Abstract
SWI/SNF chromatin-remodeling complexes have crucial roles in transcription and other chromatin-related processes. The analysis of the two members of this class in Saccharomyces cerevisiae, SWI/SNF and RSC, has heavily contributed to our understanding of these complexes. To understand the in vivo functions of SWI/SNF and RSC in an evolutionarily distant organism, we have characterized these complexes in Schizosaccharomyces pombe. Although core components are conserved between the two yeasts, the compositions of S. pombe SWI/SNF and RSC differ from their S. cerevisiae counterparts and in some ways are more similar to metazoan complexes. Furthermore, several of the conserved proteins, including actin-like proteins, are markedly different between the two yeasts with respect to their requirement for viability. Finally, phenotypic and microarray analyses identified widespread requirements for SWI/SNF and RSC on transcription including strong evidence that SWI/SNF directly represses iron-transport genes.
Changes in chromatin structure are required to facilitate fundamental nuclear processes including transcription, DNA replication, DNA repair, recombination and chromosome segregation. The restructuring of chromatin is often accomplished by ATP-dependent chromatin remodelers, which alter DNA-histone contacts to mediate nucleosomal structural alterations, removal and movement1. There are four different classes of ATP-dependent chromatin-remodeling complexes, SWI/SNF, ISWI, Ino80 and Mi-2, and each of these has at its catalytic core an ATPase subunit that belongs to the Snf2-ATPase superfamily2. The most widely studied ATP-dependent chromatin-remodeling complexes are in the SWI/SNF class, broadly conserved among eukaryotes2.
The two SWI/SNF-class remodelling complexes of S. cerevisiae, SWI/SNF, the founding member of this class, and RSC, have been shown to have vital roles in vivo3. SWI/SNF is involved in transcriptional activation4,5, telomeric and rDNA silencing6 and DNA repair7. RSC is ten-fold more abundant than SWI/SNF and, in contrast to SWI/SNF, is essential for cell viability8,9. RSC regulates transcription by RNA polymerases II and III10–14, and possibly by RNA polymerase I14,15. Furthermore, RSC has important roles throughout the cell cycle16, including functions in kinetochore function17, sister chromatid cohesion18 and DNA repair7,19. Thus, SWI/SNF and RSC function in most chromatin-related processes in S. cerevisiae.
Saccharomyces cerevisiae SWI/SNF and RSC have been extensively analyzed biochemically. Although both complexes have ATP-dependent nucleosome-remodeling activity3, they have distinct compositions. SWI/SNF contains 12 different proteins, whereas RSC contains 17 different subunits2. Snf 2, the ATPase subunit of SWI/SNF, is a paralog of Sth1 of RSC. In addition, the two complexes contain four other paralogs (Snf5, Swi3, Snf12/Swp73 and Swp82 of SWI/SNF, and Sf h1, Rsc8, Rsc6 and Rsc7 of RSC, respectively). Three other components are shared between the complexes, two of which, the actin-related proteins Arp7 and Arp9, are crucial for growth8,20. Many components of SWI/SNF and RSC contain motifs involved in different activities, including DNA binding, protein-protein interaction and recognition of acetylated histones2.
The SWI/SNF class of chromatin-remodeling complexes is evolutionarily conserved throughout eukaryotes, and they are of substantial importance in vivo3,21. The two mammalian SWI/SNF subclasses, BAF and PBAF (corresponding to SWI/SNF and RSC, respectively), are important in several cellular processes (reviewed in refs. 2,21). These complexes contain the core ATPase subunit, Brg1 (also known as Smarca4) or Brm (also known as Smarca2), plus seven or more noncatalytic subunits that are largely shared between BAF and PBAF 22,23, with the precise composition of the complex dependent on cell type and the cell cycle. Mammalian SWI/SNF can function both as a positive and negative regulator of transcription, depending on association with other proteins21,24. The importance of SWI/SNF in mammalian cells is emphasized by the finding that several mammalian SWI/SNF subunits are defined as tumor suppressors25.
Saccharomyces cerevisiae has been a valuable system for understanding the in vivo mechanisms and roles of SWI/SNF complexes. However, S. cerevisiae has major differences in chromatin structure compared to other eukaryotes. Therefore, we have embarked on studies of SWI/SNF and RSC in the distantly related yeast, S. pombe. Study of these complexes in S. pombe provides an opportunity to gain insight into their in vivo functions in an organism whose chromatin more closely resembles that of mammalian cells26. In this work, we present the purification and characterization of the S. pombe SWI/SNF and RSC complexes, thereby establishing S. pombe as a new model system for the study of chromatin-remodeling complexes and providing the resources for such analysis. We show that the two S. pombe complexes differ greatly in composition from their S. cerevisiae counterparts and in some ways are more similar to those of metazoans. Furthermore, the two actin-related proteins shared between the S. pombe SWI/SNF and RSC complexes are functionally distinct from those in the S. cerevisiae complexes as they are not required for growth in S. pombe, whereas they are essential or critical for growth in S. cerevisiae, depending on the strain background. Finally, microarray analyses and other phenotypic characterizations of S. pombe SWI/SNF and RSC mutants show that these complexes have widespread roles in transcription and strongly suggest a direct role for SWI/SNF in transcriptional repression.
RESULTS
Purification of S. pombe SWI/SNF and RSC complexes
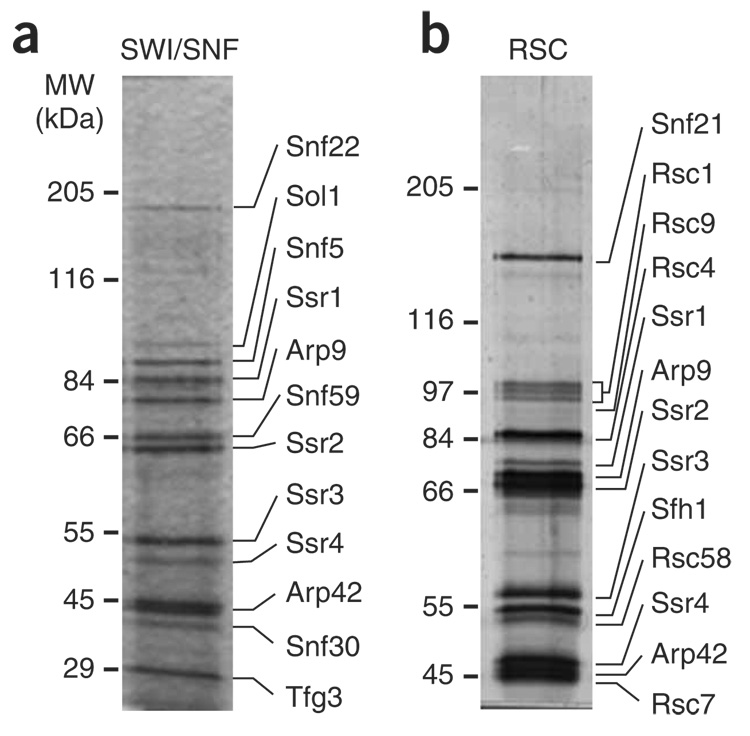
To initiate study of S. pombe SWI/SNF and RSC, we identified putative S. pombe homologs of S. cerevisiae SWI/SNF and RSC components by sequence homology (Supplementary Table 1 online). Among the candidates identified were the putative homologs of the S. cerevisiae ATPases Snf2 of SWI/SNF and Sth1 of RSC, previously named Snf21 and Snf22 (ref. 27). Although sequence homology could not clearly distinguish which S. pombe gene corresponded to which S. cerevisiae homolog, previous analysis showed that snf22+ is not essential for growth and snf21+ is essential27, suggesting that snf22+ is the SNF2 ortholog and snf21+ is the STH1 ortholog. Among the other genes identified were SPAC2F7.08c and SPCC16A11.14, putative S. pombe orthologs to S. cerevisiae SNF5 and SFH1, respectively. To purify the putative S. pombe SWI/SNF and RSC complexes, a tandem-affinity purification (TAP) sequence was fused to the 3′ end of each of these four S. pombe genes. Then, each TAP-tagged protein was purified, and the associated proteins were analyzed by SDS-PAGE and MS.
For the putative S. pombe SWI/SNF complex, purification of Snf22-TAP and Snf5-TAP yielded the identical 12-protein complex (Fig. 1a, Table 1 and data not shown). This complex also contained homologs of other S. cerevisiae SWI/SNF proteins, including Sol1 (switch one–like, a homolog of S. cerevisiae Swi1) and Tfg3 (a homolog of S. cerevisiae Taf14), strongly suggesting that this is the S. pombe SWI/SNF complex. The complex that we have identified is probably the same as an uncharacterized complex that was shown to contain Sol1 (ref. 28). Our MS analysis also identified a previously uncharacterized protein of approximately 30 kDa, which we called Snf30, and a highly divergent Swp82 homolog, called Snf59 (Table 1 and data not shown). To confirm that Snf30 and Snf59 are components of SWI/SNF, each was TAP-tagged and purified. Both SDS-PAGE and MS gave results identical to those seen for the Snf22-TAP and Snf5-TAP purifications (Table 1 and data not shown), demonstrating that Snf30 and Snf59 are both components of S. pombe SWI/SNF. In summary, the composition of the 12-subunit S. pombe SWI/SNF complex has both substantial similarities and differences with the S. cerevisiae SWI/SNF complex (Table 2).
Figure 1.
Purification of the fission yeast SWI/SNF and RSC complexes. Representative silver-stained 10% SDS-PAGE gels are shown for the SWI/SNF (a) and RSC (b) purifications. For SWI/SNF an Snf5-TAP (FWP220) purification is shown, and for RSC a Snf21-TAP (FWP218) purification is shown. Molecular weight (MW) marker positions are shown on the left. Components of each complex were identified by MS (Table 1), and the predicted assignment of a protein band to a subunit is shown.
Table 1.
The S. pombe SWI/SNF and RSC chromatin-remodeling complexes
| Protein | Systematic ID | Sizea | Sequence coverage (%)b | |||
|---|---|---|---|---|---|---|
| SWI/SNF complex | Snf22c | Snf5 | Snf59 | Snf30 | ||
| Snf22 | SPCC1620.14c | 1680 | 44 | 44 | 41 | 39 |
| Sol1 | SPBC30B4.04c | 865 | 43 | 49 | 43 | 31 |
| Snf5 | SPAC2F7.08c | 632 | 37 | 38 | 42 | 38 |
| Ssr1 | SPAC17G6.10 | 527 | 58 | 55 | 53 | 44 |
| Arp9 | SPAC1071.06 | 523 | 37 | 37 | 30 | 29 |
| Snf59 | SPBC26H8.09c | 515 | 57 | 62 | 64 | 50 |
| Ssr2 | SPAC23H3.10 | 503 | 33 | 38 | 34 | 23 |
| Arp42 | SPAC23D3.09 | 430 | 47 | 47 | 45 | 45 |
| Ssr3 | SPAC23G3.10c | 425 | 73 | 71 | 59 | 67 |
| Ssr4 | SPBP23A10.05 | 395 | 36 | 39 | 35 | 32 |
| Snf30 | SPAC23G3.07c | 274 | 37 | 42 | 33 | 32 |
| Tfg3 | SPAC22H12.02 | 241 | 34 | 37 | 39 | 30 |
| RSC complex | Snf21 | Sfh1 | Rsc7 | Rsc1 | ||
| Snf21 | SPAC1250.01 | 1199 | 58 | 48 | 58 | 64 |
| Rsc1 | SPBC4B4.03 | 803 | 57 | 47 | 53 | 51 |
| Rsc9 | SPBC1703.02 | 780 | 41 | 34 | 42 | 45 |
| Rsc4 | SPBC1734.15 | 542 | 64 | 64 | 61 | 61 |
| Ssr1 | SPAC17G6.10 | 527 | 62 | 54 | 51 | 55 |
| Arp9 | SPAC1071.06 | 523 | 43 | 40 | 30 | 34 |
| Ssr2 | SPAC23H3.10 | 503 | 33 | 32 | 35 | 38 |
| Arp42 | SPAC23D3.09 | 430 | 64 | 59 | 54 | 62 |
| Ssr3 | SPAC23G3.10c | 425 | 81 | 75 | 67 | 76 |
| Sfh1 | SPCC16A11.14 | 418 | 42 | 41 | 36 | 28 |
| Rsc58 | SPAC1F3.07c | 403 | 52 | 50 | 55 | 54 |
| Ssr4 | SPBP23A10.05 | 395 | 42 | 37 | 35 | 42 |
| Rsc7 | SPCC1281.05 | 390 | 54 | 55 | 61 | 59 |
Mass-spectrometry results showing the identification SWI/SNF and RSC components for each TAP-tagged subunit preparation.
Protein size in amino acids.
Sequence coverage (percentage of amino acids) for the respective protein.
Protein name indicates the TAP fusion used for purification.
Table 2.
Composition of S. pombe SWI/SNF and RSC complexes compared to those of S. cerevisiae and human
| S. pombe | S. cerevisiae | Human | |||
|---|---|---|---|---|---|
| SWI/SNFa | RSC | SWI/SNF | RSC | BAF | PBAF |
| Snf22 | Snf21 | Snf2 | Sth1 | BRG1 or BRM | BRG1 |
| Sol1 | Swi1 | BAF250 | |||
| Snf5 | Sfh1 | Snf5 | Sfh1 | SNF5 | SNF5 |
| Ssr1, Ssr2b | Ssr1, Ssr2 | Swi3 | Rsc8 | BAF170, BAF155 | BAF170, BAF155 |
| Ssr3 | Ssr3 | Snf12 | Rsc6 | BAF60a | BAF60a or BAF60b |
| Ssr4 | Ssr4 | ||||
| Arp42 | Arp42 | BAF53 | BAF53 | ||
| Arp9 | Arp9 | Arp9 | Arp9 | ||
| Arp7 | Arp7 | ||||
| Actin | Actin | ||||
| Tfg3 | Taf14 | ||||
| Rsc1 | Rsc1 or Rsc2 | BAF180 | |||
| Rsc4 | Rsc4 | ||||
| Rsc9 | Rsc9 | ||||
| Rsc58 | Rsc58 | ||||
| Snf59 | Rsc7 | Swp82 | Rsc7 | ||
| Snf30 | |||||
| BAF57 | BAF57 | ||||
| Rtt102 | Rtt102 | ||||
| Snf11 | |||||
| Snf6 | |||||
| Rsc3 | |||||
| Rsc30 | |||||
| Ldb7 | |||||
| Htl1 | |||||
Subunits of the respective complexes are listed in columns, with orthologous proteins grouped horizontally.
Paralogs in the same complex are grouped in the same row of the column.
To determine the composition of S. pombe RSC, we purified Snf21-TAP and Sfh1-TAP. These purifications yielded an identical 13-member complex (Fig. 1b, Table 1 and data not shown). To confirm this composition, two additional putative members of the complex, Rsc1 and Rsc7, were TAP-tagged and used to purify the complex. These purifications gave identical results to the Snf21 and Sfh1 purifications (Table 1). S. pombe RSC contains orthologs to many of the proteins in its S. cerevisiae counterpart, including the bromodomain proteins Rsc1, Rsc4 and Rsc58 (Table 2). However, S. pombe RSC also has notable differences from S. cerevisiae RSC. First, S. pombe RSC does not contain five subunits that are found in S. cerevisiae RSC but that are not conserved in metazoans, including Rsc3, which is essential for growth in S. cerevisiae10,29. In addition, S. cerevisiae RSC exists in at least two forms owing to the mutually exclusive presence of the bromodomain proteins Rsc1 or Rsc2 (ref. 30). However, S. pombe RSC has only a single Rsc1 and Rsc2 homolog, suggesting that S. pombe RSC exists in fewer forms than in S. cerevisiae.
Shizosaccharomyces pombe SWI/SNF and RSC share six components
A notable feature of the S. pombe SWI/SNF and RSC complexes is that they share six proteins, a much greater degree of overlap than the three subunits shared between the S. cerevisiae complexes (Table 2). Analysis of these shared proteins highlights similarities between the two S. pombe complexes and their metazoan counterparts (Table 2). The presence of Ssr1 and Ssr2 in both S. pombe complexes is similar to human BAF and PBAF, which share both BAF150 and BAF170 (ref. 23); however, this contrasts with complexes from S. cerevisiae, where Swi3 and Rsc8 are specific to SWI/SNF and RSC, respectively. Ssr3 is a homolog of S. cerevisiae Snf12 (in SWI/SNF) and Rsc6 (in RSC) and, similarly to its metazoan ortholog, BAF60a, is shared between SWI/SNF and RSC. Ssr4 is a member of a previously uncharacterized protein family (Pfam family PF08549) that has no apparent Saccharomyces homologs. Finally, the shared actin-related proteins, Arp42 and Arp9, differ from the Arp7–Arp9 module in S. cerevisiae SWI/SNF and RSC (Supplementary Fig. 1 online). Notably, this is the first example in which an Arp4-like protein is associated with Arp9 and not nuclear actin31. In summary, S. pombe SWI/SNF and RSC components overlap extensively, a situation distinct from S. cerevisiae and similar to the mammalian BAF and PBAF complexes.
Deletion analysis of SWI/SNF and RSC genes
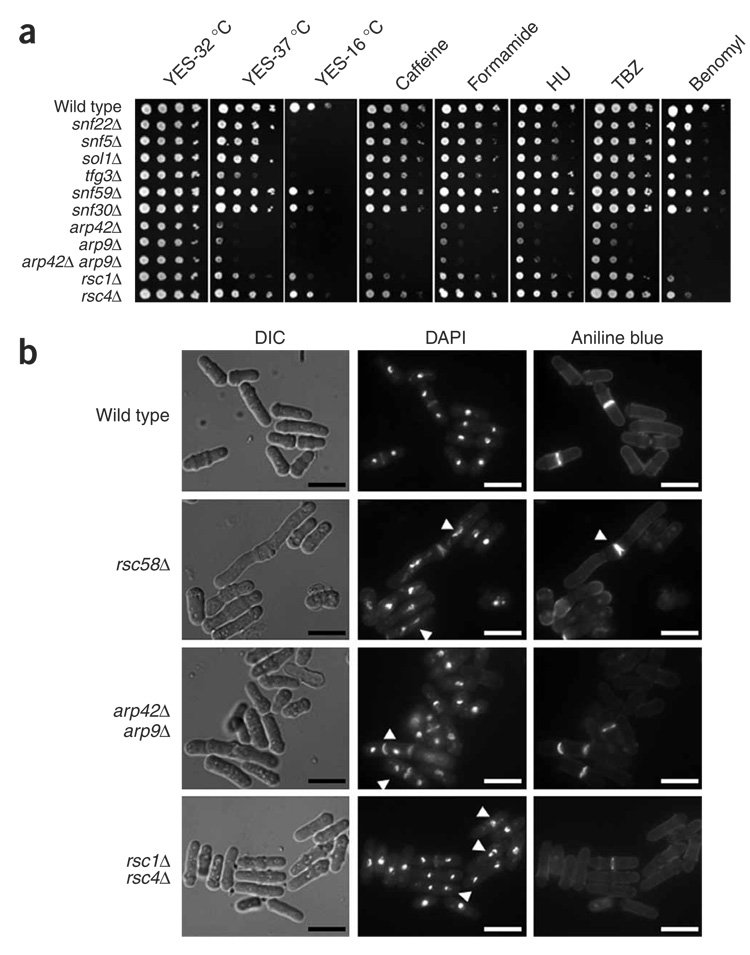
To analyze the roles of S. pombe SWI/SNF and RSC in vivo, we deleted each of the genes that encodes a subunit of the complexes (summarized in Supplementary Table 2 online). Our results show that none of the six genes encoding SWI/SNF-specific proteins are essential for viability (Fig. 2a). Thus, as in S. cerevisiae, SWI/SNF is not essential for growth. In contrast, four of the seven RSC-specific genes (snf21+, sfh1+, rsc9+ and rsc7+), as well as four of the six genes encoding shared proteins (ssr1+–ssr4+) are essential for growth, demonstrating that S. pombe RSC is required for cell viability. One unexpected discovery from the deletion analysis was that S. pombe arp42Δ, arp9Δ and arp42Δ arp9Δ mutants are viable and grow well on rich medium at 32 °C (Fig. 2a). This result contrasts with results for S. cerevisiae ARP7 and ARP9, which are essential or critical for growth depending on the genetic background20,32. The requirement for viability for other components also differed between S. pombe and S. cerevisiae (Supplementary Table 2). These notable differences between the S. cerevisiae and S. pombe complex members indicate that they mediate distinct roles in each organism.
Figure 2.
SWI/SNF and RSC mutants show a range of mutant phenotypes. (a) Growth phenotypes of the viable SWI/SNF and RSC mutants. Wild-type (FWP52), snf22Δ (FWP229), snf5Δ (FWP231), sol1Δ (FWP245), tfg3Δ (FWP246), snf59Δ (FWP247), snf30Δ (FWP248), arp42Δ (FWP239), arp9Δ (FWP240), arp42Δ arp9Δ (FWP249), rsc1Δ (FWP234) and rsc4Δ (FWP242) strains were grown in liquid YES medium to stationary phase, subjected to ten-fold serial dilutions and spotted onto solid YES medium containing caffeine, formamide, hydroxyurea (HU), thiabendazole (TBZ) or benomyl as indicated (see Methods for concentrations). The leftmost spot for each came from a culture at 1 × 108 cells ml−1, and approximately 3 µl were spotted. (b) Cell elongation, chromosomal segregation and septation defects of Arp and RSC mutants. Representative pictures for exponential phase cells of the wild type and rsc58Δ, rsc1Δ rsc4Δ and arp42Δ arp9Δ mutants are shown. Quantification (n = 100) indicated that the average cell length for each strain was as follows: wild type, 10.2 µm; rsc58Δ, 15.7 µm; rsc1Δ rsc4Δ, 11.3 µm; and arp42Δ arp9Δ, 12.1 µm. DAPI was used to visualize chromosomal DNA and aniline blue was used to stain septa60. Arrowheads highlight examples of abnormalities. Scale-bars represent 10 microns.
To gain more information about the roles of SWI/SNF and RSC, we tested the viable deletion mutants for growth defects under different conditions (Fig. 2a and Supplementary Fig. 2 online). For SWI/SNF, a subset of mutants (snf22Δ, snf5Δ, sol1Δ and tfg3Δ) showed tight cold sensitivity at 16 °C (Fig. 2a), a phenotype not seen for their S. cerevisiae counterparts. Some mutants also showed modest sensitivity to benomyl or hydroxyurea (Fig. 2a). Notably, S. pombe SWI/SNF mutants do not show certain phenotypes characteristic of S. cerevisiae SWI/SNF mutants33 (Supplementary Fig. 2). For RSC, two of the three viable mutants that are RSC specific, rsc58Δ and rsc1Δ, showed pleiotropic phenotypes, indicating roles for RSC in several cellular processes (Fig. 2a, Supplementary Fig. 2 and data not shown). In contrast, the rsc4Δ mutant showed near–wild-type growth. Microscopy of the rsc58Δ and rsc1Δ rsc4Δ mutants revealed elongated cells and apparent chromosomal segregation defects (Fig. 2b). Finally, the phenotypes observed for arp42Δ, arp9Δ and arp42Δ arp9Δ encompass those seen for the SWI/SNF and RSC mutants (for example, temperature sensitivity and caffeine sensitivity), consistent with Arp42 and Arp9 being important for both SWI/SNF and RSC functions (Fig. 2).
Arp proteins are not required for complex assembly
Given the marked difference between S. cerevisiae and S. pombe with respect to the requirement for Arp proteins in SWI/SNF and RSC, we determined the composition of the SWI/SNF and RSC complexes that assemble in the arp42Δ arp9Δ mutant, using TAP purification and MS. Our results (Supplementary Table 3 online) show that both SWI/SNF and RSC are fully assembled in an arp42Δ arp9Δ mutant, lacking only Arp42 and Arp9. The peptide numbers for each component are similar between wild-type and arp42Δ arp9Δ preparations, suggesting that the complexes are as stable in an arp42Δ arp9Δ mutant as in the wild type (Supplementary Table 3). Furthermore, no other Arps are present in these mutant complexes that might compensate for the loss of Arp42 or Arp9. We also purified both Arp42-TAP and Arp9-TAP, and showed that they are present only in SWI/SNF and RSC (Supplementary Fig. 3 online). Taken together, these results demonstrate that Arp42 and Arp9 are specifically associated with SWI/SNF and RSC but are not required for their assembly or for the essential function of RSC.
Transcriptome analysis of SWI/SNF and RSC mutants
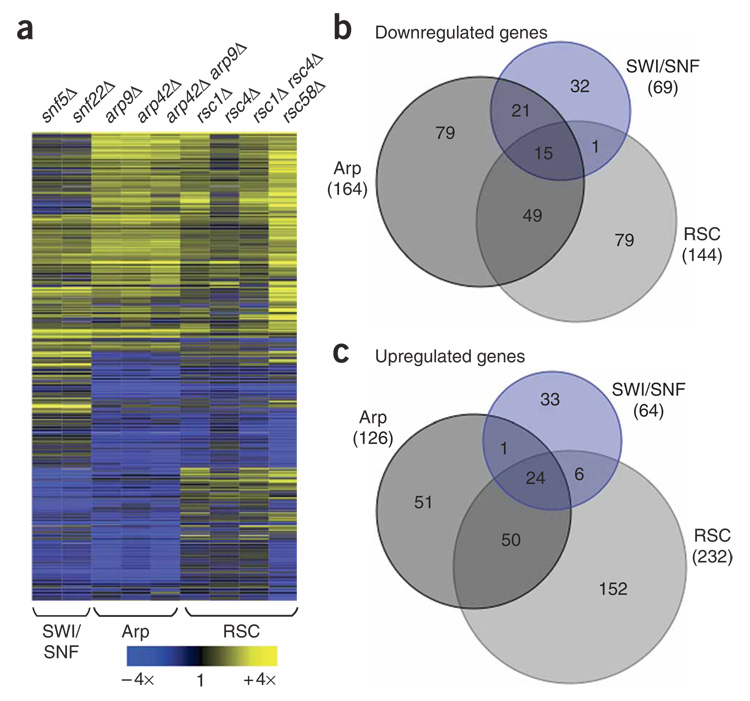
To assess the roles of SWI/SNF and RSC in gene expression, we carried out whole-genome expression analysis on nine SWI/SNF, RSC and Arp mutants. For SWI/SNF, the expression profiles for snf22Δ and snf5Δ showed almost identical patterns (Fig. 3a), with 2.6% of S. pombe genes having mRNA levels altered more than two-fold. Although an effect of this degree is similar to what is seen for S. cerevisiae SWI/SNF mutants, the gene targets are distinct between the two yeasts4,5. For RSC, rsc1Δ, rsc4Δ and rsc1Δ rsc4Δ mutants caused similar and mild effects (Fig. 3a), consistent with their growth phenotypes (Fig. 2a). Overall, expression alteration effects of more than two-fold in either direction were seen for 1.4%, 0.6%and 1.5% of S. pombe genes in the rsc1Δ, rsc4Δ and rsc1Δ rsc4Δ mutants, respectively. In contrast, in S. cerevisiae, where RSC4 is essential for growth, a rsc4 temperature-sensitive mutant caused a change in 12% of all genes13,14. For the S. pombe rsc58Δ mutant, 6.8% of all genes had mRNA levels altered more than two-fold, although the poor growth of rsc58Δ may cause indirect effects. For the shared Arp components, the expression profiles for the arp42Δ, arp9Δ and arp42Δ arp9Δ mutants showed almost identical patterns (Fig. 3a), with 5.6% of S. pombe genes being altered more than two-fold (Fig. 3b,c). As expected, we observed a significant overlap of the Arp mutants with the SWI/SNF and RSC mutant data sets (Fig. 3b,c). Some effects, however, were specific for the Arp mutants, consistent with the phenotypic analysis of these mutants and indicating modularity in the SWI/SNF and RSC complexes. This is further supported by the identification of a Rsc1 and Rsc4 submodule that seems largely dispensable for RSC function.
Figure 3.
Whole-genome expression profiling of SWI/SNF, Arp and RSC deletion mutants. (a) Transcriptome analysis for snf5Δ (FWP231), snf22Δ (FWP229), arp9Δ (FWP240), arp42Δ (FWP239), arp42Δ arp9Δ (FWP249), rsc1Δ (FWP234), rsc4Δ (FWP242), rsc1Δ rsc4Δ (FWP250) and rsc58Δ (FWP244) is shown using hierarchical clustering. A row represents a gene whose expression was altered more than two-fold in any mutant. Data are presented as the ratio of mutant to wild type (FWP165) and color coded as indicated by the key. Data from the nine mutants were grouped into SWI/SNF, Arp and RSC data sets, as indicated below, for the data analysis in b and c. The correlation values between pairs of mutants tested are as follows: snf22Δ and snf5Δ − r2 = 0.89; rsc1Δ and rsc4Δ − r2 = 0.50; rsc1Δ and rsc1Δ rsc4Δ − r2 = 0.85; rsc4Δ and rsc1Δ rsc4Δ − r2 = 0.62; arp42Δ and arp9Δ − r2 = 0.95; arp42Δ and arp42Δ arp9Δ − r2 = 0.91; and arp9Δ and arp42Δ arp9Δ − r2 = 0.87. (b,c) Venn diagrams showing the degree of overlap of genes with reduced (b) or elevated (c) mRNA levels for the SWI/SNF, Arp and RSC data sets as indicated. The total number of genes affected more than two-fold for each data set is indicated in parentheses.
RSC controls membrane and organelle development genes
To determine whether any particular biological processes were enriched in the microarray data, we analyzed genes whose expression was significantly altered in the RSC and Arp mutants for significant enrichments of any gene ontology (GO) categories34. The significance analysis of microarrays (SAM) method was used to identify significantly affected genes35. The set of genes with increased mRNA levels in RSC and Arp mutants was significantly enriched for GO categories relating to membrane (GO0016020; P-value = 6.5e−22), endoplasmic reticulum (ER) (GO0005783; P-value = 3.4e−20) and Golgi processes (GO0005794; P-value = 4.0e−7). Such GO enrichments were not observed when this analysis was restricted to the more conservative two-fold data set. Investigating a possible defect in Golgi processes, we tested the mutants for resistance to brefeldin A, a compound that inhibits protein secretion and promotes Golgi disassembly36. Indeed, strong resistance to brefeldin A was observed for the RSC and Arp mutants (Supplementary Fig. 4 online) but not for SWI/SNF mutants (data not shown), supporting the conclusion that RSC controls genes involved in this process.
Role of SWI/SNF in transcriptional activation
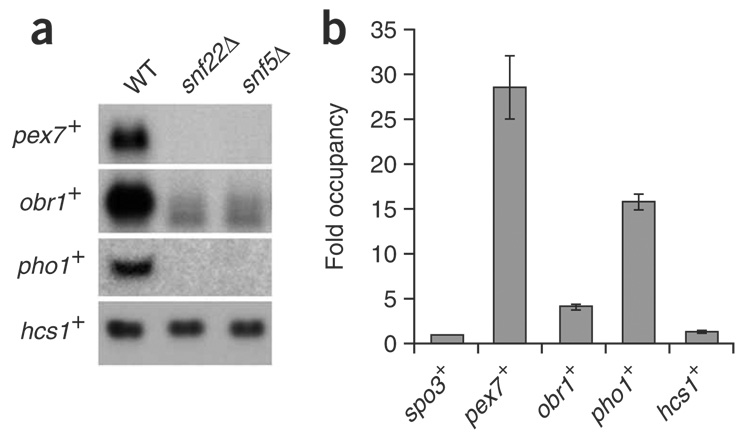
Microarray analysis identified 133 genes whose expression was altered more than two-fold in the snf22Δ and snf5Δ mutants, with approximately an equal proportion of genes up- or downregulated (64 and 69 genes, respectively; Fig. 3b,c). This result suggests that SWI/SNF is required for both activation and repression of transcription. Among the downregulated genes were pex7+, obr1+ and pho1+, whose mRNA levels were decreased 26-fold, 8-fold and 8-fold, respectively, in the snf22Δ and snf5Δ mutants. To validate the microarray data for these genes, we performed northern analysis and confirmed that SWI/SNF is required for their activation (Fig. 4a). To test whether SWI/SNF directly activates these genes, we performed chromatin immunoprecipitation (ChIP) using the Snf22-TAP construct. Our results show that Snf22-TAP is strongly and specifically associated at the pex7+, obr1+ and pho1+ promoter regions (Fig. 4b), indicating that SWI/SNF is directly required for their activation.
Figure 4.
SWI/SNF is directly required for transcriptional activation of genes. (a) Northern blot analysis of iron-uptake genes under iron-sufficient conditions. RNA was prepared from the wild type (WT, FWP165), snf22Δ (FWP229) and snf5Δ (FWP220) strains. Probes specific for the pex7+, obr1+ and pho1+ genes were used as indicated. (b) SWI/SNF is physically associated with the regulatory regions of the pex7+, obr1+ and pho1+ genes. ChIP analysis of Snf22-TAP (FWP289) cultures is shown. Occupancy levels were determined (Methods) and normalized to the spo3+ promoter region, which is set to 1. Values calculated represent the average of three independent biological replicates, and error bars indicate the s.e.m.
Direct role for SWI/SNF in transcriptional repression
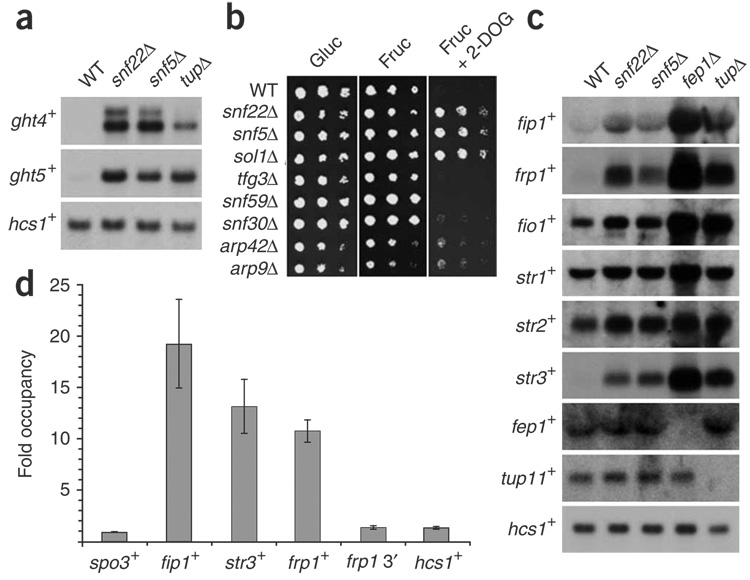
Of the 64 genes with increased mRNA levels in the SWI/SNF mutants (Fig. 3c), two gene-ontology categories were significantly enriched: hexose transporter activity (GO0015749; P-value = 9.3e−06) and iron ion transport (GO0006826; P-value = 9.2e−10; discussed below). Schizosaccharomyces pombe contains eight ght (glucose hexose transporters) genes37 and the mRNA levels for most of these genes were increased in the snf22Δ and snf5Δ mutants (Supplementary Table 4 online). To confirm the microarray data, we measured ght4+ and ght5+ mRNA levels in wild-type, snf22Δ, snf5Δ and tup11Δ tup12Δ strains. The functionally redundant Tup11 and Tup12 transcriptional co-repressors have been previously implicated in ght gene regulation38. Our results, from both northern blots (Fig. 5a) and reverse-transcription PCR (RT-PCR; data not shown) confirmed that ght4+ and ght5+ have increased mRNA levels in the snf22Δ, snf5Δ and tup11Δ tup12Δ mutants compared to wild type. Therefore, SWI/SNF represses several genes required for hexose uptake. To test whether SWI/SNF has a physiological role in hexose sugar uptake, we investigated whether SWI/SNF mutants are resistant to the nonmetabolizable glucose analog 2-deoxyglucose (2-DOG). Some S. pombe mutants resistant to 2-DOG have altered hexose transport39. Indeed, snf22Δ, snf5Δ and sol1Δ mutants show strong resistance to 1 mM 2-DOG, whereas complete growth inhibition was seen for the wild-type strain (Fig. 5b). In conclusion, SWI/SNF seems to be important for the negative regulation of hexose sugar uptake.
Figure 5.
SWI/SNF represses the transcription of hexose transport and iron-uptake genes. (a) Northern blot analysis of ght4+ and ght5+ mRNA levels under glucose-sufficient conditions. RNA was prepared from the wild type (WT, FWP165), snf22Δ (FWP229), snf5Δ (FWP220) and tup11Δ tup12Δ (tupΔ, FWP197) strains. hcs1+ served as a loading control. (b) SWI/SNF mutants show 2-DOG resistance. Ten-fold serial dilutions were spotted onto solid YES medium containing 2% (w/v) glucose (Gluc), 1 mM 2-DOG and 2% (w/v) fructose (Fruc) as indicated. Spot tests were performed as for Figure 2a. (c) Northern blot analysis of iron-uptake genes under iron-sufficient conditions. The strains used were as for a with the addition of fep1Δ (FWP253). Probes specific for the iron-uptake genes were used as indicated. (d) SWI/SNF is physically associated with the regulatory regions of iron-transport genes under repressing conditions. ChIP analysis of Snf22-TAP (FWP289) cultures is shown. Occupancy levels were determined as described for Figure 4 and in Methods.
Schizosaccharomyces pombe has both reductive and nonreductive iron-acquisition systems. The reductive system requires Fip1, Frp1 and Fio1, whereas the nonreductive system uses the siderophore–iron transporters Str1, Str2 and Str3 (reviewed in ref. 40). The expression of the six genes encoding these proteins is repressed under high-iron conditions by the repressor Fep1, which forms a complex with the co-repressors Tup11 and/or Tup12 (refs. 40,41). In the snf22Δ and snf5Δ microarrays, all six genes encoding the reductive and nonreductive components showed increased mRNA levels, although the increase was only slight for str1+ and str2+ (Supplementary Table 4). In contrast, expression of the regulators fep1+, tup11+ and tup12+ was unchanged (Supplementary Table 4). To validate the microarray data, we performed northern analysis, which confirmed that SWI/SNF is required for the repression of iron-uptake genes (Fig. 5c).
To test whether SWI/SNF directly represses the iron-uptake genes, we carried out ChIP analysis to determine whether SWI/SNF is physically associated with iron-transport gene promoters under repressing conditions. For these experiments, we used the Snf22-TAP construct. Our results show that Snf22-TAP is strongly associated at all three promoter regions tested, fip1+, str3+ and frp1+ (Fig. 5d). This association is specific, as we detected no Snf22-TAP association for several control regions (Fig. 5d). These results, taken together with the SWI/SNF mutant analysis, strongly suggest that SWI/SNF directly represses transcription of these genes.
DISCUSSION
In this work, we have presented the first purification and functional characterization of the S. pombe SWI/SNF and RSC chromatin-remodeling complexes. Our results show that these complexes have diverged substantially from their S. cerevisiae counterparts and that they have broad and crucial roles in vivo. Although several core components are conserved between the S. pombe, S. cerevisiae and metazoan complexes, the S. pombe complexes are more reminiscent of the metazoan complexes with respect to the number of components in each complex and the number of components shared between the SWI/SNF and RSC complexes. In addition, there are many notable differences between S. pombe and S. cerevisiae with respect to the requirement for particular subunits. Most notably, the two Arp proteins that are shared between SWI/SNF and RSC, which are essential or almost essential for viability in S. cerevisiae, are not required for growth in S. pombe. Finally, the classes of genes controlled by the S. pombe complexes are different from those regulated by the S. cerevisiae complexes. Our results strongly suggest that S. pombe SWI/SNF has a direct role in transcriptional repression. Taken together, these studies have comprehensively characterized the S. pombe SWI/SNF and RSC complexes and have established a foundation for a detailed understanding of their roles in transcription and chromatin structure in this organism.
An intriguing discovery from our work is the discovery that S. pombe SWI/SNF and RSC contain Arp42 and Arp9, a pairing of Arp proteins not previously observed in chromatin-remodeling complexes31. The Arp4 proteins are paired with nuclear actin within all known Arp-containing chromatin-related complexes, with the exception of SWI/SNF and RSC in S. cerevisiae31 and, now, S. pombe. Arp42 is one of two Arp4-like proteins in S. pombe (Supplementary Fig. 1); the other, Alp5, is essential for viability and is a member of the NuA4 complex42 and probably the Ino80 and Swr1 complexes. Humans also contain two Arp4 proteins, BAF53a and BAF53b, that have been shown to confer distinct properties to human SWI/SNF (BAF) complexes in the developing nervous system43,44. Thus, the functional specificity of chromatin-related complexes can be achieved through the substitution of distinct Arp4 proteins.
A related and surprising result from our studies was the finding that the loss of Arp proteins from SWI/SNF and RSC caused no substantial effect on growth under normal laboratory conditions, whereas loss of either Arp protein of S. cerevisiae SWI/SNF and RSC causes inviability or extremely poor growth, depending on the strain background20. However, similar to our findings, S. cerevisiae RSC can assemble in the absence of either Arp32. Our results can be compared to the Ino80 chromatin-remodeling complex45, which contains actin, Arp4, Arp5 and Arp8. The loss of actin or Arp4 causes inviability, whereas loss of Arp5 and Arp8 does not. Both Arp5 and Arp8, however, have crucial roles in Ino80 function, as an arp8Δ mutation causes loss of Arp4 and actin from Ino80 assembly, and either arp5Δ or arp8Δ abolishes the activity of the complex45. Therefore, the S. cerevisiae Ino80 Arps seem to have a more substantial role than the S. pombe SWI/SNF and RSC Arps. The finding that the S. pombe arp42Δ, arp9Δ and arp42Δ arp9Δ mutants possess conditional mutant phenotypes shows that they have important functions in these complexes under some circumstances. However, the conditional nature of the phenotypes suggests that their roles may be important only at specific genes, consistent with the idea that Arps may interact with histones in a way that is dependent on histone modifications31. For example, Arp4 has been shown to interact with phosphorylated H2A to recruit the NuA4 complex to sites of DNA damage46.
Another result that aligns S. pombe SWI/SNF function more closely with metazoans than S. cerevisiae is that S. pombe SWI/SNF seems to directly repress transcription at numerous genes. Although a recent study has provided evidence for a modest role for S. cerevisiae SWI/SNF in direct repression of heat-shock genes47, no case of strong repression has been identified in S. cerevisiae. One previous study suggested direct repression of the S. cerevisiae SER3 gene by SWI/SNF48; however, subsequent studies demonstrated that this regulation is indirect49. In contrast, there is substantial evidence for direct repression by mammalian SWI/SNF complexes, often by recruitment of histone deacetylase complexes21,24. We have shown that S. pombe SWI/SNF is physically associated with at least three regulatory regions where it confers strong repression. Furthermore, this repression seems to be direct, as there are no detectable RNAs produced in these regions (S.M. and J.B., unpublished results).
As in S. cerevisiae, in S. pombe both SWI/SNF and RSC are required for normal transcription of many genes, yet RSC is essential and is involved in a much broader role in transcription than is SWI/SNF. However, our results show that the requirement for particular subunits and the pattern of effects on transcription differs substantially between S. pombe and S. cerevisiae. With respect to the specific genes that are controlled by these complexes, these also differ greatly between the two yeasts. This result is consistent with a recent analysis of transcription factor binding sites between S. cerevisiae and S. bayanus that suggests that regulatory sites have diverged faster than coding regions50. Given that in many cases SWI/SNF and RSC are recruited by gene-specific regulatory proteins, such divergence would be likely to alter the genes subject to their control.
In addition to elucidating the biochemical and functional differences between the S. pombe and S. cerevisiae SWI/SNF and RSC complexes, our studies have identified several distinct biological processes in which the S. pombe complexes participate. These include glucose uptake and iron homeostasis for SWI/SNF and endoplasmic reticulum and Golgi function for RSC. Furthermore, some RSC mutants have apparent cell-cycle and chromosome-segregation defects; these phenotypes suggest that S. pombe RSC may be valuable as a model system to investigate the molecular basis for the involvement of SWI/SNF complexes in tumorigenesis in mammalian cells25. Our results have established S. pombe as an important system for the study of chromatin-remodeling complexes in gene expression.
METHODS
Schizosaccharomyces pombe strains and genetic methods
Strains used in this study are listed in Supplementary Table 5 online. The gene name ssr (SWI/SNF and RSC) has been used to name previously unannotated genes encoding products shared between SWI/SNF and RSC. Schizosaccharomyces pombe was grown at 32 °C, unless stated otherwise, in standard rich (YES) or defined (EMM) growth medium supplemented with the necessary requirements51. We carried out genetic crosses by mating strains at 29 °C onME or SPASmedia51. Diploid cells were isolated by ade6-M210 and ade6-M216 intragenic complementation followed by selection for adenine prototrophs. To test for various plate phenotypes, 2% (v/v) formamide (Fluka), 1 mM2-deoxyglucose (Sigma), 10 mM caffeine (Sigma), 10 µg ml−1 brefeldin A (Sigma), 10 µg ml−1 thiabendazole (Sigma), 10 µg ml−1 benomyl (Sigma) and 5 mM hydroxyurea (Sigma) were added individually to the growth medium. For microscopy, all images were acquired using a Nikon Eclipse TE2000-E equipped with a 100×,1.4 numerical aperture, Plan Apo objective (Nikon) and with Metamorph 6.3r7 (Molecular Devices) software. To delete genes encoding SWI/SNF and RSC components, the respective open reading frame was replaced in diploid cells with the ura4+ gene by PCR-based gene targeting as described52. In each case, ura4+ was amplified using primers of 100 bases, with 20 bases designed for amplification and 80 bases designed to direct homologous recombination to the sites flanking the coding region to be replaced (Supplementary Table 6 online). Cells were transformed as described52, except that for transformation of diploid cells, initial cultures were grown at 34 °C to prevent sporulation. Diploid transformants were screened by colony PCR for the correct gene-replacement event and candidates were confirmed by genomic DNA isolation51 and further PCR analysis. Confirmed deletion mutants were sporulated, tetrads were dissected and haploid progeny were assessed for viability. Dissected tetrads were genotyped, and the gene-replacement event was again confirmed by colony PCR. Previous analysis had conclusively shown that a SPBC1734.15 (rsc4/brd1) deletion mutant was viable53; therefore, a rsc4::ura4+ mutant was recreated in haploid cells. Double-deletion mutants were obtained by crosses. To create S. pombe TAP-fusion strains, the TAP tag was placed at the 3′ end of the respective gene by homologous recombination. Correct integration was verified by PCR. Whole-cell protein extracts51 and western blot analysis using the PAP antibody (peroxidase–anti-peroxidase; Sigma) confirmed expression of the TAP fusion protein (data not shown). Oligonucleotide primers used for the deletion of genes or epitope tagging were designed using the Pombe PCR Primer Program (PPPP), available at http://www.sanger.ac.uk/PostGenomics/S_pombe/software.
Tandem affinity purification of SWI/SNF and RSC complexes
The S. pombe SWI/SNF and RSC complexes were purified by the TAP method54. Four different SWI/SNF or RSC subunits were TAP-tagged and analyzed for each complex. For SWI/SNF, TAP fusions and purifications were done for Snf22 (SPCC1620.14c), Snf5 (SPAC2F7.08c), Snf30 (SPAC23G3.07c) and Snf59 (SPBC26H8.09c) proteins. For RSC, the Snf21 (SPAC1250.01), Sfh1 (SPCC16A11.14), Rsc7 (SPCC1281.05) and Rsc1 (SPBC4B4.03) proteins were analyzed. In addition, TAP fusions and purifications were done for Arp42 (SPAC23D3.09) and Arp9 (SPAC1071.06). TAP preparations were performed from 3-liter cultures grown at 30 °C to a cell concentration of approximately 2 × 107 cells ml−1, as previously described54, except that cells were mechanically lysed using a Krups Tipo 203 coffee grinder and dry ice. Purifications were assessed by western analysis using anti-hemagluttinin (generous gift from Brad Cairns) or anti-TAP (Open Biosystems) antibodies and by silver-stain analysis of SDS-PAGE gels using SilverQuest (Invitrogen). Total protein mixtures were precipitated with 20% (w/v) trichloroacetic acid (TCA; Sigma) before MS analysis.
Mass spectrometry analysis of complexes
For MS analysis of the purified SWI/SNF and RSC S. pombe complexes, whole-protein mixtures were digested in solution with trypsin and 10–20% was analyzed by LC-MS/MS. Peptides were separated across a 55-min gradient ranging from 7% to 30% (v/v) acetonitrile (ACN) in 0.1% (v/v) formic acid (FA) in a microcapillary (125 µm × 17 cm) column packed with C18 reverse-phase material (Magic C18AQ, 5 µm particles, 200-Å pore size, Michrom Bioresources) and on-line analyzed on a hybrid MS, either an LTQ-FT or an LTQ-Orbitrap (Thermo-Electron). For each cycle, one full MS scan acquired at high mass resolution was followed by ten MS/MS spectra on the linear ion trap from the ten most abundant ions. MS/MS spectra were searched against the S. pombe protein sequence database using the Sequest algorithm55. Peptide matches were filtered to <0.5% false positives using a target-decoy database strategy56. Final lists of proteins involved in the various complexes were obtained by subtracting protein matches found also in an untagged control sample.
Chromatin immunoprecipitation
We carried out ChIP experiments as previously described57, using the prototrophic h+ snf22+-TAP strain FWP289 and the untagged control strain FWP287. Briefly, cells were grown to mid–log phase in YES medium at 32 °C, then cross-linked in 1% (v/v) formaldehyde for 30 min. Cells were broken by bead beating, and the chromatin fraction was sheared to 200–500 bp fragments using a Bioruptor sonicator (Diagenode). For immunoprecipitations, 4 µl of anti–protein A antibody (Sigma) was coupled to 100 µl of Dynabeads (Invitrogen). ChIP DNA was quantified and occupancy levels determined by real-time PCR, using a Stratagene MX3000P. Specifically, qPCR was done in triplicate for each primer set, using a standard curve that was established by serial ten-fold dilutions of a representative input DNA. Occupancy levels were determined by dividing the relative abundance for snf22+-TAP (immunoprecipitation value over input) by that of the no-tag control. These values were then normalized to the spo3+ promoter region, which is inactive during vegetative growth58. Values calculated represent the average of three independent experiments. The primer sequences are listed in Supplementary Table 6.
Microarray experiments
Total RNA was isolated using a hot-phenol protocol as previously described59 (http://www.sanger.ac.uk/PostGenomics/S_pombe/). Between 10–20 µg of total RNA were labeled by direct incorporation of either fluorescent Cy3- or Cy5-dCTP (GE Healthcare), and the fluorescently labeled product was hybridized to S. pombe cDNA microarrays as previously described59. Microarrays were scanned using a GenePix 4000B laser scanner (Axon Instruments) and fluorescence-intensity ratios were calculated with GenePix Pro (Axon Instruments). The data were normalized using a previously described script59. At least two biological repeats were analyzed for each mutant with dye swaps. To analyze the data, repeats for every given mutant were averaged. For each complex, genes with ratios >2 or <0.5 (mutant/wild type) for at least one mutant were called up- or downregulated, respectively. The significance of overlaps between different gene lists was calculated in Gene-Spring (Agilent) by using a standard Fisher’s exact test, and the P-values were adjusted with a Bonferroni multiple testing correction. Hierarchical clustering of genes up- or downregulated in at least one mutant was performed in GeneSpring using Spearman correlation, with genes containing no data in ≤50% of the conditions being discarded. SAM35 was performed for the arp and RSC data using all available repeats, with a false-discovery rate adjusted to 1%. The correlations between different mutants were determined using the correlation function in Excel. Microarray data for the SWI/SNF and RSC mutants is shown in Supplementary Table 7 online.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank D. Helmlinger and M. Gelbart for helpful comments on the manuscript. We are grateful to C. Hoffman (Biology Department, Boston College) and S. Labbé (Department of Biochemistry, University of Sherbrooke) for providing S. pombe strains. We also thank D. Drubin and P. Silver for assistance with the microscopy analysis and use of their facilities. This work was supported by the US National Institutes of Health grant GM32967 to F.W., HG3456 to S.P.G., and a Cancer Research UK grant C9546/A6517 to J.B. B.J.M. was supported by a Post-Doctoral Research Fellowship from the New Zealand Foundation of Research Science and Technology, and S.M. was supported by a fellowship for Advanced Researchers from the Swiss National Science Foundation.
Footnotes
Accession code. Raw microarray data are available from ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-31.
References
- 1.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.van Vugt JJ, Ranes M, Campsteijn C, Logie C. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim. Biophys. Acta. 2007;1769:153–171. doi: 10.1016/j.bbaexp.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 5.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dror V, Winston F. The SWI/SNF chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:8227–8235. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and SWI/SNF ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns BR, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 9.Laurent BC, Yang X, Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol. Cell. Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angus-Hill ML, et al. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- 11.Damelin M. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- 12.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasten M, et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soutourina J, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y, Cairns BR, Kornberg RD, Laurent BC. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 19.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 21.Simone C. SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. J. Cell. Physiol. 2006;207:309–314. doi: 10.1002/jcp.20514. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 24.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by SWI/SNF complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 25.Sansam CG, Roberts CW. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle. 2006;5:621–624. doi: 10.4161/cc.5.6.2579. [DOI] [PubMed] [Google Scholar]
- 26.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 27.Yamada T, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernal G, Maldonado E. Isolation of a novel complex of the SWI/SNF family from Schizosaccharomyces pombe and its effects on in vitro transcription in nucleosome arrays. Mol. Cell. Biochem. 2007;303:131–139. doi: 10.1007/s11010-007-9465-y. [DOI] [PubMed] [Google Scholar]
- 29.Wilson B, Erdjument-Bromage H, Tempst P, Cairns BR. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics. 2006;172:795–809. doi: 10.1534/genetics.105.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns BR, et al. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, Shen X. Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 2007;19:326–330. doi: 10.1016/j.ceb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 34.Aslett M, Wood V. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turi TG, Webster P, Rose JK. Brefeldin A sensitivity and resistance in Schizosaccharomyces pombe. Isolation of multiple genes conferring resistance. J. Biol. Chem. 1994;269:24229–24236. [PubMed] [Google Scholar]
- 37.Heiland S, Radovanovic N, Hofer M, Winderickx J, Lichtenberg H. Multiple hexose transporters of Schizosaccharomyces pombe. J. Bacteriol. 2000;182:2153–2162. doi: 10.1128/jb.182.8.2153-2162.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fagerstrom-Billai F, Durand-Dubief M, Ekwall K, Wright AP. Individual subunits of the Ssn6-Tup11/12 corepressor are selectively required for repression of different target genes. Mol. Cell. Biol. 2007;27:1069–1082. doi: 10.1128/MCB.01674-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta SV, Patil VB, Velmurugan S, Lobo Z, Maitra PK. Std1, a gene involved in glucose transport in Schizosaccharomyces pombe. J. Bacteriol. 1998;180:674–679. doi: 10.1128/jb.180.3.674-679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labbe S, Pelletier B, Mercier A. Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals. 2007;20:523–537. doi: 10.1007/s10534-006-9056-5. [DOI] [PubMed] [Google Scholar]
- 41.Znaidi S, Pelletier B, Mukai Y, Labbe S. The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J. Biol. Chem. 2004;279:9462–9474. doi: 10.1074/jbc.M312787200. [DOI] [PubMed] [Google Scholar]
- 42.Minoda A, Saitoh S, Takahashi K, Toda T. BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment, and gene silencing at centromere. Mol. Biol. Cell. 2005;16:316–327. doi: 10.1091/mbc.E04-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu JI, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 45.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 46.Downs JA, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Shivaswamy S, Iyer VR. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 2008;28:2221–2234. doi: 10.1128/MCB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martens JA, Winston F. Evidence that SWI/SNF directly represses transcription in S. cerevisiae. Genes Dev. 2002;16:2231–2236. doi: 10.1101/gad.1009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 51.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 53.Aves SJ, Hindley J, Phear GA, Tongue N. A fission yeast gene mapping close to suc1 encodes a protein containing two bromodomains. Mol. Gen. Genet. 1995;248:491–498. doi: 10.1007/BF02191650. [DOI] [PubMed] [Google Scholar]
- 54.Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 55.MacCoss MJ, Wu CC, Yates JR., III Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 56.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 57.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 59.Lyne R, et al. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kippert F, Lloyd D. The aniline blue fluorochrome specifically stains the septum of both live and fixed Schizosaccharomyces pombe cells. FEMS Microbiol. Lett. 1995;132:215–219. doi: 10.1111/j.1574-6968.1995.tb07836.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.