Abstract
Translin and Trax are components of an evolutionarily conserved RNA binding complex. Deletion of Translin in yeast, Drosophila and mouse produces a dramatic loss of Trax protein indicating that its stable expression is dependent on its association with Translin. Analysis of Translin KO mice has revealed multiple behavioral abnormalities and alterations in levels of transcripts encoding synaptic proteins. A confluence of localization, biochemical and RNA trafficking studies supports the view that this complex mediates dendritic trafficking of RNAs, a process thought to play a critical role in synaptic plasticity. However, further studies are needed to define its RNA cargoes, its precise role in this process, and how its binding activity and localization are regulated. Nevertheless, there is sufficient evidence to suggest that the Translin/Trax complex be included among the cadre of RNA binding complexes, such as Staufen and CPEB, that regulate dendritic trafficking of RNA in neurons.
Keywords: RNA trafficking, dendrites, Staufen, GABA A receptors, GluR3, adenosine A2A receptors, BDNF
Introduction
Although Translin was initially identified as a protein that binds to single-stranded DNA in vitro (1), subsequent characterization of this protein indicates that it, and its partner protein, Trax (2), form a heteromeric RNA binding complex in vivo. Interest in understanding the role of this RNA binding complex in the nervous system has been driven by studies suggesting that it regulates dendritic trafficking and translation of mRNA, processes thought to play a key role in synaptic plasticity (3), and by reports that targeted deletion of Translin in mice (4,5) and Drosophila (6) produces behavioral abnormalities. As our understanding of the structure and function of the Translin/Trax complex is far from complete, this review will summarize progress in characterizing this complex and also highlight the substantial gaps in our knowledge that remain to be clarified.
Identification of Translin
Translin was identified independently by four groups that were pursuing distinct lines of research. Kasai and co-workers were seeking to identify single-stranded DNA binding proteins involved in mediating chromosomal translocations. To this end, they purified a 27 kD protein that binds to a fragment of single-stranded DNA that flanks a chromosomal translocation hotspot. They coined the name, Translin, to connote its putative involvement in chromosomal translocations (1).
Hecht and co-workers were looking for trans factors that confer translational dormancy on a set of transcripts in spermatocytes. Their search focused on identifying factors that bind to a fragment of the 3’UTR of one of these transcripts, protamine-2 mRNA, believed to contain cis sequences, referred to as Y and H elements, that mediate the delay in translation. In studies that parallel Kasai’s approach, they purified a protein complex that binds with high affinity to this RNA segment and found that it contained Translin (7).
Two other groups also identified Translin on the basis of its ability to bind to single-stranded DNA in vitro. Taira and Baraban (8) noted the presence of a robust gel-shift complex in brain extracts capable of binding to a GC rich DNA oligo, but not to its complementary strand. As this gel shift complex was highly enriched in brain compared to a wide selection of peripheral tissues, they pursued identification of this novel complex and found that it too contained Translin (9). Furthermore, Manor and co-workers identified Translin as a factor that binds to G rich single-stranded DNA sequences (10,11). Their studies revealed that Translin displays higher affinities for several G-rich sequences than for the DNA fragment used by Kasai’s group. As these higher affinity sequences were not known to be associated with translocation hotspots, these findings called into question Translin’s proposed role in chromosomal translocations.
Thus, these initial studies demonstrated that Translin is able to bind to either RNA or ssDNA with high affinity in vitro and focused attention on deciphering whether one, or possibly both, of these nucleic acids are endogenous targets of Translin in vivo. Although neither of these possibilities has been ruled out definitively, it is noteworthy that targeted deletion of Translin in either mice (4), Drosophila (12) or yeast (13) does not yield higher levels of chromosomal recombinations or susceptibility to DNA damage, findings that argue against a role for this protein in DNA protection or repair, as initially hypothesized by Kasai’s group. By default, these negative findings favor the view that Translin’s endogenous target is RNA. However, it is noteworthy that Trax binds to C1D, an activator of a DNA-dependent protein kinase implicated in DNA repair, in vitro. Furthermore, their interaction has been detected in vivo following exposure to gamma irradiation (14).
The hypothesis that Translin and Trax bind to RNA in vivo has received support from two lines of research. In one, Hecht’s group has identified several transcripts that co-precipitate with Translin from sperm cells (15,16). In the other, Finkenstadt et al. (17) sought to understand a glaring mismatch between the high levels of expression of Translin and Trax proteins in multiple peripheral tissues, as detected by immunoblot, and the negligible level of the corresponding gel-shift complex. In doing so, they found that incubation of these tissue extracts with RNAse, but not DNAse, reveals a robust gel-shift band. Thus, these findings suggest that under basal conditions the Translin complex binds to RNA and that digestion of this RNA unmasks these binding sites.
Organization of the Translin octameric complex
Initial characterization of recombinant Translin protein expressed in bacteria revealed that it forms an octameric complex which possesses nucleic acid binding activity (1). Electron microscopic visualization of these octameric complexes using negative staining revealed a ring-shaped structure with a central aperture. Examination of recombinant Translin protein from human, mouse and chicken revealed similar ring structures, however, chicken Translin appears to be composed of 10 subunits and produce rings with a larger diameter (18). Under non-reducing conditions, Translin migrates as a dimer on SDS-PAGE, suggesting that the Translin octamer may be composed of four dimers, with each dimer being held together by a disulfide bond (18,19).
The presence of a putative leucine zipper domain located near the C-terminus raised the possibility that this motif may mediate formation of dimers or assembly of dimers into octamers. Consistent with the latter view, mutation of two leucines in this region blocks formation of the octameric complex, however, Translin dimers were still present as detected on SDS-PAGE under non-reducing conditions. Of note, these point mutations in the leucine zipper domain also blocked the ability of the complex to bind to nucleic acid suggesting that formation of the octameric complex is required to confer binding activity (18). This inference is also supported by studies characterizing the composition of the Translin nucleic acid binding complex. Glycerol gradient centrifugation of either native human Translin or recombinant S. pombe Translin demonstrated that their nucleic acid binding activities sediment as octamers, while no binding activity is detected in fractions sedimenting as either dimers or tetramers (10,19). However, other studies have found that Translin dimers also possess nucleic acid binding activity, a discrepancy that may be due to the specific binding conditions used (20).
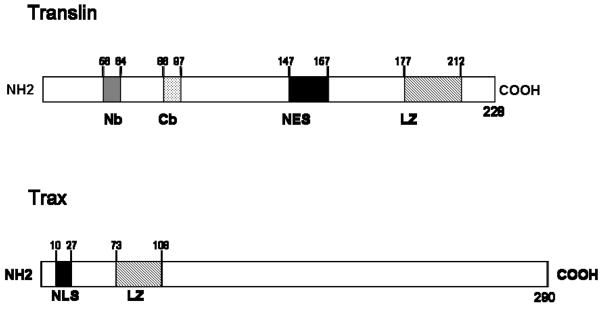
Initial insight into the portions of Translin that mediate nucleic acid binding has also been obtained by analyzing mutations in two basic regions located in the middle of the protein (Figure 1). Although mutations in either of these regions do not impair formation of the octameric complex, both decrease their RNA binding activity (18,21).
Figure 1. Translin and Trax proteins.
The location of several domains identified in Translin and Trax proteins are highlighted in these schematic diagrams. Nb and Cb, refer to basic domains located closer to the N-terminus and C-terminus, respectively, that mediate nucleic acid binding activity; NES, nuclear export signal, LZ, putative leucine zipper domain; NLS, nuclear localization signal.
Two crystallographic studies of Translin have been performed (22, 23). In both cases, Translin is present in monomeric form. Thus this approach has not provided direct insight into the organization of the octameric complex. Nevertheless, these studies have provided a detailed view of the structure of Translin monomers, which are composed of 7 alpha helices. Of note, these studies did not obtain evidence to support the existence of a leucine zipper domain inferred from sequence data. Thus, further studies are needed to assess whether the region containing the putative leucine zipper does in fact fold into that configuration in the native protein complex or not.
While crystallographic studies have not been especially informative regarding the organization of the octameric complex, EM studies performed with negative staining have yielded a clear view of its overall structure. The subunits are aligned around a central aperture in a pinwheel fashion. Reconstruction of images taken with RNA oligos bound to the structure indicate that they bind to 4 sites on one face of the complex, suggesting that every other Translin subunit makes contact with RNA (24). Thus, even though it is still unclear which domains mediate formation of the Translin octameric complex, these EM studies provide a “birds-eye” view of how this complex forms and interacts with RNA.
Interaction of Translin with Trax
Following the identification of Translin, yeast two hybrid studies were performed to find partner proteins. In addition to identifying Translin as a binding partner of itself (19), this approach also led to the identification of Trax, Translin associated protein X, as a candidate partner for Translin (2). This 33 kD protein shares substantial homology with Translin including the putative leucine zipper motif. Unlike Translin, Trax does not self-aggregate and appears to be devoid of nucleic acid binding activity when expressed alone.
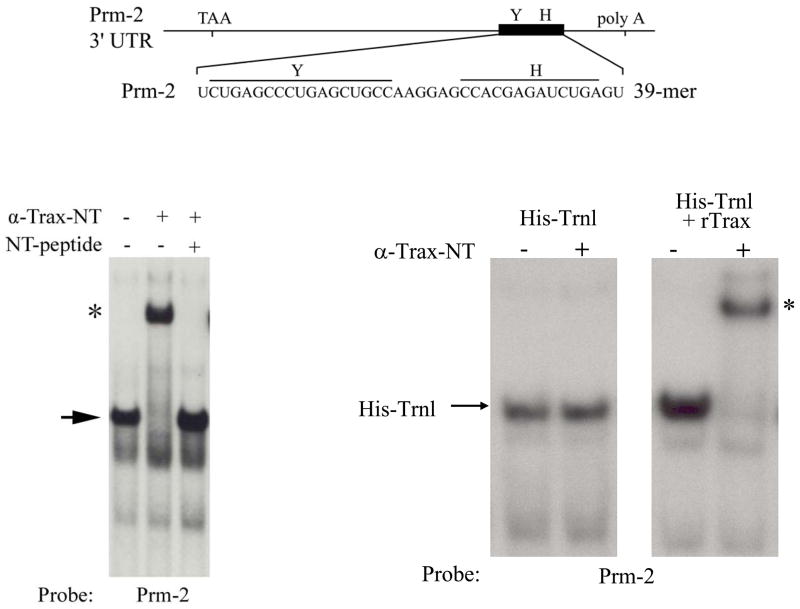
Several lines of evidence demonstrate that these proteins form a heteromeric complex under physiological conditions in vivo. First, endogenous Translin and Trax co-precipitate from brain extracts (25). Second, highly purified fractions of the Translin-containing gel-shift complex from brain also contain Trax (9). Third, this gel-shift complex is “super-shifted” by Trax antibodies confirming that the active complex contains Trax (21). Lastly, co-transfection of Translin and Trax expression constructs in 293 cells produces a gel-shift complex that co-migrates with the endogenous complex detected in brain extracts (21; Figure 2).
Figure 2. Endogenous and recombinant Translin/Trax complex.
Schematic diagram shown at the top presents the 39 nt protamine-2 (Prm-2) fragment used as a gel-shift probe. The position of Y and H elements are indicated above the sequence. Bottom left panel shows a gel-shift experiment performed on cerebellar extracts using the Prm-2 probe. The left lane shows the position of the endogenous gel-shift complex (arrow). This band is “supershifted” by incubation with Trax antibody (middle lane), but not if the antibody is blocked by pre-incubation with its antigen peptide (right lane), indicating that the band is composed of heteromeric Translin/Trax complexes. Panels at bottom right present gel-shift experiments performed on extracts of hEK293 cells transfected with either a His-Translin construct or co-transfected with His-Translin and an untagged rat Trax construct (rTrax). Arrow at left margin indicates location of the homomeric Translin binding complex detected by the Prm-2 probe. This complex is not “supershifted” by Trax antibodies (α-Trax-NT). Righthand panel shows gel-shift study performed following co-transfection of His-Translin with an untagged rat Trax construct (rTrax). Note that the complex detected is “supershifted” by incubation with Trax antibodies indicating that the gel-shift band corresponds to the heteromeric Translin/Trax complex. (Figure adapted from refs. 21 and 28 with publishers’ permission.)
While these studies demonstrate conclusively that Translin and Trax are components of endogenous heteromeric RNA binding complexes, the possibility remains that endogenous Translin homomeric complexes may also be present. To address this issue in cerebellum, Finkenstadt et al. (25) examined which portion of the gel-shift complex is “supershifted” by Trax antibodies (Figure 2). These experiments revealed that virtually all of the gel-shift band detected in cerebellar extracts was “supershifted” indicating that, at least in this tissue, heteromeric Translin/Trax compexes represent the dominant, if not the sole, configuration present. The selectivity of this Trax supershift assay has been confirmed in studies conducted on recombinant homomeric Translin and heteromeric Translin/Trax complexes generated in hEK293 cells (21; Figure 2). As expected, Translin/Trax heteromeric complexes generated by co-transfection of Translin and Trax expression plasmids are “supershifted” by incubation with Trax antibodies. In contrast, Translin homomeric complexes generated by single transfection with the Translin expression plasmid are not. Thus, it is unlikely that the “supershift” observed in cerebellar extracts is due to cross-reactivity of these Trax antibodies with Translin. However, these studies leave open the possibility that Translin homomeric complexes are present in other brain regions or in peripheral tissues.
Additional evidence for a close interaction between Translin and Trax has come, unexpectedly, from analyzing the impact of targeted deletion of Translin in mice, Drosophila and yeast (4,12,13,26). In each case, deletion of Translin is associated with either complete or nearly complete loss of Trax protein. This loss appears to be due to instability of Trax protein since normal levels of Trax mRNA are detected under these conditions. Conversely, deletion of Trax in Drosophila does not induce a similar loss of Translin protein (13). Thus, these findings indicate that Trax stability is dependent on its association with Translin, but not vice versa.
Role of Trax
Since both Translin homomeric and Translin/Trax heteromeric complexes possess nucleic acid binding activity, how do they differ? In other words, if the Translin/Trax complex is the predominant complex in vivo, what advantages does it have over the Translin homomeric complex that forms in vitro. One likely explanation has emerged from examining the subcellular localization of the complex, which is able to shuttle between the nucleus and cytoplasm (27). Specifically, Trax contains a nuclear localization signal and Translin is thought to possess a nuclear export signal (27; Figure 1). Thus, one advantage of the heteromeric complex is that these signals can operate in synergy to mediate dynamic changes in the subcellular localization of the complex. As these findings establish that these proteins can shuttle between nuclear and cytoplasmic compartments, further studies are warranted to identify how these movements are regulated.
One other possibility is that the nucleic acid binding specificity of the homomeric and heteromeric complexes may differ. However, this has not been examined rigorously. Nevertheless, even though Trax does not appear to be able to bind to nucleic acids when expressed in the absence of Translin, there is evidence that it can mediate nucleic acid binding activity in the context of the Translin/Trax complex. Introduction of mutations into two basic domains present in Translin greatly reduces the ability of Translin homomeric complexes to bind to either RNA or ssDNA without interfering with the formation of the octameric complex (18). Interestingly, co-transfection of these mutant Translin constructs with a wild type Trax construct generates heteromeric Translin/Trax constructs that display robust nucleic acid binding activity (21). Accordingly, these findings suggest that in the context of the heteromeric Translin/Trax complex, Trax may be directly involved in binding to nucleic acids. However, further studies are needed to test this hypothesis rigorously.
In summary, these studies indicate that Trax contains three functional properties: a nuclear localization signal, RNA binding activity, and the ability to interact with Translin. While the nuclear localization signal has been mapped to the extreme N-terminal portion of the protein, little is known about the location of the domains that mediate these other properties.
Binding Selectivity
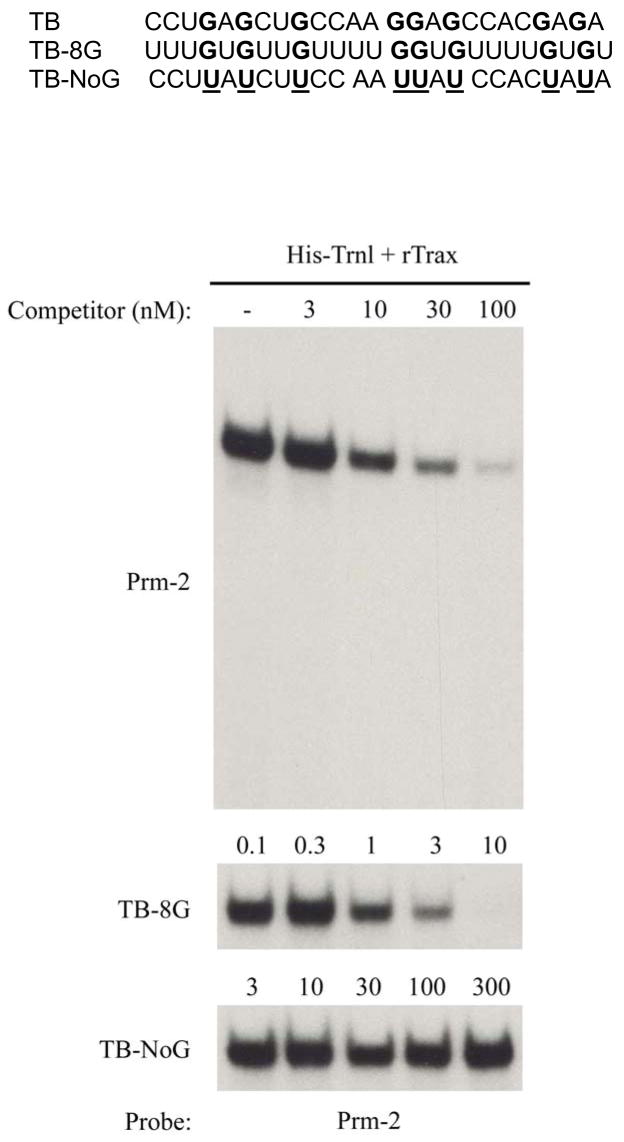
The RNA binding activity of the Translin/Trax complex is readily detectable using a probe sequence taken from the 3’UTR of protamine-2, one of the candidate targets of the complex in spermatocytes. This oligo contains two loosely defined consensus sequences referred to as Y and H elements which were thought to represent binding sites for the complex (Figure 2). However, the specific nucleotides or structural features that are needed were unclear. Systematic truncation and mutation studies have revealed that the Y and H elements per se are not necessary for binding to the complex as synthetic oligos with very dissimilar sequences also bind quite well (28). First, truncation of this 39-mer from both ends led to identification of a 25-mer central fragment that binds as well as the parent oligo, even though half of both the Y and H elements are missing. Further attempts to define the minimal features present in this 25-mer needed to confer high affinity binding indicated that G residues play a critical role. This analysis was facilitated by the observation that oligos composed of polymers of either U or A residues have little affinity for the complex. Substitution of eight G residues at the same positions found in the 25-mer protamine-2 oligo on a background of U residues generates an oligo that binds as well as the parent 25-mer (Figure 3). Thus, these eight G residues are sufficient for conferring high affinity binding to the complex. Furthermore, mutation of these G residues to U residues in the 25-mer completely abolishes its binding activity, indicating that all, or at least some, of these are also necessary. Further analysis using this approach indicated that as few as 4 G residues are sufficient to confer high affinity binding. However, there is a high degree of flexibility with regard to the spacing of these residues. Thus, while these studies indicate that the complex interacts with a high degree of selectivity for clusters of G residues, the flexible spacing precludes one from using this information to identify potential RNA targets of the complex from simple inspection of their primary sequence. Presumably, the U spacers provide a high degree of flexibility that enables the interspersed G residues to adopt the proper spacing to dock into binding sites on the complex. Perhaps, selectivity for endogenous transcripts depends on secondary structural features which confer the spacing between G residues needed to bind to the complex with high affinity. Accordingly, further studies are clearly needed to define the secondary structural features that are able to present G residues in the proper configuration for binding to the complex.
Figure 3. Binding selectivity of Translin/Trax complex.
Top panel presents the primary sequence of a series of RNA oligos used in gel-shift studies shown in bottom panels. TB, refers to the central portion of the 39-mer Prm-2 probe, which binds with high affinity to the complex. TB-8G, refers to the oligo generated by retaining the 8 G residues located in TB while replacing all the other other residues with U. In TB-NoG, the 8 G residues present in TB were replaced by U residues. The bottom panels present gel-shift experiments performed with recombinant Translin/Trax complexes generated by co-transfection of 293 cells. This panel shows competition curves obtained by preincubation of extracts with either Prm-2, TB-8G or TB-NoG. Note that TB-8G is more potent at inhibiting the gel-shift band than Prm-2. In contrast, TB-NoG is inactive even at concentrations up to 300 nM. (Figure adapted from ref. 28 with publisher’s permission.)
Clues to Function
Several lines of evidence support the hypothesis that the Translin/Trax complex mediates RNA trafficking in neurons. First, prominent Translin immunostaining is present in neuronal dendrites (25,29; Figure 4). Second, siRNA mediated knockdown of Translin impairs dendritic trafficking of BDNF mRNA, which co-precipitates with the native complex from forebrain homogenates (28). Third, earlier studies have demonstrated that an anti-sense oligo targeting a putative Y element in Cam Kinase II mRNA blocks its trafficking into dendrites (31). However, while this study is consistent with this hypothesis, it is unclear that this effect of antisense treatment is mediated by preventing the interaction of Translin or Translin/Trax complexes with this transcript. Fourth, Translin and Trax have been identified as components of a ribonucleoprotein complex containing BC1 RNA, a small non-coding RNA, implicated in regulating dendritic translation (32,33,34). Thus, taken together, these findings support the view that the Translin/Trax complex mediates RNA trafficking in dendrites. In addition, recent studies have also implicated Trax in mediating trafficking of mRNA into axons. Trax was found to be upregulated during regeneration of optic nerve, and siRNA mediated knockdown of Trax in this system both enhanced levels of GAP-43 mRNA and stimulated nerve regeneration (35).
Figure 4. Localization of Translin in Purkinje cells.
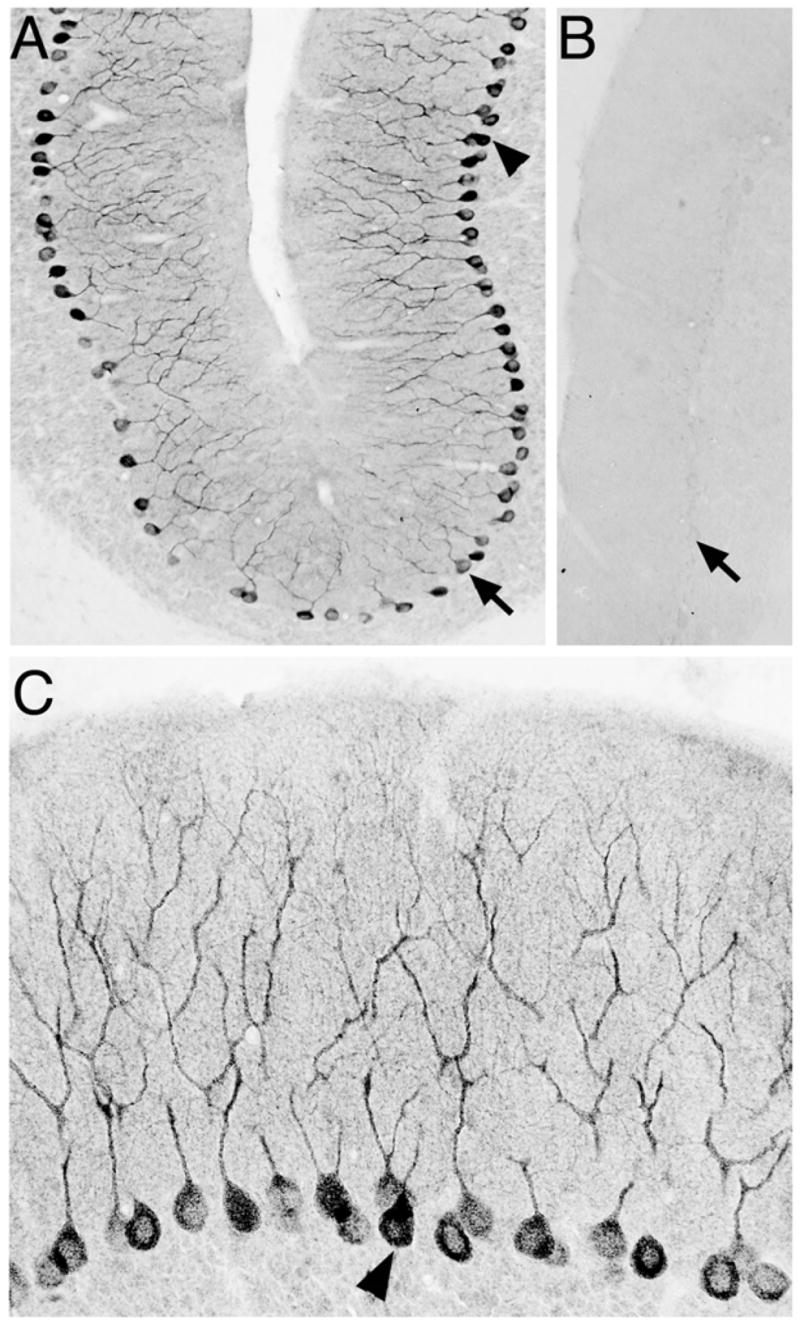
Panel A presents a low power view of Translin immunostaining in cerebellum which shows prominent, selective labeling of Purkinje cells. Panel B shows absence of staining in an adjacent section when the antibody is pre-incubated with its antigen peptide. Arrows shown in panels A and B point to a corresponding site in both sections. Panel C presents a higher power view of the area shown in upper right portion of panel A. Arrowhead in both panels point to the same pair of Purkinje neuron cell bodies. (Figure adapted from ref. 25 with publisher’s permission.)
Although a major focus of work on Translin and Trax has been aimed at elucidating their role in regulating trafficking of RNA, two recent studies indicate that these proteins also have other cellular functions. One provides evidence that the Translin/Trax complex may also regulate transcription; the other demonstrates that Trax binds to the intracellular tail of the adenosine A2A receptor and mediates its ability to suppress proliferation in PC12 cells.
A link between the Translin/Trax complex and transcription has emerged from studies aimed at identifying the components of gel-shift complexes containing a transcription factor called steroidogenic factor-1 (SF-1) in Leydig cells (36). These studies demonstrated that Translin and Trax are components of this complex and that co-transfection of Translin and/or Trax with SF-1 potentiates its ability to activate transcription. Of note, the ability of Translin to enhance SF-1 mediated transcription is not blocked by mutations that impair its RNA or ssDNA binding activity. Thus, the interaction of these proteins with SF-1 is not mediated by their interaction with nucleic acids, but appears to reflect a protein-protein interaction between either Translin or Trax and SF-1. Conceivably, their role in transcription may reflect a feedback mechanism that is coordinated with their role in RNA trafficking. For example, if the complex is unoccupied by RNA, it may enter the nucleus and then bind to SF-1 or other transcription factors to stimulate transcription of RNA cargoes that it will then shepherd into the cytoplasm.
The interaction between Trax and the adenosine A2A receptor was identified in studies aimed at understanding how this receptor suppresses proliferation of PC12 cells (37). Overexpression of the C-terminal tail of this receptor blocked this effect. Therefore, yeast two hybrid studies were used to search for proteins that bind to this tail and led to identification of Trax as a candiate interactor. Trax co-precipitates with the native A2A receptor and overexpression of Trax mimics the suppressive effect of A2A receptor stimulation on proliferation. Furthermore, antisense mediated knockdown of Trax expression blocks this effect of A2A receptor stimulation. Thus, taken together, these studies provide strong evidence that Trax mediates downstream effects of A2A receptor activation. Although it is unclear how Trax elicits this effect, these findings are particularly interesting in that they suggest that the Translin/Trax complex may be regulated via direct interaction with G protein coupled receptors. As little is known about how the activity and localization of this complex is regulated, this new information is particularly welcome.
Analysis of Translin KOs
Characterization of Translin KO mice have yielded several important findings (4,5). As mentioned above, these mice do not display increased levels of or susceptibility to chromosomal recombination events, bolstering the view that the complex targets RNA, rather than ssDNA, in vivo. Furthermore, Trax levels are undetectable in these mice. This striking deficit appears to be due to degradation of Trax protein as Trax mRNA levels remain normal. From a neuronal perspective, biochemical and behavioral analyses have revealed several potentially interesting changes. Microarray analysis of Translin KO mice has identified changes in the levels of multiple mRNAs. These include transcripts encoding GluR3, a member of the AMPA receptor family of glutamate receptors, and the alpha 1 subunit of the GABA A receptor. While these transcripts represent candidate targets of the complex, it is not clear that they bind directly to the complex as they did not co-precipitate with the complex from brain.
Behavioral characterization of these mice on a panel of tasks has identified multiple deficits. On the accelerating Rota-Rod test which provides a measure of cerebellar function, Translin KO mice fell off the apparatus sooner than wild type littermates. This deficit fits well with the strong expression of Translin in cerebellum (25). To evaluate anxiety-related behaviors, Translin KO mice were evaluated in both the elevated zero maze and light-dark exploration. In both cases, Translin KO mice spent more time in the open compartments consistent with reduced anxiety. In a variety of other tests, including Morris water, fear conditioning and acoustic startle, Translin KO mice showed abnormal responses in a gender-specific fashion, making interpretation less straightforward.
These mice also display other interesting phenotypes. First, they noted “multiple episodes of tonic-clonic seizures in both male and female Translin KO mice” during routine handling but not during behavioral testing. It is tempting to speculate that this seizure phenotype may be related to the marked decrease in expression of the alpha 1 subunit of GABA A receptors in these KO mice. Clearly, further studies aimed at examining the effect of Translin deficiency on inhibitory synaptic transmission are needed to explore these findings. Second, Translin KO mice have reduced levels of two major monoamine neurotransmitters, norepinephrine and serotonin, in several forebrain regions in both males and females. Since these neurotransmitters play critical roles in mediating anxiety, learning and seizure activity, it will also be useful to examine the cause of these changes and their role in mediating the behavioral changes identified in these mice. Third, it is noteworthy that the overall profile of biochemical and behavioral changes displayed by Translin KO mice resembles many of those reported for mice lacking FMRP. As FMRP has been implicated in regulating translation of dendritic transcripts, these parallels lend additional support to the hypothesis that the Translin/Trax complex may also regulate trafficking and/or translation of dendritic mRNAs as well. However, the significance of this superficial resemblance will need to be tested further.
Behavioral analysis of Drosophila Translin mutants has also identified an abnormal phenotype (6). These flies performed normally in tests of geotactic and phototactic responses. In addition, they displayed normal avoidance response to benzaldehyde, indicating normal olfactory function. However, Translin mutants displayed a reduced jump response to brief exposure to a high concentration of benzaldehyde, a defect that could be due to impaired motor function. Thus, further characterization of the behavioral deficiencies displayed by both mice and Drosophila lacking Translin may be very helpful in deciphering the role of this protein in the nervous system.
Genetic association between TRAX and psychiatric disorders
In humans, the TRAX gene is located within 50kb of the first exon of DISC1, a gene that was identified as the site of a chromosomal translocation associated with psychotic disorders in a Scottish pedigree (38). Heightened interest in the DISC1/TRAX locus has prompted several high resolution analyses of this region. In addition to confirming a link between DISC1 and psychiatric disorders, these studies have also yielded evidence for linkage between SNPs located within the TRAX gene and major mental disorders. For example, a high density SNP analysis of this locus detected under-representation of a haplotype defined by 2 SNPs located within intron 4 of the TRAX gene in schizophrenia (39). In addition, a SNP located immediately distal to the final TRAX exon (exon6) showed significant assocation with schizophrenia in a Taiwanese population (40) and bipolar illness in a group of families that display a high incidence of this disease (41). Furthermore, a population-based twin cohort study detected a striking over-representation of a haplotype consisting of 4 SNPs, the two located in intron 4 of TRAX and two located in the initial portion of DISC1, with schizophrenia (42). Thus, while it is certainly conceivable that SNPs located in the vicinity of TRAX may contribute to susceptibility for psychiatric diseases by affecting expression of DISC1, these data also raise the possibility that TRAX should be considered a candidate susceptibility gene for psychiatric disorders in its own right.
Summary
Although Translin was originally identified and named for its postulated role in chromosomal translocation and DNA repair, characterization of Translin and its partner protein, Trax, indicate that these proteins form a complex that binds to RNA in vivo. While initial studies suggested that this complex is highly enriched in brain, it is now clear that the Translin/Trax complex is expressed in a broad range of tissues. The available evidence suggests that this complex is one of several factors, such as CPEB and Staufen, which mediate RNA trafficking in dendrites. Studies aimed at defining the sequence features that confer high affinity binding to the complex have revealed a key role for G residues, however the precise configuration of these residues remains to be clarified. Although initial analysis of Translin KO mice has yielded several interesting phenotypes, further studies are needed to identify the cellular mechanisms that underlie these changes. Finally, genetic association studies suggest that Trax is a candidate susceptibility gene for psychiatric disorders. Accordingly, substantial progress has been made in characterizing this complex in yeast, Drosophila and mouse. However, we are only beginning to understand the organization of the complex and how it interacts with RNA. Furthermore, additional studies are needed to understand how its binding activity and subcellular localization are regulated, and to identify the set of transcripts that it binds to in vivo. As genetic association studies and analysis of Translin KO mice have linked Translin and Trax to behavioral abnormalities, progress in understanding the structure and function of this complex may help elucidate the pathophysiology of neuropsychiatric disorders.
Acknowledgments
Supported in part by NARSAD (Z.L.) and NIDA (J.M.B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosal translocations. Nat Genet. 1995;10:167–74. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 2.Aoki K, Ishida R, Kasai M. Isolation and characterization of a cDNA encoding a Translin-like protein, TRAX. FEBS Lett. 1997;401:109–112. doi: 10.1016/s0014-5793(96)01444-5. [DOI] [PubMed] [Google Scholar]
- 3.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–89. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 4.Chennathukuzhi V, Stein JM, Abel T, Donlon S, Yang S, Miller JP, Allman DM, Simmons RA, Hecht NB. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain and behavioral changes. Mol Cell Biol. 2003;23:6419–6434. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein JM, Bergman W, Fang Y, Davison L, Brensinger C, Robinson MB, Hecht NB, Abel T. Behavioral and neurochemical alterations in mice lacking the RNA binding protein Translin. J Neurosci. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suseendranathan K, Sengupta K, Rikhy R, D’Souza JS, Kokkanti M, Kulkarni MG, Kamdar R, Changede R, Sinha R, Subramanian L, Singh K, Rodrigues V, Rao BJ. Expression pattern of Drosophila translin and behavioral analyses of the mutant. Eur J Cell Biol. 2007;86:173–186. doi: 10.1016/j.ejcb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Wu XQ, Gu W, Meng X, Hecht NB. The RNA-binding protein, TB-RBP, is the mouse homologue of Translin, a recombination protein associated with chromosomal translocations. Proc Natl Acad Sci USA. 1997;94:5640–5. doi: 10.1073/pnas.94.11.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taira E, Baraban JM. Identification of a strand-specific Egr response element binding complex enriched in rat brain. J Neurochem. 1997;68:2255–2262. doi: 10.1046/j.1471-4159.1997.68062255.x. [DOI] [PubMed] [Google Scholar]
- 9.Taira E, Finkenstadt PM, Baraban JM. Identification of Translin and TRAX as components of the GS1 strand-specific DNA binding complex enriched in brain. J Neurochem. 1998;71:471–477. doi: 10.1046/j.1471-4159.1998.71020471.x. [DOI] [PubMed] [Google Scholar]
- 10.Aharoni A, Baran N, Manor H. Characterization of a multisubunit human protein which selectively binds single stranded d(GA)n and d(GT)n sequence repeats in DNA. Nucleic Acids Res. 1993;21:5221–5228. doi: 10.1093/nar/21.22.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob E, Pucshansky L, Zeruya E, Baran N, Manor H. The human protein translin specifically binds single-stranded microsatellite repeats, d(GT)n, and G-strand telomeric repeats, d(TTAGGG)n: a study of the binding parameters. J Mol Biol. 2004;344:939–50. doi: 10.1016/j.jmb.2004.09.095. [DOI] [PubMed] [Google Scholar]
- 12.Claussen M, Koch R, Jin Z-Y, Suter B. Functional characterization of Drosophila Translin and Trax. Genetics. 2006;174:1337–1347. doi: 10.1534/genetics.106.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaendling A, Ramayah S, Pryce DW, McFarlane RJ. Functional characterisation of the Schizosaccharomyces pombe homologue of the leukaemia-associated translocation breakpoint binding protein translin and its binding partner, TRAX. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2007.10.014. in press. [DOI] [PubMed] [Google Scholar]
- 14.Erdemir T, Bilican B, Oncel D, Goding CR, Yavuzer U. DNA damage-dependent interaction of the nuclear matrix protein C1D with Translin-associated factor X (TRAX) J Cell Sci. 2002;115(Pt 1):207–16. doi: 10.1242/jcs.115.1.207. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Chennathukuzhi V, Miki K, O’Brien DA, Hecht NB. Mouse testis brain RNA binding protein/Translin selectively binds to the messenger RNA of the fibrous sheath protein glyceraldehydes 3-phosphate dehydrogenase-S and suppresses its translation in vitro. Biol of Reproduction. 2003;68:853–859. doi: 10.1095/biolreprod.102.008631. [DOI] [PubMed] [Google Scholar]
- 16.Cho YS, Iguchi N, Yang J, Handel MA, Hecht NB. Meiotic messenger RNA and noncoding RNA targets of the RNA-binding protein Translin (TSN) in mouse testis. Biol of Reproduction. 2005;73:840–847. doi: 10.1095/biolreprod.105.042788. [DOI] [PubMed] [Google Scholar]
- 17.Finkenstadt PM, Jeon M, Baraban JM. Masking of the Translin/Trax complex by endogenous RNA. FEBS Lett. 2001;498:6–10. doi: 10.1016/s0014-5793(01)02470-x. [DOI] [PubMed] [Google Scholar]
- 18.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–6. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 19.Laufman O, Ben Yoseph R, Adir N, Manor H. Cloning and characterization of the S. pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res. 2005;33:4128–4139. doi: 10.1093/nar/gki727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu XQ, Xu L, Hecht NB. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 1998;26:1675–80. doi: 10.1093/nar/26.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkenstadt PM, Jeon M, Baraban JM. TRAX is a component of the Translin-containing RNA binding complex. J Neurochem. 2002;83:202–210. doi: 10.1046/j.1471-4159.2002.01158.x. [DOI] [PubMed] [Google Scholar]
- 22.Pascal JM, Chennathukuzhi VM, Hecht NB, Robertus JD. Mouse testis-brain RNA-binding protein (TB-RBP): expression, purification and crystal X-ray diffraction. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 11):1692–4. doi: 10.1107/s0907444901014548. [DOI] [PubMed] [Google Scholar]
- 23.Sugiura I, Sasaki C, Hasegawa T, Kohno T, Sugio S, Moriyama H, Kasai M, Matsuzaki T. Structure of human translin at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 4):674–9. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 24.VanLoock MS, Yu X, Kasai M, Egelman EH. Electron microscopic studies of the translin octameric ring. J Struct Biol. 2001;35:58–66. doi: 10.1006/jsbi.2001.4383. [DOI] [PubMed] [Google Scholar]
- 25.Finkenstadt PM, Kang WS, Jeon M, Tang W, Baraban JM. Somatodendritic localization of Translin, a component of the Translin/Trax RNA binding complex. J Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Cho YS, Chennathukuzhi VM, Underkoffler LA, Loomes K, Hecht NB. TRAX is post-transcriptionally regulated by its partner protein TB-RBP and both are essential for normal cell proliferation. J Biol Chem. 2004;279:12605–12614. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]
- 27.Cho YS, Chennathukuzhi VM, Handel MA, Eppig J, Hecht NB. The relative levels of Translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J Biol Chem. 2004;279:31514–23. doi: 10.1074/jbc.M401442200. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Baraban JM. High affinity binding of the Translin/jTrax complex to RNA does not require the presence of Y or H elements. Mol Brain Res. 2004;120:123–9. doi: 10.1016/j.molbrainres.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi S, Takashima A, Anzai K. The dendritic translocation of translin protein in the form of BC1 RNA protein particles in developing rat hippocampal neurons in primary culture. Biochem Biophys Res Commun. 1998;253:448–53. doi: 10.1006/bbrc.1998.9704. [DOI] [PubMed] [Google Scholar]
- 30.Chiaruttini C, Baj G, Fiorelli R, Li Z, Braiuca P, Gardossi L, Baraban J, Tongiorgi E. Translin mediates BDNF mRNA localization into dendrites: a link with neuropsychiatric disorders?. FASEB Summer Research Conference on Intracellular RNA sorting, transport & localization.2005. [Google Scholar]
- 31.Severt WL, Biber TU, Wu X, Hecht NB, DeLorenzo RJ, Jakoi ER. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J Cell Sci. 1999;112:3691–702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- 32.Muramatsu T, Ohmae A, Anzai K. BC1 RNA protein particles in mouse brain contain two y-,h-element-binding proteins, translin and a 37 kDa protein. Biochem Biophys Res Commun. 1998;247:7–11. doi: 10.1006/bbrc.1998.8657. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–41. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Iacoangeli A, Lin D, Williams K, Denman RB, Hellen CU, Tiedge H. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171:811–21. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schröer U, Volk GF, Liedtke T, Thanos S. Translin- associated factor-X (Trax) is a molecular switch of growth-associated protein (GAP)-43 that controls axonal regeneration. Eur J Neurosci. 2007;26:2169–78. doi: 10.1111/j.1460-9568.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 36.Mellon SH, Bair SR, Depoix C, Vigne J-L, Hecht NB, Brake PB. Translin coactivates steroidogenic factor-1-stimulated transcription. Mol Endocrinology. 2007;21:89–105. doi: 10.1210/me.2005-0355. [DOI] [PubMed] [Google Scholar]
- 37.Sun C-N, Cheng H-C, Chou J-L, Lee S-Y, Lin Y-W, Lai H-L, Chen H-M, Chern Y. Rescue of p53 blockage by the A2a adenosine receptor via a novel interacting protein, translin-associated protein X. Mol Pharm. 2006;70:454–466. doi: 10.1124/mol.105.021261. [DOI] [PubMed] [Google Scholar]
- 38.Roberts RC. Schizophrenia in translation: disrupted in schizophrenia (DISC1): integrating clinical and basic findings. Schizophr Bull. 2007;33:11–5. doi: 10.1093/schbul/sbl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennah W, Varilo T, Kestilä M, Paunio T, Arajärvi R, Haukka J, Parker A, Martin R, Levitzky S, Partonen T, Meyer J, Lönnqvist J, Peltonen L, Ekelund J. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–9. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 40.Hwu HG, Liu CM, Fann CS, Ou-Yang WC, Lee SF. Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry. 2003;8:445–52. doi: 10.1038/sj.mp.4001235. [DOI] [PubMed] [Google Scholar]
- 41.Curtis D, Kalsi G, Brynjolfsson J, McInnis M, O’Neill J, Smyth C, Moloney E, Murphy P, McQuillin A, Petursson H, Gurling H. Genome scan of pedigrees multiply affected with bipolar disorder provides further support for the presence of a susceptibility locus on chromosome 12q23-q24, and suggests the presence of additional loci on 1p and 1q. Psychiatr Genet. 2003;13:77–84. doi: 10.1097/01.ypg.0000056684.89558.d2. [DOI] [PubMed] [Google Scholar]
- 42.Cannon TD, Hennah W, van Erp TG, Thmpson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–13. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]