Abstract
Monitoring the sodium concentration in vivo using 23Na MRI can be an important tool for assessing the onset of tissue disorders. Practical clinical 23Na MRI methods furthermore often do not allow one to use sufficiently small voxel sizes such that only the tissue of interest is seen, but a large signal contamination can arise from sodium in synovial fluid. Here we demonstrate that applying an inversion recovery (IR) technique allows one to distinctly select either the cartilage-bound or the free sodium for visualization in an image. The results are validated both ex vivo and in vivo.
Keywords: Inversion recovery, sodium-MRI, ordered sodium, free sodium, cartilage imaging
1 Introduction
A large amount of sodium exists in living tissues, such as cartilage and the brain, which makes 23Na MRI a very promising tool for the diagnosis of cartilage pathologies, as well as, brain tumors [1] . Notably, the cartilage tissue sodium concentration has been shown to correlate directly with the decrease of the proteoglycan content, which is frequently identified with the onset of degenerative joint disease [2–8].
Normally, the signals from free sodium and the cartilage-bound or ordered sodium overlap with each other, and several methods have been developed for their separation. A number of techniques are based on the evolution under residual quadrupolar interactions. For example, in the double-quantum filter experiment [9,10], the transverse magnetization operators evolve into second rank tensors under the action of the quadrupolar interaction. These second-rank tensors can then be converted into double-quantum coherences (, which can be filtered out using phase cycling. Alternatively, in the Jeener-Broeckaert experiment the rotational properties of different rank tensors [9] are exploited to perform this selection. Both techniques were adapted to detect the filtered signal through the central transition to obtain higher signal-to-noise ratio and higher resolution [9], however they require a large phase cycle and are difficult to implement on MRI scanners since they use a large number of pulses. More recently, methods based on frequency-sweep pulses [11, 12] and quadrupolar nutation [13] were demonstrated, which exploit coherence transfer properties that depend on the quadrupolar interaction. The quadrupolar coupling itself was shown to correlate with the onset of cartilage degeneration [14, 15].
Another form of signal separation is performed by the triple quantum filtered experiments, in which the selected signal arises from quadrupolar nuclei in slow motion [16]. Slow-motion gives rise to third-rank tensors, which, upon conversion to triple-quantum coherences, can be filtered out by phase cycling.
In the current study, we demonstrate the feasibility of employing the inversion recovery (IR) sequence to selectively detect the cartilage-bound sodium signal or the free sodium signal. The study shown here depends on the significant difference between the spin-lattice relaxation rates between the two pools. In the particular case of cartilage tissue, the difference is especially large due to immobilization and larger induced quadrupolar interactions of the bound sodium. It is possible that cartilage-tissue also contains free sodium, as one could expect in analogy to experiments performed with 2H [17, 18], but we have not seen a significant fraction thereof in bulk measurements [15].
A large fraction of the signal is typically lost in 23Na MRI due to lengthy rf-pulses. The simplicity of the inversion recovery sequence makes it hence particularly attractive for 23Na MRI.
2 Results and Discussion
A hard pulse normal IR sequence yielded the T1 values for sodium in the saline solution and for sodium in cartilage as 64.4 and 18.2 ms, respectively at 11.7 T. The zero-crossing points in the relaxation curves were at 39.2 ms and 12.3 ms, respectively. The zero-crossings are slightly different if low power pulses are employed (34.5 and 6.5 ms, respectively).
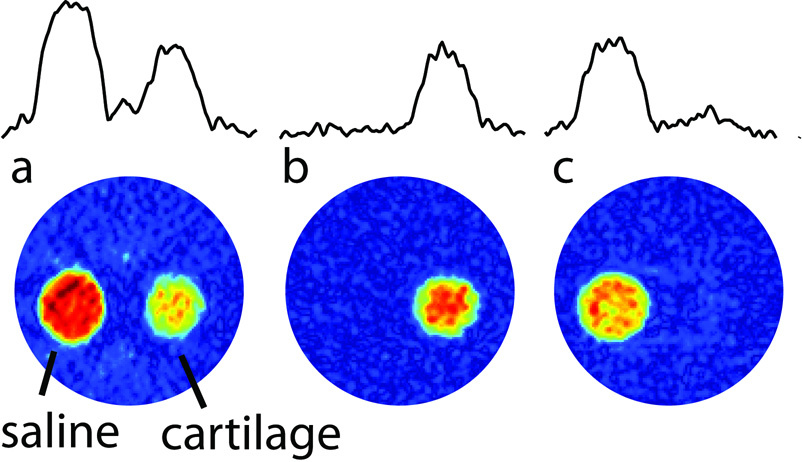
A sample containing both cartilage and saline solution was imaged using a two-dimensional projection-reconstruction image (Figure 2). By adjusting the inversion-recovery delays accordingly, one can either select the cartilage-bound sodium region, or the saline sodium in the images. In Figure 2b, the signal of cartilage-bound sodium is essentially not reduced, largely due to the big difference in T1 values between the two sodium pools. By contrast, Figure 2c shows, that the free sodium signal is reduced by approximately 30 % due to a partial saturation at the 6.5 ms inversion-recovery delay.
Figure 2.
2D images obtained from 30 1D projections using projection reconstruction. The sample consists of two capillaries aligned vertically in an NMR tube. One of the tubes contains cartilage and the other a saline solution. In (a) the single-pulse response is shown, in (b) inversion-recovery with a 34.5 ms delay was used, and in (c) an inversion-recovery delay of 6.5 ms was used. 1D projections are displayed at the top to allow one to estimate the level of suppression in these experiments.
The rf power was chosen to reflect the typical power attainable on MRI scanners (on the order of 500 Hz). The pulse durations ( 0.5 and 1ms, respectively) are still sufficiently short such that relaxation losses during the pulses are mild [19]. The sodium T1 relaxation is biexponential [16] and the signal integral after the inversion recovery sequence is modulated by
| (1) |
where , Cq is the quadrupolar coupling constant, and η the asymmetry. This biexponential relaxation mechanism will not interfere with the selection of cartilage-bound sodium, in general, since in this case, both the short and the fast components will have decayed to an appreciable extent already. In the experiment that selects the free sodium, incomplete cancellation of the bound sodium may occur, as is also seen in Figure 2c. The ratio of the rates J2/J1 approaches a maximum of four as τc → ∞. In general, however, one can minimize this effect by simply choosing the delay τ of the inversion-recovery experiment to be the zero-crossing point of Eq. [1] when selecting the free sodium signal. Some contamination of the free sodium signal may occur, however, in the presence of quadrupolar coupling. While one can adjust the optimum delay τ to compensate for such effects, a distribution of a sizable quadrupolar interaction will lead to signal contamination due to quadrupolar nutation-related effects, especially if the ratio ωrf/ωQ is small [13]. Nonetheless, In the case demonstrated here, the average quadrupoalar coupling was ωQ/2π = 370 Hz for the cartilage tissue, and a relatively clean nulling of the cartilage-bound sodium is possible in Figure 2c. Alternatively, one could adjust the pulse flip angles to partially compensate for nutation effects. In practice, however, it is expected that selecting the bound sodium over the free sodium signals will find more frequent use. In this case, quadrupolar nutation effects will be negligible.
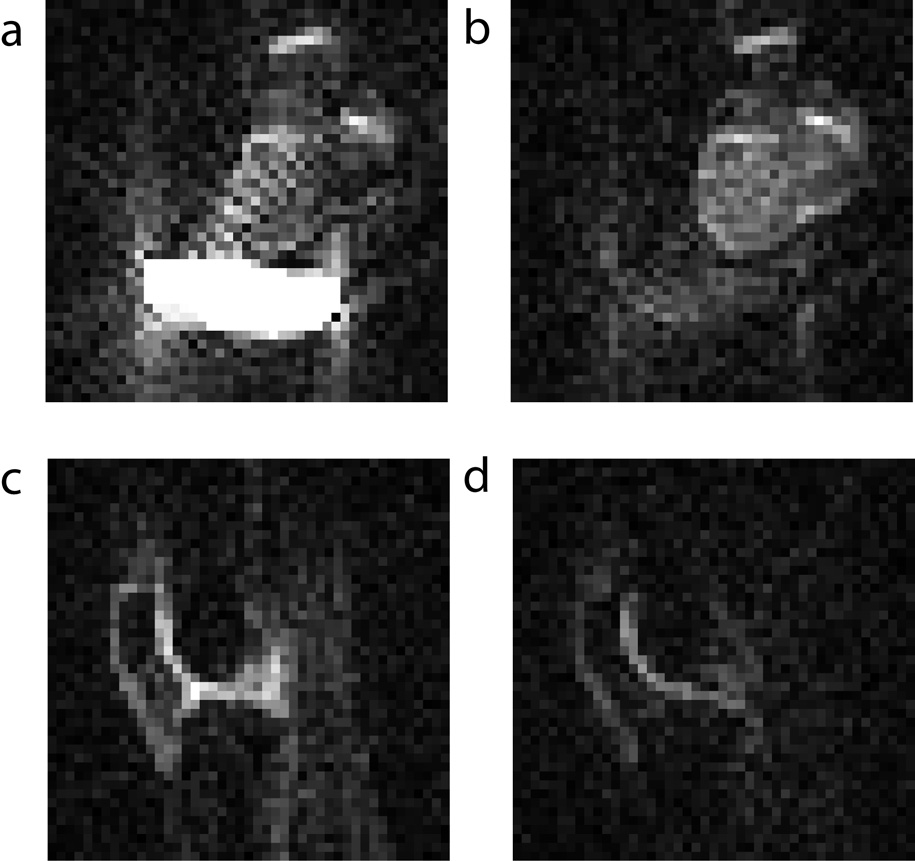
In order to demonstrate this experiment in vivo, we calibrated the optimal free-sodium suppression condition using first an ex vivo bovine knee sample together with a saline bag on a 7T scanner. Optimal suppression was found at a 40 ms inversion-recovery delay (Figures 3a and 3b). In addition, since the strong signals from the saline compartment are minimized, fewer projection-reconstruction artifacts arise in the image. This aspect can be particularly important when working with undersampled data, since the sodium images become sparser. The same images were then also performed to compare the signal suppression between a regular pulsed image, and an inversion-recovery image of a human knee (Figures 3c and 3d). The inversion-recovery sequence makes the demarcation of cartilage tissue more evident in vivo. Cross-sections showed that the signal in the cartilage region was lower by approximately 30 % in the inversion-recovery sequence. A part of this reduction comes from the fact that non-cartilage compartments no longer contribute as strongly to the signals, hence the interpretability of the 23Na signals in terms of measuring the fixed charge density in cartilage is enhanced.
Figure 3.
Axial ex vivo images of a bovine knee joint with a saline bag at 7T (a) without inversion-recovery, and (b) with an inversion-recovery delay of 40 ms. In vivo images of a human knee at 7T (a) without inversion-recovery, and (b) with an inversion-recovery delay of 40 ms.
3 Conclusions
We present here a simple method based on inversion recovery for separating the signals from the free and cartilage-bound sodium pools. Either pool can be selected in an image. The advantages of this method lie in (1) its simplicity, (2) its sensitivity, (3) its ability to selectively detect either the ordered or free sodium signal, (4) the relative robustness against flip angle errors, (5) its small phase cycle, (6) the detection of the central transition, and (7) its high sensitivity, which should make it more attractive than mutliple-quantum filtering in 23Na MRI. The presented method is especially suitable for imaging cartilage. The strength of binding and the large electric field gradients that sodium experiences near the highly-negatively charged proteoglycans lead to a particularly large difference of the relaxation rates between cartilage and synovial sodium.
4 Experimental
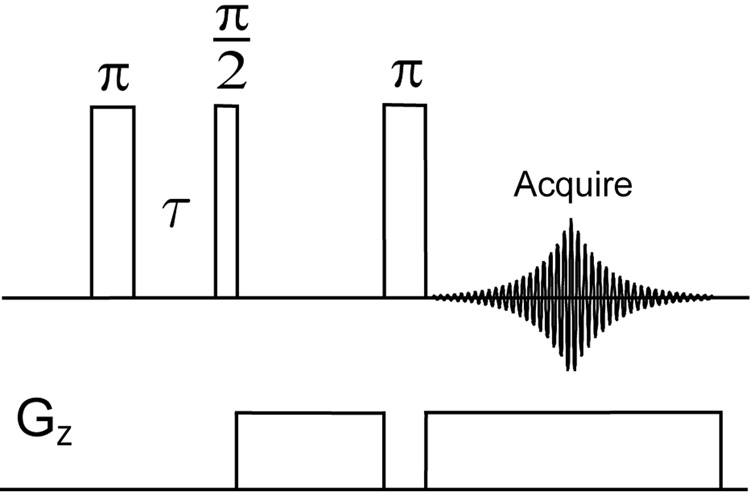
The experiments of Figure 2 were carried out on a Bruker Avance 500 MHz spectrometer (11.7 T) with a BBO probe tuned to sodium frequency. The pulse sequence used is shown in Fig. 1 We optimized the relaxation delay τ between these two pulses in order to achieve selective detection of the ordered sodium or the free sodium signal. All pulse durations were calibrated based on the free sodium signal. The rf strengths were 487 Hz for all the pulses (513.5 µs π/2 pulse duration). 2D images were obtained from 30 1D projections using projection reconstruction. The sample consists of two capillaries aligned vertically in an NMR tube. One of the tubes contains cartilage and the other a saline solution at a concentration(The saline solution was made by diluting a phosphate-buffered saline stock solution (Aldrich, pH 7.4, 137 mMNaCl) to 2/5 of original concentration). A gradient of 2 G/cm strength was applied in the transverse direction and 2048 data points were acquired to cover a spectral window of 10 kHz, with 128 transients coadded. The repetition delay was 1 s for all measurements. The first pulse was cycled between 0 and 180°.
Figure 1.
Pulse sequence of the inversion recovery sequence performed in combination with a gradient spin echo.
The experiments of Figure 3 were performed on a 7.0 T clinical MR scanner (Magnetom Tim Trio, Siemens Medical Solutions, Erlangen, Germany). A 18 cm diameter quadrature 23Na knee coil was used for all the imaging measurements, and rf irradiation with 800 Hz power was used. The image was performed with a 2D-radial sequence with a spatial resolution of 4.7 mm × 4.7 mm × 20 mm, TR/TE=500ms/3.2ms, acquisition time= 1minute. A single slice was used. The ex vivo image was performed using a bovine knee joint in axial orientation together with a 500 mL saline bag (155 mM [Na+]) to load the coil. The in vivo images were taken from a 42-year-old male volunteer (right knee) in saggital orientation. A 40 ms inversion-recovery delay yielded an optimal suppression of the saline signal and was hence used for both experiments.
The bovine cartilage and knee samples were obtained within five hours of animal sacrifice ( 4–6 months old cows) from a USDA approved slaughter house (Bierig Bros, Vineland, NJ) and then frozen at −20°C until used.
Acknowledgements
This work was supported by U.S. NSF grant CHE-0554400 and a NIH grant 1R21AR054002-01A1 and was conducted in a facility constructed with support from Research Facilities Improvement Grant Number C06 RR-16572-01 from the NCRR, NIH. NYUs NMR resources were supported by NSF Grant MRI-0116222. A.J. is a member of the New York Structural Biology Center, which is supported by the New York State Office of Science, Technology, and Academic Research and NIH Grant P41 FM66354.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohler S, Kolodny N. Sodium Magnetic Resonance Imaging and Chemical Shift Imaging. Prog. Nucl. Magn. Reson. Secptrosc. 1992;24:411–433. doi: 10.1002/mrm.1910230109. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro E, Borthakur A, Gougoutas A, Reddy R. Na-23 MRI Accurately Measures Fixed Charge Density in Articular Cartilage. Magn. Reson. Med. 2002;47:284–291. doi: 10.1002/mrm.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borthakur A, Shapiro E, Beers J, Kudchodkar S, Kneeland J, Reddy R. Sensitivity of MRI to Proteoglycan Depletion in Cartilage: Comparison of Sodium and Proton MRI. Osteoarthr. Cartilage. 2000;8:288–293. doi: 10.1053/joca.1999.0303. [DOI] [PubMed] [Google Scholar]
- 4.Wheaton A, Borthakur A, Dodge G, Kneeland B, Schumacher H, Reddy R. Sodium Magnetic Resonance Imaging of Proteoglycan Depletion in an in vivo Model of Osteoarthritis. Acad. Radiol. 2004;11:21–28. doi: 10.1016/s1076-6332(03)00574-9. [DOI] [PubMed] [Google Scholar]
- 5.Wheaton A, Borthakur A, Shapiro E, Regatte R, Akella S, Kneeland J, Reddy R. Proteoglycan loss in human knee cartilage: Quantitation with sodium MR imaging - Feasibility study. Radiology. 2004;231:900–905. doi: 10.1148/radiol.2313030521. [DOI] [PubMed] [Google Scholar]
- 6.Borthakur A, Shapiro E, Akella S, Gougoutas A, Kneeland J, Reddy R. Quantifying Sodium in the Human Wrist in vivo by using MR Imaging. Radiology. 2002;224:598–602. doi: 10.1148/radiol.2242011039. [DOI] [PubMed] [Google Scholar]
- 7.Insko E, Clayton D, Elliott M. In vivo Sodium MR Imaging of the Intervertebral Disk at 4 T. Acad. Radiol. 2002;9:800–804. doi: 10.1016/s1076-6332(03)80350-1. [DOI] [PubMed] [Google Scholar]
- 8.Reddy R, Insko E, Noyszewski E, Dandora R, Kneeland J, Leigh J. Sodium MRI of Human Articular Cartilage in vivo. Magn. Reson. Med. 1998;39:697–701. doi: 10.1002/mrm.1910390505. [DOI] [PubMed] [Google Scholar]
- 9.Kemp-Harper R, Brown S, Hughes C, Styles P, Wimperis S. 23Na NMR Methods for Selective Observation of Sodium Ions in Ordered Environments. Prog. Nucl. Magn. Reson. Spectrosc. 1997;30:157–181. [Google Scholar]
- 10.Navon G, Shinar H, Eliav U, Seo Y. Multiquantum Filters and Order in Tissues. NMR Biomed. 2001;14:112–132. doi: 10.1002/nbm.687. [DOI] [PubMed] [Google Scholar]
- 11.Ling W, Jerschow A. Selecting Ordered Environments in NMR of Spin-3/2 Nuclei via Frequency-Sweep Pulses. J. Magn. Reson. 2005;176:234–238. doi: 10.1016/j.jmr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Ling W, Jerschow A. Frequency-selective quadrupolar MRI contrast. Solid-State Nucl. Magn. Reson. Spectrosc. 2006;29:227–231. doi: 10.1016/j.ssnmr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Choy AJJ, Ling W. Selective detection of ordered sodium signals via the central transition. J. Magn. Reson. 2006;180:105–109. doi: 10.1016/j.jmr.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Shinar H, Navon G. Multinuclear NMR and microscopic MRI studies of the articular cartilage nanostructure. NMR Biomed. 2006;19:877–893. doi: 10.1002/nbm.1068. [DOI] [PubMed] [Google Scholar]
- 15.Ling W, Regatte RR, Schweitzer ME, Jerschow A. The Behavior of Ordered Sodium in Enzymatically Depleted Cartilage Tissue. Magn. Reson. Med. 2006;56:1151–1155. doi: 10.1002/mrm.21062. [DOI] [PubMed] [Google Scholar]
- 16.Jaccard G, Wimperis S, Bodenhausen G. Multiple-quantum NMR spectroscopy of S = 3/2 spins in isotropic phase: a new probe for multiexponential relaxation. J. Chem. Phys. 1986;85:6282–6293. [Google Scholar]
- 17.Shinar H, Seo Y, Ikoma K, Kusaka Y, Eliav U, Navon G. Mapping the Fiber Orientation in Articular Cartilage at Rest and Under Pressure Studied by 2H Double Quantum Filtered MRI. Magn. Reson. Med. 2002;48:322–330. doi: 10.1002/mrm.10195. [DOI] [PubMed] [Google Scholar]
- 18.Keinan-Adamsky K, Shinar H, Navon G. The effect of decalcification on the microstructure of articular cartilage assessed by 2H double quantum filtered spectroscopic MRI. MAGMA. 2005;18:231–237. doi: 10.1007/s10334-005-0009-8. [DOI] [PubMed] [Google Scholar]
- 19.van der Maarel JRC, Jesse W, Hancu I, Woessner DE. Dynamics of Spin I=3/2 under Spin-Locking Conditions in an Ordered Environment. J. Magn. Reson. 2001;151:298–313. doi: 10.1006/jmre.2001.2382. [DOI] [PubMed] [Google Scholar]