancer. In: De Lange T, et al., editors. Telomeres. 2nd edn. Cold Spring Harbor Laboratory Press; 2006. pp. 81–108. [Google Scholar]
- 26.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SB, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 28.Drummond MW, et al. Concise review: telomere biology in normal and leukemic hematopoietic stem cells. Stem Cells. 2007;25:1853–1861. doi: 10.1634/stemcells.2007-0057. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell JR, et al. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 30.Dokal I, Vulliamy T. Telomerase deficiency and human disease. In: De Lange T, et al., editors. Telomeres. Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 31.Tsakiri KD, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 33.Garcia CK, et al. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enomoto S, et al. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2626–2638. doi: 10.1091/mbc.02-02-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.IJpma AS, Greider CW. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 38.Takai H, et al. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 39.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front. Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 40.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 41.d’Adda di Fagagna F, et al. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 42.Lou Z, Chen J. Cellular senescence and DNA repair. Exp. Cell Res. 2006;312:2641–2646. doi: 10.1016/j.yexcr.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Lustig AJ, Petes TD. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. U. S. A. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenwell PW, et al. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 45.Morrow DM, et al. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 46.Ritchie KB, et al. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mallory JC, Petes TD. Protein kinase activity of tel1p and mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan SW, et al. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 2001;11:1240–1250. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- 49.Tsukamoto Y, et al. The role of the Mre11–Rad50–Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 50.Ball LG, Xiao W. Molecular basis of ataxia telangiectasia and related diseases. Acta Pharmacol. Sin. 2005;26:897–907. doi: 10.1111/j.1745-7254.2005.00165.x. [DOI] [PubMed] [Google Scholar]
- 51.O’Driscoll M, Jeggo PA. The role of double-strand break repair – insights from human genetics. Nat. Rev. Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 52.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Maser RS, DePinho RA. Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amst.) 2004;3:979–988. doi: 10.1016/j.dnarep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Zhu XD, et al. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 55.Jaco I, et al. Role of mammalian Rad54 in telomere length maintenance. Mol. Cell. Biol. 2003;23:5572–5580. doi: 10.1128/MCB.23.16.5572-5580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarsounas M, et al. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 57.Runge KW, Zakian VA. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takai H, et al. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 59.Anderson CM, et al. Tel2 mediates activation and localization of ATM/Tel1 kinase to a double-strand break. Genes Dev. 2008;22:854–859. doi: 10.1101/gad.1646208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi T, et al. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12:1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 61.Seto AG, et al. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 62.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 63.Marcand S, et al. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- 64.Taggart AKP, et al. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 65.Osterhage JL, et al. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2006;13:720–728. doi: 10.1038/nsmb1125. [DOI] [PubMed] [Google Scholar]
- 66.Wright WE, et al. Normal human telomeres are not late replicating. Exp. Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- 67.Zou Y, et al. Asynchronous replication timing of telomeres at opposite arms of mammalian chromosomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12928–12933. doi: 10.1073/pnas.0404106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 69.Jady BE, et al. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomlinson RL, et al. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcand S, et al. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teixeira MT, et al. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 73.Hemann MT, et al. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 74.Ouellette MM, et al. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 2000;275:10072–10076. doi: 10.1074/jbc.275.14.10072. [DOI] [PubMed] [Google Scholar]
- 75.Steinert S, et al. Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun. 2000;273:1095–1098. doi: 10.1006/bbrc.2000.3080. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y, et al. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sabourin M, et al. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fisher TS, et al. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 80.Goudsouzian LK, et al. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang M, et al. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hector RE, et al. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 84.Shima H, et al. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics. 2005;170:71–85. doi: 10.1534/genetics.104.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mallory JC, et al. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Repair (Amst.) 2003;2:1041–1064. doi: 10.1016/s1568-7864(03)00115-0. [DOI] [PubMed] [Google Scholar]
- 86.Tseng SF, et al. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pennock E, et al. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 88.Bianchi A, et al. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 89.Smolka MB, et al. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feldser D, et al. Ataxia telangiectasia mutated (Atm) is not required for telomerase-mediated elongation of short telomeres. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2249–2251. doi: 10.1073/pnas.0511143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakamura TM, et al. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161:1437–1452. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verdun RE, et al. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 93.Viscardi V, et al. Sudden telomere lengthening triggers a Rad53-dependent checkpoint in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:3126–3143. doi: 10.1091/mbc.E02-11-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Viscardi V, et al. MRX-dependent DNA damage response to short telomeres. Mol. Biol. Cell. 2007;18:3047–3058. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shimada K, et al. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tercero JA, et al. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 97.Bessler JB, Zakian VA. The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity. Genetics. 2004;168:1205–1218. doi: 10.1534/genetics.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hockemeyer D, et al. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hirano Y, Sugimoto K. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol. Biol. Cell. 2007;18:2026–2036. doi: 10.1091/mbc.E06-12-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Michelson RJ, et al. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larrivee M, et al. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hemann MT, Greider CW. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blackburn EH. A history of telomere biology. In: De Lange T, et al., editors. Telomeres. 2nd edn. Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]