Abstract
In all cells examined, specific endoplasmic reticulum (ER) membrane arrays are induced in response to increased levels of the ER membrane protein 3-hydroxy 3-methylglutaryl coenzyme A (HMG-CoA) reductase. In yeast, expression of Hmg1p, one of two yeast HMG-CoA reductase isozymes, induces assembly of nuclear-associated ER stacks called karmellae. Understanding the features of HMG-CoA reductase that signal karmellae biogenesis would provide useful insights into the regulation of membrane biogenesis. The HMG-CoA reductase protein consists of two domains, a multitopic membrane domain and a cytosolic catalytic domain. Previous studies had indicated that the HMG-CoA reductase membrane domain was exclusively responsible for generation of ER membrane proliferations. Surprisingly, we discovered that this conclusion was incorrect: sequences at the carboxyl terminus of HMG-CoA reductase can profoundly affect karmellae biogenesis. Specifically, truncations of Hmg1p that removed or shortened the carboxyl terminus were unable to induce karmellae assembly. This result indicated that the membrane domain of Hmg1p was not sufficient to signal for karmellae assembly. Using β-galactosidase fusions, we demonstrated that the carboxyl terminus was unlikely to simply serve as an oligomerization domain. Our working hypothesis is that a truncated or misfolded cytosolic domain prevents proper signaling for karmellae by interfering with the required tertiary structure of the membrane domain.
INTRODUCTION
Assembly of specific membranes is an essential process throughout cell growth and development, but the molecular mechanisms responsible for specific membrane biogenesis are not understood in even a single case. Specific endoplasmic reticulum (ER) membrane arrays are induced in yeast by increasing the levels of certain ER membrane proteins, such as 3-hydroxy 3-methylglutaryl coenzyme A (HMG-CoA) reductase (Wright et al., 1988), cytochrome P450 (Schunck et al., 1991), cytochrome b5 (Vergeres et al., 1993), Sec12p (Nishikawa et al., 1994), the canine ribosome receptor (Wanker et al., 1995), and Pbn1p (protease B negative 1 protein) (Naik and Jones, 1998). Analysis of these inducible membranes provides an opportunity to discover the molecular mechanisms cells use to regulate the synthesis and organization of new membrane arrays.
HMG-CoA reductase is an integral ER membrane protein that catalyzes the production of mevalonate, a key intermediate in the synthesis of sterols and nonsterol isoprenoid compounds. The structure of HMG-CoA reductase can be divided into two domains: a complex membrane-spanning domain at the amino terminus followed by a cytosolic catalytic domain (Liscum et al., 1985; Basson et al., 1986; Roitelman et al., 1991). The membrane domain of mammalian and yeast HMG-CoA reductases spans the membrane seven or eight times and is connected to the catalytic domain through a flexible linker sequence (Liscum et al., 1985; Lum et al., 1996; Roitelman et al., 1992). The membrane domain is essential for both the proliferation of ER membranes (Jingami et al., 1987; Parrish et al., 1995) and the regulated degradation of both the mammalian and yeast HMG-CoA reductases (Gil et al., 1985; Skalnik et al., 1988; Hampton et al., 1996). The cytosolic domain is responsible for catalysis and is thought to mediate protein dimerization (Edwards et al., 1985; Basson et al., 1987; Frimpong and Rodwell, 1994).
Unlike mammalian cells, which have one HMG-CoA reductase, yeast express two functional HMG-CoA reductase isozymes, Hmg1p and Hmg2p (Basson et al., 1986). Each isozyme triggers the proliferation of distinct sets of membrane arrays that reflect the localization of the particular isozyme (Koning et al., 1996). Specifically, Hmg1p triggers the formation of karmellae, which are stacked pairs of membranes associated with the nucleus (Wright et al., 1988). Hmg2p induces peripheral ER membrane stacks and short karmellae (Koning et al., 1996). The ability to respond to HMG-CoA reductase elevations by generating ER membrane arrays is not unique to yeast but occurs in all cell types that have been examined. For example, in mammalian cells, HMG-CoA reductase induces the formation of hexagonal arrays of smooth ER tubules called crystalloid ER (Chin et al., 1982; Anderson et al., 1983; Orci et al., 1984; Kochevar and Anderson, 1987). In addition, many tissues that produce sterol derivatives, such as steroid hormones, contain characteristic ER membrane arrays (Fawcett, 1981).
Results of our laboratory and others using chimeric fusion proteins indicated that the catalytic domain of HMG-CoA reductase played no role in either induction of ER membranes or controlling their morphology. For example, proteins containing the HMG-CoA reductase membrane domain fused to unrelated carboxyl-terminal sequences such as β-galactosidase (β-gal) (Skalnik et al., 1988) or Suc2His4Cp (containing a portion of the invertase protein and histidinol dehydrogenase) induced the proliferation of ER membrane arrays that were indistinguishable from the membranes generated by the wild-type HMG-CoA reductase (Parrish et al., 1995). To determine whether the membrane domain of Hmg1p was sufficient to induce karmellae, we generated truncations of Hmg1p that shortened or completely removed the carboxyl terminus. Surprisingly, although the truncated proteins were expressed at high levels, yeast did not generate karmellae in response to them. Therefore, sequences beyond the membrane domain must be playing some role in the process of signaling for karmellae. One postulated role for the carboxyl-terminal sequences is to provide an oligomerization domain that would allow HMG-CoA reductase monomers to associate with each other. This hypothesis was tested by fusing the Hmg1p membrane domain to an oligomerization-competent β-gal and to an oligomerization-incompetent truncated β-gal (Tsuneoka and Mekada, 1992). Both β-gal chimeras induced karmellae, indicating that oligomerization of the cytosolic domain was not a requirement. Our experiments suggested that the carboxyl terminus of HMG-CoA reductase was not neutral for karmellae assembly and that the role of the cytosolic domain in karmellae assembly was unlikely to be oligomerization.
MATERIALS AND METHODS
Strains and Media
The yeast strains used in this study are listed in Table 1. Strains were grown at 30°C on rich minimal medium (0.67% yeast nitrogen base without amino acids, 2% casamino acids, and 2% glucose or 2% galactose plus 3% sucrose) supplemented with the appropriate acids or nucleotide bases (Sherman et al., 1986) or complete synthetic medium lacking histidine and uracil from Bio 101 (La Jolla, CA). Solid medium contained 2% agar (Sherman et al., 1986).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| JRY527a | MATa ade2-101 his3Δ200 lys2-801 met ura3-52 |
| RWY410b | JRY527 + pAK266 |
| RWY406 | JRY527 + pAK260 |
| RWY614 | JRY527 + pDP428 |
| RWY572 | JRY527 + pCS40 |
| RWY621b | JRY527 + pCR425 |
| RWY626 | JRY527 + pCR427 |
| RWY900 | JRY527 + pRH127-3 |
| RWY1494 | JRY527 + pDP586 |
| RWY1497 | JRY527 + pDP586 + pRH127-3 |
| RWY943 | JRY527 + pDP601 |
| RWY944 | JRY527 + pDP602 |
| Plasmid | Description |
|---|---|
| pAK266b | pGAL:Hmg1p in pRS316; CEN6 URA3 |
| pAK260 | pGAL:Hmg1Δ29p in pRS316; CEN6 URA3 |
| pDP428 | pHMG1:HMG1:HA in YEp352; 2 micron origin URA3 |
| pCS40 | pHMG1:Hmg1mem:Hmg2cat in YEp352; 2 micron origin URA3 |
| pCR425b | pGAL:Hmg1mem:HMG1525–987:GFP in pRS316; CEN6 URA3 |
| pCR427 | pGAL:Hmg1mem:GFP in pRS316; CEN6 URA3 |
| pRH127-3c | pGAPDH:Hmg1 catalytic domain 2 in Yplac195; micron origin URA3 |
| pDP586 | pGAL:Hmg1p in pRS313; CEN6 HIS3 |
| pDP601 | pHMG1:Hmg1:β-galactosidase in YEp352; 2 micron origin URA3 |
| pDP602 | pHMG1:Hmg1:β-galΔ20 in YEp352; 2 micron origin URA3 |
Plasmids
To truncate Hmg1p after the membrane domain, pCS4–14, a derivative of pCS4 with an XhoI site immediately after the membrane domain coding region (Sengstag et al., 1990), was digested with XhoI and ClaI to add a synthetic double-stranded oligonucleotide with the hemagglutinin (HA) epitope and a stop codon, resulting in plasmid pDP428. The HA epitope is a nonapeptide sequence derived from the influenza HA protein (Wilson et al., 1984). This plasmid encodes the N-terminal 523 amino acids of Hmg1p (the entire membrane domain), followed by 14 amino acids that included the HA epitope and a stop codon. The following two synthetic oligonucleotides were annealed to provide the 14 amino acids: 5′-TCGAGCATACCAGTTACCCATACGATGTTCCAGATTACGC TTAACTAGTTGA-3′ and 5′-CGTCAACTAGTTAAGCGTAATCTGGAACATCGTATGGGT AACTGGTATGC-3′.
To create Hmg1:β-gal fusion proteins, pDP428 was digested with XhoI and SpeI to insert an XhoI–SpeI fragment containing wild-type lacZ (pDP598) or to insert an XhoI–SpeI fragment containing a truncated lacZ (pDP600). The full-length β-gal was obtained from the plasmid pMKITNeo-XhoI-HMGalδ5′, and the truncated β-gal was obtained from the plasmid pMKITNeo-XhoI-HMGalδ20 (both provided by Helen Cheng and Robert Simoni, Stanford University, Palo Alto, CA). These plasmids contain Syrian hamster HMG-CoA reductase fused to β-gal or a truncated β-gal missing the last 20 amino acids needed for tetramerization (Cheng et al., 1999). Both intermediate plasmids, pDP598 and pDP600, needed a correction in reading frame to encode the N-terminal 523 amino acids of Hmg1p, followed by 76 amino acids of Syrian hamster HMG-CoA reductase linker region, and then ending with the entire β-gal or β-gal missing the last 20 amino acids (β-galΔ20). The reading frame between the Hmg1p membrane domain and the hamster HMG-CoA reductase linker region was corrected by inserting a double-stranded oligonucleotide. The double-stranded oligonucleotide was prepared by annealing two oligonucleotides: 5′-TCGAACATACCAGTACG-3′ and 5′-TCGACGTACTGGTATGT-3′. The final plasmids are pDP601 with the full-length β-gal fusion and pDP602 with the truncated β-gal fusion Δ20.
The galactose-inducible HMG-CoA reductase plasmid pGAL-HMG1 (pAK266) has been described previously (Koning et al., 1996). To create a catalytically inactive HMG-CoA reductase mutant, pAK266 was digested with KpnI and then religated. This plasmid (pAK260) encodes the GAL1/10 promoter, N-terminal 957 amino acids of Hmg1p (which includes the entire membrane domain and most of the catalytic domain), followed by a 29-amino-acid gap, and then the remaining 68 amino acids of Hmg1p. The green fluorescent protein (GFP) (Chalfie et al., 1994; Chalfie, 1995; Prasher et al., 1992; Prasher, 1995) expression constructs used in these studies were derived from the plasmid pJC81 (provided by Jeff Cox and Peter Walter, University of California San Francisco, San Francisco, CA). This plasmid contains a mutant version of GFP10 in which the first two codons have been changed to encode a BamHI site. To create a membrane domain Hmg1:GFP fusion, a 1.5-kb BamHI–EagI fragment of pJC81 (containing the 710 bp GFP10 ORF, 400 bp of the ACT1 gene containing the transcriptional terminator, and 390 bp of the tet gene) was subcloned into the BamHI–EagI sites of pRS316 (Sikorski and Hieter, 1989) creating plasmid pCR415. This plasmid, pCR415, was then digested with BamHI and SmaI, treated with Klenow to fill in the ends, and religated, creating pCR426. A 2.2-kb EcoRI fragment containing pGAL-HMG1 from pJR435 (Basson et al., 1988) was ligated into the EcoRI site of pCR426, creating plasmid pCR427. This plasmid encodes an Hmg1mem:GFP fusion, consisting of the GAL1/10 promoter, N-terminal 525 amino acids of Hmg1p, followed by three linker residues and all of GFP except the initiating methionine. The cytosolic Hmg1:GFP fusion encoded by pCR425 has been described previously (Koning et al., 1996). This plasmid encodes the GAL1/10 promoter, N-terminal 987 amino acids of Hmg1p, followed by two linker residues and all of GFP except the initiating methionine.
To exchange the Hmg1p catalytic domain with the catalytic domain of Hmg2p, pA7, which contains an introduced XhoI site after the sequences encoding the Hmg1p last transmembrane domain (Sengstag et al., 1990), was digested with HindIII, treated with Klenow, and then cut with XhoI. The resulting 9-kb fragment contained the HMG1 promoter and all the membrane domain coding sequences as well as vector sequences. A 2-kb fragment that contains the Hmg2p linker region and catalytic domain coding sequences was isolated from pB7, which contains an introduced SalI site after the last transmembrane domain of Hmg2p (Sengstag et al., 1990). This fragment was isolated after digestion with EcoRI and treatment with Klenow and then digested with SalI. Ligation of the fragments produced pCS40, which encodes the HMG1 promoter, N-terminal 525 amino acids of Hmg1p, fused to the Hmg2p linker and catalytic domain.
The multicopy (2μ) plasmid pRH127–3, which expressed the Hmg1p catalytic domain under control of the constitutive glyceraldehyde-3-phosphate dehydrogenase promoter, has been previously described (Donald et al., 1997). To coexpress full-length Hmg1p with the soluble Hmg1p catalytic domain, a galactose-inducible HMG1 plasmid, pAK266, was digested with XhoI and SpeI, yielding a 4.3-kb fragment with the GAL1/10 promoter and HMG1. This fragment was ligated into the XhoI and SpeI sites of pRS313 (Sikorski and Hieter, 1989) to produce a CEN-GAL1/10-HMG1 plasmid in a vector that contains the HIS3 selectable marker (pDP586).
3,3′-Dihexyloxacarbocyanineiodide (DiOC6) Staining
DiOC6 staining was performed as described previously (Koning et al., 1993). Cells in log phase growth were stained with 10 μg/ml DiOC6 (Kodak, Rochester, NY) per 107 cells using a 1 mg/ml ethanolic stock. Stained cells were observed with conventional fluorescence optics, using a Nikon (Melville, NY) Microphot-FXA epifluorescence microscope with excitation (480 ± 20 nm) and barrier (535 ± 40 nm) filters appropriate for fluorescein.
Electron Microscopy
Preparation of cells for electron microscopy was a variation on methods described previously (Wright and Rine, 1989). Specifically, cells were grown to 1 OD600/ml, and the culture was fixed in 2% glutaraldehyde in buffer [0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 1 mM CaCl2, 1 mM MgCl2], postfixed in 2% KMnO4, and stained en bloc with 1% uranyl acetate. The cells were dehydrated through a graded ethanol series and embedded in Spurr’s resin (Spurr, 1969). Sections were stained with Reynolds’ lead citrate (Reynolds, 1963). Observations were made on a Philips (Eindhoven, The Netherlands) CM100 transmission electron microscope at 60–80 kV.
Immunofluorescence
Immunofluorescence was performed using a procedure similar to that described by Pringle et al. (1989). Log phase cells were fixed with 3.7% formaldehyde, treated with Zymolyase (United States Biological, Swampscott, MA) to partially remove their cell walls, and applied to multiwell slides (Koning et al., 1996). Antisera generated against the carboxyl terminal 15 amino acids of Hmg1p (LDII) was used at a 1:100 dilution. Kar2p antibodies were a gift of Mark Rose (Princeton University, Princeton, NJ) or Jeff Brodsky (University of Pittsburgh, Pittsburgh, PA) and were used at a 1:2000 dilution. GFP antibody was purchased from Clontech (Palo Alto, CA) and used at a 1:100 dilution. HA antisera (12CA5) was purchased from Boehringer Mannheim (Indianapolis, IN) and used at a 1:100 dilution. A 1:800 dilution of rabbit anti-β-gal antibody (Cappel Organon Technika, Durham, NC) was used after pretreatment with a yeast cell lysate to decrease nonspecific binding. In all cases, the incubation in primary antisera was for 1 h at room temperature. The antibody solution was gently aspirated away, and the cells were washed five times with TBST (25 mM Trizma base, 3 mM KCl, 140 mM NaCl, 0.05% Tween 20). Then 10 μl of blocking solution (TBST and 1% ovalbumin) was applied to each well. In all cases, 10 μl of secondary antibody was diluted in blocking solution, centrifuged for 10 min at 12,000 rpm in a microcentrifuge, and applied to the appropriate wells. The secondary antibodies used were 1:1000 goat anti-rabbit fluorescein conjugated, 1:200 goat anti-mouse fluorescein conjugated, and 1:500 goat anti-rabbit Texas Red conjugated (Cappel Organon Technika). After 45 min, the secondary antibody was washed five times with TBST. Ten microliters of a 1:1000 dilution of 1 mg/ml DAPI (Sigma, St. Louis, MO) in Tris-buffered saline was added to each well for 1 min. After one rinse with Tris-buffered saline, a drop of Citifluor (Ted Pella, Redding, CA) was applied to each well, and the slide was sealed with a coverslip and nail polish. Each experiment included a positive control using an antibody to detect tubulin (Yol1/34; Accurate Chemical, Westbury, NY) used at 1:10 dilution and a negative control lacking primary antiserum.
Protein Preparation and Immunoblotting
After growth in rich minimal medium containing 2% galactose and 3% sucrose for 12 h, the cultures were diluted into fresh medium and grown for 4–6 h until they reached 1 OD600/ml. A 16- to 18-h galactose induction was used because a high level of karmellae is observed in cells after 12 h in galactose. The cells were then pelleted for 5 min at 834 × g in a clinical centrifuge, and the cell pellets were frozen at −80°C until lysed. Strains that did not contain galactose-inducible plasmids were grown overnight in rich minimal medium containing 2% glucose medium. The next morning, the cells were diluted into fresh glucose medium and grown to 1 OD600/ml before harvesting a cell pellet. A total membrane fraction was prepared from the cells using modifications of previously described methods (Deschenes and Broach, 1987; Koning et al., 1996). The pelleted membranes were resuspended in 100 μl of lysis buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 20 mM MOPS, pH 7.4) containing protease inhibitors (2 μg/ml each N-tosyl-l-phenylalanine-chloromethyl ketone, leupeptin, pepstatin A, aprotinin) found in Complete Protease Inhibitor tablets (Boehringer Mannheim). The absorbance at 280 nm was measured for each sample in 1% SDS, and 100 A280 units were loaded on duplicate gels after heating at 65°C in 1× Laemmli sample buffer (0.03 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.005% bromphenol blue) for 10 min, followed by a 5 min centrifugation at top speed in a microcentrifuge (Laemmli, 1970). One gel was blotted to nitrocellulose and processed for immunodetection as described (Lum and Wright, 1995), and the duplicate gel was stained with colloidal blue from Novex (San Diego, CA). The duplicate gel and Western blot were digitized by scanning and analyzed using NIH Image 1.60 software as described (Lum and Wright, 1995). The same membrane preparation procedure was used to determine the stability of the Hmg1p or Hmg1 fusion protein pool by measuring the relative amount of Hmg1p immunoreactivity remaining at various time points after blocking protein synthesis with 50 μg/ml cycloheximide (CHX) (Hampton and Rine, 1994).
β-Gal Activity Assays
The strains RWY 943 and RWY 944 were assayed for β-gal activity according to the protocol of Guarente and Ptashne (1981). Briefly, log phase cells were resuspended in buffer, SDS, and chloroform. Then o-nitrophenyl-β-d-galactoside was added as the substrate for β-gal. The reaction was stopped by adding Na2CO3, and the cell debris was removed by centrifugation. The OD420 was measured, and the results were normalized to the OD600 of the culture and to the assay time. Duplicate assays were run in each case, and cell dilutions were determined to be in the linear range of the assay.
RESULTS
Truncating the Hmg1p Carboxyl Terminus Reduced or Eliminated the Protein’s Ability to Induce Karmellae
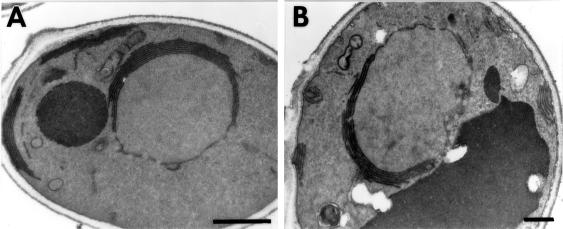
Yeast cells respond to increased levels of Hmg1p by proliferating stacked pairs of nuclear-associated membranes called karmellae (Wright et al., 1988). To determine whether the cytosolic domain of Hmg1p was required for karmellae biogenesis, we examined the organization of the ER in cells expressing wild-type or truncated Hmg1p proteins, using fluorescence microcopy (summarized in Figure 1) and electron microscopy (Figure 2). In this study Hmg1p levels were elevated by use of either a galactose-inducible promoter or a multicopy plasmid, resulting in an ∼10-fold increase in protein in both cases. As expected, karmellae were observed in 35% of the cells expressing wild-type Hmg1p (Figure 2A). Note that karmellae are never observed in 100% of the cell population in part because karmellae remain in the mother cell at mitosis (Wright et al., 1988). Surprisingly, the truncated Hmg1p, which lacked a catalytic domain (Hmg1mem:HA), failed to induce karmellae or any other type of ER membrane proliferation (Figure 2B).
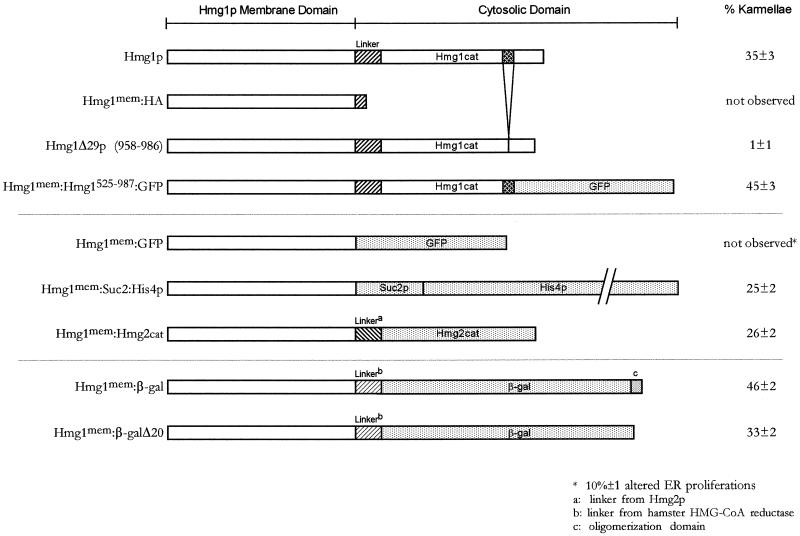
Figure 1.
Summary of karmellae levels in cells expressing intact Hmg1p or the different Hmg1 fusion proteins. The fusion proteins used in this study are diagrammed together with the percentage of karmellae observed for the strains expressing each protein. These karmellae percentages were determined by scoring cells from three independent experiments. At least 400 cells were scored per experiment.
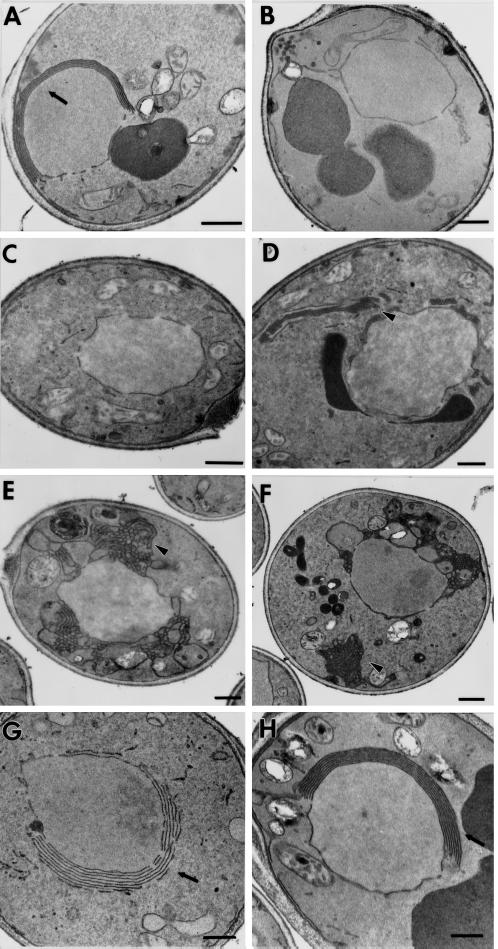
Figure 2.
The ultrastructure of the nuclear envelope in a wild-type strain and in strains expressing the Hmg1 fusion proteins was compared. (A) A wild-type strain expressing intact Hmg1p (RWY 410) contains karmellae membranes in an ordered array on the nucleus. (B) Cells expressing the truncated Hmg1:HA protein (RWY 614) lack any ER membrane proliferations. (C and D) Cells expressing Hmg1Δ29p (RWY 406) usually lack karmellae and sometimes produce extended ER networks. (E and F) Cells expressing the Hmg1mem:GFP (RWY 626) do not generate karmellae but have altered ER arrays. (G) Cells expressing the Hmg1mem:Hmg1525–987:GFP (RWY 621) generate karmellae. (H) Cells expressing the Hmg1mem:Hmg2 cat (RWY 572) generate karmellae predominantly. Occasionally, Hmg2-type membranes are found in the population of cells expressing the Hmg1mem:Hmg2cat. Arrows point to karmellae; arrowheads point to proliferated ER. Bars, 0.5 μm.
We considered the possibility that removal of the entire catalytic domain might have profound effects on the folding of the Hmg1p membrane domain. If so, less dramatic changes to the catalytic domain should allow the protein to remain karmellae competent. To test this idea, we examined the ultrastructure of cells expressing a catalytically inactive Hmg1p missing only 29 amino acids within the catalytic domain (Hmg1Δ29p) and determined that only 1% of the cell population contained karmellae (Figure 2C). In an additional 1% of the cell population, the Hmg1Δ29p induced tubular ER stacks, which resembled the mammalian crystalloid ER (Figure 2D) (Anderson et al., 1983). This dramatic reduction in karmellae was unexpected, because the catalytic domain can be completely replaced with unrelated carboxyl-terminal sequences without adversely affecting the ability of the protein to induce karmellae (Skalnik et al., 1988; Parrish et al., 1995). In addition, we had previously confirmed that the catalytic activity of Hmg1p was not required for karmellae biogenesis by examining the ultrastructure of cells expressing a catalytically inactive allele of HMG1 in which a single amino acid had been changed; this mutant Hmg1p induced karmellae at the wild-type level (Profant, unpublished observations). Thus, neither a specific protein sequence nor enzymatic activity appeared to be necessary for the protein’s ability to induce karmellae, yet even rather minor changes to the Hmg1p carboxyl terminus could interfere with karmellae assembly.
We also examined cells expressing an Hmg1 membrane:Hmg2 catalytic domain fusion, in which the Hmg2p catalytic domain replaced the Hmg1p catalytic domain (Hmg1mem:Hmg2cat). If the information in the Hmg1p membrane domain dictates the cell’s response as predicted, the Hmg1mem:Hmg2cat should generate karmellae. In addition, the catalytic domain is highly conserved between Hmg1p and Hmg2p (Basson et al., 1988), making it an unlikely source of the membrane morphology differences induced by Hmg1p versus Hmg2p. As expected, the Hmg1mem:Hmg2cat induced predominantly karmellae (Figure 2H). However, in 4% of the cell population, the Hmg1mem:Hmg2cat induced Hmg2-type membranes. Therefore, information present in the Hmg2 catalytic domain continued to exert some influence on the membranes generated even in the context of the Hmg1 membrane domain.
To investigate the role of carboxyl-terminal sequences in karmellae biogenesis further, we characterized the membrane proliferations induced by two different Hmg1:GFP fusion proteins. The first fusion included the linker sequence and a portion of the Hmg1 catalytic domain (Hmg1mem:Hmg1525–987:GFP). In the second fusion, the cytosolic domain of Hmg1p was completely replaced by GFP (Hmg1mem:GFP). Karmellae were induced by Hmg1mem:Hmg1525–987:GFP (Figure 2G). In contrast, as for the Hmg1:HA protein, karmellae were never observed in cells expressing the Hmg1mem:GFP; instead 10% of the cells assembled disorganized ER membrane stacks (Figure 2, E and F). The disorganized ER consisted of either portions of the nuclear envelope folding away from the nucleus or as peripheral membrane stacks. These observations are consistent with an essential, but unexpected, role for the carboxyl terminus in karmellae biogenesis.
Differences in Protein Amounts Did Not Account for the Inability to Induce Karmellae
Elevated levels of Hmg1p are required to induce karmellae (Wright et al., 1988). Thus, mutations that decrease the amount of Hmg1p would result in proteins that are unable to induce karmellae assembly. We determined that the steady-state levels of all of the fusion proteins were 70–94% of wild type, verifying that adequate protein was expressed to induce karmellae assembly in each case (Figure 3 and Table 2).
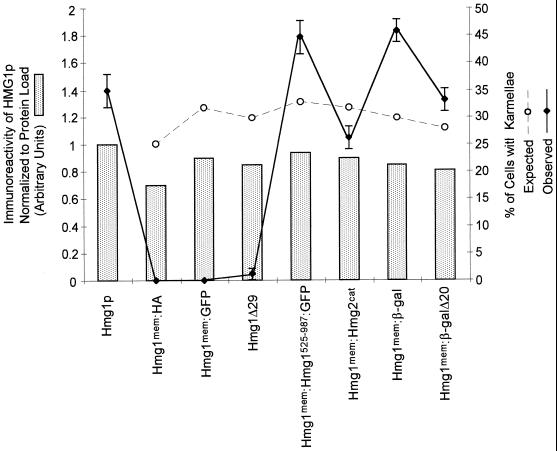
Figure 3.
Comparison of relative protein levels and ability to induce karmellae for the Hmg1 fusion proteins. The graph compares the relative protein levels of the Hmg1 fusion proteins with the wild-type Hmg1p level. The wild-type Hmg1p steady-state level was set to 1.0. For each strain, the percentage of cells in the population with karmellae was observed using DiOC6, which stains the ER. The expected percentage of cells with karmellae was determined by adjusting the observed percentage of karmellae in the wild-type Hmg1p strain for the relative HMG-CoA reductase protein level in each subsequent strain.
Table 2.
The inability to generate karmellae did not correlate with relative amount of HMG-CoA reductase protein expressed
| Protein expressed | Relative protein amount | Karmellae observed (%) | Karmellae expected (%) |
|---|---|---|---|
| wild-type Hmg1p | 1.00 | 35 | 35 |
| Hmg1mem:HA | 0.70 | 0 | 25 |
| Hmg1mem:GFP | 0.90 | 0 | 32 |
| Hmg1Δ29p | 0.85 | 1 | 30 |
| Hmg1mem:Hmg1525–987GFP | 0.94 | 45 | 33 |
| Hmg1mem:Hmg2cat | 0.90 | 26 | 32 |
| Hmg1mem:β-galactosidase | 0.85 | 46 | 30 |
| Hmg1mem:β-galΔ20 | 0.81 | 33 | 28 |
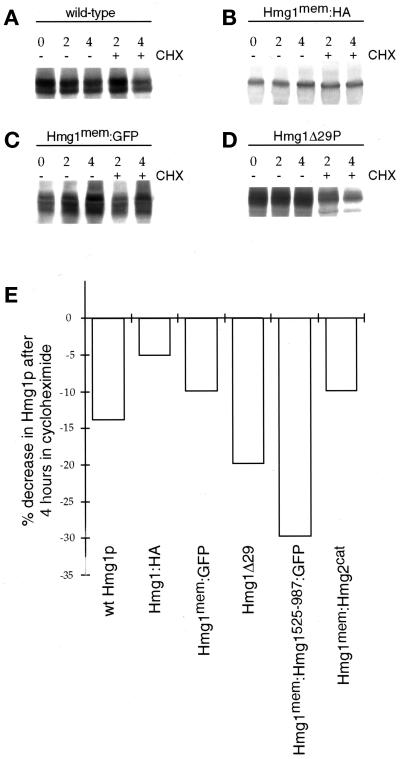
An alternate explanation for the inability of certain HMG1 constructs to generate karmellae is that Hmg1 fusion proteins that cannot induce karmellae may have decreased stability. To investigate this possibility, we compared levels of Hmg1p that remain after addition of CHX. Because CHX inhibits new protein synthesis, the amount of protein remaining at various times after treatment can be used to assess the degradation rate of the protein in question (Hampton and Rine, 1994). As expected (Hampton and Rine, 1994), wild-type Hmg1p was stable, with little degradation observed after 4 h of CHX treatment (Figure 4A). The truncated Hmg1p that lacked a cytosolic domain (Hmg1:HA) (Figure 4B) and the Hmg1Δ29p (Figure 4D) were also stable after 4 h of CHX treatment. In addition, both the Hmg1mem:GFP protein (Figure 4C) and the Hmg1mem:Hmg1525–987:GFP protein (Figure 4E) were stable after 4 h in CHX. In contrast to Hmg1p, intact Hmg2p is quite unstable, with a half-life of 60 min (Hampton and Rine, 1994). Unlike wild-type Hmg2p, the Hmg1mem:Hmg2cat protein was stable after 4 h of CHX treatment (Figure 4E), confirming that the membrane domain of the protein determines its half-life (Gil et al., 1985; Skalnik et al., 1988; Hampton and Rine, 1994). In Figure 4E, the percent decrease in protein after 4 h was plotted for the different Hmg1 fusions, showing that the fusion proteins had similar stabilities. These results eliminated the possibility that the fusions were unable to signal for karmellae because of inadequate steady-state protein levels or instability of the Hmg1p variants.
Figure 4.
Comparison of Hmg1p levels in wild-type cells and cells expressing the Hmg1 fusion proteins. Total cellular membranes were isolated and prepared for immunoblotting. At time zero, the culture was split into two flasks with one flask receiving CHX. At subsequent times (2 and 4 h), aliquots were removed for membrane isolation. (A) Intact Hmg1p (RWY 410) was detected using an antisera against the Hmg1p catalytic domain. (B) The truncated Hmg1mem:HA protein (RWY 614) was detected using the 12CA5 anti-HA antibody. (C) The Hmg1mem:GFP fusion (RWY 626) was detected using anti-GFP antisera. (D) Hmg1Δ29p (RWY 406) was detected using an antisera against the Hmg1p catalytic domain. (E) The graph compared the fusion protein levels, normalized to total protein and to designation of the zero time point level for each protein set to 1.0. The relative levels of protein were determined for all time points, and the percent decrease was calculated by comparing the 0-h protein level with the 4-h with-CHX protein level.
Alteration of the Cytosolic Domain of Hmg1p Did Not Cause Mislocalization of the Protein to the Peripheral ER
The ability of a protein to induce karmellae might depend on its specific localization within the cell (Koning et al., 1996). Because karmellae arise from the nuclear envelope, a protein capable of signaling for karmellae assembly would be expected to be localized in the nuclear envelope. Consistent with this hypothesis, increased levels of wild-type Hmg1p are present predominantly in the nuclear envelope; in contrast, increased levels of Hmg2p localize predominantly to the peripheral ER (Koning et al., 1996).
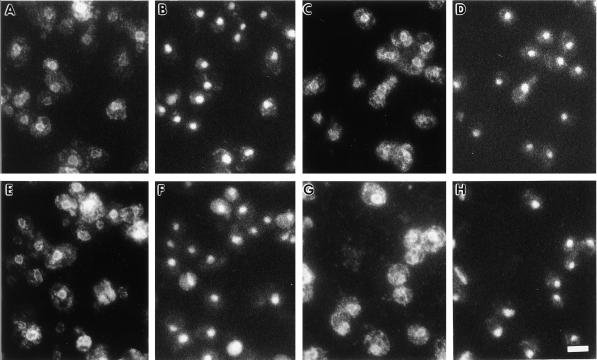
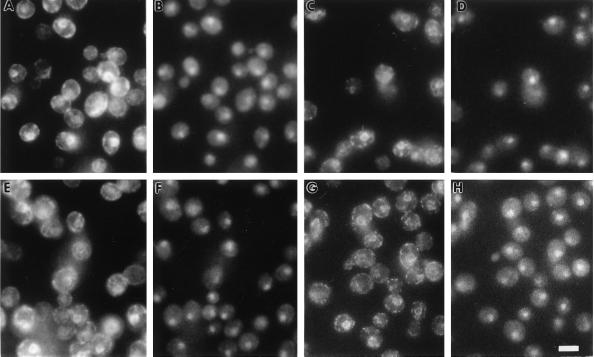
The subcellular localization of the fusion proteins was examined by immunofluorescence to assess whether alterations in the cytosolic domain of Hmg1p influenced the distribution of the protein within the ER. As expected, intact Hmg1p was present predominantly in the nuclear envelope, with increased fluorescence found in cells generating karmellae (Figure 5, A and B). The localization of the lumenal ER protein Kar2p demonstrates the pattern for proteins found throughout the ER (Rose et al., 1989; Preuss et al., 1991). In Figure 5, C and D, Kar2p staining appeared in the nuclear envelope and in a network stretching to the cell periphery. The Hmg1Δ29p had a localization pattern indistinguishable from wild-type Hmg1p (Figure 5, E and F), indicating that the inability of this protein to generate karmellae was not due to an altered distribution within the ER. The localization pattern for Kar2p in the strain expressing Hmg1Δ29p was unaltered from wild-type Kar2p localization (Figure 5, G and H).
Figure 5.
Localization of elevated levels of wild-type Hmg1p and the Hmg1 fusion proteins with altered cytosolic domains by indirect immunofluorescence. Indirect immunofluorescent localizationof Hmg1p or Kar2p is shown in A, C, E, and G. Localization of the nucleus in the same cells is shown by DAPI staining in B, D, F, and H. (A and B) Localization of elevated levels of wild-type Hmg1p was determined in cells of strain RWY 410 using an antibody to the catalytic domain of Hmg1p. (C and D) Localization of Kar2p in strain RWY 410 is shown for comparison. (E and F) Localization of elevated levels of Hmg1Δ29p in strain RWY 406 was determined by using an antibody to the catalytic domain of Hmg1p. (G and H) Localization of Kar2p in strain RWY 406 is shown for comparison. Bar, 5 μm.
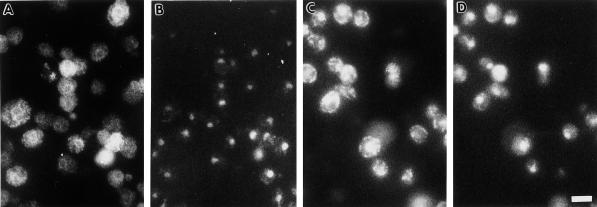
In Figure 6, the truncated Hmg1p lacking a catalytic domain (Hmg1:HA) was localized using a monoclonal antibody (12CA5) to the HA epitope. The protein was detected in the nuclear envelope and in the ER network extending throughout the cytosol (Figure 6, A and B). Thus, the truncated Hmg1:HA protein appeared to be localized throughout the ER rather than displaying a predominantly nuclear envelope pattern. The Kar2p localization pattern is shown for comparison in Figure 6, C and D. To confirm the specificity of the HA antibody, immunofluorescence was performed on a yeast strain that was not expressing an HA-tagged protein. No immunofluorescence pattern was observed in these cells, but the entire cell was faintly visible (our unpublished data).
Figure 6.
Localization of the truncated Hmg1:HA protein by indirect immunofluorescence. Indirect immunofluorescent localization of the truncated Hmg1:HA protein or Kar2p is shown in A and C. Localization of the nucleus in the same cells is shown by DAPI staining in B and D. (A and B) Localization of elevated levels of the truncated Hmg1:HA protein in strain RWY 614 was detected by using 12CA5 anti-HA antibody. (C and D) Localization of Kar2p in strain RWY 614 is shown for comparison. Bar, 5 μm.
Next, we investigated the localization patterns of the two Hmg1:GFP fusions. The Hmg1mem:Hmg1525–987:GFP protein, which generates karmellae, was localized in the nuclear envelope (Figure 7, E and F; Koning et al., 1996) similar to wild-type Hmg1p. The Hmg1mem:GFP fusion, which failed to generate karmellae, had a nuclear envelope localization pattern with some peripheral ER staining (Figure 7, A and B), again appearing similar to that of wild-type Hmg1p.
Figure 7.
Localization of the Hmg1mem:GFP fusion and the Hmg1mem:Hmg1525–987:GFP fusion by indirect immunofluorescence. Indirect immunofluorescent localization of the GFP fusion proteins or Kar2p in shown in A, C, E, and G. Localization of the nucleus in the same cells is shown by DAPI staining in B, D, F, and H. (A and B) Localization of elevated levels of the Hmg1mem:GFP protein in strain RWY 626 was determined by using a GFP antibody. (C and D) Localization of Kar2p in strain RWY 626 is shown for comparison. (E and F) Localization of elevated levels of the Hmg1mem:Hmg1525–987:GFP protein was determined in cells of strain RWY 621 using a GFP antibody. (G and H) Localization of Kar2p in strain RWY 621 is shown for comparison. Bar, 5 μm.
Taken together, the localization patterns demonstrated that the failure to assemble karmellae in response to the Hmg1 fusions was not due to a large proportion of the protein being mislocalized out of the ER or mislocalized exclusively to the peripheral ER. However, from our data we cannot rule out the possibility that subtle alterations in the localization of the protein, such as that observed for Hmg1p:HA, might influence the ability of the protein to induce karmellae.
Competition with the Soluble Catalytic Domain Reduced the Overall Level of Karmellae Generated by the Wild-Type Hmg1p
Our results suggested that the membrane domain and the cytosolic domain of Hmg1p cooperate in some way to generate karmellae. To test this idea, we examined karmellae assembly in a strain expressing both full-length Hmg1p and a truncated protein containing only the linker and the catalytic domain of Hmg1p. Because HMG-CoA reductase is thought to act as a dimer using contact sites in the catalytic domain (Edwards et al., 1985; Basson et al., 1987; Frimpong and Rodwell, 1994; Lawrence et al., 1995; Rogers et al., 1997; Tabernero et al., 1999), we hypothesized that expression of a soluble catalytic domain would interfere with the ability of the holoprotein to dimerize and to induce karmellae. It is important to note that the catalytic domain is capable of proper folding and oligomerization in the absence of attachment to the membrane domain, because it is enzymatically active,and the amount of activity parallels the protein amount (Donald et al., 1997). As expected, elevated expression of the soluble catalytic domain alone was incapable of inducing karmellae (Table 3). In the strain expressing both the holoprotein and the soluble catalytic domain, the soluble Hmg1p catalytic domain was expressed at threefold higher levels than the holoprotein (Table 3). The Hmg1p holoprotein was expressed at levels similar to strains that are not simultaneously expressing the catalytic domain. Consequently, by the law of mass action, and assuming that the proteins dimerized via carboxyl-terminal sequences, we expected that 86% of the holoprotein would be complexed with a soluble catalytic domain and that 14% of the holoprotein would form holoprotein-holoprotein dimers. Thus, if dimerization between holoproteins were important for karmellae assembly, a dramatic drop in the level of karmellae should be seen. Specifically, we would predict that the proportion of cells containing karmellae should decrease by 85% (from 41 to <6%). Unexpectedly, karmellae assembly was decreased by only 33% in the strain expressing both the holoprotein and the soluble catalytic domain (Table 3). This result suggested that carboxyl terminus–mediated dimerization between holoproteins may not be essential for karmellae assembly.
Table 3.
Expression of the soluble catalytic domain interfered with the production of karmellae without lowering the level of Hmg1p holoprotein
| Strain | Protein expressed
|
Karmellae (%)a | Relative holoprotein levelb | Relative catalytic domain levelb | |
|---|---|---|---|---|---|
| pGAL:Hmg1p | pGAPDH:Hmg1p | ||||
| RWY1494 | X | 41 ± 1 | 1.00 | ||
| RWY900 | X | Never seen | 3.60 | ||
| RWY1497 | X | X | 27 ± 1 | 1.05 | 3.30 |
The percent karmellae was determined from three independent experiments.
After adjusting for total protein loaded, the amount of holoprotein or catalytic domain was normalized to holoprotein in RWY1494.
Oligomerization of the Cytosolic Domain of Hmg1p Was Not Required for Karmellae Formation
To further test the hypothesis that oligomerization via the cytosolic domain was not required for karmellae assembly, we fused the Hmg1p membrane domain to β-gal or to a truncated β-gal missing the last 20 amino acids (β-galΔ20). Full-length β-gal forms tetramers, which are the enzymatically active form of the enzyme; dimers and monomers are not catalytically active (Fowler and Zabin, 1983). Truncated β-gal missing the last 10 amino acids is found as a monomer (Fowler and Zabin, 1966, 1968). In addition, β-gal missing the last 16 amino acids lacks enzyme activity and runs on a nondenaturing gel as a monomer (Tsuneoka and Mekada, 1992). Although we did not directly assess the oligomerization state of either β-gal fusion, the lack of β-gal activity (Table 4) exhibited by Hmg1:β-galΔ20 was consistent with this carboxyl terminus being monomeric. Additionally, because Hmg1:β-gal was enzymatically active (Table 4), the carboxyl termini of separate fusion proteins were capable of forming tetramers. If oligomerization of the cytosolic region on Hmg1p is critical for karmellae, then the Hmg1:β-galΔ20 will fail to induce karmellae.
Table 4.
Karmellae biogenesis in response to β-galactosidase fusion proteins
| Protein | Karmellae (%)a
|
β-gal activityb (U/OD/min) | |
|---|---|---|---|
| 4 h | 8 h | ||
| Hmg1:β-galactosidase | 46 ± 2 | 56 ± 2 | 236 ± 12 |
| Hmg1:β-galΔ20 | 33 ± 2 | 37 ± 2 | 0.7 ± 0.4 |
The percent karmellae was determined from three independent experiments.
β-Galactosidase activity was determined from three independent experiments, running duplicate tubes for each experiment.
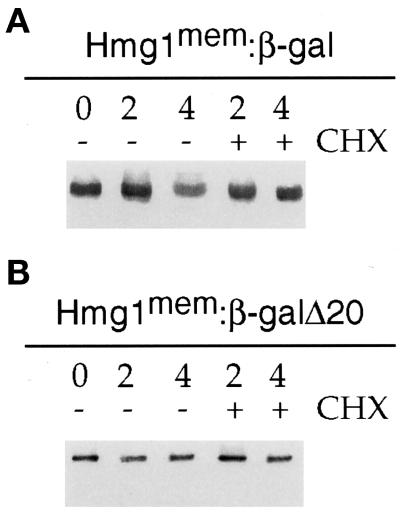
We performed immunoblots to measure expression of the β-gal fusion proteins and to confirm the stability of the proteins. Hmg1:β-gal was present at 85% and Hmg1:β-galΔ20 was present at 81% of the wild-type steady-state Hmg1p level, and both fusions were stable during the 4-h treatment with CHX (Figure 8). Both Hmg1:β-gal and Hmg1:β-gal Δ20 generated karmellae (Figure 9) and were localized predominantly to the nuclear envelope of the ER (Figure 10). Because the Hmg1:β-galΔ20 induced karmellae, oligomerization of the cytosolic domain did not appear to be required for karmellae signaling. However, cells expressing the Hmg1:β-gal generated a higher level of karmellae compared with the cells expressing the Hmg1:β-galΔ20 (Table 4).
Figure 8.
Comparison of the stability of the Hmg1:β-gal fusion proteins by CHX chase. Total cellular membranes were isolated and prepared for immunoblotting. At time zero, the culture was split into two flasks with one flask receiving CHX. At subsequent times (2 and 4 h), aliquots were removed for membrane isolation. (A) The Hmg1:β-gal protein expressed in RWY 943 was detected using an antisera generated against the membrane domain of Hmg1p. (B) The Hmg1:β-galΔ20 protein level in RWY 944 was detected using the same antisera against the Hmg1p membrane domain.
Figure 9.
Comparison of the karmellae induced by either Hmg1:β-gal or Hmg1:β-galΔ20. Cells from both strains were fixed for electron microscopy. (A) RWY 943 cells expressing Hmg1:β-gal generated karmellae. (B) RWY 944 cells expressing the Hmg1:β-galΔ20 also generated karmellae. Bars, 0.5 μm.
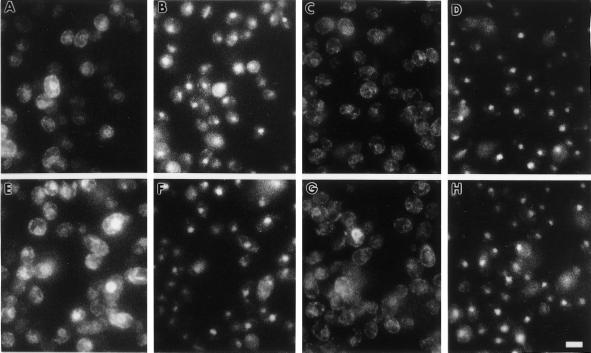
Figure 10.
Indirect immunofluorescent localization of the Hmg1:β-gal fusions or Kar2p is shown in A, C, E, and G. Localization of the nucleus in the same cells is shown by DAPI staining in B, D, F, and H. (A and B) Localization of elevated levels of Hmg1:β-gal was determined in strain RWY 943 by using anti-β-gal antibody. (C and D) Localization of Kar2p in strain RWY 943 is shown for comparison. (E and F) Localization of elevated levels of Hmg1:β-gal 20 was determined in strain RWY 944 by using the same anti-β-gal antibody. (G and H) Localization of Kar2p in strain RWY 944 is shown for comparison. Bar, 5 μm.
DISCUSSION
To explore how cells regulate the synthesis and organization of new membrane arrays, we sought to define the features of Hmg1p that are necessary for this protein to act as a signal for karmellae biogenesis. In eukaryotes, the carboxyl terminus of HMG-CoA reductase contains the cytosolic catalytic domain, and the amino terminus contains the membrane domain (Liscum et al., 1985). Although details concerning the structure and function of the HMG-CoA reductase catalytic domain are beginning to be solved (Darnay et al., 1992; Lawrence et al., 1995; Rogers et al., 1997; Tabernero et al., 1999), the HMG-CoA reductase membrane domain remains much more cryptic. In plants, the membrane domain contains two transmembrane helices (Denbow et al., 1996; Re et al., 1997), whereas in animals and fungi, it contains of a series of seven or eight transmembrane helices (Liscum et al., 1985; Basson et al., 1988; Sengstag et al., 1990; Olender and Simon, 1992; Roitelman et al., 1992). Although there is little obvious primary sequence conservation, the structural conservation of this complex membrane domain over the 1 billion years of evolution since divergence of fungi and animals (Doolittle et al., 1996; Lum et al., 1996; Feng et al., 1997) suggests that the structure has an important functional role. However, the molecular logic of tethering the catalytic activity to a membrane appears enigmatic, because prokaryotic HMG-CoA reductase proteins are soluble (Beach and Rodwell, 1989; Bischoff and Rodwell, 1996; Baltscheffsky et al., 1997; Bochar et al., 1997; Takahashi et al., 1999), and the catalytic domain of eukaryotic HMG-CoA reductase can support apparently normal growth of cells when it is freed from the membrane domain (Gil et al., 1985; Donald et al., 1997).
Although the molecular details are not yet understood, two functions can be assigned to the HMG-CoA reductase membrane domain. First, it mediates regulation of protein half-life in response to sterol levels (Skalnik et al., 1988; Chun and Simoni, 1992; Hampton and Rine, 1994; Kumagai et al., 1995; Sekler and Simoni, 1995). Second, it is required for stimulation of ER expansion in response to increased HMG-CoA reductase levels (Chin et al., 1982; Jingami et al., 1987; Wright et al., 1988; Parrish et al., 1995). For both functions, regions of the membrane domain, including transmembrane segments and the loops that connect them, have been identified as playing particularly critical roles (Gil et al., 1985; Pathak et al., 1986; Jingami et al., 1987; Hampton and Rine, 1994; Kumagai et al., 1995; Parrish et al., 1995; Sekler and Simoni, 1995; Koning et al., 1996). For example, at least some aspects of protein half-life control may function via a sterol-sensing domain located within the region of the membrane domain extending from transmembrane segments 2–6 (Lange and Steck, 1998; Osborne and Rosenfeld, 1998; Cheng et al., 1999). In yeast, the amino-terminal region before transmembrane segment 1 is important for regulated degradation (Gardner et al., 1998), and the final ER-lumenal loop between predicted seventh and eighth transmembrane domains (loop G) carries information required for the assembly of karmellae (Parrish et al., 1995). We hypothesize that loop G mediates karmellae assembly via interactions with other proteins, perhaps other HMG-CoA reductase molecules. Consequently, an obvious prediction is that alterations of HMG-CoA reductase that prevent proper folding or tertiary structure of this ER–lumenal loop would interfere with necessary protein–protein interactions, resulting in failure to induce karmellae assembly. This prediction is supported by the identification of point mutations in loop G that can nearly abolish the protein’s ability to induce karmellae without affecting the protein’s stability or steady-state levels (Profant and Wright, unpublished observations).
Studies of chimeric Hmg-CoA reductase proteins in which the carboxyl terminus was replaced with unrelated sequences suggested that the ability of this protein to induce membrane proliferation resided solely with the HMG-CoA reductase membrane domain (Skalnik et al., 1988; Parrish et al., 1995). Thus, our initial observation that removal of the carboxyl terminus prevented karmellae assembly was surprising and suggested that sequences present at the carboxyl terminus were important for membrane biogenesis. Pursuing this initial observation, we found that the inability of certain Hmg1p carboxyl-terminal mutants to induce karmellae was not due to mislocalization of the protein, an insufficient level of protein, an unstable protein, or the inability of the protein to oligomerize via its carboxyl terminus. How, then, does the carboxyl terminus exercise its effects on the ability of Hmg1p to induce karmellae? Our working hypothesis is that the conformation of the carboxyl terminus of HMG-CoA reductase affects the folding of the membrane domain, particularly loop G, in certain cases rendering it unable to achieve the conformation needed to stimulate karmellae assembly.
This hypothesis reflects the well-established observation that membrane protein function typically involves conformational changes. For example, conformational changes are required for the function of ion channels and pumps, as well as receptors for extracellular matrix proteins, hormones, neurotransmitters, light, and odorant molecules. In many cases the necessary conformational change is at least initiated by subtle alterations in one or more transmembrane-spanning regions of the protein. For example, fluorescence spectroscopy reveals that agonist binding to the β2 adrenoreceptor causes conformational changes in two of the seven transmembrane helices that comprise the membrane domain (Gether et al., 1997). These changes are then believed to alter cytoplasmic regions of the protein, allowing interactions with the associated G-protein α subunit necessary for signal transduction. Another potentially relevant example is the “inside-out” signaling of integrins (Ginsberg et al., 1992; Dedhar, 1999). In the absence of thrombin, αIIbβ3 integrin of platelets is inactive and has poor affinity for fibrinogen. Upon activation of platelets by exposure to thrombin or ADP, the αIIbβ3 integrin takes on an altered conformation that greatly increases its affinity for fibrinogen. This inside-out signaling is believed to involve changes in the αIIbβ3 integrin cytoplasmic domain that are transmitted to the extracellular domain, exposing previously hidden fibrinogen binding sites (O’Toole et al., 1994; Hughes et al., 1995; Leisner et al., 1999). Deletions or point mutations within the cytoplasmic domain can constituively activate αIIbβ3 integrin for fibrinogen binding, indicating that the intracellular and extracellular domains are “conformationally and functionally coupled” (Leisner et al., 1999). Thus, the mechanics of transmembrane signaling illustrate the ability of subtle changes in protein conformation on one side of a membrane to be transmitted across a membrane bilayer, ultimately changing the protein’s function or interactions.
We envision a similar effect of different carboxyl terminal sequences on the folding of the Hmg1p membrane domain. One possibility is that carboxyl terminal sequences that are incompatible with karmellae assembly affect the packing of the transmembrane helices so that loop G cannot properly fold. Consistent with this notion, we have been unsuccessful in using the yeast two-hybrid system to identify proteins that interact with loop G. For this analysis, it was necessary to eliminate the transmembrane sequences from the “bait” construct, so that the protein would be soluble and able to enter the nucleus to activate reporter genes. Consequently, the bait consisted solely of loop G sequences, flanked by several hydrophobic residues. Extensive searches using this construct did not reveal interacting proteins. Although the failure of the two-hybrid approach may indicate the absence of proteins that interact with loop G, it is also consistent with the idea that proper folding of loop G may be very sensitive to the status of adjacent transmembrane domains.
The ability of changes in one protein domain to affect transmembrane helix packing is well illustrated by structural studies of receptor tyrosine kinases such as the erythropoietin receptor. Upon ligand binding, the extracellular domains of the receptor homodimer undergo a dramatic shift in conformation that brings transmembrane helices more closely together, allowing activation of marker enzymes that replace the cytoplasmic kinase domains (Remy et al., 1999). The packing of or interactions between the Hmg1p transmembrane helices may have similar sensitivity to the folding status of its carboxyl terminus.
Dimerization of the catalytic domains is critical for HMG-CoA reductase activity, because the enzyme active site is formed by interactions between two monomers (Lawrence et al., 1995; Rogers et al., 1997; Tabernero et al., 1999). Recent results also support the importance of oligomerization for regulated degradation of HMG-CoA reductase, such that monomeric fusion proteins are degraded rapidly relative to fusion proteins capable of oligomerizing via their carboxyl termini (Cheng et al., 1999). Cheng and coworkers (1999) did not analyze the ability of their fusion proteins to induce crystalloid ER. However, studies of microsomal aldehyde dehydrogenase, which induces crystalloid ER similar to that induced by HMG-CoA reductase, also indicate the importance of interactions via the carboxyl termini for induction of membrane assembly (Masaki et al., 1994, 1996; Gong et al., 1996; Yamamoto et al., 1996). Cheng et al. (1999) propose that dimerization via the HMG-CoA reductase catalytic domains serves to promote or to stabilize interactions between the attached membrane domains. However, to account for the unequivocal importance of the membrane domain in regulated degradation, they propose that the key focus for regulated degradation is the dimerization status of the protein’s membrane domain. Thus, they propose that carboxyl-terminal fusions capable of oligomerization stabilize the protein by bringing the membrane domains close to one another, facilitating interactions between membrane domains that are critical for regulated degradation.
Although our results do not support an absolute requirement for dimerization via catalytic domains for karmellae assembly, they are consistent with the model proposed by Cheng et al. (1999). Thus, interactions via the carboxyl termini may increase the efficiency of membrane domain interactions, thereby increasing the efficiency of karmellae signaling. However, unlike the mammalian protein, the Hmg1p membrane domain may have sufficient affinity for one another to allow efficient interactions in the absence of cytosolic domain oligomerization, provided that the membrane domains can properly fold. If so, only the carboxyl terminus fusions that resulted in altered membrane domain folding would prevent karmellae assembly by preventing the necessary interactions between membrane domains. Our data predict that these proteins include the construct lacking a carboxyl-terminal domain (Hmg1mem:HA), as well as the protein with a truncated catalytic domain (Hmg1memΔ29p), or in which the carboxyl terminus is replaced by GFP (Hmg1mem:GFP).
It is interesting to note that the Hmg1mem:GFP fusion could not induce karmellae, although the Hmg1mem:β-galΔ20 was able to do so. Both of these fusion proteins are predicted to be unable to dimerize via their carboxyl termini, yet one induced karmellae and the other did not. Although this result supports the conclusion that oligomerization via the carboxyl terminus is not essential for karmellae assembly, it raises questions about how the proteins differ. A possible explanation for the different abilities of Hmg1mem:GFP and Hmg1mem:β-galΔ20 to induce karmellae is the absence of a linker sequence in Hmg1mem:GFP. In this protein, the cytosolic domain (GFP) immediately follows the membrane domain, whereas the cytosolic domain of Hmg1mem:β-galΔ20 is separated from the membrane by a 143-amino-acid linker sequence. Thus, sufficient separation of the membrane and catalytic domains may be important for allowing each domain to achieve its proper conformation. Consistent with this idea, a fusion protein in which the GFP domain was widely separated from the catalytic domain (Hmg1mem:Hmg1595–987:GFP) was capable of generating karmellae. Separation of the membrane and catalytic domain is not sufficient, however, because Hmg1memΔ29p has a linker sequence but is unable to induce karmellae assembly.
Recently, another view that emphasizes protein quality has emerged to explain different types of ER proliferations that occur in response to mutant and wild-type cytochrome p450. When expressed in Saccharomyces. cerevisiae, a wild-type cytochrome P450 of Candida maltosa generates tubular stacks of ER throughout the cytoplasm that appear distinct from karmellae (Zimmer et al., 1995). In contrast, mutant forms of Candida maltosa cytochrome P450 expressed in S. cerevisiae lead to the proliferation of karmellae-like structures (Zimmer et al., 1995). Because the mutant cytochrome P450 forms have lower protein stability than the wild-type P450 enzyme, these authors favor a model in which a quality control system sorts the mutant P450 forms into a stacked ER subcompartment for degradation rather than the subcompartment responsible for tubular expansion (Zimmer et al., 1995). In our study, the Hmg1p variants that failed to induce karmellae did not have lower protein stability than the wild-type Hmg1p, making it unlikely that karmellae are assembled as degradative compartments to remove an unstable protein.
Our ultimate goal is to understand the molecular events that underlie karmellae assembly as a model for how cells modulate membrane biogenesis in response to changing physiological demands. Characterizing the features of Hmg1p that are critical for induction of karmellae is an important step in achieving that goal. Although the molecular details remain to be discovered, our results suggest that proper folding of the Hmg1p membrane domain is a critical first step in the karmellae signaling process and indicate that this folding may be influenced by sequences at the carboxyl terminus. Our results also rule out models that suggest that oligomerization of a membrane protein is sufficient to induce alterations in membrane assembly (Gong et al., 1996). Consequently, identifying trans-acting factors that respond to the Hmg1p membrane domain will be essential for learning how this protein stimulates the dramatic changes in membrane biogenesis and organization that accompany karmellae assembly.
ACKNOWLEDGMENTS
We thank the members of the Wright laboratory, particularly Mark Parrish, for their helpful advice and discussions. In addition, we thank Helen Cheng for providing the wild-type lacZ plasmid (pMKITNeo-XhoI-HMGal δ5′) and the lacZΔ20 plasmid (pMKITNeo-XhoI-HMGalδ20), Randy Hampton for providing the glyceraldehyde-3-phosphate dehydrogenase-HMG1-CAT plasmid (pRH127–3), Jeff Cox and Peter Walter for providing a plasmid containing GFP coding sequences, Mark Rose for providing Kar2p antiserum, and Jeff Brodsky for providing additional Kar2p antiserum. This work was supported by National Institutes of Health grant GM45726 to R.W.
Abbreviations:
- β-gal
β-galactosidase
- CHX
cycloheximide
- CoA
coenzyme A
- DiOC6
3,3′-dihexyloxacarbocyanine iodide
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- HMG
3-hydroxy 3-methylglutaryl
REFERENCES
- Anderson RG, Orci L, Brown MS, Garcia-Segura LM, Goldstein JL. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J Cell Sci. 1983;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]
- Baltscheffsky M, Brosche M, Hultman T, Lundvik L, Nyren P, Sakai-Nore Y, Severin A, Strid A. A 3-hydroxy-3-methylglutaryl-CoA lyase gene in the photosynthetic bacterium Rhodospirillum rubrum. Biochim Biophys Acta. 1997;1337:113–122. doi: 10.1016/s0167-4838(96)00158-6. [DOI] [PubMed] [Google Scholar]
- Basson ME, Moore RL, O’Rear J, Rine J. Identifying mutations in duplicated functions in Saccharomyces cerevisiae: recessive mutations in HMG-CoA reductase genes. Genetics. 1987;117:645–655. doi: 10.1093/genetics/117.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson ME, Thorsness M, Finer-Moore J, Stroud RM, Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl CoA reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988;8:3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson ME, Thorsness M, Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1986;83:5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach MJ, Rodwell VW. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl CoA reductase. J Bacteriol. 1989;171:2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff KM, Rodwell VW. 3-Hydroxy-3-methylglutaryl-CoA reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J Bacteriol. 1996;178:19–23. doi: 10.1128/jb.178.1.19-23.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Brown JR, Doolittle WF, Klenk HP, Lam W, Schenk ME, Stauffacher CV, Rodwell VW. 3-Hydroxy-3-methylglutaryl CoA reductase of Sulfolobus solfataricus: DNA sequence, phylogeny, expression in Escherichia coli of the hmgA gene, and purification and kinetic characterization of the gene product. J Bacteriol. 1997;179:3632–3638. doi: 10.1128/jb.179.11.3632-3638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Cheng HH, Xu L, Kumagai H, Simoni RD. Oligomerization state influences the degradation rate of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1999;274:17171–17178. doi: 10.1074/jbc.274.24.17171. [DOI] [PubMed] [Google Scholar]
- Chin DJ, Luskey KL, Anderson RGW, Faust JR, Goldstein JL, Brown MS. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold elevation in 3-hydroxy-3-methylglutaryl CoA reductase. Proc Natl Acad Sci USA. 1982;79:1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun KT, Simoni RD. The role of the membrane domain in the regulated degradation of 3-hydroxy-3-methylglutaryl CoA reductase. J Biol Chem. 1992;267:4236–4246. [PubMed] [Google Scholar]
- Darnay BG, Wang Y, Rodwell VW. Identification of the catalytically important histidine of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1992;267:15064–15070. [PubMed] [Google Scholar]
- Dedhar S. Integrins and signal transduction. Curr Opin Hematol. 1999;6:37–43. doi: 10.1097/00062752-199901000-00007. [DOI] [PubMed] [Google Scholar]
- Denbow CJ, Lang S, Cramer CL. The N-terminal domain of tomato 3-hydroxy-3-methylglutaryl-CoA reductases. Sequence, microsomal targeting, and glycosylation. J Biol Chem. 1996;271:9710–9715. doi: 10.1074/jbc.271.16.9710. [DOI] [PubMed] [Google Scholar]
- Deschenes RJ, Broach JR. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol Cell Biol. 1987;7:2344–2351. doi: 10.1128/mcb.7.7.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald KA, Hampton RY, Fritz IB. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl CoA reductase on squalene synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 1997;63:3341–3344. doi: 10.1128/aem.63.9.3341-3344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF, Tsang S, Cho G, Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Kempner ES, Lan S-F, Erickson SK. Functional size of rat hepatic 3-hydroxy-3-methylglutaryl CoA reductase as determined by radiation inactivation. J Biol Chem. 1985;260:10278–10282. [PubMed] [Google Scholar]
- Fawcett DW. The Cell. Philadelphia: W.B. Saunders; 1981. pp. 301–368. [Google Scholar]
- Feng DF, Cho G, Doolittle RF. Determining divergence times with a protein clock: update and reevaluation [see comments] Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler AV, Zabin I. Colinearity of beta-galactosidase with its gene by immunological detection of incomplete polypeptide chains. Science. 1966;154:1027–1029. doi: 10.1126/science.154.3752.1027. [DOI] [PubMed] [Google Scholar]
- Fowler AV, Zabin I. Beta-galactosidase: immunological studies of nonsense, missense and deletion mutants. J Mol Biol. 1968;33:35–47. doi: 10.1016/0022-2836(68)90279-9. [DOI] [PubMed] [Google Scholar]
- Fowler AV, Zabin I. Purification, structure, and properties of hybrid beta-galactosidase proteins. J Biol Chem. 1983;258:14354–14358. [PubMed] [Google Scholar]
- Frimpong K, Rodwell VW. The active site of hamster 3-hydroxy-3-methylglutaryl-CoA reductase resides at the subunit interface and incorporates catalytically essential acidic residues from separate polypeptides. J Biol Chem. 1994;269:1217–1221. [PubMed] [Google Scholar]
- Gardner R, Cronin S, Leader B, Rine J, Hampton R, Leder B. Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1998;9:2611–2626. doi: 10.1091/mbc.9.9.2611. (erratum [1999]. 10[3]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U, Lin S, Ghanouni P, Ballesteros JA, Weinstein H, Kobilka BK. Agonists induce conformational changes in transmembrane domains III and VI of the beta2 adrenoceptor. EMBO J. 1997;16:6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G, Faust JR, Chin DJ, Goldstein JL, Brown MS. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41:249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Ginsberg MH, Du X, Plow EF. Inside-out integrin signaling. Curr Opin Cell Biol. 1992;4:766–771. doi: 10.1016/0955-0674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW. Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc Natl Acad Sci USA. 1996;93:2219–2223. doi: 10.1073/pnas.93.5.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Koning A, Wright R, Rine J. In vivo examination of membrane protein localization and degradation with GFP. Proc Natl Acad Sci USA. 1996;93:828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PE, O’Toole TE, Ylanne J, Shattil SJ, Ginsberg MH. The conserved membrane-proximal region of an integrin cytoplasmic domain specifies ligand binding affinity. J Biol Chem. 1995;270:12411–12417. doi: 10.1074/jbc.270.21.12411. [DOI] [PubMed] [Google Scholar]
- Jingami H, Brown MS, Goldstein JL, Anderson RG, Luskey KL. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl CoA reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J Cell Biol. 1987;104:1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochevar D, Anderson RGW. Purified crystalloid endoplasmic reticulum from UT-1 cells contains multiple proteins in addition to 3-hydroxy-3-methylglutaryl CoA reductase. J Biol Chem. 1987;262:10321–10326. [PubMed] [Google Scholar]
- Koning AJ, Lum PY, Williams JM, Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Chun KT, Simoni RD. Molecular dissection of the role of the membrane domain in the regulated degradation of 3-hydroxy-3-methylglutaryl CoA reductase. J Biol Chem. 1995;270:19107–19113. doi: 10.1074/jbc.270.32.19107. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lange Y, Steck TL. Four cholesterol-sensing proteins. Curr Opin Struct Biol. 1998;8:435–439. doi: 10.1016/s0959-440x(98)80119-x. [DOI] [PubMed] [Google Scholar]
- Lawrence CM, Rodwell VW, Stauffacher CV. Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 angstrom resolution. Science. 1995;268:1758–1762. doi: 10.1126/science.7792601. [DOI] [PubMed] [Google Scholar]
- Leisner TM, Wencel-Drake JD, Wang W, Lam SC. Bidirectional transmembrane modulation of integrin alphaIIbbeta3 conformations. J Biol Chem. 1999;274:12945–12949. doi: 10.1074/jbc.274.18.12945. [DOI] [PubMed] [Google Scholar]
- Liscum L, Finer-Moore J, Stroud RM, Luskey KL, Brown MS, Goldstein JL. Domain structure of 3-hydroxy-3-methylglutaryl CoA reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985;260:522–530. [PubMed] [Google Scholar]
- Lum PY, Edwards S, Wright R. Molecular, functional, and evolutionary characterization of the gene encoding HMG-CoA reductase in the fission yeast, Schizosaccharomyces pombe. Yeast. 1996;12:1107–1124. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1107::AID-YEA992%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lum PY, Wright R. Degradation of HMG CoA reductase-induced membranes in the fission yeast. Schizosaccharomyces pombe. J Cell Biol. 1995;131:81–94. doi: 10.1083/jcb.131.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J Cell Biol. 1994;126:1407–1420. doi: 10.1083/jcb.126.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Membrane topology and retention of microsomal aldehyde dehydrogenase in the endoplasmic reticulum. J Biol Chem. 1996;271:16939–16944. doi: 10.1074/jbc.271.28.16939. [DOI] [PubMed] [Google Scholar]
- Naik RR, Jones EW. The PBN1 gene of Saccharomyces cerevisiae: an essential gene that is required for the posttranslational processing of the protease B precursor. Genetics. 1998;149:1277–1292. doi: 10.1093/genetics/149.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Hirata A, Nakano A. Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol Biol Cell. 1994;5:1129–1143. doi: 10.1091/mbc.5.10.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender EH, Simon RD. The intracellular targeting and membrane topology of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1992;267:4223–4235. [PubMed] [Google Scholar]
- Orci L, Brown MS, Goldstein JL, Garcia-Segura LM, Anderson RG. Increase in membrane cholesterol: a possible trigger for degradation of HMG CoA reductase and crystalloid endoplasmic reticulum in UT-1 cells. Cell. 1984;36:835–845. doi: 10.1016/0092-8674(84)90033-3. [DOI] [PubMed] [Google Scholar]
- Osborne TF, Rosenfeld JM. Related membrane domains in proteins of sterol sensing and cell signaling provide a glimpse of treasures still buried within the dynamic realm of intracellular metabolic regulation. Curr Opin Lipidol. 1998;9:137–140. doi: 10.1097/00041433-199804000-00010. [DOI] [PubMed] [Google Scholar]
- O’Toole TE, Katagiri Y, Faull RJ, Peter K, Tamura R, Quaranta V, Loftus JC, Shattil SJ, Ginsberg MH. Integrin cytoplasmic domains mediate inside-out signal transduction. J Cell Biol. 1994;124:1047–1059. doi: 10.1083/jcb.124.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish ML, Sengstag C, Rine JD, Wright RL. Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol Biol Cell. 1995;6:1535–1547. doi: 10.1091/mbc.6.11.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RK, Luskey KL, Anderson RG. Biogenesis of the crystalloid endoplasmic reticulum in UT-1 cells: evidence that newly formed endoplasmic reticulum emerges from the nuclear envelope. J Cell Biol. 1986;102:2158–2168. doi: 10.1083/jcb.102.6.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasher DC. Using GFP to see the light. Trends Genet. 1995;11:320–323. doi: 10.1016/s0168-9525(00)89090-3. [DOI] [PubMed] [Google Scholar]
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Re EB, Brugger S, Learned M. Genetic and biochemical analysis of the transmembrane domain of Arabidopsis 3-hydroxy-3-methylglutaryl CoA reductase. J Cell Biochem. 1997;65:443–459. [PubMed] [Google Scholar]
- Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KS, Rodwell VW, Geiger P. Active form of Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl CoA reductase. Biochem Mol Med. 1997;61:114–120. doi: 10.1006/bmme.1997.2596. [DOI] [PubMed] [Google Scholar]
- Roitelman J, Bar-Nun S, Inoue S, Simoni RD. Involvement of calcium in the mevalonate-accelerated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1991;266:16085–16091. [PubMed] [Google Scholar]
- Roitelman J, Olender EH, Bar-Nun S, Dunn JWA, Simoni RD. Immunolgical evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol. 1992;5:959–973. doi: 10.1083/jcb.117.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Mistra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Schunck W, Vogel F, Gross B, Kargel E, Mauersberger S, Kopke K, Gengnagel C, Muller H. Comparision of two cytochromes P-450 from Candida maltosa: primary structures, substrate specificities and effects of their expression in Saccharomyces cerevisiae on the proliferation of the endoplasmic reticulum. Eur J Cell Biol. 1991;55:336–345. [PubMed] [Google Scholar]
- Sekler MS, Simoni RD. Mutation in the lumenal part of the membrane domain of HMG-CoA reductase alters its regulated degradation. Biochem Biophys Res Commun. 1995;206:186–193. doi: 10.1006/bbrc.1995.1026. [DOI] [PubMed] [Google Scholar]
- Sengstag C, Stirling C, Schekman R, Rine J. Genetic and biochemical evaluation of eukaryotic membrane protein topology: the polytopic structure of S. cerevisiae HMG-CoA reductase. Mol Cell Biol. 1990;10:672–680. doi: 10.1128/mcb.10.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A uniform set of multipurpose shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalnik DG, Narita H, Kent C, Simoni RD. The membrane domain of 3-hydroxy-3-methylglutaryl CoA reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto β-galactosidase. J Biol Chem. 1988;263:6836–6841. [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Tabernero L, Bochar DA, Rodwell VW, Stauffacher CV. Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1999;96:7167–7171. doi: 10.1073/pnas.96.13.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kuzuyama T, Seto H. Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl CoA reductase, a key enzyme involved in biosynthesis of terpenoids. J Bacteriol. 1999;181:1256–1263. doi: 10.1128/jb.181.4.1256-1263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka M, Mekada E. Degradation of a nuclear-localized protein in mammalian COS cells, using Escherichia coli beta-galactosidase as a model protein. J Biol Chem. 1992;267:9107–9111. [PubMed] [Google Scholar]
- Vergeres G, Yen TSB, Aggeler J, Lausier J, Waskell L. A model system for studying membrane biogenesis: overexpression of cytochrome b5 in yeast results in marked proliferation of the intracellular membrane. J Cell Sci. 1993;106:249–259. doi: 10.1242/jcs.106.1.249. [DOI] [PubMed] [Google Scholar]
- Wanker EE, Sun Y, Savitz AJ, Meyer DI. Functional characterization of the 180-kD ribosome receptor in vivo. J Cell Biol. 1995;130:29–39. doi: 10.1083/jcb.130.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Wright R, Basson M, D’Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Rine J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Masaki R, Tashiro Y. Formation of crystalloid endoplasmic reticulum in COS cells upon overexpression of microsomal aldehyde dehydrogenase by cDNA transfection. J Cell Sci. 1996;109:1727–1738. doi: 10.1242/jcs.109.7.1727. [DOI] [PubMed] [Google Scholar]
- Zimmer KP, Matsuda I, Matsuura T, Mori M, Colombo JP, Fahimi HD, Koch HG, Ullrich K, Harms E. Ultrastructural, immunocytochemical and stereological investigation of hepatocytes in a patient with the mutation of the ornithine transcarbamylase gene. Eur J Cell Biol. 1995;67:73–83. [PubMed] [Google Scholar]