Abstract
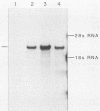
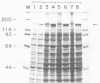
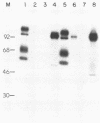
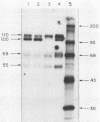
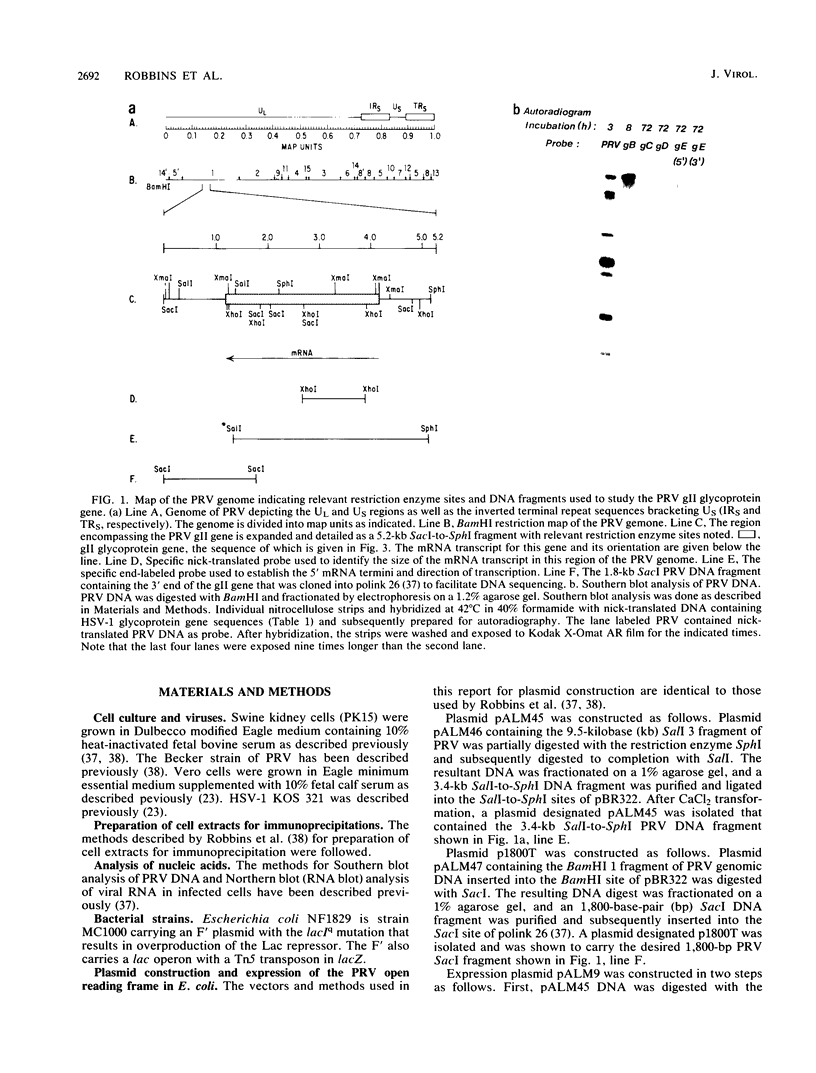
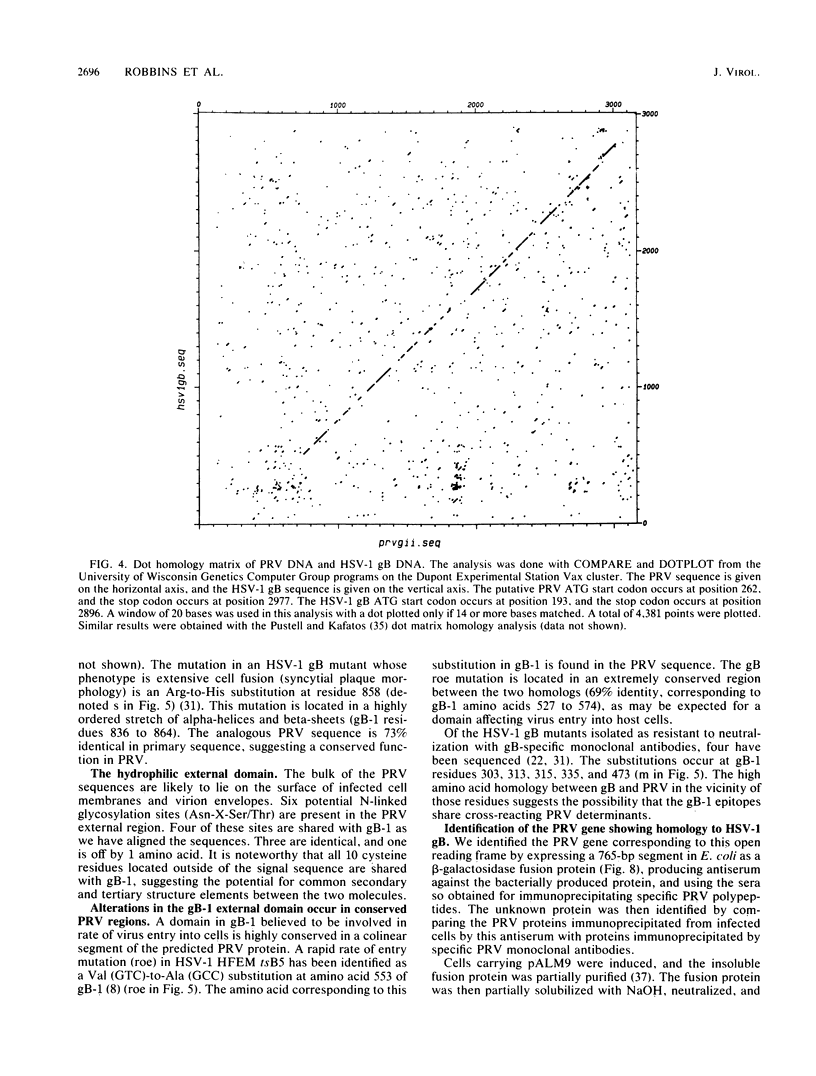
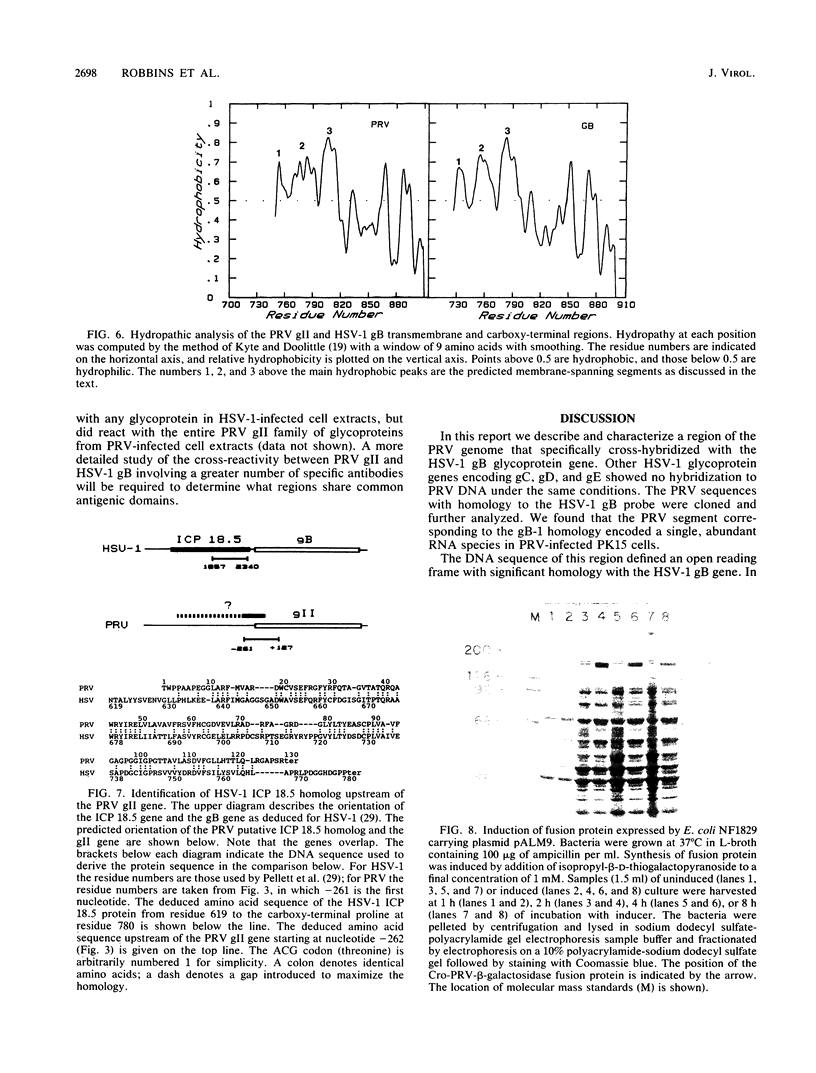
We have looked for conserved DNA sequences between four herpes simplex virus type 1 (HSV-1) glycoprotein genes encoding gB, gC, gD, and gE and pseudorabies virus (PRV) DNA, HSV-1 DNA fragments representing these four glycoprotein-coding sequences were hybridized to restriction enzyme fragments of PRV DNA by the Southern blot procedure. Specific hybridization was observed only when HSV-1 gB DNA was used as probe. This region of hybridization was localized to a 5.2-kilobase (kb) region mapping at approximately 0.15 map units on the PRV genome. Northern blot (RNA blot) analysis, with a 1.2-kb probe derived from this segment, revealed a predominant hybridizing RNA species of approximately 3 kb in PRV-infected PK15 cells. DNA sequence analysis of the region corresponding to this RNA revealed a single large open reading frame with significant nucleotide homology with the gB gene of HSV-1 KOS 321. In addition, the beginning of the sequenced PRV region also contained the end of an open reading frame with amino acid homology to HSV-1 ICP 18.5, a protein that may be involved in viral glycoprotein transport. This sequence partially overlaps the PRV gB homolog coding sequence. We have shown that the PRV gene with homology to HSV-1 gB encoded the gII glycoprotein gene by expressing a 765-base-pair segment of the PRV open reading frame in Escherichia coli as a protein fused to beta-galactosidase. Antiserum, raised in rabbits, against this fusion protein immunoprecipitated a specific family of PRV glycoproteins of apparent molecular mass 110, 68, and 55 kilodaltons that have been identified as the gII family of glycoproteins. Analysis of the predicted amino acid sequence indicated that the PRV gII protein shares 50% amino acid homology with the aligned HSV-1 gB protein. All 10 cysteine residues located outside of the signal sequence, as well as 4 of 6 potential N-linked glycosylation sites, were conserved between the two proteins. The primary protein sequence for HSV-1 gB regions known to be involved in the rate of virus entry into the cells and cell-cell fusion, as well as regions known to be associated with monoclonal antibody resistance, were highly homologous with the PRV protein sequence. Furthermore, monospecific antibody made against PRV gII immunoprecipitated HSV-1 gB from infected cells. Taken together, these findings suggest significant conservation of structure and function between the two proteins and may indicate a common evolutionary history.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., DeMarchi J., Pendrys J., Veach R. A., Kaplan A. S. Proteins specified by the short unique region of the genome of pseudorabies virus play a role in the release of virions from certain cells. J Virol. 1986 Jan;57(1):191–196. doi: 10.1128/jvi.57.1.191-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Kaplan A. S. Synthesis of proteins in cells infected with herpesvirus. V. Viral glycoproteins. Virology. 1970 Jun;41(2):265–273. doi: 10.1016/0042-6822(70)90078-4. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984 Aug;137(1):185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Claesson-Welsh L., Spear P. G. Oligomerization of herpes simplex virus glycoprotein B. J Virol. 1986 Nov;60(2):803–806. doi: 10.1128/jvi.60.2.803-806.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson C. M., Hosler B. A., Respess R. A., Waters D. J., Thorley-Lawson D. A. Cross-reactivity between herpes simplex virus glycoprotein B and a 63,000-dalton varicella-zoster virus envelope glycoprotein. J Virol. 1985 Oct;56(1):333–336. doi: 10.1128/jvi.56.1.333-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl H., Ben-Porat T., Ehrlicher L., Habermehl K. O., Kaplan A. S. Characterization of the envelope proteins of pseudorabies virus. J Virol. 1984 Nov;52(2):583–590. doi: 10.1128/jvi.52.2.583-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland T. C., Homa F. L., Marlin S. D., Levine M., Glorioso J. Herpes simplex virus type 1 glycoprotein C-negative mutants exhibit multiple phenotypes, including secretion of truncated glycoproteins. J Virol. 1984 Nov;52(2):566–574. doi: 10.1128/jvi.52.2.566-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killington R. A., Yeo J., Honess R., Watson D. H., Duncan B. E., Halliburton I. W., Mumford J. Comparative analyses of the proteins and antigens of five herpesviruses. J Gen Virol. 1977 Nov;37(2):297–310. doi: 10.1099/0022-1317-37-2-297. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee G. T., Para M. F., Spear P. G. Location of the structural genes for glycoproteins gD and gE and for other polypeptides in the S component of herpes simplex virus type 1 DNA. J Virol. 1982 Jul;43(1):41–49. doi: 10.1128/jvi.43.1.41-49.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukàcs N., Thiel H. J., Mettenleiter T. C., Rziha H. J. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J Virol. 1985 Jan;53(1):166–173. doi: 10.1128/jvi.53.1.166-173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin S. D., Highlander S. L., Holland T. C., Levine M., Glorioso J. C. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J Virol. 1986 Jul;59(1):142–153. doi: 10.1128/jvi.59.1.142-153.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin S. D., Holland T. C., Levine M., Glorioso J. C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985 Jan;53(1):128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mettenleiter T. C., Lukacs N., Rziha H. J. Mapping of the structural gene of pseudorabies virus glycoprotein A and identification of two non-glycosylated precursor polypeptides. J Virol. 1985 Jan;53(1):52–57. doi: 10.1128/jvi.53.1.52-57.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T. C., Lukàcs N., Thiel H. J., Schreurs C., Rziha H. J. Location of the structural gene of pseudorabies virus glycoprotein complex gII. Virology. 1986 Jul 15;152(1):66–75. doi: 10.1016/0042-6822(86)90372-7. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachl C., Burke R. L., Stuve L. L., Sanchez-Pescador L., Van Nest G., Masiarz F., Dina D. Expression of cell-associated and secreted forms of herpes simplex virus type 1 glycoprotein gB in mammalian cells. J Virol. 1987 Feb;61(2):315–325. doi: 10.1128/jvi.61.2.315-325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Jenkins F. J., Ackermann M., Sarmiento M., Roizman B. Transcription initiation sites and nucleotide sequence of a herpes simplex virus 1 gene conserved in the Epstein-Barr virus genome and reported to affect the transport of viral glycoproteins. J Virol. 1986 Dec;60(3):1134–1140. doi: 10.1128/jvi.60.3.1134-1140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Kousoulas K. G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985 Jan;53(1):243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Petrovskis E. A., Timmins J. G., Armentrout M. A., Marchioli C. C., Yancey R. J., Jr, Post L. E. DNA sequence of the gene for pseudorabies virus gp50, a glycoprotein without N-linked glycosylation. J Virol. 1986 Aug;59(2):216–223. doi: 10.1128/jvi.59.2.216-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovskis E. A., Timmins J. G., Post L. E. Use of lambda gt11 to isolate genes for two pseudorabies virus glycoproteins with homology to herpes simplex virus and varicella-zoster virus glycoproteins. J Virol. 1986 Oct;60(1):185–193. doi: 10.1128/jvi.60.1.185-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea T. J., Timmins J. G., Long G. W., Post L. E. Mapping and sequence of the gene for the pseudorabies virus glycoprotein which accumulates in the medium of infected cells. J Virol. 1985 Apr;54(1):21–29. doi: 10.1128/jvi.54.1.21-29.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Watson R. J., Whealy M. E., Hays W. W., Enquist L. W. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex virus type 1 and type 2 glycoprotein C. J Virol. 1986 May;58(2):339–347. doi: 10.1128/jvi.58.2.339-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins A. K., Weis J. H., Enquist L. W., Watson R. J. Construction of E. coli expression plasmid libraries: localization of a pseudorabies virus glycoprotein gene. J Mol Appl Genet. 1984;2(5):485–496. [PubMed] [Google Scholar]
- Robbins A. K., Whealy M. E., Watson R. J., Enquist L. W. Pseudorabies virus gene encoding glycoprotein gIII is not essential for growth in tissue culture. J Virol. 1986 Sep;59(3):635–645. doi: 10.1128/jvi.59.3.635-645.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden B. W., Kinchington P. R., Powell K. L., Halliburton I. W. Antigenic and biochemical analysis of gB of herpes simplex virus type 1 and type 2 and of cross-reacting glycoproteins induced by bovine mammillitis virus and equine herpesvirus type 1. J Gen Virol. 1985 Feb;66(Pt 2):231–247. doi: 10.1099/0022-1317-66-2-231. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Straus S. E., Aulakh H. S., Ruyechan W. T., Hay J., Casey T. A., Vande Woude G. F., Owens J., Smith H. A. Structure of varicella-zoster virus DNA. J Virol. 1981 Nov;40(2):516–525. doi: 10.1128/jvi.40.2.516-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuve L. L., Brown-Shimer S., Pachl C., Najarian R., Dina D., Burke R. L. Structure and expression of the herpes simplex virus type 2 glycoprotein gB gene. J Virol. 1987 Feb;61(2):326–335. doi: 10.1128/jvi.61.2.326-335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen M. W., Wathen L. M. Isolation, characterization, and physical mapping of a pseudorabies virus mutant containing antigenically altered gp50. J Virol. 1984 Jul;51(1):57–62. doi: 10.1128/jvi.51.1.57-62.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]