Abstract
Background
We present the first population genetic analysis of homologous loci from two sympatric human malaria parasite populations sharing the same human hosts, using full-length sequences of ama1 genes from Plasmodium vivax and P. falciparum collected in the Venezuelan Amazon.
Methodology/Principal Findings
Significant differences between the two species were found in genetic diversity at the ama1 locus, with 18 distinct haplotypes identified among the 73 Pvama1 sequences obtained, compared to 6 unique haplotypes from 30 Pfama1 sequences, giving overall diversity estimates of h = 0.9091, and h = 0.538 respectively. Levels of recombination were also found to differ between the species, with P. falciparum exhibiting very little recombination across the 1.77kb sequence. In contrast, analysis of patterns of nucleotide substitutions provided evidence that polymorphisms in the ama1 gene of both species are maintained by balancing selection, particularly in domain I. The two distinct population structures observed are unlikely to result from different selective forces acting upon the two species, which share both human and mosquito hosts in this setting. Rather, the highly structured P. falciparum population appears to be the result of a population bottleneck, while the much less structured P. vivax population is likely to be derived from an ancient pool of diversity, as reflected in a larger estimate of effective population size for this species. Greatly reduced mosquito transmission in 1997, due to low rainfall prior to the second survey, was associated with far fewer P. falciparum infections, but an increase in P. vivax infections, probably due to hypnozoite activation.
Conclusions/Significance
The relevance of these findings to putative competitive interactions between these two important human pathogen species is discussed. These results highlight the need for future control interventions to employ strategies targeting each of the parasite species present in endemic areas.
Introduction
Plasmodium vivax infection causes 132–391 million cases each year worldwide, with a significant, but under-reported burden of severe malaria requiring hospitalisation [1]. Vivax malaria is endemic in both tropical and some temperate regions and it is estimated that 2.6 billion people are at risk from infection each year [2]. P. vivax is sympatric with P. falciparum in many regions often with P. ovale and/or P. malariae also [3]. The majority of laboratory and field studies of human malaria have been concerned with P. falciparum, due to the development of methods for continuous in vitro culture of this species, and the large number of genetic markers available [1], [4]. However the current elucidation of the P. vivax genome, and the recent development of improved in vitro drug sensitivity testing protocols [5] are cause for optimism regarding future research into this important human pathogen.
Control regimes targeting any particular malaria species must also consider knock-on effects this may have on other sympatric Plasmodium species. The effects a circulating patent species may have on liver-stage or sub-patent blood-stage infection by other species remains unclear. Working in Vanuatu, Maitland et al. [6] found that although the individual prevalence of P. vivax and P. falciparum fluctuated, the overall burden of malaria remained constant, suggesting that there was direct interaction between the two species. In regions where more than one species co-circulate, it has also been observed that fewer than expected mixed infections occur [6], [7], [8]. A suggested explanation for these observations is that an erythrocytic P. falciparum infection actively suppresses subsequent P. vivax infections that emerge from the liver [7] through non-specific effectors such as cytokines, and cross-species immune responses [7], [9]. When this suppression is lifted, either through drug treatment or host-mediated responses, there can be a swift expansion of P. vivax within the host [10]. Although a particular species, such as P. falciparum, may be reduced in prevalence or even eradicated in a particular setting, other malaria species, such as P. vivax, may survive the intervention through the persistence of dormant liver-stage hypnozoites, and subsequently proliferate through the human population to occupy the niche vacated by the targeted species. Thus although mortality due to P. falciparum may be reduced by such an intervention, malaria morbidity may continue; merely the species-specific dynamics within the region have been altered. In the light of drug resistance in P. vivax, [11], [12] and also possibly P. malariae [13], [14], control measures for the future that include novel drug and vaccine development should therefore consider all local sympatric Plasmodium spp. [3], [15].
To be effective in areas with multiple Plasmodium species, vaccine-based interventions would ideally generate protective responses against clinical episodes from each species present. Much vaccine development has focused on those antigens expressed during the merozoite stage, the form that invades circulating erythrocytes and reticulocytes during clinical stages of infection. Merozoite antigens are under considerable investigation as potential vaccine candidates despite extensive sequence variation within and between populations, as there is some evidence that antibodies raised to one variant are cross-reactive with other variants from the same species [16], [17]. Whether this cross-reactivity also indicates cross-isolate protection in subsequent infection with different variants is not known. A current vaccine candidate for both P. falciparum and P. vivax is the merozoite-expressed apical membrane antigen (AMA1). Widespread variation of the genes encoding AMA1 proteins from different populations and in more than one species have been reported, providing evidence that this gene, ama1, is under significant selection pressures in natural populations [15], [18], [19]. A number of studies have investigated polymorphism within Pfama1 to assist development of anti-AMA1 vaccines able to elicit broad protective responses [18], [20], and this has lead to the development of multiple Pfama1 antigens for simultaneous vaccination in order to overcome this diversity [17]. Analyses of diversity have also been carried out Pvama1, but these have used parasite isolates collected at hospital clinics, rather than cross-sectional P. vivax population samples [3], [15], [21]. There are no studies that examine ama1 diversity simultaneously in both species in a setting where P. vivax and P. falciparum are sympatric.
We have been engaged in analysis of genetic diversity in malaria parasite populations circulating among forest-dwelling communities along the Padamo River basin in southern Venezuela. These studies have shown that the P. falciparum population in this area exhibits inter-genic linkage disequilibrium, and is dominated by a small number of multi-locus genotypes, whereas the P. vivax population appears more diverse [22], [23], [24]. Here we present full-length sequence analysis of Pvama1 and Pfama1 from cross-sectional surveys of malaria parasites co-circulating among the human population between 1995 and 1997. We use these homologous sequence datasets to directly compare genetic structure between the two parasite populations, to contrast selective forces acting upon the two genes, and to examine any evidence of direct interplay between the two species in this setting of mesoendemic, sympatric transmission among two geographically defined parasite populations sharing the same human hosts.
Materials and Methods
Ethical approval for the original survey was obtained from the Ethics committee of the Venezuelan Ministry of Health, and the Ethics Committee of the London School of Hygiene & Tropical Medicine in 1995. The study protocol was also approved by CAICET (Centro Amazónico de Investigación y Control de Enfermedades Tropicales); the Dirección de Malariologa y Saneamiento Ambiental and the Regional Health Service, each institution providing a written statement of approval and support. Oral and written informed consent was obtained from the community leaders once the study was explained to the assembled community population, one community at a time.
Sample collection
Between October 1995 and November 1997, blood samples and census data from all individuals in nine villages along the Padamo River, Amazon Basin, Venezuela, were collected in each of two cross-sectional surveys [23]. Due to the low P. falciparum prevalence found in 1997, the boundaries of the survey were extended in the second survey to include an additional large village. More than 90% of the surveyed population gave informed consent to participate: 708 individuals in 1996 and 945 in 1997. Blood samples from all participants were obtained for microscopy and collected onto filter paper for malarial genetic analyses. Individuals shown to have malaria parasites were treated accordingly, as described [23].
DNA extraction
DNA was extracted from all available filter paper blood-spots corresponding to slide-positive samples as previously described [25]. Sufficient P. falciparum DNA samples for amplification and sequencing were not available from the 1997 survey. Samples that did not easily or consistently amplify were re-extracted using a commercially available kit (Qiagen) and this DNA source was used instead.
Pvama1 and Pfama1 amplification and sequencing
Pvama1-specific primers were designed based on the published P. vivax sequence (GENBANK accession L27504) to amplify 1663 bp in a nested PCR approach. Previously published Pfama1-specific primers [19] were used in a nested PCR to amplify 1770 bp of the Pfama1 gene. PCR positive samples were re-amplified in a 50 µl nest 2 reaction which was prepared for direct sequencing by ethanol precipitation into 20 µl. New sequencing primers for Pvama1 were designed based on L27504. Previously published sequencing primers for Pfama1 [19] were used in conjunction with amplification primers to determine full-length sequences (Perkin-Elmer BigDye 3.1). Sequencing reactions were cleaned by precipitation in 0.3 M NaAc, 125 mM EDTA and 2.2 volumes of ethanol, and analysed on an ABI prism 3730 automated capillary sequencer. Amplification and sequencing primers used in this study are presented in Table 1, with the PCR conditions used.
Table 1. PCR primers and conditions.
| Nest 1 | Nest 2 | |||
| Pvama1 1663 bp | Pvama1_F16 | 5′ gcg gtt act tcc acc cc 3′ | Pvama1_F29 | 5′ gca aac caa atc gct gcc 3′ |
| Pvama1_R613 | 5′ gcg tgg tgt ggg agg ccc 3′ | Pvama1_R598 | 5′ gcc tcg ggg tcg agc atc tcg tc 3′ | |
| Nest 1 PCR: | 94°C3 min×1 cycle, (94°C1 min−46°C1 min−72°C2.5 min)×40 cycles, 72°C10 min×1 | Nest 2 PCR: | 94°C3 min×1 cycle, (94°C1 min−57°C1 min–72°C2.5 min)×40 cycles, 72°C10 min×1 | |
| Fwd sequencing: | Rev sequencing | |||
| Pvama1_F142 | aga att cca gct gga aga tg | Pvama1_R138 | ttc cac ttc tgc atc ttc ccc | |
| Pvama1_F301 | cgt aaa aat tta gga aac gcc | Pvama1_R293 | aca ttt ttg ctc aaa tac acc | |
| Pvama1_F441 | ccc ctg cag cat ata taa aga c | Pvama1_R442 | ggg gaa atc ccg gtc tac ttc | |
| Pfama1 1770 bp | Pfama1_F5* | 5′ tgc gta tta tta ttg agc 3′ | Pfama1_F10* | 5′ gag cgc ctt tga gtt tac 3′ |
| Pfama1_R613* | 5′ gtg ttg tat gtg atg ctc 3′ | Pfama1_R598* | 5′ gcc tca gga tct aac att tca tc 3′ | |
| Nest 1 PCR: | 94°C3 min×1 cycle, (94°C1 min−46°C1 min−72°C2.5 min)×40 cycles, 72°C10 min×1 | Nest 2 PCR: | 94°C3 min×1 cycle, (94°C1 min−60°C1 min−72°C2.5 min)×40 cycles, 72°C10 min×1 | |
| Fwd sequencing: | Rev sequencing: | |||
| Pfama1_F143 | gac ttc cat cag gga aat gtc c | Pfama1_R188 | gag gtt ctg ttg gag gaa aag c | |
| Pfama1_F312 | cgg att atg ggt cga tgg aaa ttg | Pfama1_R350 | tta ggt tga tcc gaa gca ctc aa | |
| Pfama1_F485 | gac agt tta aaa tgc cca tgt gc | Pfama1_R492 | cac atg ggc att tta aac tgt c | |
(Polley & Conway, 2001).
Editing and assembling of sequence products
Each sequence from the same isolate was assembled and initially edited into a full-length contig with at least double coverage using SeqMan™II, and EditSeq™ (DNAStar Inc, Madison, WI). Secondary editing was done to confirm polymorphisms only occurring once (singletons) within the population using MegAlign™ (DNAStar Inc). Sequencing was repeated to confirm singletons and for isolates that did not have full-length double reads. A single ama1 contig could not be determined for two isolates (one P. falciparum and one P. vivax). DNA was re-extracted and each was fully processed independently. In both cases, the two DNA extractions each yielded two different sequences that were already represented in the population, making them unlikely to be PCR artefacts, but rather indicating the infections were comprised of more than one genotype of that respective species. Confirmed single genotype data was exported as a PAUP alignment for statistical analyses, computed using DnaSP v.4.10 software.
Within population analyses
In the 1997 survey, an extra village to the south of the main survey region, Koshirowetheri, was included for data collection and contributed a further 250 participants. To analyse data between the two years on a like-for-like basis, sequences from Koshirowetheri were at first excluded. Sequences of Pvama1 from the original survey region from 1997 and those from Koshirowetheri in 1997 were subsequently compared by tests of departure from neutrality and analysis of within-population diversity to identify any significant differences between them that would preclude all Pvama1 sequences obtained in the 1997 survey being treated as a single dataset.
The binomial probability was used to test whether the number of different haplotypes, and the relative frequency of the most common haplotype in each case, was significantly different between the Pfama1 and Pvama1 datasets. As 6 distinct Pfama1 haplotypes (successes) were described in 30 sequences (trials) the probability of success in this dataset is 0.20. The probability that the observed number of successes in the Pvama1 dataset was within a 2-sided 95% confidence interval of 0.20 was then calculated. Similarly, as 20 of 30 Pfama1 sequences were identical, we tested whether the frequency of the most common Pvama1 haplotype was significantly different to 0.667.
Tests to determine any significant departure of variation from neutrality using Tajima's D [26] and Fu and Li's D* and F* [27] indices were performed in DNAsp v. 4.10 [28] on each gene population as a whole and via a sliding window approach. Tajima's D tests the departure from neutral by comparing the estimations of nucleotide diversity (θ) derived from the average pairwise diversity (π) and the total number of polymorphic sites. The Fu and Li tests identify departures from neutral patterns of nucleotide substitutions as deviations between the estimates of θ derived from the number of phylogenies compared to either the total number of mutations (D*) or the average pairwise diversity (F*). Analysis of recombination and linkage disequilibrium was performed to calculate the minimum number of recombination events within each of the Pvama1 and Pfama1 sequences [29] and to give an estimation of the recombination parameter, C [30]. The indices of linkage disequilibrium, D' [31] and R2 [32] were also determined and their relationship with distance between sites was plotted.
Between population analyses
The Mc Donald-Kreitman test, using either P. cynomolgi (accession number X86099) or P. knowlesi (accession numbers AF298218, M58317, M61097) for outgroup comparisons with the P. vivax populations, and P. reickenowi (accession number AJ252087) for comparisons with P. falciparum, allows for determination of the ratios of synonymous and nonsynonymous changes between and within species. Between-population divergence of the Pvama1 data presented here and previously published Pvama1 data for Domain I (DI) from global sites [15], [33], [34], [35] was determined using the θ-estimator of Wright's fixation index (FST) to determine the relative contribution of the differences observed between each population to the overall diversity seen using Arlequin v.3.11 software [36]. As only two previously published studies [15], [35] allow comparisons across the whole Pvama1 ectodomain, FST values in our dataset were determined across the whole gene and for each of the domains separately. Mean FST values for the Venezuelan Pfama1 sequences were compared with two published Pfama1 data sets [18], [19] that each includes all three domains, allowing determination of mean FST values for the full gene and each of the three domains separately.
Results
Parasite samples
In the first survey, light microscopy determined 40 P. vivax, 32 P. falciparum, 16 P. malariae and 2 mixed infections (one P. vivax with P. malariae and one P. vivax with P. falciparum), out of 708 individuals. In the second survey of 945 individuals, microscopy determined 92 P. vivax, 12 P. falciparum, 20 P. malariae and 3 mixed infections of P. vivax with P. malariae. The overall malaria prevalence did not vary between surveys being 12.7% in 1995/6 and 13.4% in 1997. However, vivax malaria comprised a significantly greater proportion of malaria infections in the second survey (O.R. 3.39, 95% C.I. 1.83–6.29; P<0.0001), whereas falciparum was significantly less common (O.R. 0.180, 95% C.I. 0.079–0.394; P<0.0001). Full-length ama1 contigs with double sequence reads were obtained for 73 P. vivax and 30 P. falciparum isolates (GenBank accession nos. EU346015-EU346087 for Pvama1, and EU332414-EU332443 for Pfama1). All sequences encoded cysteine residues in previously described positions. Neither Pvama1 nor Pfama1 populations contained sequences identical to their respective reference strains; Sal-1 (gene ID Pv092275 from http://www.PlasmoDB.org) and 3D7 (accession number XM_001347979).
Within population analyses
Diversity among Pvama1 sequences was first analysed within comparable groups: those from the 9 villages surveyed in 1995/6 (28 sequences), those collected from these 9 villages in the second survey (12 sequences), and sequences collected from Koshirowetheri village in the second survey (33 sequences). Diversity of the Pfama1 gene was analysed in sequences from the 9 villages in the first survey only (30 sequences).
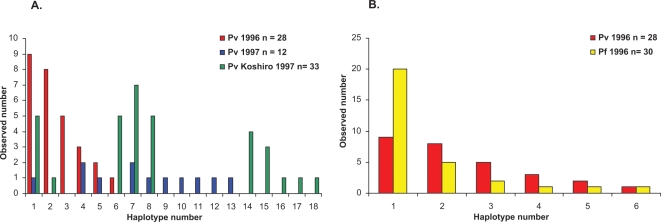
Genetic diversity, as measured by h, is greater among the Pvama1 sequences than among the Pfama1 sequences (Table 2). This remains true when isolates from Koshirowetheri are excluded from the analysis, supporting the conclusion that there is a significant increase in the circulating diversity of P. vivax at the Pvama1 locus between the two years (Figure 1). Of the 73 Pvama1 sequences obtained, there were 18 unique haplotypes, compared to 6 unique haplotypes from 30 Pfama1 sequences (2-sided binomial probability, P = 0.308). However, among the 30 Pfama1 sequences 20 were of the most common haplotype, whereas among the 73 Pvama1 sequences 15 were of the most common haplotype (P<0.001). The overall diversity estimates for Pvama1 and Pfama1 (h = 0.9091, h = 0.538 respectively) are consistent with our previous studies from this region using rif, Pfglurp, Pfmsp1 and Pfmsp2 genes for P. falciparum, and using Pvmsp3α for P. vivax [22], [23], and demonstrate greater diversity in the P. vivax population.
Table 2. Within-population analyses of Pvama1 and Pfama1 from the Venezuelan Amazon.
| P. falciparum ama1 | P. vivax ama1 | |||
| 1996 | Villages 1–9 in 1996 | Villages 1–9 in 1997 | 1997 Koshirowetheri | |
| Number of sequences | 30 | 28 | 12 | 33 |
| Number of haplotypes | ||||
| Whole gene | 6 | 6 | 10 | 10 |
| Domain I | 5 | 5 | 6 | 9 |
| Domain II | 2 | 3 | 4 | 3 |
| Domain III | 3 | 2 | 3 | 2 |
| Gene diversity, h | ||||
| Whole gene | 0.54 | 0.80 | 0.97 | 0.89 |
| Domain I | 0.45 | 0.78 | 0.68 | 0.86 |
| Domain II | 0.37 | 0.58 | 0.74 | 0.48 |
| Domain III | 0.42 | 0.50 | 0.59 | 0.41 |
| Tajima's D | 1.73927* | 1.00507** | −0.48752 | 1.82661 |
| Fu and Li's D* | 1.72279*** | 0.71243** | −0.53615 | 1.72312*** |
| Fu and Li's F* | 2.03574*** | 0.94666** | −0.59625 | 2.07362*** |
| Recombination parameter, C (Between adjacent sites) | 0.001 (0.000) | 10.0 (0.0061) | 14.7 (0.0090) | 16.9 (0.0103) |
| Effective population size, Ne † | 4.2×102 | 4.2×106 | 6.1×106 | 7.0×106 |
| Minimum Recombination events, Rm | 3 | 1 | 4 | 8 |
* = 0.10>P>0.05; ** = P>0.10; *** = P<0.02.
= Effective population size, Ne, = C/4r, where r is the recombination rate and r of P. falciparum is used for all, approximately 6×10−7, as the recombination rate for P. vivax has not yet been determined (Joy et al., 2006).
Figure 1. Graphical representation of the distributions of the Pvama1 and Pfama1 allele frequencies from the Venezuelan Amazon.
A. Distribution of 18 Pvama1 haplotypes across surveys in 1996 and 1997. Sequences from the additional village sampled in 1997 (Koshiro) are presented separately from those of the villages sampled in both surveys. B. Haplotype frequency distribution for Pfama1 and Pvama1 from the 1996 survey only.
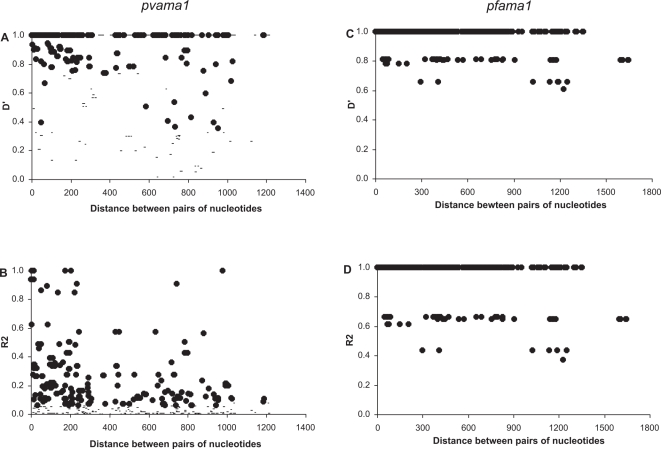
The frequency of recombination within the Pvama1 population is relatively high compared to the predicted frequency within the Pfama1 population (10 minimum recombination events within 73 Pvama1 sequences vs. 3 in 30 Pfama1 sequences; Table 3), yet both of these are low compared to estimates from other regions. Among 23 Pvama1 sequences from Sri Lanka, the estimated minimum number of recombination events was 9 [15]. For Pfama1, analysis of 50 isolates from Nigeria and 51 from Thailand provided estimates of the minimum number of recombination events of 25 and 16 respectively [18], [19]. Our linkage disequilibrium analysis indicates declining inter-site linkage with increasing nucleotide distance within the Pvama1 sequences, in contrast to the significant linkage across the whole of the Pfama1 sequences (Figure 2.). The very low value of the Pfama1 recombination parameter, C = 0.001, and the observed maintenance of significant linkage suggests very little meiotic recombination occurs in this gene in our population, supporting the view that clonal propagation of P. falciparum has occurred over a significant period of time [23]. This is supported by our estimation of en effective population size for P. vivax that is 4 orders of magnitude higher than that for P. falciparum (Table 2).
Table 3. McDonald-Kreitman analyses of Pvama1 and Pfama1.
| Region | Polymorphic changes within species | Polymorphic changes within species | Fixed differences between species | Fixed differences between species | Neutrality index1 | Fisher's exact test2 | |
| Synonymous | Nonsynonymous | Synonymous | Nonsynonymous | ||||
| P. vivax vs | Whole | 6 | 21 | 119 | 69 | 6.04 | 0.000095** |
| P. cynomolgi | DI | 3 | 11 | 35 | 19 | 6.75 | 0.0057* |
| DII | 2 | 2 | 21 | 11 | 1.91 | 0.61 | |
| DIII | 0 | 2 | 12 | 10 | - | 0.48 | |
| P. vivax vs | Whole | 9 | 22 | 130 | 82 | 3.88 | 0.00086** |
| P. knowlesi | DI | 4 | 12 | 37 | 21 | 5.29 | 0.0095* |
| DII | 2 | 2 | 20 | 21 | 1.67 | 1.00 | |
| DIII | 1 | 2 | 15 | 3 | 10.00 | 0.13 | |
| P. falciparum vs | Whole | 0 | 29 | 19 | 53 | - | 0.0013* |
| P. reickenowi | DI | 0 | 14 | 4 | 10 | - | 0.098 |
| DII | 0 | 5 | 3 | 4 | - | 0.20 | |
| DIII | 0 | 3 | 1 | 5 | - | 1.00 |
= Neutrality index indicates the extent to which the levels of amino acid polymorphism depart from the expected in the neutral model.
= Fisher's exact test.
*0.001<P<0.01; ** P<0.001.
Figure 2. Linkage disequilibrium indices of Pvama1 and Pfama1.
Linkage disequilibrium plots of D' and R2 for Pvama1 (Graphs A and B) and Pfama1 (Graphs C and D). Sites with significant linkage (P<0.05) are shown as solid circles; non-significant sites are shown as for Pvama1 only. No non-significant sites for Pfama1 were found.
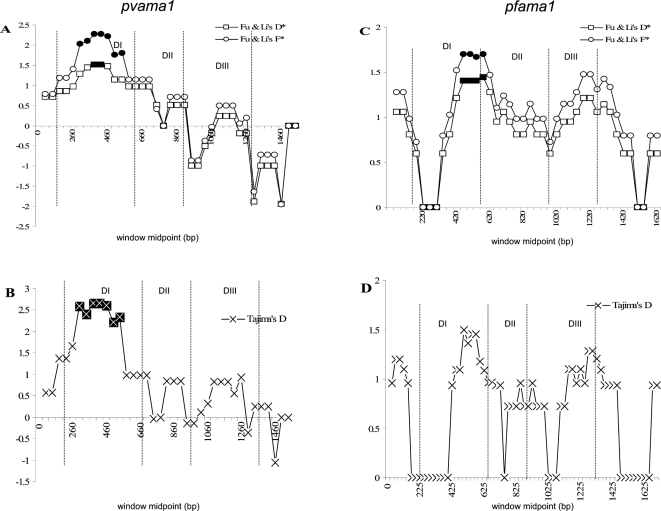
Evidence of selective signals in the two sequence sets was provided by tests for neutrality of polymorphisms. Across the two full-length genes, significant departures from neutrality were only found for Pfama1, with Fu and Li's D* and F* analyses providing parameter values of 1.72279 and 2.03574 (P<0.02 in each case) respectively. For pvama1, sequences from the 1997 survey gave significant values for Tajima's D and Fu and Li's F* across domain 1 only (data not shown). Sliding window analyses were also performed to identify any departures from neutral patterns of nucleotide substitution across smaller regions within both genes. Plots of each of these analyses, with the individual domains highlighted, are shown in Figure 3. As with other studies [15], [33], [34], a region of DI of Pvama1 within this Venezuelan population shows a significantly positive departure from neutral substitution patterns, indicating that balancing selection may be maintaining alleles of this domain. However, unlike previous studies [15], no evidence was found of balancing or directional selection upon DII or DIII in Pvama1 sequences from this Venezuelan population, and the values for these two domains are effectively zero (minimally positive and minimally negative in many cases). Like the Pvama1 data, Pfama1 showed a significant positive departure from neutrality in DI. The trend across the other two domains matches published Nigerian and Thai data of Polley et al. [18], [19], in that evidence of allele maintenance by balancing selection was stronger in DIII than within DII, although this was not significant in our study population.
Figure 3. Sliding window plots of Tajima's D and Fu and Li's F* for Pvama1 and Pfama1.
A and B – Pvama1; C and D – Pfama1. Midpoints where the sequence significantly departs from zero (P<0.05) are shown as solid points. Domains, DI, DII, DIII, are marked.
Between population analyses
We also examined the two sequence sets for evidence of selection using the McDonald-Kreitman test, which compares the relative frequency of non-synonymous nucleotide substitutions within the species, to the relative frequency of fixed differences between this and a related species. Data are summarised in Table 3, and support results from the tests of neutrality, as alleles within these Venezuelan Pvama1 and Pfama1 populations, particularly within DI, have significantly more non-synonymous substitutions than expected from comparison with related species. Thus diversifying selection is maintaining ama1 allelic diversity in both P. falciparum and P. vivax in our population.
There are 12, 4 and 3 Pvama1 haplotypes within this studied Venezuelan population across DI, DII and DIII, respectively. When this Venezuelan data is added to previously published data [15], [33], [34], [35], there are 90 described haplotypes across DI, 11 of which are unique to Venezuela; 8 haplotypes across DII, 1 unique to Venezuela; 4 haplotypes across DIII, none unique to Venezuela. This observed greater overlap of haplotypes across DII and DIII between samples from geographically diverse regions may be due simply to the scarcity of polymorphic sites in these smaller domains, exacerbated by the fact that only two other studies are available for comparison across these domains [15], [35]. Inter-population comparison of Pvama1 sequences from all available studies shows that the majority of diversity is within each population and not among populations, as indicated by low FST values (Tables 4 and 5).
Table 4. Geographical differences in Pvama1 sequences across DI only.
| Venezuelaa | |||||||||||||||
| Venezuelab | 0.140 | Venezuelab | |||||||||||||
| Venezuelac | 0.059 | 0.824 | Venezuela1 | ||||||||||||
| Sri Lanka2 | 0.125 | Sri Lanka | (23) | ||||||||||||
| Thailand3 | 0.135 | 0.122 | Thailand | (8) | |||||||||||
| ADS4 | 0.170 | 0.152 | 0.139 | ADS | (21) | ||||||||||
| Africa4 | 0.134 | 0.003 | 0.104 | 0.156 | Africa | (5) | |||||||||
| China4 | 0.092 | 0.075 | 0.020 | 0.096 | 0.047 | China | (7) | ||||||||
| India4 | 0.085 | 0.034 | 0.081 | 0.126 | 0.044 | 0.032 | India | (18) | |||||||
| Indonesia4 | 0.072 | 0.043 | 0.059 | 0.116 | 0.050 | 0.000 | −0.011 | Indonesia | (5) | ||||||
| Morong4 | 0.146 | 0.128 | 0.107 | 0.155 | 0.115 | 0.084 | 0.099 | 0.078 | Morong | (110) | |||||
| Palawan4 | 0.163 | 0.143 | 0.139 | −0.012 | 0.146 | 0.091 | 0.117 | 0.094 | 0.155 | Palawan | (17) | ||||
| PNG4 | 0.107 | 0.097 | 0.045 | 0.120 | 0.029 | 0.016 | 0.050 | 0.022 | 0.102 | 0.112 | PNG | (23) | |||
| Solomons4 | 0.131 | 0.123 | −0.007 | 0.151 | 0.097 | 0.048 | 0.075 | 0.051 | 0.125 | 0.142 | 0.009 | Solomons | (7) | ||
| Thailand4 | 0.074 | −0.008 | −0.007 | 0.112 | −0.020 | 0.000 | −0.014 | −0.034 | 0.078 | 0.102 | 0.007 | −0.024 | Thailand (6) | ||
| India (33)5 | 0.109 | 0.066 | 0.089 | 0.137 | 0.050 | 0.039 | 0.011 | 0.023 | 0.109 | 0.128 | 0.054 | 0.083 | 0.009 |
Pairwise FST values between Venezuelan sequences presented here and other sites only are highlighted. a = 1996 villages, N = 28; b = 1997 villages, N = 12; c = Koshirowetheri 1997 only, N = 33; 1 = all Venezuela data irrespective of village, N = 73; 2 = [15]; 3 = [33]; 4 = [34]; 5 = [35]; Sample sizes available are shown in parenthesis after each location; Significant difference (P<0.05) indicated by bold underline. Slightly negative numbers, indicated by italics, are biologically meaningless and are equivalent to FST = 0.000.
Table 5. Geographical differences in Pvama1 and Pfama1 sequences across all domains.
| P. vivax ama1 pairwise FST estimates | Venezuela (73) 1 | P. falciparum ama1 pairwise FST estimates | Venezuela (30) 2 | ||
| Sri Lanka (23) 2 | Nigeria (50) 4 | ||||
| Whole | 0.074 | Whole | 0.258 | ||
| DI | 0.120 | DI | 0.265 | ||
| DII | 0.185 | DII | 0.192 | ||
| DIII | 0.322 | Sri Lanka | DIII | 0.240 | Nigeria |
| India 3 | Thailand (51) 5 | ||||
| Whole (33) | 0.091 | 0.048 | Whole | 0.273 | 0.019 |
| DI (33) | 0.109 | 0.058 | DI | 0.286 | 0.039 |
| DII (34) | 0.034 | 0.223 | DII | 0.344 | 0.086 |
| DIII (34) | 0.349 | −0.035 | DIII | 0.276 | 0.056 |
Pairwise FST values between Venezuelan sequences and other sites are highlighted. Sample sizes are in parenthesis. 1 = all Venezuela Pvama1data from both 1996 and 1997; 2 = all Venezuela Pfama1 data from 1996 only; 2 = [15]; 3 = [35] one sequence was ignored in the whole gene and DI analysis as it was not full length; 4 = [19]; 5 = [18]. Significant difference (P<0.05) indicated by bold text. Slightly negative numbers, indicated by italics, are biologically meaningless and are equivalent to FST = 0.000.
The number of Pfama1 haplotypes observed in the Venezuelan dataset is 6, 2 and 3 across DI, DII and DIII, respectively. When added to previously published data from Nigeria and Thailand [18], [19], there are 58 haplotypes across DI, 5 of which are unique to Venezuela; 26 haplotypes across DII, none unique to Venezuela; 17 haplotypes across DIII, none of them unique to Venezuela. Pairwise inter-population comparisons show there is a significant diversity between population pairs, particularly across DI, with generally higher FST values than for Pvama1 (Table 5).
Discussion
We have analysed diversity in full length gene sequences encoding the apical membrane antigen (AMA1), from 73 P. vivax and 30 P. falciparum isolates collected in the same cross-sectional surveys of an isolated human population. To our knowledge, this is the first such detailed analysis of two homologous genes in sympatric Plasmodium parasite populations. The majority of people present at the time of the surveys agreed to provide blood samples for analysis, irrespective of malaria status, and so these surveys represent a good sample of both co-circulating P. vivax and P. falciparum [22], [23]. This is an important strength of our study, compared to analyses of symptomatic cases that have presented passively at health facilities with undefined catchment areas.
Previous studies had suggested that P. vivax and P. falciparum circulating in the Padamo exhibited very different population structures, but a direct test of this hypothesis required detailed analysis of homologous genes in the two species populations, so that the magnitude and effect of selection would be as similar as possible in the two datasets. AMA1 is a well-characterised parasite protein in both species, with homologues in all Plasmodium spp. examined, and is currently being tested as a vaccine candidate for P. falciparum. AMA1 contributes to the reorientation of merozoites, and is located at the tight junction during ingress of the erythrocyte. AMA1 also has a role in sporozoite invasion of hepatocytes [37], [38], [39], [40].
Genetic diversity was found to be significantly higher in the P. vivax population, and indices of intra-population recombination were significantly lower in the P. falciparum population. In contrast, indices of selective diversification on the two gene populations were similar, suggesting that differences in functional and immunological constraints upon them could not explain differences in gene diversity, and the frequency of recombination. We therefore conclude that whereas the P. vivax population is comprised of a diverse gene pool that frequently undergoes recombination, the P. falciparum population exhibits restricted diversity and appears to propagate most commonly by clonal expansion, with minimal opportunities for recombination. These findings are wholly consistent with the observation of stable multi-locus genotypes supported by high levels of inter-genic linkage disequilibrium among the P. falciparum population in the Padamo. As argued previously, this strongly suggests that this parasite population has passed through an evolutionary bottleneck which has both restricted genetic diversity and reduced opportunities for recombination within the population [23]. In contrast, we found no evidence that the P. vivax population has also passed through such a bottleneck. Evidence of a similar difference in structure between these two species has been observed in a study of 11 isolates of each from Papua New Guinea [41]. Interestingly, our earlier analysis of var gene sequences in the Padamo P. falciparum population indicated that these genes partially transcended the clonal structure observed at other loci, supporting the view that this gene family recombines, and evolves, at a higher frequency than do other loci in P. falciparum [42].
The majority of diversity seen among the Venezuelan ama1 sequences is in DI of both P. vivax and P. falciparum and, in accordance with other global studies, this domain accounts for the majority of distinct haplotypes within a population. However, the polymorphisms generating this diversity do not demonstrate neutral patterns of substitution. The pattern of polymorphisms seen in DI show a significant positive departure from neutral, suggesting they are under biological functional constraints as well as being targets of host immune mechanisms. Evidence of such ‘balancing selection’ in DI of ama1 across global isolates suggests this region is a target of natural host immunity, and supports the development of this domain as a potential vaccine target in both P. vivax and P. falciparum.
The importance of DII is less clear. In this Venezuelan population, there is no evidence to suggest balancing selection has contributed to the pattern of polymorphic substitution neither in DII nor in DIII of either species. Previous data confirms this finding for Pfama1 [18], [19], but this is contradictory to available Pvama1 data [15]. However, the Pvama1 sequences from this smaller Sri Lanka dataset (N = 23) were derived from clinical malaria presentations, and it is unknown how those alleles seen compare to the full repertoire of alleles present within the circulating P. vivax population from that region. Such differences in sampling procedure may also explain the neutral pattern of substitution across DIII observed among the Venezuelan Pvama1 sequences, in contrast to DIII from other regions.
Low sample numbers may have inhibited our analyses of the Venezuelan Pfama1 sequences by reducing the power needed to identify significant changes or signals of selection beyond those identified in DI. Only 30 Pfama1 sequences were obtained, all from the first survey in 1996. Yet, as only 33 individuals were slide positive for P. falciparum in this year (one of which was a mixed infection with P. malariae), under-representation is unlikely. The potential lack of power within Pfama1 is shown in DIII. Although the trend follows that previously described, there is no significant evidence to conclude this domain is also under balancing selection.
There is a marked difference in the prevalence of P. vivax infections between the two surveys. The second survey in 1997 followed a period of low rainfall which effectively interrupted mosquito-borne transmission for that season [23]. P. falciparum prevalence (slide positive) dropped from 4.7% to 1.3% but P. vivax increased from 5.5% in 1996 to 9.8% in 1997. P. malariae prevalence remained similar (2.3% and 2.1% respectively) as did the overall burden of malaria −12.7% in 1996 to 13.44% in 1997. In the Venezuelan Amazon, both P. vivax and P. falciparum are transmitted by the predominant vector species, Anopheles darlingi, and an interruption of transmission would affect both parasite species equally.
We suggest the increase of prevalence of P. vivax regardless of reduced transmission is a result of new merozoites being released from dormant hypnozoites (relapse). In the presence of active P. falciparum infection, such emergent P. vivax blood-stages may be prevented from establishing an infection by cross-species regulation and suppression [7], [9]. The presence of circulating P. falciparum may have effectively limited P. vivax infection in the previous (wet) year, but during a season with greatly reduced transmission, these new merozoites are able to expand significantly within their individual hosts. It has usually been assumed that genotypes seen as a result of relapses will always be representative of those genotypes previously seen, i.e. those of a primary infection [43], [44]. If true, then even though the actual numbers of P. vivax increased from one survey to the next in this Venezuelan study, the genetic profiles between the two samples would be expected to be similar. We found a greater apparent diversity in Pvama1 sequences from the 1997 survey, with whole gene diversity h = 0.913 compared to h = 0.796 in 1996. This may be partially due to the lower number of isolates obtained in the first survey, and given the large effective population size we have estimated for P. vivax, we cannot be certain our two cross-sectional samples were sufficiently representative to permit comparison of P. vivax diversity between surveys. Pairwise FST values show small but significant differences between Pvama1 sequence sets between these two years across the whole gene (9.1%) and across the polymorphic region of DI only (6.3%). We cannot determine whether any apparent increase in diversity in the second survey is due to relapse from heterologous hypnozoites, as we did not successfully amplify Pvama1 from any sample pairs of individuals that were P. vivax positive in both 1996 and 1997 surveys (there were 3 such people). Further work on pvama1 diversity in larger samples from this population would be required to compare with recent findings from Thailand, India and Myanmar which suggest that relapse of P. vivax in these regions is usually from activation of heterologous hypnozoites [45].
Of the four Plasmodium species that infect humans, P. vivax and P. falciparum are the two that are most often sympatric. Despite this, control regimes have usually targeted only P. falciparum in isolation, and in many cases the contribution of P. vivax to the local and global malarial burden has been ignored [1]. Substantial differences in the level of genetic diversity between these two species in the Padamo are consistent with one of them, P. falciparum, having passed through an evolutionary bottleneck either at the time of the introduction of this species to Venezuela from Europe and Africa or due to more recent constraints on recombination, whereas the P. vivax population displays evidence of a distinct recent evolutionary history in which diversity is maintained, and recombination is common. Whereas the former species displays characteristics of an epidemic population structure, P. vivax exhibits a structure consistent with that of an established endemic pathogen. Our data thus demonstrate that these two human pathogens adopt different strategies to survive in this setting of mesoendemic transmission in the Venezuelan Amazon, and as a result differ in their susceptibility to changing environmental conditions, and seasonal fluctuations in vector abundance. Future control regimes and interventions therefore need to include strategies that will target each of the parasite populations present, and so aim to reduce the overall malarial burden.
Acknowledgments
We thank Spencer Polley for helpful discussions and comments on the manuscript. Study participants gave informed consent for genetic studies to be carried out on the parasite isolates, at the time they were collected.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was not externally funded. CJS is supported by the UK Health Protection Agency.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006;22:353–358. doi: 10.1016/j.pt.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller I, Zimmerman PA, Reeder JC. Plasmodium malariae and Plasmodium ovale - the ‘bashful’ malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui L, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, et al. Genetic diversity and multiple infections of Plasmodium vivax malaria in Western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- 5.Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, et al. Chloroquine Resistant Plasmodium vivax: In Vitro Characterisation and Association with Molecular Polymorphisms. PLoS ONE. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maitland K, Williams TN, Bennett S, Newbold CI, Peto TE, et al. The interaction between Plasmodium falciparum and P. vivax in children on Espiritu Santo island, Vanuatu. Trans R Soc Trop Med Hyg. 1996;90:614–620. doi: 10.1016/s0035-9203(96)90406-x. [DOI] [PubMed] [Google Scholar]
- 7.Maitland K, Williams TN, Newbold CI. Plasmodium vivax and P. falciparum: Biological interactions and the possibility of cross-species immunity. Parasitol Today. 1997;13:227–231. doi: 10.1016/s0169-4758(97)01061-2. [DOI] [PubMed] [Google Scholar]
- 8.Smith T, Genton B, Baea K, Gibson N, Narara A, et al. Prospective risk of morbidity in relation to malaria infection in an area of high endemicity of multiple species of Plasmodium. Am J Trop Med Hyg. 2001;64:262–267. doi: 10.4269/ajtmh.2001.64.262. [DOI] [PubMed] [Google Scholar]
- 9.Bruce MC, Day KP. Cross-species regulation of malaria parasitaemia in the human host. Curr Opin Microbiol. 2002;5:431–437. doi: 10.1016/s1369-5274(02)00348-x. [DOI] [PubMed] [Google Scholar]
- 10.Mason DP, Krudsood S, Wilairatana P, Viriyavejakul P, Silachamroon U, et al. Can treatment of P. vivax lead to a unexpected appearance of falciparum malaria? Southeast Asian J Trop Med Public Health. 2001;32:57–63. [PMC free article] [PubMed] [Google Scholar]
- 11.Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–4083. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet. 1989;2:1183–1184. doi: 10.1016/s0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- 13.Maguire JD, Sumawinata IW, Masbar S, Laksana B, Prodjodipuro P, et al. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet. 2002;360:58–60. doi: 10.1016/S0140-6736(02)09336-4. [DOI] [PubMed] [Google Scholar]
- 14.Tan-ariya P, Pasuralertsakul S. First report of in vitro susceptibility of Plasmodium malariae Thai isolates to chloroquine. Southeast Asian J Trop Med Public Health. 1994;25:784–787. [PubMed] [Google Scholar]
- 15.Gunasekera AM, Wickramarachchi T, Neafsey DE, Ganguli I, Perera L, et al. Genetic diversity and selection at the Plasmodium vivax apical membrane antigen-1 (PvAMA-1) locus in a Sri Lankan population. Mol Biol Evol. 2007;24:939–947. doi: 10.1093/molbev/msm013. [DOI] [PubMed] [Google Scholar]
- 16.Franks S, Baton L, Tetteh K, Tongren E, Dewin D, et al. Genetic diversity and antigenic polymorphism in Plasmodium falciparum: extensive serological cross-reactivity between allelic variants of merozoite surface protein 2. Infect Immun. 2003;71:3485–3495. doi: 10.1128/IAI.71.6.3485-3495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun. 2008;76:2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, et al. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 21.Grynberg P, Fernandes Fontes CJ, Braga EM. Association between particular polymorphic residues on apical membrane antigen 1 (AMA-1) and platelet levels in patients with vivax malaria. Clin Microbiol Infect. 2007;13:1089–1094. doi: 10.1111/j.1469-0691.2007.01815.x. [DOI] [PubMed] [Google Scholar]
- 22.Ord R, Polley S, Tami A, Sutherland CJ. High sequence diversity and evidence of balancing selection in the Pvmsp3alpha gene of Plasmodium vivax in the Venezuelan Amazon. Mol Biochem Parasitol. 2005;144:86–93. doi: 10.1016/j.molbiopara.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Tami A, Grundmann H, Sutherland C, McBride JS, Cavanagh DR, et al. Restricted genetic and antigenic diversity of Plasmodium falciparum under mesoendemic transmission in the Venezuelan Amazon. Parasitology. 2002;124:569–581. doi: 10.1017/s0031182002001713. [DOI] [PubMed] [Google Scholar]
- 24.Ord RL. Genetic Diversity of Plasmodium vivax and Plasmodium falciparum co-circulating among the same hosts in the Venezuelan Amazon. London: London School of Hygiene and Tropical Medicine, University of London.; 2008. p. 249. [Google Scholar]
- 25.Sutherland CJ, Alloueche A, Curtis J, Drakeley CJ, Ord R, et al. Gambian children successfully treated with chloroquine can harbor and transmit Plasmodium falciparum gametocytes carrying resistance genes. Am J Trop Med Hyg. 2002;67:578–585. doi: 10.4269/ajtmh.2002.67.578. [DOI] [PubMed] [Google Scholar]
- 26.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 29.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genet Res. 1987;50:245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- 31.Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill WG, Roberts A. Linkage disequilibrium in finite populations. Theoretical and Applied Genetics. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Q, Saul A. Sequence analysis of the apical membrane antigen I (AMA-1) of Plasmodium vivax. Mol Biochem Parasitol. 1994;65:183–187. doi: 10.1016/0166-6851(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 34.Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, et al. Plasmodium vivax synonymous substitution frequencies, evolution and population structure deduced from diversity in AMA 1 and MSP 1 genes. Mol Biochem Parasitol. 2000;108:53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- 35.Rajesh V, Elamaran M, Vidya S, Gowrishankar M, Kochar D, et al. Plasmodium vivax: Genetic diversity of the apical membrane antigen-1 (AMA-1) in isolates from India. Exp Parasitol. 2007;116:252–256. doi: 10.1016/j.exppara.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 37.Hehl AB, Lekutis C, Grigg ME, Bradley PJ, Dubremetz JF, et al. Toxoplasma gondii homologue of plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect Immun. 2000;68:7078–7086. doi: 10.1128/iai.68.12.7078-7086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect Immun. 2004;72:154–158. doi: 10.1128/IAI.72.1.154-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24:74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem. 2004;279:9490–9496. doi: 10.1074/jbc.M311331200. [DOI] [PubMed] [Google Scholar]
- 41.Mueller I, Kaiok J, Reeder JC, Cortes A. The population structure of Plasmodium falciparum and Plasmodium vivax during an epidemic of malaria in the Eastern Highlands of Papua New Guinea. Am J Trop Med Hyg. 2002;67:459–464. doi: 10.4269/ajtmh.2002.67.459. [DOI] [PubMed] [Google Scholar]
- 42.Tami A, Ord R, Targett GA, Sutherland CJ. Sympatric Plasmodium falciparum isolates from Venezuela have structured var gene repertoires. Malar J. 2003;2:7. doi: 10.1186/1475-2875-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirchgatter K, del Portillo HA. Molecular analysis of Plasmodium vivax relapses using the MSP1 molecule as a genetic marker. J Infect Dis. 1998;177:511–515. doi: 10.1086/517389. [DOI] [PubMed] [Google Scholar]
- 44.Craig AA, Kain KC. Molecular analysis of strains of Plasmodium vivax from paired primary and relapse infections. J Infect Dis. 1996;174:373–379. doi: 10.1093/infdis/174.2.373. [DOI] [PubMed] [Google Scholar]
- 45.Imwong M, Snounou G, Pukrittayakamee S, Tanomsing N, Kim JR, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 46.Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]