Abstract
ARNO is a member of a family of guanine-nucleotide exchange factors with specificity for the ADP-ribosylation factor (ARF) GTPases. ARNO possesses a central catalytic domain with homology to yeast Sec7p and an adjacent C-terminal pleckstrin homology (PH) domain. We have previously shown that ARNO localizes to the plasma membrane in vivo and efficiently catalyzes ARF6 nucleotide exchange in vitro. In addition to a role in endocytosis, ARF6 has also been shown to regulate assembly of the actin cytoskeleton. To determine whether ARNO is an upstream regulator of ARF6 in vivo, we examined the distribution of actin in HeLa cells overexpressing ARNO. We found that, while expression of ARNO leads to disassembly of actin stress fibers, it does not result in obvious changes in cell morphology. However, treatment of ARNO transfectants with the PKC agonist phorbol 12-myristate 13-acetate results in the dramatic redistribution of ARNO, ARF6, and actin into membrane protrusions resembling lamellipodia. This process requires ARF activation, as actin rearrangement does not occur in cells expressing a catalytically inactive ARNO mutant. PKC phosphorylates ARNO at a site immediately C-terminal to its PH domain. However, mutation of this site had no effect on the ability of ARNO to regulate actin rearrangement, suggesting that phosphorylation of ARNO by PKC does not positively regulate its activity. Finally, we demonstrate that an ARNO mutant lacking the C-terminal PH domain no longer mediates cytoskeletal reorganization, indicating a role for this domain in appropriate membrane localization. Taken together, these data suggest that ARNO represents an important link between cell surface receptors, ARF6, and the actin cytoskeleton.
INTRODUCTION
The actin cytoskeleton of animal cells plays an active role in a large number of cellular functions, such as cell shape change, formation of stress fibers and focal adhesions, cell motility, membrane ruffling, cytokinesis, cell-to-cell adhesion, and endocytosis (Nobes and Hall, 1995b; Hall, 1998). To accomplish these functions, the actin cytoskeleton is capable of rapid remodeling in response to a diverse array of extracellular signals. Examination of the signaling pathways that translate signals originating at the cell surface into actin reorganization led to the finding that members of the Rho-related family of GTPases, which include Cdc42, Rac1, and RhoA, could regulate the formation of distinct actin structures. In fibroblasts, activation of Cdc42 and Rac1 results in the formation of filopodia and lamellipodia, respectively, while RhoA is linked to the formation of stress fibers and focal contacts (Ridley and Hall, 1992; Ridley et al. 1992; Kozma et al., 1995; Nobes and Hall, 1995a). Each of these GTPases can be activated by specific membrane receptors, including receptor tyrosine kinases and G protein-coupled receptors (Kozma et al., 1995; Nobes and Hall, 1995a). These findings illustrate the existence of distinct signaling pathways exist that modulate the formation of unique actin-based structures.
The ADP-ribosylation factors (ARFs)1 are also members of the Ras superfamily. In contrast to the Rho family proteins, ARFs have traditionally been thought to play a role in the regulation of intracellular membrane trafficking (Donaldson and Klausner, 1994). ARF1, which is the best understood of the six mammalian ARFs, plays a key role in the secretory pathway. Activation of ARF1 results in its recruitment from the cytosol to the Golgi complex where it mediates the binding of coat proteins and adaptins to Golgi membranes (reviewed by Donaldson and Klausner, 1994; Boman and Kahn, 1995). In contrast to ARF1, ARF6 localizes to the plasma membrane and endosomes where it may modulate some aspects of endocytosis (D’Souza-Schorey, et al., 1995; Peters et al., 1995; Cavenaugh et al., 1996; Radhakrishna and Donaldson, 1997; Yang et al., 1998). However, recent work has suggested that ARF6, like Rho GTPases, may also be capable of regulating actin structure and function (Radhakrishna et al., 1996; D’Souza-Schorey et al., 1997). In HeLa cells overexpressing wild-type ARF6, aluminum fluoride treatment results in the rapid redistribution of cortical actin into plasma membrane protrusions resembling lamellipodia (Radhakrishna et al., 1996). These structures, while enriched in several actin-associated molecules (e.g., gelsolin, talin, FAK), are nevertheless morphologically distinct from the rearrangements stimulated by Rac1 and RhoA.
The specific cellular effectors that mediate ARF6 function are poorly understood. ARFs 1, 5, and 6 are potent activators of phospholipase D (PLD) (Brown et al., 1993; Massenburg et al., 1994). Once activated, PLD generates phosphatidic acid, and it has been suggested that the localized production of phosphatidic acid may play a key role in both vesicle formation (Ktistakis et al., 1996) and actin reorganization (Ha and Exton, 1993; Cross et al., 1996). Another potential target of ARF6 is POR1, a previously described Rac1 interacting protein whose function is required for membrane ruffling (Van Aelst, et al., 1996; D’Souza-Schorey et al., 1997). POR1 has also recently been shown to interact with GTP-bound ARF6, and POR1 mutants are capable of inhibiting ARF6-dependent actin remodeling (D’Souza-Schorey et al., 1997). These data suggest that POR1 represents a common downstream effector of ARF6 and Rac1 and may indicate that the cytoskeletal rearrangements regulated by these GTP-binding proteins are coordinated.
Like other GTPases, the cycling of ARFs between GDP- and GTP-bound states is aided by a number of regulatory proteins, including GTPase-activating proteins (Cukierman et al., 1995) and guanine-nucleotide exchange factors (GEFs) (Schimmoller et al., 1997). A number of GEFs, which promote binding of GTP to ARF by facilitating the release of GDP, have been identified (Chardin et al., 1996; Peyroche et al., 1996; Klarlund et al., 1997; Meacci et al., 1997; Morinaga et al., 1997; Sata et al., 1998). Although different in size and sequence, all share an ∼200-amino acid catalytic domain, referred to as the Sec7 domain. The specific role of any of these regulators in the diverse processes regulated by ARF proteins is largely unknown.
A subfamily of ARF GEFs, comprised of the proteins ARNO, cytohesin-1 and GRP1, is distinguished by a unique domain structure consisting of an N-terminal coiled-coil domain, a central Sec7 domain, and a C-terminal pleckstrin homology (PH) domain that seems to be important for membrane recruitment through interaction with inositol phospholipids (Chardin et al., 1996; Klarlund et al., 1997; Meacci et al., 1997). ARNO was originally characterized as an exchange factor for ARF1 (Chardin et al., 1996). However, we recently reported that, in vitro, ARNO can catalyze nucleotide exchange on both ARFs 1 and 6 (Frank et al., 1998) and have subsequently found that it exhibits exchange activity for all ARFs (our unpublished observations). Given the highly conserved nature of the Sec7 domain, it seems likely that all of the ARF GEFs will behave promiscuously when assayed in vitro, and that the function of specific GEFs in vivo will be determined by their subcellular localization. We previously reported that, in BHK cells, ARNO is not associated with Golgi or ER membranes, as would be expected of an ARF1 GEF, but rather is enriched in plasma membrane fractions. Immunofluorescence microscopy revealed extensive overlap in the distribution of ARNO and ARF6 at the plasma membrane (Frank et al., 1998), further suggesting that ARNO catalyzes exchange on ARF6 in vivo.
We have now extended our previous studies by assessing the function of ARNO in intact cells. Work in other systems reveals that overexpression of GEFs frequently recapitulates the phenotype observed in cells expressing activated forms of their target G proteins. For example, Tiam1 encodes a GEF for Rac1 and, similar to constitutively activated (V12)Rac1, overexpression of Tiam1 in fibroblasts induces membrane ruffling (Michiels et al., 1995). We hypothesized that if ARNO is, in fact, an ARF6 nucleotide exchange factor, its overexpression should generate a phenotype similar to that of activated ARF6. Here we report that, like ARF6, ARNO regulates the structure of the cortical actin cytoskeleton. Importantly, ARNO-dependent actin remodeling requires ARNO catalytic activity and the concurrent activation of protein kinase C (PKC), indicating that the intracellular signaling pathways regulated by ARNO and PKC are coordinated. We demonstrate that an ARNO mutant lacking the C-terminal PH domain no longer mediates cytoskeletal reorganization, suggesting a role for this domain in appropriate intracellular targeting. Our data provide the first evidence of a specific physiological role for ARNO and suggest that its regulated recruitment to distinct plasma membrane sites may provide an essential link between extracellular signals and the activation of ARF6.
MATERIALS AND METHODS
Cells, Reagents, and Antibodies
HeLa cells were grown in DMEM supplemented with 10% FCS, antibiotics, and 2 mM l-glutamine. For the studies described, the following antibodies were used: anti-myc monoclonal antibody 9E10, anti-HA polyclonal 16B12 (BabCo, Berkeley, CA) anti-giantin rabbit polyclonal antibody (Seelig et al., 1994), donkey anti-rabbit IgG coupled to Texas red (Jackson ImmunoResearch, West Grove, PA), donkey anti-mouse IgG coupled to Cy2 (Jackson ImmunoResearch), and goat anti-mouse IgG coupled to horseradish peroxidase (Southern Biotechnology, Birmingham, AL). The rabbit polyclonal antibody to ARNO has been described previously (Frank et al., 1998). For use in immunofluorescence microscopy anti-ARNO antisera were purified by passage over a GST-ARNOΔ1–53 (deletion of amino acids Met1 to Asn53) affinity column. GST-ARNOΔ1–53 was used for affinity purification since antibodies binding to the N-terminal coiled-coil domain of ARNO cross-reacted with the intermediate filament protein vimentin when assayed by indirect immunofluorescence (our unpublished observations). All other chemicals were purchased from Sigma Chemical (St. Louis, MO).
Plasmids and Transient Transfections
To generate a wild-type tagged ARNO construct, native ARNO cDNA was amplified by PCR using a noncoding primer that included the c-myc epitope sequence. ARNO point mutants were generated using wild-type myc-tagged ARNO as a template and overlapping mutant primers following the protocol outlined in the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). To generate ARNOΔPH, native ARNO cDNA was amplified by PCR using a coding primer that included the c-myc epitope sequence and a noncoding primer in which Leu269 is replaced by a stop codon. Wild-type ARNO myc-tagged at either the N- or C terminus functioned indistinguishably in inducing cytoskeletal rearrangments. The wild-type and various mutant constructs were all subcloned into the cytomegalovirus-based mammalian expression vector pCB7 (Brewer, 1994). For production of recombinant His6-tagged proteins, wild-type ARNO and ARNO(E156K) were subcloned into the bacterial expression vector pQE8 (Qiagen, Santa Clarita, CA). Sequences of all constructs were confirmed by restriction digests and DNA sequencing.
For transient transfections, HeLa cells grown on glass coverslips (5 × 104 cells/well) were transfected using calcium-phosphate precipitation. Thirty to 36 h after transfection, cells were fixed with 2% formaldehyde in PBS, washed twice with PBS, and then incubated with the appropriate antibodies diluted in blocking buffer (PBS/10% normal goat serum/0.2% saponin). For actin staining, rhodamine-phalloidin (Molecular Probes, Eugene, OR) was combined with the secondary antibody. Images were obtained using a Nikon Microphot fluorescence microscope (Nikon, Melville, NY) equipped with a DEI 750 Optronics Engineering (Goleta, CA) video camera and IP Lab Spectrum software (Vienna, VA).
Infection of HeLa Cells with Recombinant Adenovirus and Quantitative Immunoblotting
Recombinant adenovirus encoding either wild-type or the E156K ARNO mutant under the control of a tetracycline-repressible promoter was produced in collaboration with Yoram Altschuler and Keith Mostov (Department of Anatomy, University of California, San Francisco) and used to infect HeLa cells stably expressing the tetracycline transactivator (kindly provided by Dr. Sandra Schmid, Scripps Institute, San Diego, CA). Cells were infected in the absence of tetracycline at an m.o.i of 10 for 3 h, at which point ARNO expression was readily detectable by immunofluorescence microscopy. Cells were then lysed in SDS sample buffer and subjected to SDS-PAGE, along with a set of recombinant His6-ARNO standards of known concentration, and a sample from noninfected cells. Gels were then transferred to nitrocellulose and immunoblotted with anti-ARNO antiserum. Quantitation of immunoblots was performed using IP-Labgel software.
GEF Assays
Nucleotide exchange assays were performed as previously described (Chardin et al., 1996). Briefly, recombinant, myristoylated ARF6 was diluted to a final concentration of 1 μM into reaction buffer containing 50 mM HEPES, pH 7.5, 1 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, 4 μM [35S]GTPγS (∼3 × 106 cpm), and 1.5 mg/ml azolectin vesicles. Reactions were initiated by addition of 100 nM His6-ARNO.
In Vivo Phosphorylation Analysis
HeLa cells were transfected as above. Thirty six to 48 h after transfection, cells were washed twice with 5 ml in DMEM without sodium phosphate and incubated twice for 1 h in the same medium. Labeling was for 4.5 h in 2.5 ml of medium supplemented with 1.25 mCi of [32P]orthophosphate (Dupont New England Nuclear, Boston, MA). If required, phorbol 12-myristate 13-acetate (PMA) (1 μM) was added for 30 min at the end of this labeling period. The cells were then scraped in 0.5 ml/dish SDS lysis buffer (50 mM Tris, pH 8.1, 100 mM NaCl, 5 mM EDTA, 0.5% SDS), boiled for 3 min, and then diluted 1:1 with Triton dilution buffer (100 mM Tris, pH 8.6, 100 mM NaCl, 5 mM EDTA, 2.5% Triton X-100). After preclearing, supernatants were incubated with anti-ARNO antiserum covalently coupled to protein A-Sepharose for 1 h at room temperature. Samples were resolved by 10% SDS-PAGE, transferred to nitrocellulose, and analyzed by autoradiography. For analysis of expression levels, blots were subsequently probed with the anti-myc monoclonal antibody 9E10.
In Vitro PKC Phosphorylation Assay
Purified His6-ARNO (3 μg) was incubated for 30 min at 37°C in the presence or absence of 20 ng of rat brain PKC (Biomol, Plymouth Meeting, PA) in 5 mM Tris-HCl buffer, pH 7.5, containing 2.5 μM PMA, 0.125 mg/ml phosphatidylserine, 100 μM ATP, 25 μM CaCl2, 1.25 mM MgCl2, 0.0075% Triton X-100, and 0.25 μCi/μl [γ-32P]ATP (3000 Ci/mmol). When specified, the PKC-specific inhibitor, bis-indolylmaleimide (BIM) (Calbiochem, San Diego, CA) was used at 10 μM.
RESULTS
Effects of ARNO Expression on the Actin Cytoskeleton
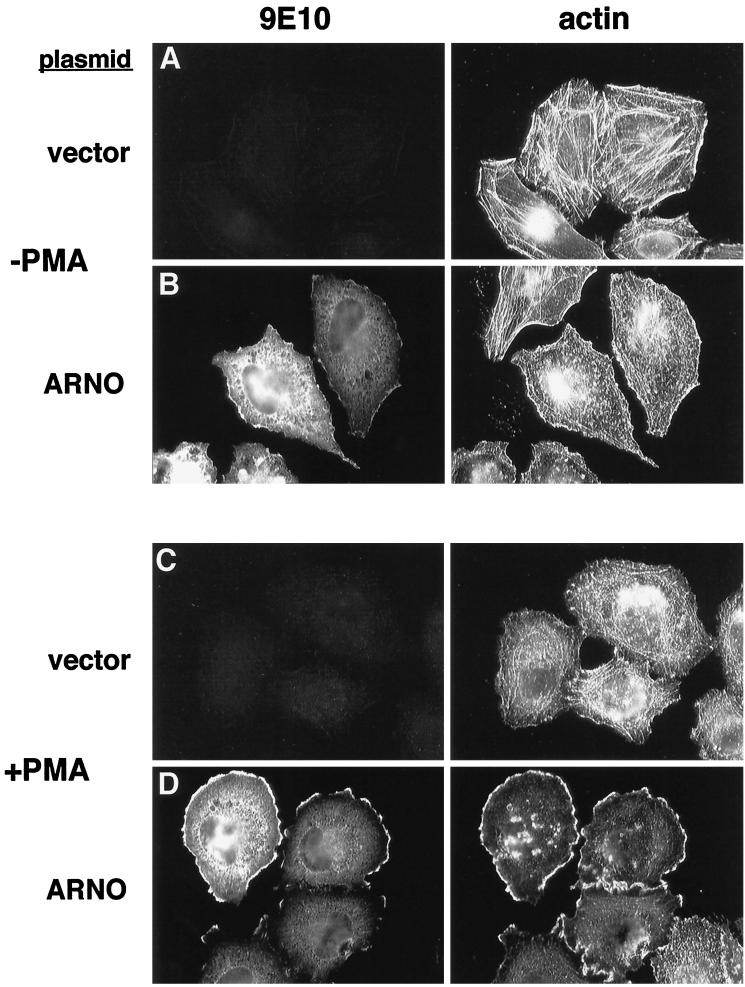
Because ARF6 has been shown to modulate the organization of the actin cytoskeleton (Radhakrishna, et al., 1996; D’Souza-Schorey et al., 1997), we examined actin distribution in HeLa cells expressing an epitope-tagged form of the ARF exchange factor ARNO. Transiently transfected cells were double-labeled with the monoclonal antibody 9E10 to detect myc-ARNO and with rhodamine-phalloidin to label F-actin. In mock-transfected cells, the most prominent feature of the actin cytoskeleton was a network of stress fibers that were present in all cells and in most cases traversed the entire length of the cell (Figure 1A). As we have previously described, the distribution of ARNO in transfected cells was primarily cytosolic, but with a readily detectable component in the plasma membrane at the margins of the cell (Figure 1B, left). Examination of actin distribution in ARNO transfectants revealed a dramatic loss of stress fibers and the appearance of a more punctate pattern of actin staining (Figure 1B, right). This phenotype was only observed in ARNO-expressing cells, and not in their nontransfected neighbors, indicating that the loss of stress fibers is a consequence of ARNO expression and not an artifact of the transfection procedure.
Figure 1.
ARNO regulates cytoskeletal reorganization in HeLa cells treated with PMA. HeLa cells on coverslips were transiently transfected with empty vector (A and C) or vector encoding epitope-tagged ARNO (B and D). Cells were treated for 30 min without (A and B) or with 1 μM PMA (C and D), and then fixed in 2% formaldehyde. ARNO was detected by labeling with the monoclonal antibody 9E10 (anti-myc epitope) followed by Cy2-labeled donkey anti-mouse IgG (left panels). Cells were costained with rhodamine-labeled phalloidin to visualize filamentous actin (right panels).
Examination of the ARNO amino acid sequence revealed the presence of a consensus PKC phosphorylation site at the C terminus, immediately adjacent to the PH domain. Because PKC is known to modulate assembly of the actin cytoskeleton (Downey, et al., 1992; Apgar, 1995), we tested the effect of PKC activation on the distribution of both ARNO and actin by treating cells with the phorbol ester PMA (1 μM) for 30 min at 37°C. PMA treatment of mock-transfected HeLa cells resulted in a loss of stress fibers and the appearance of a more punctate actin staining pattern (Figure 1C), similar to that observed in cells expressing ARNO in the absence of PMA. However, in ARNO-expressing cells, PMA treatment resulted in a dramatic redistribution of both ARNO and actin to the cell periphery in structures resembling lamellipodia (Figure 1D). These structures, which occurred in more than 70% of ARNO transfectants, most resembled those observed in cells expressing wild-type ARF6 treated with aluminum fluoride (our unpublished results and Radhakrishna et al., 1996). However, they tended to be less protrusive and more lamellar in appearance. Interestingly, these structures were distinct from the actin-rich microspikes previously described in CHO cells expressing constitutively activated ARF6 (D’Souza-Schorey et al., 1997). It is likely that PKC activation modulates the activity of other signaling molecules in addition to ARF6. Nevertheless, the strict dependency on ARNO expression in regulating cytoskeletal rearrangements indicates that activation of an ARF6 signaling pathway is central to this process. These protrusions are also distinct from those observed in cells expressing activated Rac1 alleles in which actin folds were distributed over the entire surface of the cells (Radhakrishna et al., 1996); rather, these structures are restricted to the lateral margins of the cell where they contact the substrate. ARNO-dependent actin redistribution occurred rapidly (5 min) and became more pronounced with time (our unpublished results). Finally, this phenotype was observed in cells over a wide range of expression levels. Using adenovirus-mediated gene transfer, which results in ARNO expression in every cell at equivalent levels, we found that actin reorganization was observed in cells within hours of infection where expression levels were 20- to 40-fold over that of the endogenous protein (as determined by quantitative Western blotting [our unpublished results]). Taken together, these results suggest that ARNO coordinates with signals generated by PKC to regulate actin architecture.
ARF6 Colocalizes with ARNO in Lamellipodia
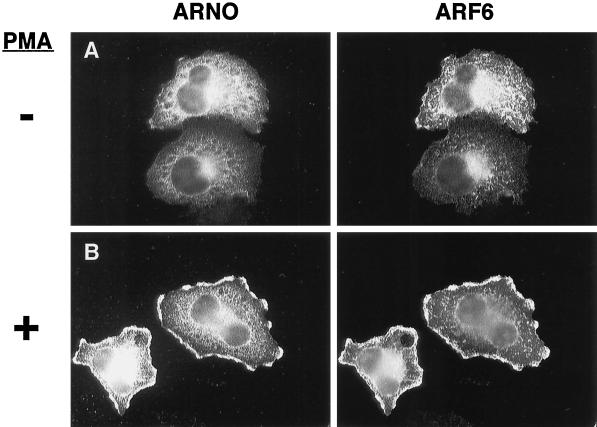
We have previously described the colocalization of ARNO and ARF6 at discrete plasma membrane sites in BHK cells (Frank et al., 1998). As alluded to above, the actin protrusions induced by PMA treatment of ARNO-expressing cells bore a strong resemblance to those previously observed in cells expressing ARF6. It would be expected therefore that these structures should also contain ARF6. Unfortunately, using a variety of ARF6-specific antibodies, we were unable to detect the endogenous protein by standard immunofluorescence protocols. Therefore, to compare the intracellular distribution of ARNO and ARF6, ARNO was coexpressed in HeLa cells with an hemagglutinin (HA)-tagged ARF6 construct, and their localization was assayed using an affinity-purified polyclonal anti-ARNO antibody and a monoclonal anti-HA antibody (16B12). As previously described (Peters et al., 1995; Radhakrishna and Donaldson, 1997), ARF6 localizes primarily to endosomal structures and the plasma membrane (Figure 2A, right). In cotransfected cells, ARNO exhibits some overlap with ARF6 on internal punctate structures (Figure 2A, left), suggesting a possible role for ARNO function at these sites. Most importantly, PMA treatment results in the rapid redistribution of ARNO to plasma membrane protrusions (Figure 2B, left), which is accompanied by a simultaneous redistribution of ARF6 to the same sites (Figure 2B, right). We have found that PMA treatment of cells transfected with ARF6 alone results in a similar redistribution, albeit at lower frequency, suggesting that this pathway can be activated by endogenous ARNO (our unpublished results).
Figure 2.
ARNO and ARF6 colocalize in structures resembling lamellipodia after PMA treatment. HeLa cells cultured on coverslips were cotransfected with expression vectors encoding ARNO and HA-ARF6. Cells were treated 48 h later for 30 min without (A) or with 1 μM PMA (B), fixed, and double labeled with an affinity-purified polyclonal anti-ARNO antibody (ARNO) and a mouse anti-HA antibody (ARF6).
Formation of Lamellipodia Requires ARNO Catalytic Activity
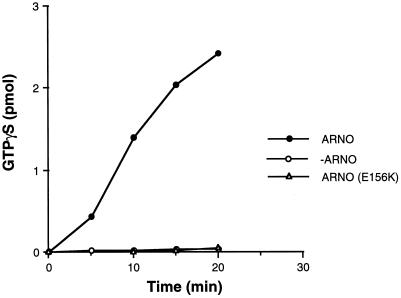
As described above, formation of lamellipodia required treatment of cells with PMA and was observed only in cells expressing elevated levels of ARNO, suggesting that the nucleotide exchange activity of ARNO was necessary to this process. To test this hypothesis, we constructed a catalytically inactive ARNO mutant by substitution of a conserved glutamic acid (E156) with lysine. An analogous substitution was first identified as a recessive mutation in the Arabidopsis Sec7 family member EMB30 (Shevell et al., 1994). Recent solution of the crystal structure of the ARNO Sec7 domain indicates that E156 is located within the catalytic site, and the substitution of this residue has been shown to abrogate ARNO-catalyzed ARF1 nucleotide exchange (Cherfils et al., 1998; Mossessova, et al., 1998). To be certain that this mutation also inhibits ARF6 nucleotide exchange, we compared the ability of wild-type ARNO and ARNO(E156K) to catalyze nucleotide exchange on ARF6, in vitro. As shown in Figure 3, incubation of recombinant myristoylated ARF6 with 100 nM ARNO increased [35S]GTPγS binding by more than 18-fold compared with ARF6 in buffer alone. In contrast, no stimulation of ARF6 nucleotide exchange was observed in the presence of an equivalent concentration of ARNO(E156K), indicating that it is indeed catalytically inactive.
Figure 3.
A Sec7 domain mutant is incapable of catalyzing nucleotide exchange on ARF6. Myristoylated ARF6 (1 μM) was incubated at 37°C in the absence (open circles) or presence of either 100 nM recombinant His6-ARNO (closed circles) or His6-E156K (open triangles) as described in MATERIALS AND METHODS. Data are the average of two simultaneous determinations and are representative of three separate experiments.
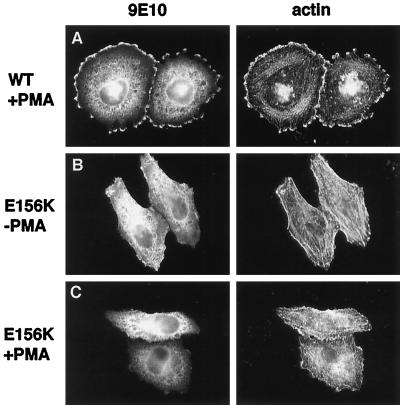
When expressed in HeLa cells, ARNO(E156K) exhibited a distribution similar to that of wild-type ARNO (Figure 4B, left). Interestingly, expression of ARNO(E156K) did not result in the reduction of stress fibers seen in cells expressing the wild-type protein (Figure 4B, right). Most importantly, PMA treatment of ARNO(E156K)-expressing cells did not result in the formation of lamellipodial extensions (Figure 4C). These data indicate that ARNO’s nucleotide exchange activity, leading to ARF6 activation, is required for the cytoskeletal rearrangements seen in ARNO-expressing cells. Moreover, because this construct contains an intact PH domain, which may bind and sequester phosphoinositides that are important regulators of cytoskeletal assembly, these findings demonstrate that the observed phenotypic effects are not simply due to expression of the PH domain alone.
Figure 4.
Catalytically inactive ARNO does not support cytoskeletal rearrangements in response to PMA treatment. HeLa cells transfected with plasmid encoding myc-tagged wild-type ARNO (A) or ARNO/E156K (B and C) were incubated in the presence (A and C) or absence (B) of PMA, fixed, and labeled for immunofluorescence as in Figure 1. Note the presence of intact stress fibers in panel B.
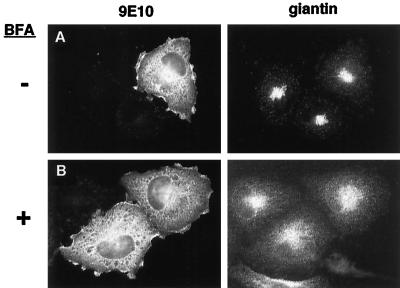
ARNO Function Is Not Sensitive to Brefeldin A In Vivo
The fungal metabolite brefeldin A (BFA) inhibits ARF1 nucleotide exchange activity in Golgi membrane preparations (Donaldson et al., 1992; Helms and Rothman, 1992; Randazzo et al., 1993) and results in the rapid redistribution of resident Golgi proteins to the ER in intact cells (Doms et al., 1989; Lippincott-Schwartz, et al., 1989). However, ARNO catalytic activity is unaffected by BFA in vitro (Chardin et al., 1996; Frank et al., 1998), and we suggested previously that this was consistent with a role for ARNO in ARF6 function since, unlike ARF1, the activation of ARF6 in vivo has been reported to be unaffected by BFA (Peters et al., 1995; Cavenaugh et al., 1996; Radhakrishna, et al., 1996; Radhakrishna and Donaldson, 1997). Nevertheless, it has been suggested that ARNO may complex with other proteins or intracellular factors that confer BFA sensitivity (Chardin, et al., 1996). To test this possibility, we assessed the ability of BFA to inhibit ARNO-dependent cytoskeletal rearrangements. HeLa cells expressing ARNO were pretreated with 10 μg/ml BFA for 15 min followed by addition of PMA for an additional 30 min. BFA had no effect on the redistribution of either ARNO (Figure 5) or actin (our unpublished results) after PMA treatment. As a positive control for BFA action, we stained the same cells with antibodies to the cis-Golgi marker giantin. Consistent with its ability to inhibit Golgi-associated exchange activity, BFA treatment results in a shift in the intracellular distribution of giantin from a punctate Golgi staining pattern (Figure 5A, right) to a more diffuse distribution (Figure 5B, right). Therefore, in keeping with previous in vitro findings, ARNO function does not appear to be sensitive to BFA in vivo and differs from Golgi-associated exchange factor(s) in this manner. Moreover, these data indicate that ARNO does not confer a BFA-resistant phenotype on Golgi membranes in vivo, even at elevated levels of expression, further supporting the hypothesis that ARNO does not function at the Golgi in vivo.
Figure 5.
BFA does not inhibit actin reorganization in cells overexpressing ARNO. Cells transiently transfected with myc-tagged wild-type ARNO were incubated in medium without (A) or with (B) 10 μg/ml BFA for 40 min. PMA was included for the final 30 min of incubation. Cells were costained with the anti-myc antibody 9E10 (left panels) and rabbit polyclonal antibody recognizing the Golgi marker giantin (right panels).
ARNO Is a Substrate for PKC In Vivo and In Vitro
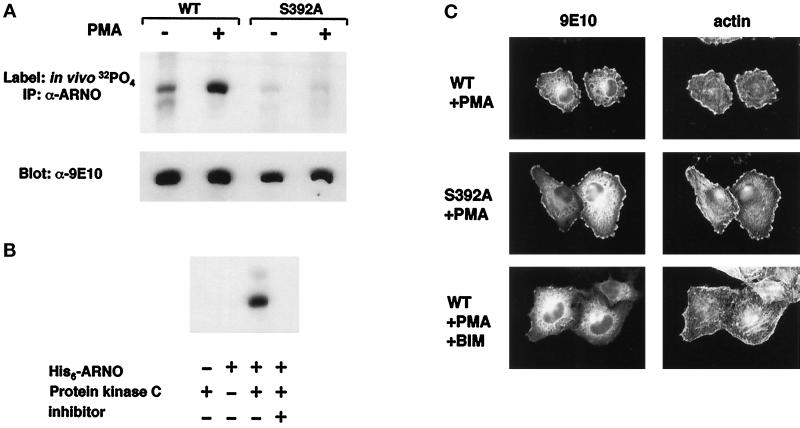
Activation of PKC by the short-term addition of phorbol esters is known to result in the translocation of PKC isoforms from cytosol to membrane and rapid phosphorylation of PKC substrates. To determine whether PKC activation resulted in ARNO phosphorylation, we immunoprecipitated myc-ARNO from HeLa cells labeled in vivo with [32P]orthophosphate. Equivalent amounts of ARNO were immunoprecipitated from control and PMA-treated cells, as determined by Western blotting of immunoprecipitates (Figure 6A, lower panel). However, while a low level of ARNO phosphorylation was observed even in the absence of phorbol ester, [32P] incorporation was increased approximately sixfold in PMA-treated cells relative to nontreated controls (Figure 6A, upper panel).
Figure 6.
PKC-dependent phosphorylation of ARNO does not positively regulate its ability to induce lamellipodia. A, HeLa cells expressing myc-tagged wild-type ARNO or ARNO(S392A) were metabolically labeled with [32P]orthophosphate for 4.5 h. During the final 30 min PMA (+) or DMSO (−) were added to culture medium. The cells were then lysed and immunoprecipitated with ARNO antiserum as described in MATERIALS AND METHODS. The resulting immune complexes were transferred to nitrocellulose membranes and analyzed by autoradiography (upper panel) followed by immunoblotting with anti-myc antibody (lower panel). B, Purified His6-ARNO was incubated with or without purified PKC for 30 min at 37°C, as described in MATERIALS AND METHODS. The PKC inhibitor BIM was included in one reaction at 10 μM. C, HeLa cells transfected with plasmid encoding myc-tagged wild-type ARNO or ARNO(S392A) were incubated with PMA, fixed, and labeled for immunofluorescence as in Figure 1. BIM was added to some cultures 10 min before the addition of PMA.
The consensus motifs for PKC phosphorylation have been defined as (S/T)X(K/R) or (K/R/)XX(S/T), with greater preference for serine over threonine (Pearson and Kemp, 1991). C-terminal alignment of ARNO family members reveals potential PKC phosphorylation sites in ARNO and cytohesin-1, but not GRP1, suggesting that these proteins may be differentially regulated (Table 1). To determine whether the PKC site of ARNO, KRISVK (amino acids 389–394), is the primary site of ARNO phosphorylation in vivo, we substituted the serine at position 392 with alanine (S392A) and introduced this construct into HeLa cells. While expressed at levels comparable to the wild-type protein (Figure 6A, lower panel), no phosphorylation of ARNO(S392A) was observed after PMA addition, demonstrating that this site is utilized in vivo. Because it was formally possible that ARNO phosphorylation in vivo could result from a kinase cascade initiated by PKC, we also ascertained whether PKC could directly phosphorylate purified ARNO in vitro. As shown in Figure 6B, incubation of recombinant ARNO with rat brain PKC, containing a mixture of α, β, and γ isoforms, resulted in readily detectable phosphorylation of ARNO. This phosphorylation was not observed in the absence of PKC and was inhibited by the addition of the PKC-specific inhibitor BIM, indicating that PKC and not a contaminating kinase in the preparation was responsible for the phosphorylation.
Table 1.
C-terminal alignment of ARNO family members
| ARNO (human) | DPFYEMLAARKKKRISVKKKQEQP |
| Cytohesin-1 (human) | DPFYEMLAARKKKVSSTKRH |
| GRP1 (mouse) | DPFYDMLATRKRRIANKK |
C-terminal alignment of ARNO family members with potential phosphorylation sites underlined. Serine residue in bold corresponds to PKC phosphorylation site used in vivo.
The PKC-mediated phosphorylation of ARNO and its apparent correlation with actin redistribution suggested that these two events may be causally linked. To test this hypothesis, we assayed the ability of cells expressing ARNO(S392A) to undergo cytoskeletal remodeling in response to PMA treatment. However, the phenotype of cells expressing ARNO(S392A) was indistinguishable from that of cells expressing wild-type ARNO (Figure 6C). This finding indicates that, while ARNO is a substrate for PKC in vivo, phosphorylation of ARNO does not provide a positive regulatory signal needed for cytoskeletal reorganization.
This observation also suggests that ARNO-dependent actin remodeling requires additional PKC target(s) other than ARNO. One possibility, for which there is considerable experimental support, is the phosphorylation by PKC of proteins directly regulating actin assembly such as the myristoylated alanine-rich C kinase substrate, which has been shown to regulate membrane ruffling and cell spreading (Myat et al., 1997). Alternatively, PMA may be binding and activating a diacylglycerol-binding protein other than a PKC family member. Interestingly these include Rho family exchange factors such as Vav, FGD1, and Lfc, which contain diacylglycerol-binding zinc fingers that regulate their function (Cerione and Zheng, 1996). To distinguish between these possibilities, cells were treated with BIM, which specifically inhibits PKC isoenzymes by competing with enzyme-bound ATP. We found that pretreatment of ARNO-expressing cells with BIM resulted in a complete inhibition of actin redistribution in the presence of PMA (Figure 6C). This clearly demonstrates a role for PKC in ARNO-regulated cytoskeletal rearrangements and indicates that phosphorylation of substrates other than ARNO is required for this process.
The C-Terminal PH Domain Is Required for Formation of Lamellipodia by ARNO
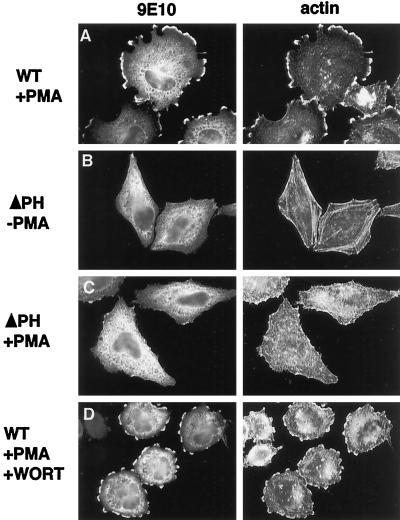
The association of ARNO with phospholipid vesicles in vitro is greatly enhanced by the presence of phosphatidylinositol 4,5-bisphosphate (PtIns (4,5)P2) in vesicle preparations (Chardin et al., 1996). This enhanced recruitment does not appear to regulate the catalytic activity of ARNO, but instead serves to concentrate the protein at the membrane surface where it interacts with ARF (Paris et al., 1997). To investigate whether the C-terminal PH domain of ARNO is required for actin remodeling, HeLa cells were transfected with a truncated ARNO construct lacking this region (ΔPH). Unlike the wild-type protein, the ΔPH mutant did not appear to concentrate at the lateral margins of cells, even in cells expressing high levels of the mutant protein (Figure 7B, left). Also, like the catalytically inactive mutant ARNO(E156K), expression of ARNO(ΔPH) did not appear to affect stress fiber morphology in non-PMA–treated cells (Figure 7B, right). Finally, no rearrangement of cortical actin was observed after treatment of cells expressing ARNO(ΔPH) with PMA (Figure 7C). These data provide evidence that ARNO-mediated actin remodeling requires its appropriate intracellular targeting, determined, at least in part, by its PH domain.
Figure 7.
Cytoskeletal reorganization requires the ARNO PH domain, but not D3 phosphoinositides. HeLa cells transfected with plasmid encoding myc-tagged wild-type ARNO (A and D) or ARNOΔPH (B and C) were incubated with (A, C, and D) or without PMA (B), fixed, and labeled for immunofluorescence as in Figure 1. To some cultures (D) 100 nM wortmannin was added 15 min before the addition of PMA.
ARNO-induced Actin Rearrangements Are not Sensitive to Wortmannin
PH domains have been subdivided into classes, based on their affinity for specific polyphosphoinositides (Rameh et al., 1997). Work with both ARNO and its close homolog GRP1 indicates that these proteins preferentially bind phosphatidylinositol 3,4,5-bisphosphate (PtIns(3,4,5)P3) in vitro (Rameh et al., 1997; Klarlund et al., 1998; Venkateswarlu et al. 1998). To determine whether the production of D3-phosphoinositides was required for ARNO function in vivo, HeLa cells expressing ARNO were pretreated for 20 min with the phosphoinositide 3-kinase (PI 3-kinase) inhibitor wortmannin (100 nM). As shown in Figure 7D, wortmannin had no detectable effect on the ability of PMA to induce actin rearrangements in these cells. Because not all isoforms of PI 3-kinase are sensitive to wortmannin, we further tested the effects of PMA treatment on cells that had been serum starved for 24 h. As observed in wortmannin-treated cells, PMA induced actin reorganization that was indistinguishable from that seen in control cells (our unpublished results). Although PI 3-kinase activity has been reported to be stimulated by PKC agonists in platelets (Hartwig et al., 1996), that activity was found to be wortmannin sensitive. Therefore, taken together, our data strongly suggest that activation of ARNO in vivo does not require synthesis of D3 phosphoinositides.
DISCUSSION
ARNO, cytohesin-1, and GRP-1 are members of a newly described subfamily of proteins that promote guanine nucleotide exchange on ARF (Chardin et al., 1996; Klarlund, et al., 1997; Meacci et al., 1997). Based on in vitro assays, ARNO was initially characterized as an exchange factor for ARF1 (Chardin et al., 1996), and this has resulted in the repeated suggestion in the literature that ARNO would function upstream of ARF1 in the formation of vesicles that bud from Golgi membranes. However, we have previously shown that ARNO and its homolog cytohesin-1 can mediate nucleotide exchange on both ARF1 and ARF6 (Frank et al., 1998) and have subsequently found that they exhibit exchange activity for all ARFs (our unpublished results). A mechanistic explanation for this result can be found in the recent observation that the switch 1 and switch 2 regions of ARF1 are the primary sites of interaction with the ARNO Sec7 domain (Mossessova et al., 1998). Hydroxyl-radical footprint analysis of the ARNO/ARF1 complex revealed that the 26 amino acids in these regions that contact the Sec7 domain are completely conserved among ARFs 1–5 and 88% identical in ARF6 (Mossessova et al., 1998). Combined with the high degree of conservation among active site residues in the Sec7 domain, these findings suggest that there is little specificity in the interaction between Sec7 domains and individual ARFs and that specificity is likely to be achieved by targeting of the exchange factor to the correct subcellular location. Determinants for targeting (and/or regulation) are presumably located in regions of these proteins outside the Sec7 domain.
Specificity of ARNO for ARF6 in Vivo
The ARF proteins are grouped into three classes on the basis of size, gene structure, and amino acid identity: ARFs 1, 2, and 3 (181 amino acids) form class I; ARFs 4 and 5 (180 amino acids) form class II; and ARF6 (175 amino acids) constitutes class 3 (Welsh et al., 1994). It is clear from recent work that ARF6 has evolved distinct intracellular functions. In contrast to ARF1, which associates with the Golgi complex, ARF6 has been localized to the plasma membrane and the endosomal sytsem and has been shown to modulate some aspects of endocytosis. Furthermore, and central to the findings of this paper, ARF6 is unique in its ability to regulate the structure of the cortical actin cytoskeleton. This morphological fingerprint provided a simple biological readout that allowed us to determine whether ARNO was a specific exchange factor for ARF6 in vivo. We had previously shown, both by subcellular fractionation and immunofluorescence microscopy, that ARNO is localized to the plasma membrane where it overlapped in distribution with ARF6. In this paper we provide several lines of evidence indicating that ARNO is a specific exchange factor for ARF6 in vivo. First, ARNO expression in HeLa cells induces morphological changes in the actin cytoskeleton similiar to those observed in cells expressing GTP-bound ARF6 (Figure 1 and Radhakrishna et al. 1996). Although these actin rearrangements require concurrent activation of PKC, they are completely dependent on ARNO catalytic activity (Figure 4). Second, we find that expression of an ARNO protein lacking its PH domain no longer undergoes translocation to the plasma membrane after PMA treatment and is unable to mediate cytoskeletal rearrangements. This indicates that overexpression of the Sec7 domain alone is not sufficient to induce activation of ARF6 and suggests that the in vivo specificity of ARNO for ARF6 is, in large part, determined by its specific intracellular targeting. Finally, ARNO-induced actin remodeling is completely resistant to BFA (Figure 5). This is consistent with previous observations that, unlike ARF1 (Donaldson et al., 1992; Helms and Rothman, 1992; Randazzo, et al., 1993), the activation of ARF6 in cells is unaffected by BFA (Peters, et al., 1995; Cavenaugh et al., 1996; Radhakrishna et al., 1996; Radhakrishna and Donaldson, 1997). In contrast, other members of the Sec7 family have been identified that are sensitive to BFA in vitro, including yeast Sec7p (Sata et al., 1998), the yeast proteins Gea1p and Gea2p (Peyroche et al., 1996), and a mammalian homolog of Sec7p (Morinaga et al., 1997). It is therefore likely that one or more of these proteins are responsible for the BFA-sensitive exchange activity in Golgi membranes. Consistent with this hypothesis, both Gea1p and Sec7p have been shown to regulate the movement of vesicles between ER and Golgi compartments in yeast (Achstetter et al., 1988; Peyroche et al., 1996).
The unique ability of ARF6 to regulate cytoskeletal reorganization suggests that it must interact with a unique subset of ARF effectors. One molecule that has been shown to a play a role in ARF6 cytoskeletal remodeling is POR1. POR1 has been shown to interact with GTP-bound forms of both ARF6 and Rac1 in vitro, and truncated mutants of POR1 inhibit cytoskeletal rearrangements induced by activated alleles of both ARF6 and Rac1 in intact cells (Van Aelst et al., 1996; D’Souza-Shorey et al., 1997). Significantly, a dominant inhibitory ARF6 mutant does not affect Rac1-induced cytoskeletal changes, nor does dominant inhibitory Rac1 inhibit ARF6-induced remodeling, indicating that these two GTPases operate on distinct pathways that may converge at POR1. The precise role of POR1 in regulation of cytoskeletal organization is not yet known.
Another potential downstream effector common to both ARF6 and PKC is PLD. PKCα and -β can directly activate PLD1 by a mechanism that is independent of its catalytic activity and appears to be mediated by the PKC-regulatory domain (Conricode et al., 1994; Singer et al., 1996). Similiarly, ARF has been shown to stimulate PLD activity, and this is synergistic with PKC-mediated stimulation of the enzyme (Singer et al., 1996). Therefore, PMA treatment of ARNO transfectants may lead to the rapid up-regulation of intracellular PLD activity, resulting from the simultaneous activation of ARF6 and PKC pathways. It is worth pointing out that a novel PLD isoform PLD2 has recently been isolated. PLD2 may be regulated by ARF (Lopez et al., 1998) and has been localized to the plasma membrane and endosomes where it modifies cortical actin structure (Colley et al., 1997). It will be of interest to determine whether activation of PLD2 may play a role in ARNO/ARF6-induced actin rearrangements.
Relationship of ARNO to PKC
Although expression of ARNO alone was sufficient to induce disassembly of filamentous actin, we found that reassembly of actin into structures resembling lamellipodia required an additional stimulus, provided in this case by PKC activation. This observation indicates that activation of ARF6, like that of the Rho-family GTPases, is regulated by signal transduction pathways emanating from the plasma membrane. In a variety of cells, PKC lies upstream of pathways that regulate the formation of actin-containing membrane ruffles and lamellipodia (Nobes and Hall, 1995a; Zigmond, 1996). However, the signaling pathways that connect PKC to actin restructuring are poorly defined. ARNO is itself a substrate for PKC both in vivo and in vitro (Figure 6, A and B). In HeLa cells, PMA treatment induces a sixfold increase in ARNO phosphorylation. However, we found that an ARNO mutant (S392A) that is not phosphorylated in vivo produced a phenotype that was indistinguishable from that of wild-type ARNO (Figure 6C), indicating that neither the catalytic activity nor membrane recruitment of ARNO is positively regulated by phosphorylation. It is worth noting that, while ARNO and cytohesin-1 both contain PKC sites at their C termini, GRP1 has an asparagine in place of the phosphorylatable serine (Table 1). While the tissue distribution of GRP1 has not been reported, ARNO and cytohesin-1 reveal distinct expression patterns (Kolanus et al., 1996). Whereas ARNO is widely expressed, cytohesin-1 expression appears to be limited to hematopoietic cells. Therefore, one possibility is that PKC phosphorylation may regulate a tissue-specific function unique to particular ARNO family members.
Role of the PH Domain in ARNO Function
As described above, it is likely that determinants outside the Sec7 domain are required for targeting of ARNO to its site of function. An obvious candidate for this targeting function is the C-terminal PH domain. Although PH domains may mediate some protein-protein interactions such as binding Gβγ subunits (Inglese et al., 1995; Mahadevan et al., 1995), it is generally thought that they function in the recruitment of proteins to membrane surfaces through their interaction with phosphoinositides (Lemmon et al., 1997).
Striking similarities exist between the domain organization of ARNO, in which the catalytic Sec7 domain is immediately followed by a PH domain, and that of the Dbl family of GEFs (Cerione and Zheng, 1996). These proteins, which are exchange factors for the Rho-family GTPases, invariably contain a PH domain immediately C-terminal to the Dbl-homology (DH) catalytic domain. In several cases, PH domains have been shown to mediate the targeting of DH domains to the appropriate subcellular location (Zheng et al., 1996; Michiels et al., 1997).
ARNO has been shown to bind avidly through its PH domain to liposomes containing PtIns(4, 5)P2 (Chardin et al., 1996). This association promotes nucleotide exchange by increasing the local concentration of ARNO at membrane surfaces where it interacts with myristoylated ARF. When assayed in the absence of lipids, a truncated ARNO mutant lacking the PH domain can catalyze nucleotide exchange in vitro as efficiently as the wild-type molecule (Paris et al., 1997). We found that a similar mutant (ΔPH) was unable to support actin remodeling when expressed in HeLa cells, even at very high expression levels (Figure 7). This result illustrates that ARNO-regulated cytoskeletal rearrangments do not result from nonspecific activation of ARF6 by overexpression of a Sec7 domain-containing protein and reflects the need for the PH domain in achieving a sufficient concentration of ARNO at specific membrane surfaces. Importantly, our studies suggest that the PH domain-dependent recruitment of ARNO lies downstream of signaling pathways that activate PKC. Our results support a recent study that found that insulin treatment of 3T3 L1 adipocytes mediated the rapid translocation of ARNO from a cytosolic to plasma membrane-bound pool (Venkateswarlu et al., 1998). It will be important to determine whether PKC lies downstream of insulin-signaling pathways leading to membrane recruitment of ARNO.
PH domains have been subdivided into classes based on their affinity for different polyphosphoinositide species (Rameh et al., 1997). Recent work indicates that ARNO and its homolog GRP1 exhibits a 50- to 100-fold higher affinity for PtIns(3,4,5)P3 than for either PtIns(4,5)P2, or PtIns(3,4)P2 in vitro (Klarlund et al., 1998; Venkateswarlu et al., 1998). This is consistent with the observation by Venkateswarlu et al. that the insulin-dependent translocation of ARNO to the plasma membrane is inhibited by wortmannin. Similiarly, the recruitment of cytohesin-1 to membranes after T cell receptor ligation requires PI-3 kinase activity (Nagel et al., 1998). This differs with our finding that wortmannin does not inhibit ARNO-regulated actin rearrangements. It is worth noting that, while ARNO may bind preferentially to PtIns(3,4,5)P3 in vitro, PtIns(4,5)P2 is present at greater than a 40-fold excess to PtIns(3,4,5)P3 in vivo (Auger et al., 1989). Therefore, ARNO should be capable of associating with both these lipids in vivo, and its binding to one or the other may depend on the particular intracellular signaling pathway activated. Alternatively, a single pathway may exist in which the PI-3 kinase- sensitive step lies downstream of receptor ligation and upstream of PKC. Interestingly, an analogous situation exists in the activation of the GTPase Rac1. For example, PI-3 kinase is required for the activation of Rac by binding of agonists to tyrosine kinases, but is not required for activation of Rac by PMA (Zigmond, 1996).
ARFs and Signal Transduction
A number of recent reports have suggested that ARF proteins may be activated by cell surface receptors, and there is growing evidence of a role for ARF in agonist regulation of PLD in vivo (Houle et al., 1995; Rumenapp et al., 1995; Shome et al., 1997; Mitchell et al., 1998). In Rat-1 fibroblasts expressing human insulin receptors, membrane-associated ARF exchange activity is markedly stimulated by insulin, and coimmunoprecipitation of ARF with insulin receptor increases after receptor ligation (Shome et al., 1997). Similarly, treatment of HL-60 cells with the chemotatic peptide f-met-leu-phe results in an increase in membrane-associated ARF and GTPγS-stimulated PLD activity (Houle et al., 1995). Moreover, ARF6 has been shown to play an important role in the regulation of chromaffin granule exocytosis (Galas et al., 1997; Caumont et al., 1998). ARF6 is associated with granule membranes in resting cells, but undergoes translocation to the plasma membrane after treatment of cells with secretagogues. Importantly, granule exocytosis was shown to be inhibited by synthetic peptides corresponding to the ARF6 (but not ARF1) N-terminal domain.
Finally, cytohesin-1, the first ARNO family member to be isolated, has been shown to lie downstream of T-cell receptor signaling pathways that result in increased adhesiveness of the integrin αLβ2 (Kolanus et al., 1996). Overexpression of the cytohesin-1 PH domain, but not that of Vav, βARK, or Ras-GTPase-activating protein was found to inhibit induction of adhesion, indicating that cytohesin was an important component of the pathway. The data presented here suggest that, analogous to ARNO, cytohesin-1 may function by inducing ARF6-dependent reorganization of cortical actin. This is intriguing since the actin-based cell surface clustering of integrins has been proposed as a mechanism of modulating integrin avidity. Taken together, these data suggest that members of the ARNO/cytohesin/GRP1 family of ARF exchange factors link signal transduction pathways to reorganization of cortical actin in a manner similar to, and possibly integrated with, the Rho family GTPases.
ACKNOWLEDGMENTS
We thank Drs. Yoram Altschuler and Keith Mostov (University of California, San Francisco) for help with production of adenoviruses, and Dr. Sandra Schmid (Scripps) for HeLa cells expressing the tetracycline transactivator. We also thank Victor Hsu, Jeff Settleman, Bobby Cherayil, Steen Hansen, and Lorraine Santy for critical reading of the manuscript. This work was supported by National Institutes of Health grants AI-32991 and DK-33506 and a gift from the Good Samaritan Foundation to J.E.C. S.R.F. was supported in part by of a predoctoral training grant to the Harvard Medical School Program in Immunology. We also thank Dr. Dennis Brown for the use of the imaging facilities of the morphology core of the Massachusetts General Hospital Renal Unit Program Project funded by grant DK-38452.
Abbreviations used:
- ARF
ADP-ribosylation factor
- BFA
brefeldin A
- BIM
bis-indolylmaleimide
- GEF
guanine nucleotide exchange factor
- GTPγS
guanosine 5′-(γ-thio)triphosphate
- HA
hemagglutinin
- PH
pleckstrin homology
- PI 3-kinase
phosphoinositide 3-kinase
- PLD
phospholipase D
- PtIns(4
5)P2, PtIns(3,4)P2, and PtIns(3,4,5)P3, phosphatidylinositol 4,5-bisphosphate, 3,4-bisphosphate and 3,4,5-trisphosphate
REFERENCES
- Achstetter T, Franzusoff A, Schekman R. SEC7 encodes an unusual, high molecular weight protein required for membrane traffic from the yeast Golgi apparatus. J Biol Chem. 1988;263:11711–11717. [PubMed] [Google Scholar]
- Apgar JR. Activation of protein kinase C in rat basophilic leukemia cells stimulates increased production of phosphatidylinositol 4-phosphate and phophatidylinositol 4,5-bisphosphate: correlation with actin polymerization. Mol Biol Cell. 1995;6:97–108. doi: 10.1091/mbc.6.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- Boman AL, Kahn RA. Arf proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- Brewer CB. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. In: Roth MG, editor. Protein Expression in Animal Cells. Vol. 43. San Diego, CA: Academic Press; 1994. , 379. [DOI] [PubMed] [Google Scholar]
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- Caumont AS, Galas MC, Vitale N, Aunis D, Bader MF. Regulated exocytosis in chromaffin cells. Translocation of ARF6 stimulates a plasma membrane-associated phospholipase D. J Biol Chem. 1998;273:1373–1379. doi: 10.1074/jbc.273.3.1373. [DOI] [PubMed] [Google Scholar]
- Cavenaugh MM, Whitney JA, Carroll K, Zhang C-J, Boman A, Rosenwald AG, Mellman I, Kahn RA. Intracellular distribution of ARF proteins in mammalian cells: ARF6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson C, Chabre M. A human exchange factor for ARF contains Sec7 and pleckstrin homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Beraud-Dufour S, Antonny B, Chardin P. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature. 1998;392:101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung T-C, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Conricode KM, Smith JL, Burns DJ, Exton JH. Phospholipase D activation in fibroblast membranes by the alpha and beta isoforms of protein kinase C. FEBS Lett. 1994;342:149–153. doi: 10.1016/0014-5793(94)80490-7. [DOI] [PubMed] [Google Scholar]
- Cross MJ, Roberts S, Ridley AJ, Hodgkin MN, Stewart A, Claesson-Welsh L, Wakelam MJO. Stimulation of actin stress fibre formation mediated by activation of phospholipase D. Curr Biol. 1996;6:588–597. doi: 10.1016/s0960-9822(02)00545-6. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Boshans RL, McDonough M, Stahl PD, Van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Li G, Colombo MI, Stahl PD. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- Doms RW, Russ G, Yewdell JW. Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J Cell Biol. 1989;109:61–72. doi: 10.1083/jcb.109.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Klausner RD. ARF: a key regulatory switch in membrane traffic and organelle structure. Curr Opin Cell Biol. 1994;6:527–532. doi: 10.1016/0955-0674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Downey GP, Chan CK, Lea P, Takai A, Grinstein S. Phorbol ester-induced actin assembly in neutrophils: role of protein kinase C. J Cell Biol. 1992;116:695–706. doi: 10.1083/jcb.116.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation 6. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- Galas MC, Helms JB, Vitale N, Thierse D, Aunis D, Bader MF. Regulated exocytosis in chromaffin cells. A potential role for a secretory granule-associated ARF6 protein. J Biol Chem. 1997;272:2788–2793. doi: 10.1074/jbc.272.5.2788. [DOI] [PubMed] [Google Scholar]
- Ha KS, Exton JH. Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J Cell Biol. 1993;123:1789–1796. doi: 10.1083/jcb.123.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Kung S, Kovacsovics T, Janmey PA, Cantley LC, Stossel TP, Toker A. D3 phosphoinositides and outside-in integrin signaling by glycoprotein IIb-IIIa mediate platelet actin assembly and filopodial extension induced by phorbol 12-myristate 13-acetate. J Biol Chem. 1996;271:32986–32993. doi: 10.1074/jbc.271.51.32986. [DOI] [PubMed] [Google Scholar]
- Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyzes exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Houle MG, Kahn RA, Naccache PH, Bourgoin S. ADP- ribosylation factor translocation correlates with potentiation of GTP-γ S-stimulated phospholipase D activity in membrane fractions of HL-60 cells. J Biol Chem. 1995;270:22795–22800. doi: 10.1074/jbc.270.39.22795. [DOI] [PubMed] [Google Scholar]
- Inglese J, Koch WJ, Touhara K, Lefkowitz RJ. Gβγ interactions with PH domains and Ras-MAPK signaling pathways. Trends Biochem Sci. 1995;20:151–156. doi: 10.1016/s0968-0004(00)88992-6. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Rameh LE, Cantley LC, Buxton JM, Holik JJ, Sakelis C, Patki V, Corvera S, Czech MP. Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. αLβ2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis N, Brown HA, Waters MG, Sternweiss PC, Roth MG. Evidence that phospholipase D mediates ADP ribosylation factor- dependent formation of Golgi coated vesicles. J Cell Biol. 1996;108:2169–2181. doi: 10.1083/jcb.134.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Falasca M, Ferguson KM, Schlessinger J. Regulatory recruitment of signalling molecules to the cell membrane by pleckstrin-homology domains. Trends Cell Biol. 1997;7:237–242. doi: 10.1016/S0962-8924(97)01065-9. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez I, Arnold RS, Lambeth JD. Cloning and initial characterization of a human phospholipase D2 (hPLD2). ADP-ribosylation factor regulates hPLD2. J Biol Chem. 1998;273:12846–12852. doi: 10.1074/jbc.273.21.12846. [DOI] [PubMed] [Google Scholar]
- Mahadevan D, et al. Structural studies on the PH domains of Dbl, Sos1, IRS-1, and βARK1 and their differential binding to G βγ subunits. Biochemistry. 1995;34:9111–9117. doi: 10.1021/bi00028a021. [DOI] [PubMed] [Google Scholar]
- Massenburg D, Han JS, Liyanage M, Patton WA, Rhee SG, Moss J, Vaughan M. Activation of rat brain phospholipase D by ADP- ribosylation factors 1,5, and 6: separation of ADP-ribosylation factor- dependent and oleate-dependent enzymes. Proc Natl Acad Sci USA. 1994;91:11718–11722. doi: 10.1073/pnas.91.24.11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacci E, Tsai S-C, Adamik R, Moss J, Vaughan M. Cytohesin-1, a cytosolic guanine nucleotide exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels F, Habets GGM, Stam JC, van der Kammen RA, Collard JG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- Mitchell R, McCulloch D, Lutz E, Johnson M, MacKenzie C, Fennell M, Fink G, Zhou W, Sealfon SC. Rhodopsin-family receptors associate with small G proteins to activate phospholipase D. Nature. 1998;392:411–414. doi: 10.1038/32937. [DOI] [PubMed] [Google Scholar]
- Morinaga N, Moss J, Vaughan M. Cloning and expression of a cDNA encoding a bovine brain brefeldin A-sensitive guanine nucleotide- exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:12926–12931. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Gulbis JM, Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human ARNO and analysis of the interaction with ARF GTPase. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- Myat MM, Anderson S, Allen LA, Aderem A. MARCKS regulates membrane ruffling and cell spreading. Curr Biol. 1997;7:611–614. doi: 10.1016/s0960-9822(06)00262-4. [DOI] [PubMed] [Google Scholar]
- Nagel W, Zeitlmann L, Schilcher P, Geiger C, Kolanus J, Kolanus W. Phosphoinositide 3-OH kinase activates the β2 integrin adhesion pathway and induces membrane recruitment of cytohesin-1. J Biol Chem. 1998;273:14853–14861. doi: 10.1074/jbc.273.24.14853. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995a;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995b;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. Role of protein-phospholipid interactions in the activation of ARF1 by the guanine nucleotide exchange factor ARNO. J Biol Chem. 1997;272:22221–22226. doi: 10.1074/jbc.272.35.22221. [DOI] [PubMed] [Google Scholar]
- Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD. Overexpression of wild-type and mutant ARF1 and ARF6: distinct pertubations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG. ARF6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Klausner RD, Donaldson JG. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, et al. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Yang YC, Rulka C, Kahn RA. Activation of ADP-ribosylation factor by Golgi membranes: evidence for a brefeldin A and protease sensitive activating factor on Golgi membranes. J Biol Chem. 1993;268:9555–9563. [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Rumenapp U, Geiszt M, Wahn F, Schmidt M, Jakobs KH. Evidence for ADP-ribosylation-factor-mediated activation of phospholipase D by m3 muscarinic acetylcholine receptor. Eur J Biochem. 1995;234:240–244. doi: 10.1111/j.1432-1033.1995.240_c.x. [DOI] [PubMed] [Google Scholar]
- Sata M, Donaldson JG, Moss J, Vaughan M. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc Natl Acad Sci USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoller F, Itin C, Pfeffer S. Vesicle traffic: get your coat! Curr Biol. 1997;7:235–237. doi: 10.1016/s0960-9822(06)00109-6. [DOI] [PubMed] [Google Scholar]
- Seelig HP, Schranz P, Schroter H, Wiemann C, Griffiths G, Renz M. Molecular genetic analyses of a 376-kilodalton Golgi complex membrane protein (giantin) [retracted in Mol Cell Biol 1995 Jan;15(1):591] Mol Cell Biol, 1994;14:2564–2576. doi: 10.1128/mcb.14.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shevell DE, Leu W-M, Gillmor CS, Xia G, Feldmann KA, Chua N-H. EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Shome K, Vasudevan C, Romero G. ARF proteins mediate insulin-dependent activation of phospholipase D. Curr Biol. 1997;7:387–396. doi: 10.1016/s0960-9822(06)00186-2. [DOI] [PubMed] [Google Scholar]
- Singer WD, Brown HA, Jiang X, Sternweis PC. Regulation of phospholipase D by protein kinase C is synergistic with ADP-ribosylation factor and independent of protein kinase activity. J Biol Chem. 1996;271:4504–4510. doi: 10.1074/jbc.271.8.4504. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- Welsh CF, Moss J, Vaughan M. ADP-ribosylation factors: a family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol Cell Biochem. 1994;138:157–166. doi: 10.1007/BF00928458. [DOI] [PubMed] [Google Scholar]
- Yang CZ, Heimberg H, D’Souza-Schorey C, Mueckler MM, Stahl PD. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J Biol Chem. 1998;273:4006–4011. doi: 10.1074/jbc.273.7.4006. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zangrilli D, Cerione RA, Eva A. The pleckstrin homology domain mediates transformation by oncogenic dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Signal transduction and actin filament organization. Curr Opin Cell Biol. 1996;8:66–73. doi: 10.1016/s0955-0674(96)80050-0. [DOI] [PubMed] [Google Scholar]