Abstract
Background: Second internal transcribed spacer (ITS2) has proven to contain useful biological information at higher taxonomic levels. Objectives: This study was carried out to unravel the biological information in the ITS2 region of An. culicifacies and the internal relationships between the five species of Anopheles culicifacies. Methodology: In achieving these objectives, twenty two ITS2 sequences (~370bp) of An. culicifacies species were retrieved from GenBank and secondary structures were generated. For the refinement of the primary structures, i.e. nucleotide sequence of ITS2 sequences, generated secondary structures were used. The improved ITS2 primary structures sequences were then aligned and used for the construction of phylogenetic trees. Results and discussions: ITS2 secondary structures of culicifacies closely resembled near universal eukaryotes secondary structure and had three helices, and the structures of helix II and distal region of helix III of ITS2 of An. culicifacies were strikingly similar to those regions of other organisms strengthening possible involvement of these regions in rRNA biogenesis. Phylogenetic analysis of improved ITS2 sequences revealed two main clades one representing sibling B, C and E and A and D in the other. Conclusions: Near sequence identity of ITS2 regions of the members in a particular clade indicate that this region is undergoing parallel evolution to perform clade specific RNA biogenesis. The divergence of certain isolates of An. culicifacies from main clades in phylogenetic analyses suggests the possible existence of camouflaged sub-species within the complex of culicifacies. Using the fixed nucleotide differences, we estimate that these two clades have diverged nearly 3.3 million years ago, while the sibling species in clade 2 are under less evolutionary pressure, which may have evolved much later than the members in clade 1.
Keywords: Anopheles culicifacies, malaria vector, internal transcribed spacer 2, secondary structure, phylogenetic analysis
Background
Anopheles culicifacies Giles sensu lato is a major vector of malaria in Indian subcontinent and Sri Lanka [1]. In India, five sibling species of the An. culicifacies complex have been identified, species A and B [2], species C [3], species D [4], and species E [5] based on polytene chromosome banding pattern and mitotic karyotyping. Polytene chromosome studies using Xab, 2g1+h1, of Sri Lankan An. culicifacies display all specimens tested belong to sibling species B [2]. Investigations carried out with wild caught mosquitoes of An. culicifacies in Sri Lanka and the DNA probes Rp36, Rp217 and Rp234 derived from a genomic library of species B revealed non existence of species A in Sri Lanka [1]. Most recently, using mitotic Y chromosome polymorphisms, An. culicifacies species B and E were detected [6].
Behavioural differences influence whether the species are efficient or poor vectors in a species complex [7]. Vector incrimination studies on An. culicifacies revealed that A, C, D and E are considered efficient vectors whereas species B is regarded as a poor vector of malaria [5]. Correct identification of malaria vector is essential for targeted malaria control. The discrimination of members of the An. culicifacies species complex is based on cytogenetic methods and has several limitations. In contrast, the rDNA-PCR approach is widely used in the discrimination of cryptic anopheline species [8]. DNA sequences evolving in concert are identical in panmictic populations, but divergent between reproductively isolated populations or species and are therefore ideal targets for diagnostic purposes [7]. The internal transcribed spacer (ITS2) of nuclear rDNA sequences have been used for discrimination of closely related anopheline mosquitoes in the An. gambiae complex Giles [9], An. funestus, Giles complex [7] and An. dirus complex [10].
Ribosomal DNA has been used extensively and very successfully for phylogenetic analysis of both closely and distantly related organisms [11]. The spacers (ITS1 and ITS2) evolve at a faster rate than the coding sequences, therefore, these sequences are good candidates for the analysis of phylogenetic relationships between members of a species complex [11]. In addition, further information regarding species relatedness and intraspecies variation could be obtained by studying the functional folding patterns (secondary structures) of ITS2 [12]. Inclusion of conserved rDNA coding regions (5.8S & 28S) to ITS2 has often been the practice in such analysis to maintain the integrity of the ITS2 secondary structure in variety of closely and distantly related organisms [13]. Secondary structure can be used to aid comparative alignment of ITS2 sequences [12]. The objectives of the current study were to understand the internal relationships between the five sibling species and the biological information of the ITS2 region in An. culicifacies using the primary and secondary structures of the ITS2 rDNA entered in GenBank.
Methodology
Data retrieval
Twenty two ITS2 sequences of An. culicifacies sibling species (A, B, C, D, E and An. Culicifacies that are unassigned to a particular species) were retrieved using ITS2 sequence of An. culicifacies B (AY167747/ SL-B) and Blast N tools of NCBI (http://www.ncbi.nlm.nih.gov/) to which following search criteria was set; Database: Nucleotide collection (nr/nt), Organism: An. culicifacies and Entrez query: ITS2. The retrieved sequences are listed in Table 1 (see supplementary material).
Multiple sequence alignment and analyses
Multiple alignments of retrieved nucleotide sequences were carried out using CLUSTAL W of BioEdit version 5.0.6 (North Carolina State University, Department of Microbiology). Entire length (~ 370 bp) of ITS2 sequences of An. culicifacies species was used for this purpose. Manual refinement was carried out to achieve maximum similarities amongst the sequences in the alignment. Sequence identity was calculated with the same software package using “Sequence Identity Matrix” from the “Alignment” menu.
Phylogenetic analyses
The phylogenetic analysis was performed using multiple aligned sequences with a neighbor-joining distance method of Phylip software version 3.2 for Windows. TREEVIEW was used to visualize trees. Bootstrap confidence intervals were calculated by 100 heuristic search replicates.
ITS2 secondary structure prediction
The ITS2 of An. culicifacies sibling species, flanked by 10bp of 5.8S rRNA and 23bp of 28S rRNA [13] were used to generate secondary structure and to predict the probable accessibility sites (loops) for transcleaving ribozymes in the latter region [14] using the Sribo program in Sfold (http://sfold.wadsworth.org/index.pl).
Results and discussion
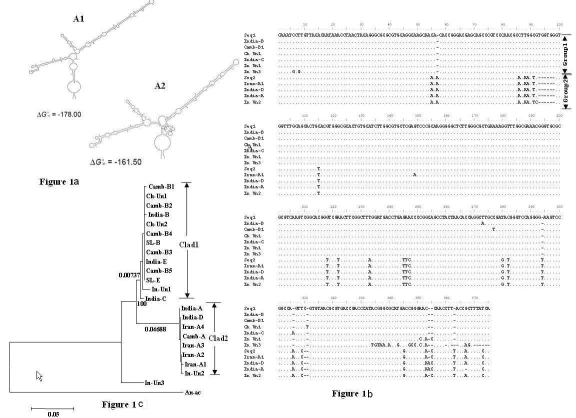
The species diversity and genetic structure of mosquitoes belonging to the culicifacies complex was investigated using the internal transcribed spacer 2 (ITS2) of ribosomal DNA (rDNA). Sequence analysis revealed (i) An. culicifacies species could be broadly divided into 2 groups on the basis of sequence similarity (i) 13 insertion or deletions, 11 transversions and 7 transitions amongst the member of An. culicifacies species (ii) considerable sequence divergence between these two groups and strict sequence conservation within the members of a given group (iv) ITS2 region of species SLB, SLE, Camb2/b3/b4/b5, IndiaE and ChUn2 of clade 1 were identical, while CamA, IranA2, Iran3 and IranA4 of clade 2 were also identical (Figure 1b). The finding of identical sequences in fast evolving ITS2 regions of the isolates from different geographical locales with eco-climatic conditions somewhat intriguing and may imply this region of these sibling species is under high evolutionary pressure to perform the group-specific functions.
Figure 1.
Proposed secondary structure of ITS2 regions of Group 1/Clade 1 (A1) and Group 2/Clade 2 (A2). Multiple alignment of the ITS2 sequences of An. culicifacies, derived from the retrieved and improved GenBank sequences using secondary structure analyses. Seq1 (SLB, SLE, Camb2, Camb3, Camb4, Camb5, ChUn2 and India E) and Seq2 (Iran A2, IranA3, Iran A4 and Camb A) (b), Table 1 (see supplementary material)). Phylogenetic tree of An. Culicifacies species based on the sequences of the ITS2 of rDNA cistron. Numerical numbers at the nodes are bootstrap values. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes. Numerical numbers at horizontal lines are node lengths (c).
The evolutionary conserved sub portion of ITS is imperative for the proper positioning of multimolecular rRNA transcript prior to processing it to unimolecular RNA transcripts. Further, secondary structure of ITS is valuable to improve the nucleotide alignments for the correct phylogenetic comparison of a given group of organisms [12]. Thus, secondary structures of the An. culicifacies species ITS2 were generated using sfold [14] and a group-specific secondary structures were seen (Figure 1a). Nucleotide polymorphism/s that changed the ITS2 secondary structure remarkably with respect to its group-specific canonical secondary structure was corrected. The 370 bp of the 22 An. culicifacies of improved ITS2 sequences were used to construct phylogenetic relationship and Anopheles aconitus sequence was used as an out group (Figure 1b, Table 1 (under supplementary material)). On the basis of the analysis, two distinct clades with confidence bootstrap value 100 were identified consisting sibling species B, C and E in one group and species A and D in the other (Figure 1c). This finding is congruent with the data derived from the previous studies on cytogenetic, repetitive DNA and D3 region of the 28s rDNA of the An. culicifacies members [15,16,17]. ITS2 regions of culicificaes Un In3 was found to be the most diverse having sequence identity/similarity in the range of 86.2% to 93.4% with the other sibling species of the complex and stayed in a separate phylogenetic position in phylogenetic tree without mixing with the counterpart members in clade 1 and 2 (Figure 1c). ITS2 repeats of an organism are reported to undergo concerted evolution and subsequent poorly understood homogenization process renders them to be identical over very short evolutionary time [11]. Therefore, if an isolates to be a member of in culicificaes complex, it is required to demonstrate a nearly identical primary structure to the other member of the complex. The primary and the secondary structure analyses of Un In3, ITS2 exhibited the presence of perfect 5.8S, relatively conserved regions of ITS2 , near universal eukaryotes secondary structure etc, are the requirement for ITS2 to be functional. Therefore, taken together these facts it is tempting to speculate whether this isolate is truly culicifacies or it represents another species/subspecies of anopheles that is camouflaged within the complex of culicifacies.
As the node length of clad 1 is greater than the clad 2, it seems that ITS2 region of the sibling species in clade 2 (node length 0.04688) is under a less selective pressure and evolving faster compared to the species in clade 1 (node length 0.00737) after splitting from a common ancestor (Figure 1c). In addition, node length of ITS2 phylogenetic tree implies sibling species of clade 1 may have evolved earlier than the sibling species in clade 2. Further, since these sibling species are reproductively isolated and near sequence identity in the ITS2 region of members in a particular clad without any possibility of cross breeding implies that the ITS2 regions of members are undergoing parallel evolution to perform clade specific RNA biogenesis. Alternatively, near identicalness of sequences in ITS2 regions within the members of clade 2 suggest the divergence of ITS2 sequences of clade 2 from clade 1 may have occurred prior to split into sibling species.
The fact that the members of the complex are the result of recent speciation events could be hypothesized by high degree of sequence conservation in ITS2. The time of divergence of these two clades can be roughly estimated using fixed nucleotide differences in ITS2 together with an average of the estimates of the rate of the ITS clock of 2.4% per million years within the malanogaster Group and 2.2% per million years between malanogaster and virilis Group of Drosophila [18]. These two clades of An. culicifacies are estimated to have diverged nearly 3.3 million years ago.
Although the involvement of ITS2 region in the biogenesis of mature rRNA is yet to be fully understood, structural integrity of the ITS2 regions has been shown an essential requirement for the correct processing of mature RNA in such process [19,20]. The secondary structure analyses of ITS2 regions of culicifacies complex revealed insertions/deletions, transitions and transversions within the members in group 1 and 2, however, these changes preserve the integrity/identity of the ITS2 secondary structure of the members in a particular group. The ITS2 secondary structure of culicifacies complex was closely resembled to near universal eukaryotes secondary structure and deemed to have three helices of which the structure of helices I, II and distal portion of helix III of culicifacies clade 1 and 2 were highly conserved. Structural identity of ITS2 in members in the two clades indicates the functional identity in rRNA processing. The secondary structures of the regions that involved in basal 10 pairing of helix II and 18 pairing of the distal region of helix III of ITS2 of An. culicifacies were strikingly similar to those regions of other organisms with a high purine content, particularity guanine with signature of YGGY, which is presumably importance for processing [11]. The cut sites for transcript processing in yeast and mammals have been known or proposed to be in the structurally conserved distal region of the helix III [19,20]. Indeed, potential cleavage site with probability of one was observed distal end of helix III. As in other organism, helix II of ITS 2 of An. culicifacies is short and lacks in any branching and the bulges near the base and middle of the helix possesses to have pyrimidine-purine and pyrimidine-pyrimidine misatches, respectively (Figure 1a). The significance of the structural conservation of helix II and its pyrimidine-pyrimidine bulge regions of ITS2 in rRNA processing yet to be ascertained. The secondary structure of helix I of culicifacies clade I and 2 was nearly identical and helix III, being longest helix in ITS2 of culicifacies as in other organism the branching of proximal region only seen in clade 2.
Conclusion
The resemblance of ITS2 secondary structure of An. culicifacies complex to the near universal eukaryotes ITS2 secondary structure suggests the functional similarity of An. culicifacies rRNA biogenesis to other eukaryotes. The presence of two distinct clads in phylogenetic analyses of ITS2 region indicated the presence of two genetically distinct groups in of An. culicifacies complex, supporting cytogenetic [5] rDNA probe [16] and allele- specific polymerase chain reaction studies [17]. The presence of very distinct ITS2 regions in An. culicifacies isolates suggest the possible existence of camouflaged sub-species within the complex of culicifacies. Finally, near sequence identity of ITS2 regions of the members in a particular clad indicate that this region is undergoing parallel evolution to perform clad specific RNA biogenesis.
Supplementary material
Acknowledgments
We thank scientists who sequenced insect ITS2 regions of An. culicifacies species, and deposited them in publicly available databases. We also thank the scientists who developed Molecular Biology Software programs and Bioinformatics tools used in this study.
References
- 1.De Silva BGDNK, et al. Indian J Malariol. 1998;35:1. [PubMed] [Google Scholar]
- 2.Green CA, Miles SJ. J Trop Med Hyg. 1980;83:75. [PubMed] [Google Scholar]
- 3.Subbarao SK, et al. Ann Entomol Soc Am. 1983;76:985. [Google Scholar]
- 4.Vasantha K, et al. Ann Entomol Soc Am. 1991;8:531. [Google Scholar]
- 5.Kar I, et al. J Med Entomol. 1999;36:595. doi: 10.1093/jmedent/36.5.595. [DOI] [PubMed] [Google Scholar]
- 6.Surendran SN, et al. Med Vet Entomol. 2000;14:437. doi: 10.1046/j.1365-2915.2000.00264.x. [DOI] [PubMed] [Google Scholar]
- 7.Besansky NJ. Parassitologia. 1999;41:97. [PubMed] [Google Scholar]
- 8.Collins FH, Paskewitz SM. Insect Mol Biol. 1996;5:1. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- 9.Paskewitz SM. Insect Mol Biol. 1993;2:247. doi: 10.1111/j.1365-2583.1994.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, et al. J Am Mosq Control Assoc. 1998;14:385. [PubMed] [Google Scholar]
- 11.Coleman AW. Nucleic Acids Res. 2007;35:3322. doi: 10.1093/nar/gkm233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman W. Trends in Genet. 2003;19:370. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 13.Severini C, et al. Insect Mol Biol. 1996;5:181. doi: 10.1111/j.1365-2583.1996.tb00052.x. [DOI] [PubMed] [Google Scholar]
- 14.Ding Y, et al. Nucleic Acids Res. 2004;32:W135. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subbarao SK, et al. Trans R Soc Trop Med Hyg. 1992;86:613. doi: 10.1016/0035-9203(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 16.Gunasekara MB, et al. Bull Entomol Res. 1996;85:345. [Google Scholar]
- 17.Singh OP, et al. J Biosci. 2004;29:275. doi: 10.1007/BF02702609. [DOI] [PubMed] [Google Scholar]
- 18.Schlotterer C, et al. Mol Biol Evol. 1994;11:513. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- 19.Van Nues RW, et al. J Mol Biol. 1995;250:24. doi: 10.1006/jmbi.1995.0355. [DOI] [PubMed] [Google Scholar]
- 20.Joseph N, et al. Nucleic Acids Res. 1999;27:4533. doi: 10.1093/nar/27.23.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.