Abstract
The majority of nucleus-encoded chloroplast proteins are targeted to the organelle by direct binding to two membrane-bound GTPase receptors, Toc34 and Toc159. The GTPase activities of the receptors are implicated in two key import activities, preprotein binding and driving membrane translocation, but their precise functions have not been defined. We use a combination of in vivo and in vitro approaches to study the role of the Toc159 receptor in the import reaction. We show that atToc159-A864R, a receptor with reduced GTPase activity, can fully complement a lethal insertion mutation in the ATTOC159 gene. Surprisingly, the atToc159-A864R receptor increases the rate of protein import relative to wild-type receptor in isolated chloroplasts by stabilizing the formation of a GTP-dependent preprotein binding intermediate. These data favor a model in which the atToc159 receptor acts as part of a GTP-regulated switch for preprotein recognition at the TOC translocon.
Introduction
Plastids (e.g., chloroplasts) are a diverse group of interrelated organelles that have evolved to perform numerous and diverse metabolic processes within all plant cells, including essential roles in photosynthesis, amino acid and lipid synthesis, and cell signaling (Neuhaus and Emes, 2000). The biogenesis of these organelles and their differentiation into specialized forms in different tissues relies on the import of thousands of nuclear-encoded proteins from the cytoplasm (Jarvis, 2004; Richly and Leister, 2004). Protein import is mediated by several translocation systems, designated translocons, which reside within the outer and inner envelope membranes of the organelles (Bedard and Jarvis, 2005; Gutensohn et al., 2006; Kessler and Schnell, 2006). The majority of plastid proteins are synthesized with N-terminal cleavable transit peptides (Bruce, 2001), and their import is mediated by the TOC–TIC translocon system. The translocon at the outer envelope membrane of chloroplasts (TOC) recognizes the intrinsic transit peptides of plastid preproteins at the organelle surface and initiates the import process by transferring the preprotein to a protein-conducting channel (Kessler and Schnell, 2006; Li et al., 2007; Rounds et al., 2007). The Toc complex physically associates with the translocon at the inner envelope membrane of chloroplasts (TIC) to facilitate simultaneous transport of proteins across the double membrane of the envelope (Gutensohn et al., 2006).
The core of the TOC translocon consists of two receptor GTPases, Toc159 and Toc34, which associate with a membrane channel that contains the β-barrel protein, Toc75 (Li et al., 2007; Rounds et al., 2007). The gene families encoding these proteins are essential in arabidopsis (Jarvis et al., 1998; Bauer et al., 2000; Baldwin et al., 2005), underscoring their central roles in the import process. Toc34 consists of a cytosolically exposed GTPase domain (G-domain) that is anchored to the outer membrane by a simple, C-terminal α-helical transmembrane segment. Toc159 contains a homologous, cytosolic G-domain, but its C-terminal membrane anchor domain (M-domain) is larger (∼50 kD) and more complex. Furthermore, it contains an N-terminal acidic domain that is highly variable among members of the Toc159 gene family and is proposed to contribute to transit peptide specificity of the translocon (Ivanova et al., 2004; Kubis et al., 2004).
Multiple lines of evidence indicate that Toc34 and Toc159 play key roles in regulating protein targeting and plastid biogenesis. First, the two receptors mediate preprotein recognition at the chloroplast surface by binding directly to transit peptides (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997; Sveshnikova et al., 2000; Schleiff et al., 2002; Smith et al., 2004). The import of bound preproteins is inhibited by nonhydrolyzable GTP analogues, indicating that GTP hydrolysis at the TOC regulates transfer of the preprotein into the protein-conducting channel (Olsen and Keegstra, 1992; Ma et al., 1996; Young et al., 1999). The GTPase domains of the two receptors are involved in transit peptide binding, and binding stimulates the GTPase activities of the receptors, providing a link between preprotein recognition and GTPase activity (Jelic et al., 2002, 2003; Smith et al., 2004). As such, Toc34 and Toc159 are hypothesized to act as a coordinated GTPase switch that controls protein import into the organelle.
Second, members of the Toc159 and Toc34 gene families in arabidopsis have been shown to form structurally and functionally distinct translocon complexes that constitute distinct pathways for protein import into plastids. Genetic and biochemical analyses indicate that different members of the arabidopsis Toc159 (atToc159) and Toc34 (atToc33) families function in the import of different classes of preproteins that are required for specialized (e.g., photosynthesis) and housekeeping functions of the organelles (Jarvis et al., 1998; Bauer et al., 2000; Kubis et al., 2003, 2004; Ivanova et al., 2004). Mutations in genes encoding specific atToc159 and atToc33 family members interfere with plastid differentiation at different stages. These studies demonstrate an important role for the TOC GTPase receptors in coordinating protein trafficking with the changes in gene expression that accompany plant development.
Despite the central role for the TOC GTPases in regulating protein import, the molecular mechanism by which their GTPase activities control preprotein recognition and translocation remain undefined. Two major models have been proposed as working hypotheses for TOC receptor function. In one model, Toc159 and Toc34 form a GTP regulated gate that controls access of preproteins to the TOC channel, thereby maintaining the fidelity of import and regulating the specificity of translocons for distinct classes of preproteins (Seedorf et al., 1995; Kessler and Schnell, 2004). Although this model includes roles for both GTPases in preprotein binding at the translocon, Toc159 is proposed to function primarily in defining the selectivity of the translocon, whereas Toc34 is envisioned to function in conjunction with Toc159 to initiate transfer of the preprotein to the Toc channel. This proposal is supported by covalent cross-linking studies that demonstrate that Toc159 forms the primary contact with transit peptides during preprotein binding (Perry and Keegstra, 1994; Ma et al., 1996). Furthermore, the atToc159 family members appear to largely dictate the specificity of different TOC complexes for distinct classes of preproteins (Hiltbrunner et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; Smith et al., 2004), consistent with a role for this receptor family in regulating initial preprotein recognition. In contrast, the two Toc34 receptors in arabidopsis appear to be largely functionally redundant in vivo (Jarvis et al., 1998).
A second model proposes that Toc159 GTPase activity provides the energetic driving force for preprotein translocation through the Toc channel (Schleiff et al., 2003; Becker et al., 2004). In this scenario, Toc34 is proposed to function primarily in preprotein recognition and Toc159 is envisioned to function downstream as a repetitive GTP-driven motor that forces the preprotein through the Toc channel. This model is based on the observation that reconstituted proteoliposomes containing Toc75 and a fragment of Toc159, corresponding to its G- and M-domains, can support a minimum level of GTP-dependent preprotein translocation (Schleiff et al., 2003).
The proposals for TOC GTPase function are largely in agreement on the critical role of the functional interactions between the two GTPases and their involvement in GTP-regulated preprotein recognition at the Toc complex. However, the models present very distinct hypotheses for the molecular mechanism of TOC GTPase function during import. The major differences between these models center on the role of Toc159 and its GTPase activity in preprotein recognition and translocation. Toc159 is alternatively proposed to function early in import as the primary preprotein receptor or late in import as the driving force for membrane translocation. One major issue in defining the specific role for Toc159 in TOC complex function is the lack of in vivo analysis of its GTPase function. The models based on in vitro biochemical analysis of Toc159 also are qualified by the fact that they rely on the use of purified or overexpressed fragments of the receptor that lack one or more of the domains found in the full-length native protein.
In this study, we aimed to more clearly define the role of Toc159 GTPase activity in TOC translocon function using in vivo genetic approaches combined with in vitro import assays using intact chloroplasts with full-length receptor. To this end, we took advantage of an arabidopsis mutant, ppi2, which lacks expression of atToc159, the major Toc159 family member expressed in chloroplasts (Bauer et al., 2000). We expressed atToc159-A864R, a mutated form of atToc159 that has significantly reduced GTPase activity, in ppi2 and examined the effects of the mutation on protein import and chloroplast biogenesis. Remarkably, the atToc159-A864R receptor was able to fully complement the ppi2 lethal phenotype. Furthermore, chloroplasts containing the atToc159-A864R mutant exhibited an increased rate of protein import relative to wild-type chloroplasts. Biochemical analysis indicated that the atToc159-A864R receptor increases the efficiency of preprotein targeting to chloroplasts. Our studies demonstrate that the nucleotide state of Toc159 regulates the formation of a stable preprotein binding intermediate, and thereby plays a regulatory role in preprotein recognition at the TOC translocon.
Results
Expression of an atToc159 GTPase mutant can complement the ppi2 lethal phenotype
Our strategy for assessing the role of atToc159 GTPase activity centered on expressing the atToc159-A864R mutant in stable transformants of the atToc159 null mutant, ppi2, and investigating the effects on chloroplast biogenesis and protein import. AtToc159-A864R contains a single point mutation that converts a conserved alanine to an arginine in the G1 motif (P-loop) of atToc159. Previous studies showed that the Escherichia coli–expressed G-domain of atToc159-A864R binds GTP and GDP with equal or slightly higher affinities compared with wild-type receptor, but its GTPase activity is reduced to ∼10% of that of atToc159 (Smith et al., 2002). To confirm that the activities of the full-length receptors are comparable to those measured with isolated G-domains, we purified full-length wild-type atToc159 and atToc159-A864R from a reticulocyte lysate in vitro translation system using immunoprecipitation and measured their activities using a standard GTPase assay. Fig. S1 A (available at http://www.jcb.org/cgi/content/full/jcb.200803034/DC1) shows that the hydrolytic activity of atToc159-A864R (KM = 1.9 ± 0.30 μM; kcat = 0.0042 ± 0.0001 min−1) is ∼30% of that measured for wild-type atToc159 (KM = 3.2 ± 0.34 μM; kcat = 0.014 ± 0.0022 min−1). The hydrolytic activity of wild-type atToc159 is similar to that measured for purified, soluble atToc159 G-domain (KM = 9.0 ± 1.35 μM; kcat = 0.019 ± 0.0015 min−1), indicating that the activities of the full-length and isolated G-domains are comparable. Furthermore, these data demonstrate that immunoprecipitation does not significantly affect the enzymatic activity of the receptor. As negative controls, we included two additional receptor G-domain constructs in the assay, atToc159-G1m and atToc33G-G1m. These receptors carry triple mutations within the G1 motif of their GTP-binding sites and previously were shown to lack GTP binding and hydrolytic activities (Bauer et al., 2002; Smith et al., 2002; Wallas et al., 2003). No measurable hydrolytic activity was detected in either case (Fig. S1 B). Together, the data are consistent with the results using the purified G-domains and indicate that the atToc159-A864R mutant has a significantly lower hydrolytic rate than wild-type receptor.
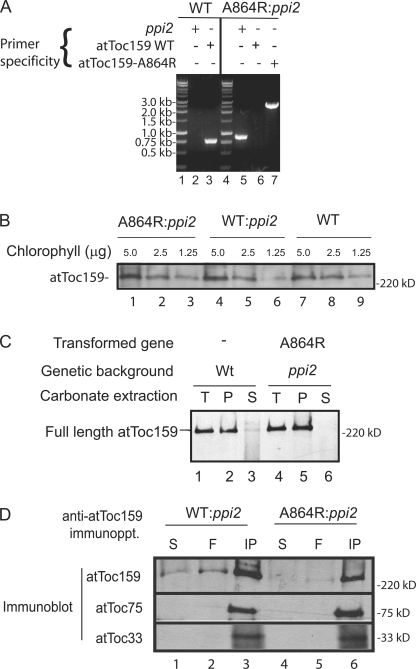
A transformation vector encoding atToc159-A864R under control of the cauliflower mosaic virus 35S promoter (CaMV35S) was transformed into heterozygous ppi2 plants. Independent transformants were segregated to select for plants homozygous for both ppi2 and CaMV35S∷atTOC159-A864R (A864R:ppi2). The genotypes of the transformed plants were confirmed by PCR amplification of the wild-type authentic atTOC159 gene, the ppi2 T-DNA marker, and the CaMV35S:atTOC159-A864R transgene (Fig. 1 A). To confirm that the transgene was expressed in the ppi2 background, we performed immunoblots of plant extracts with anti-atToc159 serum. A ppi2 line expressing CaMV35S∷atTOC159 (WT:ppi2) that was previously generated was used as a control (Bauer et al., 2002). Full-length atToc159 was present in wild-type and WT:ppi2, whereas a full-length protein corresponding to atToc159-A864R was detected in A864R:ppi2 plants (Fig. 1 B). Immunoblots of serial dilutions of extracts from these plants indicates that receptor expression in WT:ppi2 and A864R:ppi2 plants is similar, and both transgenes are expressed at levels only moderately higher than authentic atToc159 in wild-type (WT) plants. Alkaline extraction of membrane fractions from wild-type and A864R:ppi2 plants indicated that atToc159-A864R was membrane integrated, consistent with correct targeting to the outer envelope membrane (Fig. 1 C). Moreover, atToc75 and atToc33 coimmunoprecipitated with atToc159 and atToc159-A864R from detergent-soluble extracts, demonstrating that the mutation did not detectably affect receptor targeting or TOC complex assembly (Fig. 1 D).
Figure 1.
Arabidopsis ppi2 homozygous plants expressing atToc159 and atToc159-A864R. Heterozygous ppi2 plants were transformed with CaMV35S∷atToc159 and CaMV35S∷atToc159-A864R binary vectors encoding atToc159 and atToc159-A864R, respectively. (A) Homozygous ppi2 plants carrying the CaMV35S∷atToc159-A864R transgene (A864R:ppi2) were identified by PCR genotyping using primers specific for the ppi2 allele or the wild-type atTOC159 gene (atToc159 WT). (B) Serial dilutions of protein extracts from A864R:ppi2, WT:ppi2, and wild-type (WT) plants were immunoblotted with anti-atToc159 serum. (C) Isolated chloroplasts (T) from wild-type and A864R:ppi2 plants were extracted with alkaline carbonate, pH 11.5, and separated into membrane pellet (P) and soluble (S) fractions. The fractions were resolved by SDS-PAGE and immunoblotted with anti-atToc159 serum. (D) Detergent-solubilized chloroplasts from WT:ppi2 and A864R:ppi2 plants were subjected to immunoprecipitation with anti-atToc159 serum. 10% of the total extracts (S) and unbound fractions (F) or the total fractions from the immunoprecipitates (IP) were resolved by SDS-PAGE and immunoblotted with antisera to atToc159, atToc75, and atToc33.
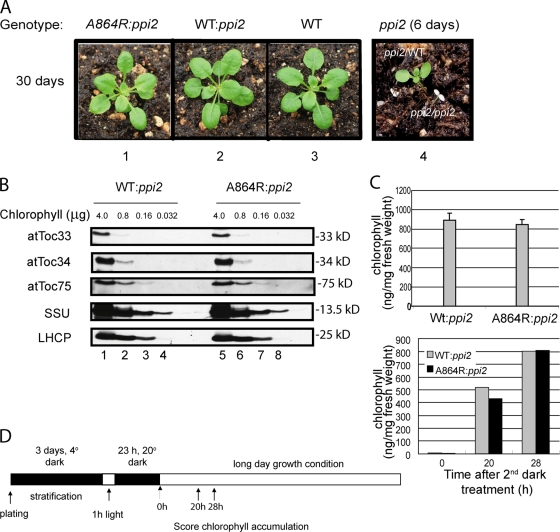
The homozygous ppi2 mutant is albino and seedling lethal, resulting in plants that die shortly after germination on soil (Fig. 2 A) (Bauer et al., 2000). Remarkably, the atToc159-A864R gene complemented the ppi2 mutation. A864R:ppi2 plants were visibly indistinguishable from WT:ppi2 or wild-type plants at all stages of development under normal growth conditions (Fig. 2 A). Furthermore, the levels of chlorophyll (Fig. 2 C) and representative chloroplast proteins, including other TOC components (Fig. 2 B), were indistinguishable from wild-type plants.
Figure 2.
The atTOC159-A864R gene complements the ppi2 lethal phenotype. (A) Visible phenotypes of plants with the indicated genetic backgrounds expressing atToc159-A864R (A864R:ppi2) or wild-type atToc159 (WT:ppi2). Panel 4 contains 6-d-old seedlings from ppi2/WT or ppi2/ppi2 plants for comparison. (B) Serial dilutions of protein extracts from WT:ppi2 or A864R:ppi2 plants were resolved by SDS-PAGE and immunoblotted with antisera to the proteins indicated at the left of each panel. (C) Chlorophyll content of WT:ppi2 and A864R:ppi2 plants at 14 d after germination. The data in the graph represent the mean of three independent experiments. (D) Chlorophyll content of WT:ppi2 and A864R:ppi2 plants (right) that were subjected to a de-etiolation protocol (left) to assess the effects of the mutation on chloroplast biogenesis.
To investigate if the atToc159-A864R mutation might have a more subtle effect that was not apparent under normal growth conditions, we performed a de-etiolation experiment (Fankhauser and Casal, 2004) to more closely examine chloroplast biogenesis in the complemented plants (Fig. 2 D). The atToc159 receptor has previously been shown to be required for the development of chloroplasts from undifferentiated plastids in the majority of green tissues (Bauer et al., 2000). Therefore, we tested whether the atToc159-A864R mutation might delay this developmental process by partially disrupting the protein import process. In this experiment, the seeds were stratified in the dark at 4°C and exposed to a short pulse of light to initiate germination. The germinating seeds were equilibrated in the dark at 20°C, resulting in etiolated (nongreen) seedlings that lack fully developed chloroplasts. The seedlings were then transferred to light to trigger photomorphogenesis and chloroplast development. We monitored chlorophyll accumulation at 20 and 28 h after transferring the plants to light. As shown in Fig. 2 D, WT:ppi2 and A864R:ppi2 plants accumulated chlorophyll at similar rates. These data indicate that the atToc159-A864R mutation does not disrupt chloroplast biogenesis and can fully complement ppi2.
AtToc159-A864R chloroplasts have an increased rate of import relative to wild-type chloroplasts
To further understand the role of atToc159 GTPase activity, we examined the levels of preprotein import in chloroplasts isolated from wild-type, WT:ppi2, and A864R:ppi2 plants. For these studies, we modified our chloroplast isolation procedure to minimize the degradation of atToc159 based on published procedures (Brock et al., 1993; Schulz et al., 2004). Our standard isolation procedures for pea and arabidopsis chloroplasts result in proteolytic cleavage of the amino-terminal A-domain of Toc159, yielding an 86-kD receptor fragment (Bolter et al., 1998; Chen et al., 2000). We found that modification of the standard isolation methods based on published procedures (Brock et al., 1993; Schulz et al., 2004) resulted in a dramatic decrease in atToc159 degradation (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200803034/DC1). Comparison of the import capabilities of these chloroplasts also demonstrates a ∼2-fold increase in the efficiency of import (Fig. S2, B and C). All subsequent experiments were performed with chloroplasts isolated using this procedure.
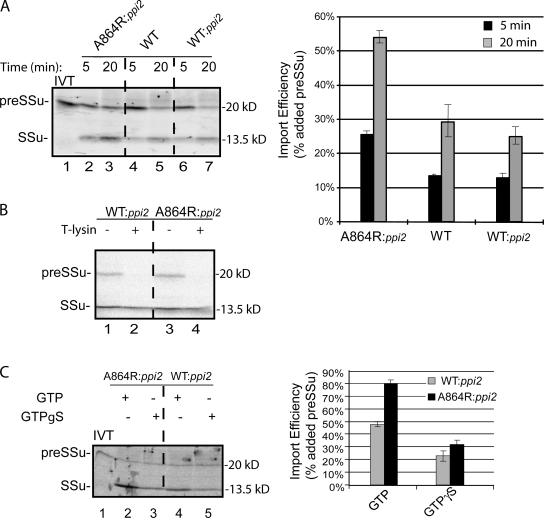
Isolated chloroplasts were incubated with [35S]-labeled chloroplast preprotein, preSSu, in the presence of 1 mM GTP and 2 mM ATP to promote import for 5 or 20 min at 25°C (Fig. 3 A). Import into chloroplasts from wild-type and WT:ppi2 plants were indistinguishable, demonstrating that expression of wild-type atToc159 under a non-native promoter does not measurably affect the import process. Surprisingly, the levels of preSSu import were significantly higher in atToc159-A864R chloroplasts compared with the two controls at each time point. The accumulation of SSu in atToc159-A864R chloroplasts was approximately double both wild-type or ppi2 chloroplasts expressing wild-type atToc159 (Fig. 3 A). Thermolysin treatment of the atToc159-A864R chloroplasts following the import reaction removed bound preSSu but did not digest the processed form (Fig. 3 B), demonstrating that the mature SSu represented completely imported protein and not a partially imported substrate. Interestingly, import into atToc159-A864R chloroplasts remained sensitive to the GTP analogue, GTPγS (Fig. 3 C). Import into both wild-type and atToc159-A864R chloroplasts was reduced by ∼60% in the presence of the analogue. Therefore, GTP hydrolysis within the TOC translocon is still required for the import reaction, even when atToc159 hydrolytic activity is reduced by 70%. The inhibition by the analogue could be due to an effect on the residual activity of atToc159-A864R or on atToc33.
Figure 3.
The levels of protein import into atToc159-A864R chloroplasts are increased compared with wild-type chloroplasts. (A) Isolated chloroplasts from WT, WT:ppi2, and A864R:ppi2 plants were incubated with in vitro–translated [35S]preSSu under import conditions for the times indicated. The graph presents quantification of triplicate experiments from import of preSSu at the 5-min and 20-min time points. (B) Chloroplasts from preSSu 20-min import reactions in A were treated with 100 μg/ml thermolysin on ice for 30 min to discriminate between bound and fully imported protein. (C) Effect of GTPγS on protein import into WT:ppi2 and A864R:ppi2 chloroplasts. Chloroplasts were incubated under standard import conditions for 20 min in the presence of GTP or GTPγS. The graph presents quantification of preSSu import from triplicate experiments. The position of preSSu and imported mature SSu are indicated at the left of each panel. IVT, 10% of in vitro–translated [35S]preSSu added to each import reaction. Dashed lines indicate that the figures were generated from different regions of the same SDS-PAGE gel using samples from the same experiment.
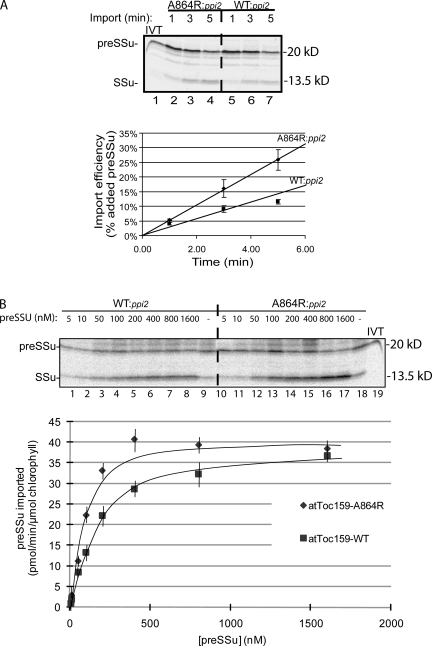
To examine the nature of the increased levels of import observed in atToc159-A864R chloroplasts, we examined import rates by conducting a time course of preSSu import. The results in Fig. 3 A and subsequent studies (unpublished data) indicated that preSSu import was linear with time between 0 and 7 min and leveled off beyond that time period. Therefore, we limited our import time course to between 0 and 5 min. Consistent with the increased SSu accumulation, the atToc159-A864R chloroplasts exhibited a nearly twofold higher import rate compared with the wild-type chloroplast controls (Fig. 4 A). To analyze whether the higher rate of import into atToc159-A864R chloroplasts represented an increase in the affinity of TOC translocons for preSSu (i.e., increased import rate constants) or an overall increase in the maximum import rate (i.e., turnover number), we performed a kinetic analysis of import using purified E. coli–expressed [35S]-preSSu (Fig. 4 B). The maximum import rates calculated from the kinetic data for atToc159-A8684R (49 ± 1.0 pmol preSSu/min/μmol chlorophyll) and atToc159 (41 ± 0.75 pmol preSSu/min/μmol chlorophyll) chloroplasts were similar. Therefore, the increased rate of preSSu does not appear to result from an overall increase in turnover number. In contrast, the import rate constants (apparent Km) for atToc159-A8684R (158 ± 18 nM) and atToc159 (224 ± 11 nM) chloroplasts were significantly different. The lower apparent Km for atToc159-A8684R chloroplasts suggests that the TOC translocons have a higher apparent affinity for preSSu relative to wild-type translocons, accounting for the increased rate of preSSu import under substrate-limiting conditions.
Figure 4.
The atToc159-A864R chloroplasts exhibit an increased rate of protein import. (A) In vitro–translated [35S]preSSu was imported into WT:ppi2 and A864R:ppi2 chloroplasts for the times indicated. The experimental conditions were the same as described in Fig. 3. Quantification of the data from triplicate experiments is presented in the graph. (B) Michaelis-Menten plot of the import of increasing concentrations of purified, E. coli–expressed [35S]preSSu into WT:ppi2 and A864R:ppi2 chloroplasts.
The atToc159-A864R receptor enhances preprotein binding to TOC translocons
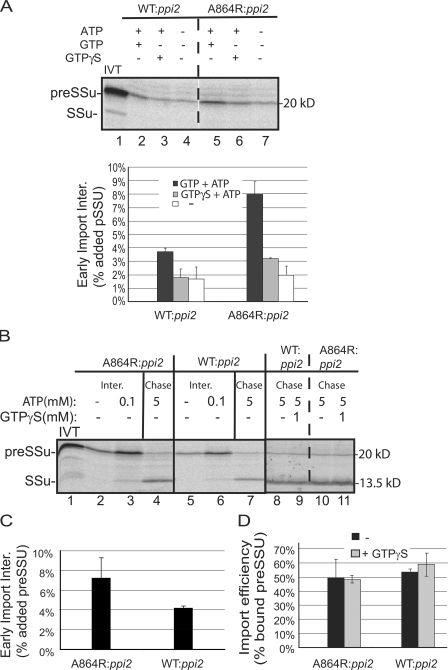
GTPγS has previously been shown to inhibit the early stages of protein import by preventing the formation of an early import intermediate that has inserted into the TOC channel, but remains largely exposed at the chloroplast surface and therefore has not completed translocation across the membrane (Schnell and Blobel, 1993; Kessler et al., 1994; Young et al., 1999; Inoue and Akita, 2008). We examined the formation of the early intermediate in wild-type and atToc159-A864R chloroplasts in more detail to determine if the mutation affected insertion into the outer membrane channel or subsequent translocation across the envelope. Isolated chloroplasts were incubated with [35S]-preSSu in the presence of GTP and low levels of ATP (0.1 mM) to promote formation of the early import intermediate. The levels of early import intermediate in atToc159-A864R chloroplasts were nearly twofold higher than in chloroplasts from WT:ppi2 plants (Fig. 5 A, compare lanes 2 and 5 and graph). These results demonstrate that the reduction of atToc159 GTPase activity actually enhances, rather than reduces, the amount of early import intermediate. Consistent with its effects on import, GTPγS inhibits the formation of the early intermediate (Fig. 5 A). These results confirm that the initiation of outer membrane translocation is not disrupted by inhibiting atToc159 GTPase activity.
Figure 5.
Preprotein translocation across the outer membrane is not affected by decreasing atToc159 GTPase activity. (A) In vitro–translated [35S]preSSu was incubated with isolated chloroplasts from WT:ppi2 or A864R:ppi2 plants in the presence of 0.1 mM ATP and 0.1 mM GTP to initiate translocation across the outer membrane (early import intermediate), but preclude complete translocation across the envelope. Where indicated, 1 mM GTPγS was substituted for GTP. The graph presents the quantification of results from triplicate experiments. (B) WT:ppi2 and A864R:ppi2 chloroplasts containing bound early import intermediates (Inter.; lanes 3 and 6) were reisolated and resuspended under protein import conditions in the presence of 5 mM ATP (Chase; lanes 4, 7, and 8–11). Where indicated, 1 mM GTPγS was added to the chase reactions (lanes 9 and 11). (C) Quantification of the levels of early import intermediate formed in B from triplicate experiments. (D) Quantification of the effects of GTPγS on the chase of the early intermediate (B) from triplicate experiments. Dashed lines indicate that the figures were generated from different regions of the same SDS-PAGE gel using samples from the same experiment.
To determine whether subsequent translocation of the early import intermediate across the chloroplast envelope is affected by atToc159-A864R, we examined the complete translocation of the intermediate by chasing it into chloroplasts using high concentrations of ATP (5 mM). Although the levels of early intermediate are higher in mutant chloroplasts (Fig. 5 B, lanes 3 and 4; Fig. 5 C), ∼50% of the bound intermediate can be chased into chloroplasts in both atToc159-A864R and wild-type chloroplasts (Fig. 5 B, compare lanes 4 and 7; Fig. 5 D, black bars). In contrast to formation of the early import intermediate, complete translocation from the intermediate stage does not appear to require additional GTP hydrolysis at either TOC receptor because the inclusion of GTPγS in the chase experiment had no effect on import of the intermediate (Fig. 5 B, compare lanes 8–9 and 10–11; Fig. 5 D, gray bars). These results demonstrate that the intermediate in both wild-type and mutant chloroplasts represents a productive intermediate and that membrane translocation after initial insertion in the TOC channel is not disrupted by inhibiting TOC GTPase activity.
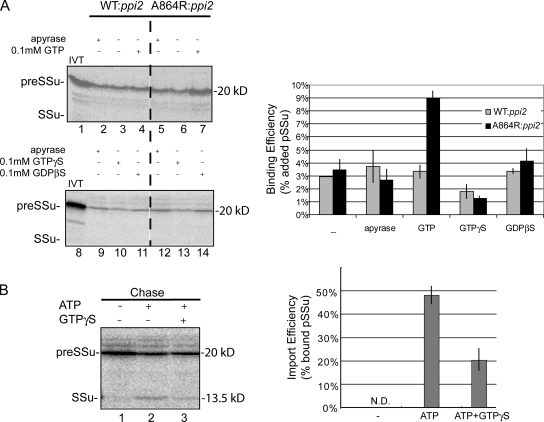
Low levels of preprotein binding to the TOC GTPase receptors have been observed in the absence of energy or in the presence of GTP alone (Perry and Keegstra, 1994; Ma et al., 1996). This prompted us to examine the initial binding step to determine its role in the increased levels of early intermediate formation and protein import observed in atToc159-A864R chloroplasts. Consistent with previous observations, wild-type chloroplasts exhibit low levels of preSSu binding in the absence of added nucleoside triphosphates (NTPs) or in the presence of the NTP hydrolytic enzyme, apyrase (Fig. 6 A). The addition of GTP or the nonhydrolyzable GDP analogue, GDPβS, do not measurably increase or decrease binding relative to the no-energy controls (Fig. 6 A and graph). The addition of the slowly hydrolyzable GTP analogue, GTPγS, reduces the basal levels of binding by nearly 50%, suggesting that forcing both TOC GTPases into their GTP bound forms inhibits the stable interaction of the preprotein with the receptors.
Figure 6.
atToc159-A864R promotes GTP-dependent binding to the TOC translocon. (A) Energy-depleted, in vitro–translated [35S]preSSu was incubated with isolated chloroplasts from WT:ppi2 or A864R:ppi2 plants in the absence of added nucleotide (−), in the presence of apyrase or in the presence of 0.1 mM of the indicated nucleotide. Chloroplasts were reisolated and bound preSSu was analyzed by SDS-PAGE and phosphorimaging. The graph presents the quantification of results from triplicate experiments. (B) atToc159-A864R chloroplasts were incubated with [35S]preSSu in the presence of 0.1 mM GTP to stimulate binding. The chloroplasts were reisolated and resuspended in the absence of added nucleotide (−), in the presence of 5 mM ATP or 5 mM ATP and 1 mM GTPγS and incubated under standard import conditions (Chase). The graph presents quantification of imported preSSu from triplicate experiments. Dashed lines indicate that the figures were generated from different regions of the same SDS-PAGE gel using samples from the same experiment.
PreSSu binding to atToc159-A864R chloroplasts in the absence of energy or in the presence of GDPβS (Fig. 6 A, lanes 5 and 6 and graph) is indistinguishable from wild-type chloroplasts. GTPγS reduces binding to atToc159-A864R to levels similar to those observed with wild-type chloroplasts (Fig. 6 A, compare lanes 12 and 13). In contrast, GTP increases the level of binding to atToc159-A864R chloroplasts approximately threefold over the no-energy control (Fig. 6 A, compare lanes 4 and 7, and graph). To ensure that the GTP-dependent binding represents a productive intermediate, we tested whether it could be chased into chloroplasts by re-isolating the chloroplasts and adding sufficient ATP to support complete envelope translocation (Fig. 6 B, lane 2 and graph). Approximately 50% of the bound preprotein can be imported and processed to its mature form, indicating that the bound precursor represents an intermediate on the import pathway. These data indicate that the GTP-bound form of atToc159 functions as the active component in stable preprotein binding to the TOC translocon. The increased binding in the presence of GTP likely results from the stabilization of the initial binding of preprotein to the TOC translocon that previously was observed only when stabilized by chemical cross-linkers (Perry and Keegstra, 1994; Ma et al., 1996). The increased binding can account for the increase in the import rates observed in atToc159-A864R chloroplasts.
In contrast to complete translocation of preproteins from the early import intermediate stage, GTPγS inhibits translocation and processing of the GTP-dependent intermediate (Fig. 6 B, compare lanes 2 and 3 and graph). The reduction of binding by GTPγS in both the wild-type and atToc159-A864R chloroplasts suggests that stable preprotein binding is blocked when both atToc159 and atToc33 are locked in their GTP-bound forms. Furthermore, it is consistent with the hypothesis that both receptor GTPases participate in the initial binding of preproteins to the translocon.
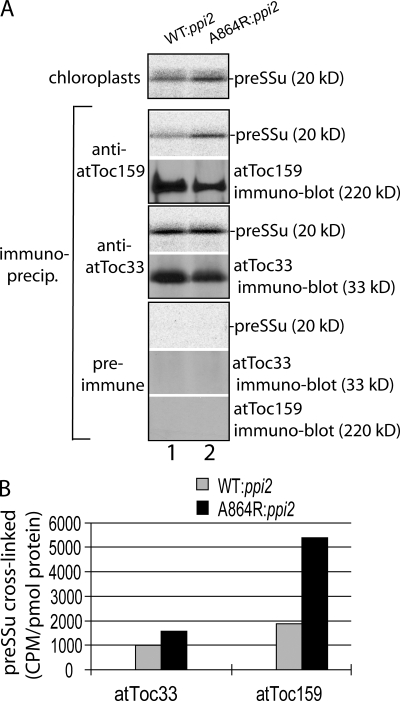
As a final step in our analysis, we examined the components involved in the GTP-dependent binding observed with atToc159-A864R chloroplasts. To this end, we used the homobifunctional cysteine cross-linker, DTME (dithio-bismaleimidoethane), to cross-link bound preSSu to isolated chloroplasts in the presence of GTP. As previously observed, preSSu binding to atToc159-A864R chloroplasts was significantly higher than the wild-type controls (Fig. 7 A, top). Consistent with this observation, approximately three times the levels of preSSu coimmunoprecipitated with atToc159-A864R compared with wild-type atToc159 from detergent solubilized chloroplasts (Fig. 7, A and B). The levels of preSSu that coimmunoprecipitated with atToc33 were increased by ∼50% in atToc159-A864R chloroplasts. We conclude that the stable, GTP-dependent binding to TOC translocons occurs due to increased binding to the GTP-bound form of atToc159 that is predominant in atToc159-A864R chloroplasts.
Figure 7.
GTP-dependent binding of preSSu to TOC translocons is mediated by atToc159. (A) Energy-depleted, in vitro–translated [35S]preSSu was incubated with isolated chloroplasts from WT:ppi2 or A864R:ppi2 plants in the presence of 0.1 mM GTP to promote GTP-dependent binding. The homo-bifunctional cross-linker DTME was added to cross-link the bound preprotein to TOC components. After quenching the cross-linker, the chloroplasts were detergent solubilized and proteins immunoprecipitated with atToc159 or atToc33 affinity-purified antibodies or preimmune serum. The immunoprecipitates were analyzed by SDS-PAGE and phosphorimaging to quantify coimmunoprecipitated [35S]preSSu or quantitative immunoblotting to detect atToc159 or atToc33. (B) Quantification of the levels of preSSu coimmunoprecipitating with atToc33 or atToc159 from WT:ppi2 or A864R:ppi2 chloroplasts. The quantification is from the single experiment presented in A. The data are representative of three replicate experiments.
Discussion
In this paper, we report the characterization of a novel intermediate in the pathway of protein import into chloroplasts. Identification of the intermediate was made possible by increasing the GTP-bound form of the atToc159 receptor using the atToc159-A864R mutant that binds but does not efficiently hydrolyze GTP. In the presence of GTP, atToc159-A864R promotes stable binding of preprotein to the TOC translocon (Fig. 6). This results in an apparent increase in the affinity of the translocon for preSSu (Fig. 4). The bound preprotein is efficiently chased into chloroplasts under import conditions, indicating that it represents a productive step in the import process. The GTP-dependent binding observed in atToc159-A864R chloroplasts results in an increased rate of protein import (Figs. 3 and 4) and accounts for the ability of the mutant receptor to complement the atToc159 null mutant, ppi2 (Figs. 1 and 2). On the basis of these results, we conclude that GTP-dependent binding represents an early step in docking of the preprotein to the TOC translocon.
Previous studies have shown that GTP alone supports low levels of preprotein binding. However, the nature of this binding has been difficult to define because the levels were similar to, or only slightly higher than, binding observed in the absence of added NTP (Olsen and Keegstra, 1992; Young et al., 1999) and were captured only by covalent cross-linking (Perry and Keegstra, 1994; Ma et al., 1996). The atToc159-A864R receptor increases GTP-dependent preprotein binding, apparently by stabilizing the GTP-bound form of the receptor. In contrast to the early import intermediate, the bound preprotein appears not to have initiated membrane translocation because import from the bound state requires GTP hydrolysis (Fig. 6), whereas complete translocation of the early import intermediate does not (Fig. 5) (Young et al., 1999). Furthermore, the early import intermediate requires low concentrations of ATP (Olsen et al., 1989; Olsen and Keegstra, 1992; Schnell and Blobel, 1993). Therefore, the GTP-dependent binding intermediate represents a stage in import that precedes preprotein insertion in the TOC channel and likely corresponds to the initial interaction of preproteins with the translocon.
This work and numerous previous studies clearly have established that GTP hydrolysis regulates the initiation of translocation across the outer membrane (Olsen and Keegstra, 1992; Kessler et al., 1994; Ma et al., 1996; Kouranov and Schnell, 1997; Young et al., 1999). Although atToc159-A864R promotes preprotein binding to the translocon, it does not appear to affect membrane translocation. Nevertheless, GTP analogues inhibit subsequent insertion of the preproteins into the TOC channel (i.e., formation of the early import intermediate) (Fig. 6). These observations are consistent with a role for GTP hydrolysis in initiating membrane translocation, but suggest that this occurs downstream of the GTP-dependent binding step and likely involves the atToc33 GTPase. The results favor models in which GTP-binding and hydrolysis at atToc159 are primarily involved in preprotein targeting to the translocon and are not strictly required for initiating membrane translocation (Li et al., 2007; Rounds et al., 2007). This hypothesis is consistent with several other observations. Pea chloroplasts that have been proteolyzed to selectively remove the amino-terminal A-domain and the G-domain of psToc159, but retain intact psToc34 and psToc75, exhibit reduced preprotein binding (Chen et al., 2000). Nonetheless, these chloroplasts remain import competent, albeit with reduced import rates. Importantly, import in these proteolyzed chloroplasts is still sensitive to nonhydrolyzable analogues, demonstrating a direct role for psToc34 GTPase activity in initiating membrane translocation (Chen et al., 2000). Moreover, an atToc159 deletion construct, consisting solely of the M-domain, can partially complement the arabidopsis ppi2 mutant when expressed in stable transgenic plants (Lee et al., 2003).
Although our data support a primary role for Toc159 in targeting preproteins to the translocon, the data also demonstrate that the initial binding of preproteins involves the concerted activities of both atToc159 and atToc33. AtToc33 and atToc159 both covalently cross-link to the preprotein at the GTP-dependent docking stage (Fig. 7). This is consistent with previous label-transfer cross-linking experiments in which both psToc159 and psToc34 were shown to interact with preproteins during initial binding (Kouranov et al., 1999). Together, these results indicate that both GTPases participate in this early binding step. Although decreasing GTP hydrolysis at atToc159 and therefore favoring its GTP-bound state appears to increase the stability of preprotein binding, the fact that GTPγS inhibits binding suggests that locking both atToc159 and atToc33 in their GTP-bound forms inhibits binding. These data suggest that atToc159 and atToc33 are not in the same nucleotide-bound state (i.e., GTP bound) during this initial interaction.
The accumulated evidence suggests that the Toc159 family of receptors constitute the key determinants of preprotein binding specificity at TOC translocons. Arabidopsis contains four atToc159 family members, and atToc159 and two other family members, atToc132 and atToc120, form separate translocons that have been shown to bind distinct preproteins in vitro and to mediate distinct import pathways in vivo (Hiltbrunner et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; Smith et al., 2004). In contrast, the two Toc34 proteins in arabidopsis, atToc33 and atToc34, exhibit modest selectivity in preprotein binding studies, and their functions are redundant in vivo (Jarvis et al., 1998; Kubis et al., 2003; Constan et al., 2004). The different import pathways generated by the Toc159 receptors appear to be critical for plastid biogenesis (e.g., chloroplast formation) at various stages of plant development. The observation that the GTP-bound form of atToc159 promotes preprotein binding suggests that atToc159 GTPase activity regulates the initial docking of preproteins to the translocon and likely plays a key role in selective preprotein targeting. In this scenario, productive preprotein binding to the GTP-bound form of Toc159 would promote the formation of a stable preprotein–TOC complex that includes binding to Toc34. Subsequent GTP binding and hydrolysis at Toc34 would result in preprotein dissociation from the receptors and transfer to the TOC channel, thereby initiating the translocation reaction. We propose that GTP hydrolysis at Toc159 resets the translocon to a prebinding state following translocation. The prebinding state would prevent translocation in the absence of productive preprotein binding. This mechanism would allow the different Toc159 family members to selectively target preproteins to specific translocons and preclude the binding and translocation of preproteins from other import pathways. Preprotein binding and import in atToc159-A864R chloroplasts are more efficient because the GTP-bound form of the receptor would stabilize the import-competent state of the translocon, thereby bypassing the prebinding state of the translocon.
A previous reconstitution study suggested that Toc159 GTPase activity was the major driving force for membrane translocation (Schleiff et al., 2003). In that work, translocation of a chloroplast preprotein was observed in artificial proteoliposomes that were reconstituted with renatured psToc75 and a fragment of psToc159 containing its G- and M-domains. Our studies demonstrate that inhibition of atToc159 GTPase activity does not inhibit membrane translocation in vivo. These data are inconsistent with a function for Toc159 as a translocation motor. However, Toc159 GTPase activity might contribute to dissociation of the preproteins from the receptor complex and transfer to the Toc channel, thereby facilitating insertion in the channel. In the reconstituted system, the activity might be sufficient to allow some transport across the membrane in the absence of Toc34. In this model, the functions of the Toc159 and Toc34 GTPase activities within native translocons are partially overlapping, with Toc159 GTPase activity primarily involved in preprotein recognition and Toc34 GTPase activity primarily involved in the initiation of translocation. Together, it is clear that cooperative activities of the Toc159 and Toc34 GTPases are required for translocon function in vivo. Future studies in which Toc34 GTPase activity can be manipulated independently and in combination with Toc159 should provide more detailed insights into the role of GTP in regulating the import process.
Materials and methods
Plant materials and intact chloroplast isolation
Arabidopsis thaliana seedlings were grown on agar plates containing 0.5× Murashige and Skoog growth medium (MS medium), 1% sucrose under long-day condition at 23°C for 14 d. To isolate intact chloroplasts, a grinding method was adapted as described previously (Brock et al., 1993; Schulz et al., 2004). In brief, plants were homogenized in ice-cold GB buffer (50 mM Hepes-KOH, pH 7.5, 2 mM EDTA, 1 mM MnCl2, 1 mM MgCl2, 330 mM sorbitol, 100 mM ascorbic acid, 0.05% protease inhibitor cocktail [PIC; Sigma-Aldrich], and 0.25% wt/vol BSA) and filtered through two layers of miracloth. The chloroplasts were collected by centrifugation for 8 min at 1,000 g at 4°C. The chloroplasts were resuspended in 5 ml of GB buffer and applied to a Percoll step gradient (35%/85% Percoll in GB buffer). After centrifugation at 7,000 g for 15 min at 4°C, the intact chloroplasts were collected from the 35%/85% Percoll interface and washed once by diluting into 50 ml ice-cold HS buffer (50 mM Hepes-KOH, pH 8.0, and 330 mM sorbitol). The chloroplasts were collected by centrifugation at 1,000 g for 6 min at 4°C and resuspended in 300 μl HS buffer and kept on ice.
Toc159 mutations and deletion constructs
The atToc159-A864R point mutation was previously introduced into pET21d-atToc159 using the PCR-based overlap extension technique (Smith et al., 2002). A C-terminal 6-histidine tag containing a TEV protease site was introduced into the construct for convenience. For expression of atToc159-A864R-TEV-his in arabidopsis, the coding region was cloned into the XbaI–EcoRI sites of the binary vector, pSMB (Mylne and Botella, 1998) to yield pSMB-CaMV35S:atToc159-A864R.
Arabidopsis transformation and the detection of transgene expression
The homozygous ppi2 plants expressing wild-type atToc159 were described previously (Bauer et al., 2002). The pSMB-CaMV35S:atToc159-A864R construct was introduced into Arabidopsis thaliana (ecotype Wassilewskija) ppi2 heterozygous plants via Agrobacterium tumefaciens–mediated transformation by the floral dip method (Clough and Bent, 1998; Chen et al., 2002). Transformed plants were selected by resistance to BASTA (glufosinate ammonium), which is encoded by the transgene-linked marker, and the presence of the transgene was confirmed by PCR of genomic DNA.
For detecting transgene expression, isolated chloroplasts were analyzed directly by SDS-PAGE and immunoblot analysis. For atToc159-A864R chloroplast membrane localization, wild-type or atToc159-A864R chloroplasts were isolated from 2-wk-old plate-grown plants, followed by alkaline carbonate treatment (Li and Schnell, 2006). The proteins from soluble and membrane fractions were dissolved in SDS-PAGE sample buffer, resolved by SDS-PAGE, and immunoblotted using atToc159 antibody.
De-etiolation experiment
The de-etiolation experiment was performed as previously described (Fankhauser and Casal, 2004). In brief, plate-sown seeds were kept in the dark for 3 d at 4°C (stratification) and treated with a 1-h exposure to white light. Plants were germinated in the dark at 20°C for 23 h. Etiolated seedlings were then placed under long-day growth conditions to initiate photomorphogenesis and chloroplast development. Chlorophyll accumulation was monitored by the method of Arnon (Arnon, 1949) at 20 and 28 h after transferring the plants to light.
Coimmunoprecipitation of TOC core complex proteins
Isolated intact chloroplasts corresponding to 200 μg chlorophyll from atToc159 or atToc159-A864R transformed plants were dissolved in 50 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 1% Triton X-100 containing 1% protease inhibitor cocktail at 4°C for 30 min with constant gentle shaking. After a 30-min centrifugation at 18,000 g, the supernatant was removed and incubated with 2 μg/ml affinity-purified atToc159 antibody overnight. The sample was incubated with 20 μl packed protein A–Sepharose beads (CL-4B; GE Healthcare) for 2 h. The solution was removed and the beads were washed three times with 500 μl washing buffer (50 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and 0.1% PIC). The bound protein was eluted directly into SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting with atToc159 antiserum and affinity-purified atToc75 and atToc33 antibodies.
For immunoprecipitation under denaturing conditions, chloroplasts (150 μg chlorophyll) were dissolved in 2% SDS containing 2% PIC by vigorous mixing. The samples were heated for 5 min at 97°C and cooled to room temperature. The samples were diluted with 720 μl dilution buffer (50 mM Tris-HCl, pH 7.5, 1.1% Triton X-100, 167 mM NaCl, 5.6 mM EDTA, and 1% PIC) and allowed to incubate on ice for 5 min. After a 30-min centrifugation at 18,000 g at 4°C, the supernatant was divided into two equal volumes and incubated with 2 μg/ml affinity-purified atToc159 A-domain antibody or atToc33 antibody for 16 h. 20 μl of packed protein A–Sepharose was added to each sample and incubated for 2 h at 4°C with constant rotation. The bound samples were washed and analyzed by SDS-PAGE.
GTP hydrolysis
GTP hydrolysis was measured using a modified method (Liang et al., 2000). atToc159, atToc159-A864R, atToc159G1m, and atToc33G1m were in vitro translated using the TNT coupled T7 reticulocyte system (Promega) and labeled by [35S]methionine. 50 μl of each translation mixture was diluted into 500 μl HNa buffer (50 mM Hepes-KOH, pH 7.5, and 150 mM NaCl) containing 0.5% Triton X-100 and 0.5% PIC. AtToc159 antibody (for atToc159 and atToc159 mutant proteins) or His6-specific antibody (for His6-tagged soluble atToc33 G1 protein) was added to a 2 μg/ml final concentration and the mixtures were incubated overnight at 4°C with constant rotation followed by the addition of 20 μl protein A–Sepharose beads for 2 h. The beads were recovered and washed five times with 500 μl HNa buffer containing 0.1% Triton X-100. The beads were incubated with 10 mM ATP in HNa buffer for 30 min at 4°C. The atToc159G protein was expressed in E. coli and purified as described previously (Smith et al., 2002). GTP hydrolysis reactions were performed on Sepharose-bound protein as previously described (Liang et al., 2000). The immunoprecipitated proteins were analyzed by SDS-PAGE and phosphorimaging to determine the amount of immunoprecipitated GTPase used in each assay. The amounts were calculated based on the specific radioactivity of the in vitro translation products.
Chloroplast import, binding, and translocation
Chloroplasts corresponding to 10 μg chlorophyll or 25 μg of chlorophyll were used for import or binding reactions, respectively, as described previously (Chen et al., 2002). For the chase experiments, chloroplasts (200 μg chlorophyll) containing bound preprotein or the early import intermediate reactions were generated as above. Chloroplasts recovered after isolation over the 35% Percoll cushion were washed once and resuspended in import buffer. Preprotein translocation was initiated with the addition of 5 mM ATP and samples were incubated at 26°C for the times indicated. Where indicated, chloroplasts were first incubated for 5 min in the dark with 0.1 mM GTPγS. All samples were resolved by SDS-PAGE and analyzed by phosphorimaging. Equivalent amounts of chloroplasts based on chlorophyll content were loaded in all lanes. Where necessary, radioactivity from in vitro translation products was normalized to reflect differing number of methionine residues.
The kinetics of preSSu import were measured using increasing concentrations of purified E. coli expressed [35S]-preSSu. The [35S]-preSSu was denatured with 6 M urea and diluted into import buffer before the addition of chloroplasts to initiate the reaction. The maximum import velocity and apparent Km for import were calculated from by plotting vo vs. vo/[S], where vo = rate of preSSu import and [S] = total concentration of preSSu. The maximum import velocity and apparent Km were determined from the y-axis intercept and slope of the plot, respectively.
Cross-linking reactions
Chloroplast binding reactions were performed using intact chloroplasts corresponding to 200 μg of chlorophyll as described above. After dilution in ice-cold HS buffer and re-isolation through a 35% Percoll cushion, chloroplasts containing bound precursors were washed twice with HS buffer. The chloroplasts were resuspended in 475 μl HS buffer and 25 μl of DMSO-dissolved DTME (dithio-bismaleimidoethane; Thermo Fisher Scientific) was added to a final concentration of 2.5 mM. Cross-linking reactions were conducted on ice in the dark for 45 min. Glycine (50 mM final concentration in HS buffer) was then added to quench the reactions. Chloroplasts were pelleted and washed once with HS buffer before further manipulation. Immunoprecipitation reactions were performed under denaturing conditions as described above, and the amounts of precipitated receptors and bound preSSu were determined by quantitative immunoblotting and phosphorimager analysis, respectively.
Online supplemental material
Fig. S1 shows Michaelis-Menten (Fig. S1 A) and rate (Fig. S1 B) analysis for GTP hydrolysis by full-length atToc159 and atToc159-A864R compared with the isolated atToc159 G-domain and atToc159 mutants known to lack GTPase activity. Fig. S2 shows the reduced degradation of atToc159 and increased protein import efficiency of chloroplasts isolated using the optimized purification procedure. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803034/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Matthew Smith (Wilfred Laurier University, Canada), and Ming Li and Caleb Rounds (University of Massachusetts, Amherst, MA) for their technical assistance.
This work was supported by National Institutes of Health (NIH) grant GM61893 to D.J. Schnell and by the Swiss National Science Foundation (SNF) grant 3100AO-109667 to F. Kessler.
Abbreviations used in this paper: A-domain, acidic domain; G-domain, GTPase domain; M-domain, membrane domain; PIC, protease inhibitor cocktail; preSSu, precursor to the small subunit of rubisco; SSu, mature small subunit of rubisco; TIC, translocon at the inner chloroplast membrane; TOC, translocon at the outer chloroplast membrane.
References
- Arnon, D.I. 1949. Copper enzymes in isolated chloroplasts. Polyphenooxidase in Beta vulgaris. Plant Physiol. 24:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, A., A. Wardle, R. Patel, P. Dudley, S.K. Park, D. Twell, K. Inoue, and P. Jarvis. 2005. A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 138:715–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J., K. Chen, A. Hiltbunner, E. Wehrli, M. Eugster, D. Schnell, and F. Kessler. 2000. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 403:203–207. [DOI] [PubMed] [Google Scholar]
- Bauer, J., A. Hiltbrunner, P. Weibel, P.A. Vidi, M. Alvarez-Huerta, M.D. Smith, D.J. Schnell, and F. Kessler. 2002. Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 159:845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T., M. Jelic, A. Vojta, A. Radunz, J. Soll, and E. Schleiff. 2004. Preprotein recognition by the Toc complex. EMBO J. 23:520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard, J., and P. Jarvis. 2005. Recognition and envelope translocation of chloroplast preproteins. J. Exp. Bot. 56:2287–2320. [DOI] [PubMed] [Google Scholar]
- Bolter, B., T. May, and J. Soll. 1998. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441:59–62. [DOI] [PubMed] [Google Scholar]
- Brock, I.W., L. Hazell, D. Michl, W.S. Nielsen, B.L. Moller, R.G. Herrmann, R.B. Klosgen, and C. Robinson. 1993. Precursors of one integral and five lumenal thylakoid proteins are imported by isolated pea and barley thylakoids: optimisation of in vitro assays. Plant Mol. Biol. 23:717–725. [DOI] [PubMed] [Google Scholar]
- Bruce, B.D. 2001. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta. 1541:2–21. [DOI] [PubMed] [Google Scholar]
- Chen, K., X. Chen, and D.J. Schnell. 2000. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 122:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., M.D. Smith, L. Fitzpatrick, and D.J. Schnell. 2002. In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell. 14:641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and A.F. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. [DOI] [PubMed] [Google Scholar]
- Constan, D., R. Patel, K. Keegstra, and P. Jarvis. 2004. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38:93–106. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and J.J. Casal. 2004. Phenotypic characterization of a photomorphogenic mutant. Plant J. 39:747–760. [DOI] [PubMed] [Google Scholar]
- Gutensohn, M., E. Fan, S. Frielingsdorf, P. Hanner, B. Hou, B. Hust, and R.B. Klösgen. 2006. Toc, Tic, Tat et al.: structure and function of protein transport machineries in chloroplasts. J. Plant Physiol. 163:333–347. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., K. Grunig, M. Alvarez-Huerta, S. Infanger, J. Bauer, and F. Kessler. 2004. AtToc90, a new GTP-binding component of the Arabidopsis chloroplast protein import machinery. Plant Mol. Biol. 54:427–440. [DOI] [PubMed] [Google Scholar]
- Inoue, H., and M. Akita. 2008. Three sets of translocation intermediates are formed during the early stage of protein import into chloroplasts. J. Biol. Chem. 283:7491–7502. [DOI] [PubMed] [Google Scholar]
- Ivanova, Y., M.D. Smith, K. Chen, and D.J. Schnell. 2004. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell. 15:3379–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P. 2004. Organellar proteomics: chloroplasts in the spotlight. Curr. Biol. 14:R317–R319. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., L.-J. Chen, H. Li, C.A. Peto, C. Fankhauser, and J. Chory. 1998. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 282:100–103. [DOI] [PubMed] [Google Scholar]
- Jelic, M., N. Sveshnikova, M. Motzkus, P. Horth, J. Soll, and E. Schleiff. 2002. The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol. Chem. 383:1875–1883. [DOI] [PubMed] [Google Scholar]
- Jelic, M., J. Soll, and E. Schleiff. 2003. Two Toc34 homologues with different properties. Biochemistry. 42:5906–5916. [DOI] [PubMed] [Google Scholar]
- Kessler, F., and D.J. Schnell. 2004. Chloroplast protein import: solve the GTPase riddle for entry. Trends Cell Biol. 14:334–338. [DOI] [PubMed] [Google Scholar]
- Kessler, F., and D.J. Schnell. 2006. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 7:248–257. [DOI] [PubMed] [Google Scholar]
- Kessler, F., G. Blobel, H.A. Patel, and D.J. Schnell. 1994. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 266:1035–1039. [DOI] [PubMed] [Google Scholar]
- Kouranov, A., and D.J. Schnell. 1997. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139:1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., H. Wang, and D.J. Schnell. 1999. Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274:25181–25186. [DOI] [PubMed] [Google Scholar]
- Kubis, S., A. Baldwin, R. Patel, A. Razzaq, P. Dupree, K. Lilley, J. Kurth, D. Leister, and P. Jarvis. 2003. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell. 15:1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis, S., R. Patel, J. Combe, J. Bedard, S. Kovacheva, K. Lilley, A. Biehl, D. Leister, G. Rios, C. Koncz, and P. Jarvis. 2004. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 16:2059–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.H., S.J. Kim, Y.J. Lee, J.B. Jin, and I. Hwang. 2003. The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 278:36794–36805. [DOI] [PubMed] [Google Scholar]
- Li, H.M., M.M. Kesavulu, P.H. Su, Y.H. Yeh, and C.D. Hsiao. 2007. Toc GTPases. J. Biomed. Sci. 14:505–508. [DOI] [PubMed] [Google Scholar]
- Li, M., and D.J. Schnell. 2006. Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J. Cell Biol. 175:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z., T. Mather, and G. Li. 2000. GTPase mechanism and function: new insights from systematic mutational analysis of the phosphate-binding loop residue Ala30 of Rab5. Biochem. J. 346:501–508. [PMC free article] [PubMed] [Google Scholar]
- Ma, Y., A. Kouranov, S. LaSala, and D.J. Schnell. 1996. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne, J., and J.R. Botella. 1998. Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol. Biol. Rep. 16:257–262. [Google Scholar]
- Neuhaus, H.E., and M.J. Emes. 2000. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:111–140. [DOI] [PubMed] [Google Scholar]
- Olsen, L.J., and K. Keegstra. 1992. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267:433–439. [PubMed] [Google Scholar]
- Olsen, L.J., S.M. Theg, B.R. Selman, and K. Keegstra. 1989. ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 264:6724–6729. [PubMed] [Google Scholar]
- Perry, S.E., and K. Keegstra. 1994. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 6:93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., and D. Leister. 2004. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 329:11–16. [DOI] [PubMed] [Google Scholar]
- Rounds, C., F. Wang, and D.J. Schnell. 2007. The toc machinery of the protein import apparatus of chloroplasts. In The Enzymes: Molecular Machines involved in Protein Transport across Cellular Membranes. Vol. 25. F. Tamanoi, R.E. Dalbey, and C.M. Koehler, editors. Academic Press. 415–438.
- Schleiff, E., J. Soll, N. Sveshnikova, R. Tien, S. Wright, C. Dabney-Smith, C. Subramanian, and B.D. Bruce. 2002. Structural and guanosine triphosphate/diphosphate requirements for transit peptide recognition by the cytosolic domain of the chloroplast outer envelope receptor, toc34. Biochemistry. 41:1934–1946. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., M. Jelic, and J. Soll. 2003. A GTP-driven motor moves proteins across the outer envelope of chloroplasts. Proc. Natl. Acad. Sci. USA. 100:4604–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell, D.J., and G. Blobel. 1993. Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. J. Cell Biol. 120:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, A., J. Knoetzel, H.V. Scheller, and A. Mant. 2004. Uptake of a fluorescent dye as a swift and simple indicator of organelle intactness: import-competent chloroplasts from soil-grown Arabidopsis. J. Histochem. Cytochem. 52:701–704. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., K. Waegemann, and J. Soll. 1995. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 7:401–411. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., A. Hiltbrunner, F. Kessler, and D.J. Schnell. 2002. The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 159:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.D., C.M. Rounds, F. Wang, K. Chen, M. Afitlhile, and D.J. Schnell. 2004. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165:323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveshnikova, N., J. Soll, and E. Schleiff. 2000. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. USA. 97:4973–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallas, T.R., M.D. Smith, S. Sanchez-Nieto, and D.J. Schnell. 2003. The roles of toc34 and toc75 in targeting the toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 278:44289–44297. [DOI] [PubMed] [Google Scholar]
- Young, M.E., K. Keegstra, and J.E. Froehlich. 1999. GTP promotes the formation of early-import intermediates but is not required during the translocation step of protein import into chloroplasts. Plant Physiol. 121:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.