Summary
The extracellular matrix (ECM) protein F-spondin mediates axon guidance during neuronal development. Its N-terminal domain, termed the reelin-N domain, is conserved in F-spondins, reelins and other ECM proteins. In this study, a recombinant human reelin-N domain has been expressed, purified and shown to bind heparin. The crystal structure of the reelin-N domain resolved to 2.0 Å reveals a variant immunoglobulin (Ig)-like fold and potential heparin-binding sites. Substantial conformational variations even in secondary structure are observed between the two chemically identical reelin-N domains in one crystallographic asymmetric unit. The variations may result from extensive, highly specific interactions across the interface of the two reelin-N domains. The calculated values of buried surface area and the interface's shape complementarity are consistent with the formation of a weak dimer. The homophilic asymmetric dimer can potentially offer advantages in binding to ligands such as glycosaminoglycans (GAGs), which may in turn bridge the two reelin-N domains and stabilize the dimer.
Keywords: ECM protein F-spondin, reelin-N domain, heparin-binding, axon guidance
Introduction
In the formation of functional neuronal networks, the guidance of axons to specific target neurons is a critical step in the development of the nervous system. Although it is a highly complex process, neural patterning involves a limited number of guidance molecules. These molecules can guide axons either by promoting or inhibiting outgrowth to ensure the extension of axons along a correct pathway 1. F-spondin, a secreted extracellular matrix (ECM) protein, plays a dual role in patterning axonal trajectory in the embryonic spinal cord by promoting outgrowth of commissural axons on one hand and inhibiting outgrowth of motor axons and migration of neural crest cells on the other hand 2. F-spondin was originally identified in the rat embryo floor plate, a cell-group implicated in the control of neural cell patterning and axonal growth in the developing vertebrate nervous system. The protein expression level is high at the time when axons start extending 3. Further studies established that F-spondin has an important function in the outgrowth of sensory 3, commissural 4 and hippocampal neurons during development 5. Secreted by cells within the floor plate F-spondin is proteolytically processed into fragments that differentially bind to the floor plate cells or the basement membrane 2. The portion of the molecule that binds to floor plate cells repels commissural axons and prevents their migration into the floor plate. Other portions of F-spondin associate with the basement membrane where they support growth cone migration. The coordinated activity of two fragments from a single guidance molecule, F-spondin, makes it a local combinatorial guidance cue for commissural axons that cross the midline 2. F-spondin also reportedly functions to promote the differentiation of progenitor cells into neurons and to prevent the migration of neural crest cells into the sclerotome 6; 7.
F-spondin may also have a role in the pathogenesis of Alzheimer's disease since it binds to amyloid precursor protein (APP) and prevents its cleavage by α- or β-secretases, which in turn prevents the production of Aβ peptides by γ-secretase 8; 9. The protection of APP by F-spondin reportedly involves the formation of a tripartite complex that also includes apoE receptor 2 9.
The function of F-spondin is not restricted to the nervous system. In normal adult tissue, F-spondin is expressed in various organs including the lung, ovary, small intestine, and kidney 10. This protein, also named vascular smooth muscle cell growth-promoting factor, has been isolated from the fluid of the adult bovine ovarian follicle where it reportedly regulates angiogenesis 11. In vitro experiments revealed that F-spondin specifically inhibits integrin αvβ3-mediated spreading of human umbilical vein endothelial cells. It also inhibits angiogenesis in an in vitro tube forming assay and an in vivo rat corneal assay 11. Thus, F-spondin may affect angiogenesis through direct effects on both endothelial and smooth muscle cells. Interestingly, some of the sites of F-spondin expression and deposition in the embryo, including the floor plate and paranotochordal area, are devoid of blood vessels 3; 7, suggesting that F-spondin may contribute to the inhibition of angiogenesis at these sites.
Like many large ECM proteins, F-spondin has multiple domains in concatenate (Figure 1A). The entire human F-spondin is composed of 807 amino acids. The N-terminal domain is a globular domain (∼1-175) with a sequence identity of 30% to the N-terminal domain of human reelin. For this reason, in the literature, the N-terminal domain of F-spondin is designated as the reelin domain while the N-terminal domain of reelin is called the F-spondin domain. For clarity, we suggest this conserved domain be named reelin-N domain. The second domain of F-spondin, sometimes referred to as the spondin domain, contains two distinctive conserved sequence repeats that can also be found in other spondin family members, including mindin 1 and 2, and M-spondin. These two conserved sequence repeats are generally designated “fs1 domain” and “fs2-domain”, and they may well represent repeats of a single novel structural fold. Six thrombospondin type 1 repeats (TSR) can be recognized between residues P440-C807 of human F-spondin. Therefore, F-spondin is also considered a member of TSR superfamily 12. Several TSR-containing proteins have been shown to support neurite outgrowth. The TSRs may mediate the interaction of F-spondin with ECM and neuronal growth cones 3; 13. The solution structures of the first and fourth TSRs of F-spondin have been reported 14, and are similar to the crystal structure of TSR of thromboposnin-1 15.
Figure 1.
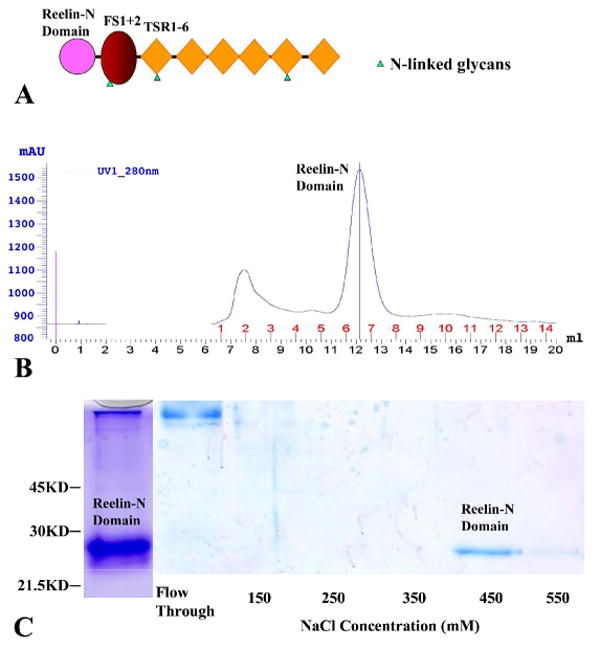
The Reelin-N Domain Binds Heparin. (A) A schematic diagram of the domain structure of F-spondin showing the relative position of the reelin-N domain, the spondin domain (FS1+2), which includes two sequential repeats designated fs1 and fs2, and the six TSRs. (B) Size-exclusion chromatography of the recombinant reelin-N domain. The recombinant protein was first purified using Ni-affinity chromatography. Six protein standards were used to calibrate the column in a separated run (data not shown). The molecular mass of the protein from the principal peak was calculated to be 21.0 kDa based on the calibration. (C) SDS PAGE of the reelin-N domain after Ni-affinity chromatography (left lane) and heparin-Sepharose affinity chromatography. The initial buffer included 100 mM NaCl and 20 mM HEPES at pH 7.8. Whereas the majority of the reelin-N domain was eluted from the heparin-Sepharose column in buffer containing 450 mM NaCl, some protein was also present in the 550 mM NaCl eluate.
The reelin-N domain is present not only in F-spondin and reelin, but also in Amphi-spondin and a hypothesized protein F10E7.4 of C.elegans. Though the reelin-N domain is part of a proteolytic fragment of F-spondin, which includes the reelin-N and spondin domains, that supports axon growth 2, the function of the individual reelin-N domain as well as its structure remain largely unknown. Here, we present the crystal structure of the reelin-N domain of human F-spondin and show that it binds heparin. The reelin-N domain has a β-sandwich core structure. Two molecules within a crystallographic asymmetric unit display substantial conformational variation, which is associated with their extensive interaction across their interface. Potential heparin-binding sites have been mapped on individual reelin-N domains. Additionally, a tentative low-affinity reelin-N domain dimer was proposed to potentially increase heparin binding, and the heparin-binding in turn may stabilize the weak dimer. The structural and functional properties of F-spondin's reelin-N domain that are revealed in this study pave the way for further exploration into the functions of F-spondin at molecular and cellular levels.
Results
Structure determination and heparin-binding analysis
A recombinant human reelin-N domain of F-spondin was expressed in Drosophila Schneider 2 (S2) cells and purified to homogeneity using His-tag immobilized metal affinity chromatography and size-exclusion chromatography. The elution profile of the size-exclusion chromatography of the reelin-N domain showed a single peak (Figure 1B). Based on a calibration curve (data not shown), the apparent molecular weight of the eluted reelin-N domain is estimated to be 21.0 kDa. The calculated protein molecular weight from its sequences is 20.5 kDa, including N-terminal (RSPWPG) and C-terminal (TGHHHHHH) vector-derived sequences. These data suggest that the reelin-N domain is a monomer in solution.
Crystals of Se-Met labeled protein were prepared for anomalous diffraction data collection to 2.8 Å at the Structural Biology Center (SBC) at the Advanced Photon Source (APS) at Argonne National Laboratory (ANL) (Table I). The anomalous signal obtained was too weak to locate the Se site correctly at this resolution. There is only one methionine residue in the domain and we suspected that this residue might reside on a mobile loop, unable to contribute enough anomalous signal. Based on secondary structure prediction (data not shown) that suggested that the reelin-N domain is a β strand dominated structure, an additional methionine was introduced in a likely buried position (Leu82) on one β strand. The Se-Met labeled L82M mutant crystal was then prepared in the same way as the native protein. One SAD (single wavelength anomalous diffraction) data set was collected at Se-peak (2.8 Å) (Table I). Despite the fact that the anomalous signal was only good to about 5.5 Å, the Se sites were readily located using the program SHELXD 16 incorporated in HKL2000 17. There are four Se sites, suggesting two molecules in one asymmetric unit, consistent with an estimated solvent content of 47%. The structure was subsequently resolved and refined with a native data set of 2.0 Å as described in detail in the Experimental Methods.
Table 1. Crystallographic Statistics.
| Data collection | Native | SelMet | SelMet
(L82/M) |
|---|---|---|---|
| Space group | P43212 | P43212 | P43212 |
| Unit Cell (Å, °) | a=b=58.61, c=219.54 | a=b=58.53, c=217.90 | a=b=58.54, c=218.92 |
| MW Da (residue) | 188061 | ||
| Mol (AU) | 2 | 2 | 2 |
| SeMet (AU) | 2 | 4 | |
| Wavelength(Å) | 0.97934 | 0.97904(peak) | 0.97907(peak) |
| Resolution(Å) | 2.0 | 2.8 | 2.8 |
| Number of unique reflections | 26722 | 97583 | 96933 |
| Redundancy | 6.7 | 11.8 | 11.7 |
| Completeness (%) | 99.0 (95.3) 2 | 97.1 (87.1) 4 | 95.7 (75.5) 4 |
| Rmerge (%) | 7.2 (41.9) 2 | 17.5 (63.8) 4 | 13.3 (61.4) 4 |
| I/σ(I) | 33.4 (3.5) 2 | 15.3 (2.0) 4 | 19.7 (2.0) 4 |
| Phasing | |||
| RCullis (anomalous) (%) | 99 | 95 | |
| Figure of merit (%) | 10.6 | ||
| Refinement | |||
| Resolution | 40.1-2.0 | ||
| Reflections (work/test) | 25408/1346 | ||
| Rcrystal/Rfree (%) | 20.6/24.1 | ||
| Rms deviation from ideal geometry | 0.018/1.76 | ||
| No. of atoms (Protein/HETATM) | 2287/153 | ||
| Mean B-value (Å2) (mainchain/sidechain) | 47.59/50.41 | ||
| Ramachandran plot statistic (%) | |||
| Residues in most favored/
additional allowed/ generously allowed regions. |
87.0
13.0 0.0 |
||
Not including cloning artifact and His tag;
(Last resolution bin, 2.00-2.07 Å);
Including Bijvoet pairs;
(Last resolution bin, 2.80-2.90 Å)
Many ECM proteins bind heparin to mediate their adhesive function. A heparin Sepharose column was used for full-length F-spondin purification 10; 11, suggesting the presence of heparin-binding site(s) within the molecule. TSR5 and TSR6 of F-spondin were found to be able to bind ECM 18, possibly through glycosaminoglycans (GAG) 15. To determine whether or not the reelin-N domain binds heparin, we have performed heparin-binding studies of reelin-N domain using affinity chromatography. The recombinant reelin-N protein does bind to the heparin-Sepharose column in buffer containing 0.10 M NaCl and can be eluted with buffer containing 0.45 M NaCl (Figure 1C). This experiment suggests that reelin-N domain of F-spondin has a relatively high affinity for heparin.
Overview of reelin-N structure
There are two reelin-N molecules in one asymmetric unit, which are not related by a proper pseudo-dyad. As predicted, reelin-N belongs to β structure (Figure 2). The core of the reelin-N domain is composed of β strands, A, B, E, and D on one sheet and A′ (or A0), G, F, C, D″ and D′ on the opposite sheet, accounting for 120 residues. Topologically it appears to fall into a variant immunoglobulin (Ig) fold. Any member of the immunoglobulin superfamily (IgSF) usually has conserved ABE and GFC sheet, with additional edge strands varying from one molecule to another 19. For instance, the variable domain of an antibody is well known to have ABED and A′GFCC′C″ sheets, whereas VCAM-1 (vascular cell adhesion molecule-1) domain 2 has ABE and GFCC′ sheets 20. In the following three aspects, reelin-N domain is regarded as a unique Ig fold. First, it has D′ and D″ strands 21, instead of C′ and C″ strands, next to the C strand on the edge of GFC sheet. Secondly, the A strand starts at Phe61 (Figure 3). Before the A strand, there is a mini A0 (or A0′) strand next to the G0 (or G0′) strand (Figure 2). The construct actually begins at Phe29 after the signal peptide. The first 15 amino acid residues are mostly disordered in the crystal. Starting from Cys44, the following 14 residues (including the mini A0 or A0′ strand), along with the elongated CD, D′D″ and FG loops, form a distinct additional layer lying on the GFCD″D′ sheet (Figure 2). As will be elaborated below, the conformation of this additional layer differs remarkably between the two reelin-N domains in the asymmetric unit. Thirdly, the reelin-N domain does not have the canonical disulfide bond between the B and F strands, but instead has two highly conserved disulfide bonds near the N- and C-termini that help define and stabilize the globular reelin-N domain (Figures 2 and 3). The first disulfide bond (Cys44-Cys128) tethers the N-terminus to the D′D″ loop, whereas the second disulfide bond (Cys156-Cys182) locks the C-terminus to the beginning of the F strand (Figure 2). The 12 C-terminal residues after Cys182 in the recombinant protein are mostly disordered.
Figure 2.

The Structure of the Reelin-N Domain. The stereo views of ribbon drawings of monomer A (A) and B (B) on the left side and their topology schemes on the right side. In the ribbon drawings the β strands in the front β sheet are colored in green while those in the back β sheet are colored in cyan. The secondary structures that form additional layer (see text) and represent dramatic conformational change between the two monomers are colored in dark yellow. A segment on the CD loop of monomer A that forms two hydrogen bonds to the A0 strand is drawn in thick dark yellow line. The four cysteine residues that form two disulfide bonds as well as some of positively charged residues that may involve in heparin-binding are drawing in ball-and-stick format. Figure 2 and 4 were prepared using the program MOLSCRIPT 38.
Figure 3.

Multiple Sequence Alignment of Reelin-N Domains across Different Species. Based on the reelin-N domain structures of the two monomers, the secondary structures are indicated with arrows (β strands) and rods (α helices) below the appropriate sequences, respectively. Identical residues are highlighted in red and highly similar residues in brown. The sequences used in the alignment include human F-spondin (gi:110347423), rat F-spondin (gi:544353), chicken F-spondin (gi:45382337), Xenopus F-spondin (gi:544354), zebrafish F-spondin-1 (gi:18859413), zebrafish F-spondin-2 (gi:2529227), amphiF-spondin (gi:3319874), Drosophila melanogaster F-spondin (gi: 19922086), human reelin (gi:20139420), rat reelin (gi:149046582) and C.elegans hypothetical protein F10E7.4 (gi:32564253). The first 14 N-termnal residues (FSDETLDKVPKSEG) and the last 10 C-terminal residues (DSTFDGVTDK) in the expression construct of the human F-spondin reelin-N domain in this study were not used for the alignment. Most of these residues are disordered in the structure. The two highly conserved disulfide bonds are marked in lines connecting paired cysteines. For structural determination, the site of mutation of L82/M is marked with “ * ”.
The top five hits for reelin-N domain in Dali structural homology search (http://www2.ebi.ac.uk/dali/) find the β-sandwich core of the large domain of chloroplast cytochrome f (PDB code:1HCZ), the calf-2 domain from integrin αvβ3 (PDB code:1JV2), the C. elegans p53-like (Cep-1) DNA-binding domain (PDB code:1T4W), the receptor-binding domain of α2-macroglobulin (PDB code:1AYO) and the fibrinogen-binding domain of MSCRAMM (microbial surface components recognizing adhesive matrix molecules), clumping factor A (PDB code:1N67). These protein fragments all have the same variant Ig-like topology as does the reelin-N domain, namely to have unique D′ and D″ strands at the edge of GFC β sheet. This variant Ig-fold was termed DEv-IgG fold and thought to adopt diverse functions during evolution 21. Notably, in the structural alignments of the reelin-N domain with other structural homologues, only the core DEv-IgG fold can be readily superposed. The residues of the reelin-N domain contributing to the hydrophobic core of the DEv-IgG fold include residues Tyr78, Val80 and Leu82 of the B strand, Phe93 and Leu95 of the C strand, Val145, Trp147 and Ala149 of the E strand, as well as Leu159 and Ala161 of the F strand (Figure 3). All of these residues are either identical or highly conserved in the family members that contain the reelin-N domain (Figure 3). Based on this sequence conservation, we predict that the features of the reelin-N domain as a DEv-IgG fold should be conserved among them. However, the additional layer in front of the GFCD″D′ β sheet and the two highly conserved disulfide bonds described above have no counterpart in these homologues, suggesting the uniqueness of reelin-N domain structure.
The substantial conformational variation of reelin-N domain
One striking feature of the reelin-N domain structure is the substantial conformational differences between the two chemically identical molecules in one asymmetric unit as depicted in the stereoviews of Figures 2A and 2B. The two monomers can be superimposed with an rmsd (root mean square deviation) value of 0.88 Å, in which most of the core structure of the domain (Thr59-Glu100, A111-S162, total 92 Cα atoms from each monomer) can be aligned. The N-terminal region (up to Tyr58), CD loop and the C-terminal region (after S162) are totally out of the structural alignment with rmsd values of 4.97 Å (12 Cα), 2.03 Å (4 Cα) and 2.92 Å (22 Cα), respectively. The N-terminal variable region together with the elongated CD, D′D″ and FG loops actually form an additional layer in front of the GFCD″D′ sheet and account for its dramatic variation between the two molecules (Figure 2). Meanwhile, the plasticity of the additional layer seems to be restrained by the two highly conserved disulfide bonds that lock the two termini to the reelin-N domain core.
After the first disulfide bond (Figure 2A), monomer A starts with a short irregular β strand (A0′), which is parallel to another short strand (G0′) on the FG loop. The tracing of the domain then crosses over to the back β sheet, forming an edge strand, the A strand. The A strand is the first strand of the reelin-N domain core structure. There are a total of 17 amino acid residues from the Cys44 of the first disulfide bond to the beginning of the A strand. This N-terminal region is rather variable in both the primary sequences of reelin-N domain members and the secondary structure (Figure 3). The connection between the anti-parallel A and B strands on the back β sheet is rather long (10 residues) and it swings toward the G strand in the front β sheet, forming a short A′ strand parallel to the G strand. The cross-sheet secondary structure has been observed in many molecules classified as V-type fold in IgSF members. The secondary structures of the C, D, D′ D″, E and F strands are largely conserved between the two molecules in the asymmetrical unit. In monomer A, the G strand seems to split into two short segments, as G0′ and G. The short G0′ is unique and it is no longer anti-parallel to the F strand. Instead, it is bent down towards the A0′ strand, forming a mini parallel β ribbon in front of the D′D″CFG β sheet. Meanwhile, the CD loop at the bottom of domain is also moved up, forming two mainchain-mainchain hydrogen bonds to the A0′ strand. The interactions were not adequate to define a strand that is anti-parallel to A0′. The portion of the loop involved in the interactions is instead highlighted as a thick yellow line in Figure 2A. The final G strand is like a regular end strand found in IgSF domains, which is parallel to the F strand.
In monomer B (Figure 2B), the N-terminal variable region starts from a short α-helix (α1) packed onto the front β sheet. It is followed by a short β strand A0 that is parallel to the G0 strand at the edge of the front β sheet. Possibly due to the A0 strand and its spatial restriction, there is no A′ formed parallel to the G strand as found in monomer A. After the A and B strands, the secondary structures of the C, D, D′ D″, E and F strands are largely similar to those in monomer A except for the CD loop, which is not moved towards the beginning of the N-terminal variable region. The split edge strands, G0 and G are both anti-parallel to the F strand as the final secondary elements before the C-terminal disulfide bond.
Conformational variation at this scale is rare in one asymmetric unit. It seems to result from the strong interaction between the two monomers, which are packed mostly with their additional layers contacting each other without a proper pseudo-dyad (Figure 4A). Across the asymmetric interface between monomers A and B, salt-bridges and hydrogen bonds dominate their interactions while hydrophobic residues are largely absent (Figure 4B). Given the monomeric state of the reelin-N domain in solution (Figure 1B), and the lack of core hydrophobic interaction between two molecules, one would assume that the asymmetric conformation of two monomers may have resulted from crystal packing that causes the large conformational changes between the two molecules as described early. However, considering the large number of interactions across their interface, it will be intriguing to explore the possibility that the two reelin-N monomers in such an asymmetric conformation may present a weak dimer.
Figure 4.

The Asymmetric Reelin-N Domain Dimer and its Interface. (A) A ribbon diagram drawing of the asymmetric reelin-N domain dimer in a crystallographic asymmetric unit. The color codes for the secondary structures of the two monomers and individual residues are the same as those in Figure 2. The disulfide bond forming cysteine residues and some of the potential heparin-binding residues shown in Figure 2 are also drawing in ball-and-stick format in this figure for comparing molecules' orientations in the two figures. (B) A stereo view of the interface of reelin-N domain dimer. The residues involved in the high-specific interaction are drawn in the ball-and-stick format. The interactions across the interface are very asymmetric for the homodimer.
Discussion
The reelin-N domain of F-spondin assumes a variant Ig-like fold in its core structure while its N-terminal variable region and long CD, D′D″ and FG loops form a unique additional layer. The conformational difference between the two reelin-N domains within an asymmetric unit is substantial and is closely associated with their extensive interaction through primarily their additional layers (Figure 5A). Across their asymmetric interface there are at least 15 pairwise interactions including seven salt bridges (Figure 5B). The residues contributing to the salt bridges, such as Arg91, Glu134, Arg139 and Arg167 are highly conserved within F-spondins (Figure 3). There are also other highly specific and conserved hydrogen bonds such as those involving Asn121, Gln123, Thr151 and Gln165. The buried surface area between the two reelin-N domains is as high as 1781 A2, well within the range of being physiologically significant 22. Moreover, the interface's shape complementarity 23, the measurement of geometric match between two juxtaposed surfaces, is as high as 0.65, a value within the range of antibody-antigen interface complementarities 23. These data all imply that the observed asymmetric reelin-N domain dimer may represent a physiological entity, rather than a pure crystallographic packing artifact. However, the interface without extensive hydrophobic interactions is unlikely to hold the homophilic dimer in solution (Figure 1B). The monomeric reelin-N domain found in the size-exclusion chromatography could result from low affinity bonding and a fast off rate. Highly specific and low-affinity homophilic or heterophilic interactions are commonly found in adhesion molecules, which play important roles in molecular recognition and adhesion 24; 25; 26. One similar and well characterized interface is the one between CD2 and CD58, which has extensive salt links and hydrogen bonds and only a small patch of hydrophobic interactions 27. The heterophilic adhesion dimer of CD2/CD58 is well characterized with low affinity and very fast Koff and Kon28; 29.
Figure 5.

The Surface Potentials and Mapping of Heparin-binding Sites. (A) The electrostatic potential surface representation of the two individual reelin-N domains, monomer A (left) and monomer B (right). Some positively charged residues that may form heparin-binding sites are mapped to their surface to show their relative orientation to those in Figure 2. (B) The electrostatic potential surface representation of the reelin-N domain dimer. The two orientations of the dimer is related by a 160 degrees of rotation along the horizontal axis. The first one is similar to that shown in Figure 4A. Some potential heparin-binding residues from two monomers are mapped to the dimer surface. Figure 5 was prepared with the program GRASP 39.
Considering that its buried interface area and the interface's shape complementarity are both significantly larger than that of the CD2-CD58 heterodimer 27, we propose that the asymmetric reelin-N domain dimeric assembly found in the structure (Figure 4A) represents a weak dimer. The question is why the reelin-N domain can potentially form such a weak dimer. It is conceivable that such a dimer can bridge intercellular or cell-matrix interaction through two F-spondin molecules. Another possibility is that a dimer has the advantage of providing multivalency, which may increase binding strength and/or combine the binding activities of different domains 30. The roles of oligomerization of thrombospondins and other ECM proteins have been studied extensively 30. The importance of multivalence is also true for individual domains of these ECM proteins. Recently, we have shown that heparins can bind TSPN domains either individually or as dimers for more extensive interactions 31.
Our experimental data (Figure 1C) indicate that the reelin-N domain is capable of binding heparin with relatively high affinity. The reelin-N domain structure revealed in this study now allows us to map the possible binding sites on the domain for heparin. A cluster of positively-charged residues is regarded as a good indicator of a heparin-docking site, which involves mostly ionic interactions between heparin's sulfate groups and the protein's positively charged residues 32. A highly conserved sequence motif (R138RRTR, see Figure 3) is located on the D″E loop and the beginning of the D″ strand, which are on the top of reelin-N domain (Figure 2). This sequence motif creates the single largest positively charged patch on the molecular surface of the reelin-N domain and probably functions as the primary heparin-binding site (Figure 5A). Other residues that are likely to be engaged are Lys166 and Arg167, which are on the FG0 (or FG0′) loop, and also located on the top of the reelin-N domain (Figures. 2 and 5A). Despite the fact that the FG loop changes its conformation dramatically from one molecule to the other (Figure 2), Lys166 and Arg167 are always positioned near the proposed primary heparin-binding site, and therefore should enhance heparin binding. In molecule A, these two residues, together with Arg46, form the second positively charged patch on the protein's surface (Figures. 2A and 5A). The distance between this site and the primary binding site is about 16 Å, a distance that corresponds to about one turn of the heparin right-handed helix. The two binding sites with such a separation help to bind heparin collectively on one side of the helical molecule. In molecule B, Lys166 and Arg167 move closer to the primary binding site, forming an extended primary positively-charged patch (Figure. 2B and 5A). Therefore, Lys166 and Arg167 seem to function in coordination with the prominent positively-charged sequence motif (R138RRTR), irrespective of the conformational variation.
Interestingly, the potential asymmetric reelin-N dimer seems to bring the heparin-binding sites from two individual monomers together to create a groove that is fully lined with positively charged residues, with a second extended positively charged patch on the other side of the dimer (Figure 5B). On one hand, an extensive heparin-binding site can certainly increase heparin-binding affinity. On the other hand, a heparin sitting along one of these extended binding sites could externally bridge two reelin-N domains and help to stabilize the asymmetric dimer. The scenario seems to be reminiscent of what has been observed in the two dimerization modes induced by heparin binding of the N-terminal domain of thrombospondin-1 31. However, the potential role of the weak asymmetric reelin-N domain dimer formation in binding to heparin, and possible other ligands, remains to be explored and verified.
The functions of the GAG-binding site of F-spondin or its reelin-N domain have not been fully established. The GAG binding activity of the reelin-N domain of F-spondin may serve to anchor F-spondin to ECM in such a way that the spondin domain and the TSRs are available for interactions with cells or possibly visa verse. The orientation of the TSRs in such a scenario could be important for the appropriate guidance of the growth cones of commissural axons. This orientation may also facilitate the proteolytic processing of F-spondin that releases the TSRs from the intact molecule 2. After proteolysis, the remaining fragment, designated the reespo domain, would contain the reelin-N domain and the spondin domain 4. The reespo domain reportedly promotes the outgrowth of sensory neurons 4. The reelin-N and spondin domains of F-spondin also mediate binding to APP 8; 9. Like the reelin-N domain of F-spondin, APP binds to GAGs 33. These data raise the possibility that the GAG binding activity of the reelin-N domain of F-spondin may facilitate its interaction with APP by co-localizing the two proteins on the cell surface.
Materials and Methods
Preparation of the Recombinant Reelin-N domain
A cDNA pool was prepared from a mixture of human spleen, placeta and liver poly(A)+ RNA (Stratagene) using a reverse transcription kit (GIBCO). A recombinant version of the human reelin-N domain (amino acids F29-K194) was prepared by PCR using the cDNA pool as the template. The reelin-N domain was prepared using the forward primer fs101f (GAT GAT CCC GGG TTC TCC GAC GAG ACC CTG) and the reverse primer fs103r (ATG ATG ACC GGT TTT GTC AGT CAC CCC ATC). To introduce an extra methionine for improving structure phasing power as discussed below, a site-directed mutant (L82M) was introduced by overlapping PCR using a pair of primers, CC AGC TAC CGC GTA ACA ATG TCA GCT GCT CCT CCC TCC (forward, fs105f) and GGA GGG AGG AGC AGC TGA CAT TGT TAC GCG GTA GCT GG (reverse, fs106r). The PCR products were sequenced and cloned between the SmaI and the AgeI sites of the vector pMT/BiP V5-HisA (Invitrogen, Carlsbad, CA) for expression in Drosophila S2 cell. The recombinant proteins include the vector-derived sequence RSPWPG at the N-terminal and the sequence TGHHHHHH at the C-terminal.
The vector transfection and cell selection, as well as the reelin-N domain expression and purification were performed as described previously 34. To label the proteins with selenomethionine (Se-Met), the cells were grown to high density (∼ 1×107/ml) in media (Hyclone) and then transferred to methionine-free medium for 4 hours. Se-Met (Sigma) was subsequently added to the medium to a final concentration of 400 mg/l. The cultures were monitored for cell viability and harvested between three and five days. The Se occupancy was estimated to be about 90% based on mass spectral analysis. All proteins were further purified by HPLC in protein buffer containing 200 mM NaCl and 20 mM HEPES at pH 7.8.
Crystallization and X-ray Diffraction Data Collection
The purified protein was concentrated to about 10-12 mg/ml for crystallization with the vapor diffusion hanging drop method. Protein crystals grew from the buffer containing 0.2 M ammonium sulfate, 30% (w/v) PEG 4000 and 0.1 M sodium citrate tribasic dehydrate at pH 5.6. The crystals that were produced had a cubic form of a dimension of about 30μm on edge. Crystals of the Se-Met labeled reelin-N domain and the labeled mutant reelin-N domain (L82/M) were also produced under the same crystallization condition, and eventually used for initial phasing.
Diffraction data sets were collected from pre-frozen crystals at 100°K at 19ID/SBC/APS/ANL beamline. For structure resolution, initial extensive heavy atom derivative screening was unsuccessful. The crystals generally diffracted to 2.5 Å to 3.5 Å, however, one 2.0 Å resolution data set was obtained during screening and it was used for phase extension and the final structural refinement (Table I).
Se-Met labeled protein and crystal were then prepared. One SAD data set (2.8 Å) was collected at the Se peak (Table I). The anomalous signal obtained was very weak and not good enough to located Se site(s) correctly. The sole methionine residue of the domain was suspected to be on a possible mobile loop. A secondary structure prediction suggested the reelin-N domain was a β strand dominated structure. We decided to introduce a methionine to replace a likely buried residue on a β strand; the residue L82 was judged to be a suitable candidate. Then the Se-Met labeled mutant reelin-N domain (L82/M) protein and crystals were prepared in the same way as the unlabeled protein, and one SAD data set was collected at Se-peak (2.8Å) (Table I). Although anomalous signal was only good to about 5.5 Å, four Se sites were located from this data set for the Se-labeled mutant reelin-N domain (L82/M) crystal with the program SHELXD 16, and an initial phasing helped finding two Se sites for the first Se-labeled reelin-N domain crystal. They suggested that there are two monomers in one asymmetric unit, consistent with an estimation that was based on solvent content.
The above two SAD data sets were used together with native data for phasing and phase extension 35. A map after density modification 35(2.0 Å) revealed several β strands arranged in two β sandwich-like structures though the maps were rather noisy. Within each β sandwich-like structure, there was a Se-site, indicating the mutated L82/M position. The mutated position greatly helped the initial tracing from the noisy maps. After manual model building and sequence fitting using the program Coot 36, about 70% of the structure (including two monomers) was built. It was noticed at the early stages of model building, that the two monomers seemed to be quite different at the edge strands and some loops. No map averaging was applied and two monomers were built individually.
The final cycles of model building and refinement were performed using the 2.0Å native data set with the programs Coot 36 and Refmac37 (Table 1). In the final structural model, the mainchain densities of two monomers were well connected except breaks between T53 and Y58 of monomer A and between E100 and E107 of monomer B.
Heparin affinity chromatography
Heparin-Sepharose affinity chromatography was used to analyze the heparin-binding function of reelin-N domain. A heparin-Sepharose column was prepared by packing a Poly-Prep chromatography column (Bio-Rad) with heparin-Sepharose CL-6B beads (Pharmacia Biotech Inc.). The column was then equilibrated with 20 mM HEPES, pH 7.5, containing 100 mM NaCl, and the reelin-N domain after Ni-affinity chromatography was loaded onto the column at 4°C. The bound protein was eluted from the column using a step-wise salt gradient of 150 mM, 250 mM, 350 mM, 450 mM and 550 mM NaCl in 20 mM HEPES (pH 7.8). Elution of 2 ml was collected for each step and probed using SDS-PAGE.
Acknowledgments
We thank Florence Poy for help in cDNA pool preparation and Qunyan Lu in protein purification. This work was support by a grant (HL49081 and HL68003) from the National Heart, Lung and Blood Institute of the National Institutes of Health to JHW and JL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Codes: The coordinates and structure factors of human reelin-N domain have been deposited in the PDB data bank under the access code 3COO.
References
- 1.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science (New York, NY) 1996;274:1123–33. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 2.Zisman S, Marom K, Avraham O, Rinsky-Halivni L, Gai U, Kligun G, Tzarfaty-Majar V, Suzuki T, Klar A. Proteolysis and membrane capture of F-spondin generates combinatorial guidance cues from a single molecule. The Journal of cell biology. 2007;178:1237–49. doi: 10.1083/jcb.200702184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klar A, Baldassare M, Jessell TM. F-spondin: a gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell. 1992;69:95–110. doi: 10.1016/0092-8674(92)90121-r. [DOI] [PubMed] [Google Scholar]
- 4.Burstyn-Cohen T, Tzarfaty V, Frumkin A, Feinstein Y, Stoeckli E, Klar A. F-Spondin is required for accurate pathfinding of commissural axons at the floor plate. Neuron. 1999;23:233–46. doi: 10.1016/s0896-6273(00)80776-x. [DOI] [PubMed] [Google Scholar]
- 5.Feinstein Y, Borrell V, Garcia C, Burstyn-Cohen T, Tzarfaty V, Frumkin A, Nose A, Okamoto H, Higashijima S, Soriano E, Klar A. F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development (Cambridge, England) 1999;126:3637–48. doi: 10.1242/dev.126.16.3637. [DOI] [PubMed] [Google Scholar]
- 6.Schubert D, Klar A, Park M, Dargusch R, Fischer WH. F-spondin promotes nerve precursor differentiation. Journal of neurochemistry. 2006;96:444–53. doi: 10.1111/j.1471-4159.2005.03563.x. [DOI] [PubMed] [Google Scholar]
- 7.Debby-Brafman A, Burstyn-Cohen T, Klar A, Kalcheim C. F-Spondin, expressed in somite regions avoided by neural crest cells, mediates inhibition of distinct somite domains to neural crest migration. Neuron. 1999;22:475–88. doi: 10.1016/s0896-6273(00)80703-5. [DOI] [PubMed] [Google Scholar]
- 8.Ho A, Sèudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2548–53. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoe HS, Wessner D, Beffert U, Becker AG, Matsuoka Y, Rebeck GW. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Molecular and cellular biology. 2005;25:9259–68. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyamoto K, Morishita Y, Yamazaki M, Minamino N, Kangawa K, Matsuo H, Mizutani T, Yamada K, Minegishi T. Isolation and characterization of vascular smooth muscle cell growth promoting factor from bovine ovarian follicular fluid and its cDNA cloning from bovine and human ovary. Archives of biochemistry and biophysics. 2001;390:93–100. doi: 10.1006/abbi.2001.2367. [DOI] [PubMed] [Google Scholar]
- 11.Terai Y, Abe M, Miyamoto K, Koike M, Yamasaki M, Ueda M, Ueki M, Sato Y. Vascular smooth muscle cell growth-promoting factor/F-spondin inhibits angiogenesis via the blockade of integrin alphavbeta3 on vascular endothelial cells. Journal of cellular physiology. 2001;188:394–402. doi: 10.1002/jcp.1122. [DOI] [PubMed] [Google Scholar]
- 12.Adams JC, Tucker RP. The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Developmental dynamics : an official publication of the American Association of Anatomists. 2000;218:280–99. doi: 10.1002/(SICI)1097-0177(200006)218:2<280::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Tzarfati-Majar V, Burstyn-Cohen T, Klar A. F-spondin is a contact-repellent molecule for embryonic motor neurons. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4722–7. doi: 10.1073/pnas.081062398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pèaèakkèonen K, Tossavainen H, Permi P, Rakkolainen H, Rauvala H, Raulo E, Kilpelèainen I, Gèuntert P. Solution structures of the first and fourth TSR domains of F-spondin. Proteins. 2006;64:665–72. doi: 10.1002/prot.21030. [DOI] [PubMed] [Google Scholar]
- 15.Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang JH. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. The Journal of cell biology. 2002;159:373–82. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta crystallographica Section D, Biological crystallography. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter JCW, Sweet RM, editors. Methods in Enzymology Macromolecular Crystallography, Part A. Vol. 276. Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 18.Tzarfaty-Majar V, Lâopez-Alemany R, Feinstein Y, Gombau L, Goldshmidt O, Soriano E, Muänoz-Câanoves P, Klar A. Plasmin-mediated release of the guidance molecule F-spondin from the extracellular matrix. The Journal of biological chemistry. 2001;276:28233–41. doi: 10.1074/jbc.M102585200. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Springer TA. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunological reviews. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang JH, Pepinsky RB, Stehle T, Liu JH, Karpusas M, Browning B, Osborn L. The crystal structure of an N-terminal two-domain fragment of vascular cell adhesion molecule 1 (VCAM-1): a cyclic peptide based on the domain 1 C-D loop can inhibit VCAM-1-alpha 4 integrin interaction. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5714–8. doi: 10.1073/pnas.92.12.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hèoèok M, Narayana SV. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. The EMBO journal. 2002;21:6660–72. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of molecular biology. 1999;285:2177–98. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. Journal of molecular biology. 1993;234:946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 24.Wang J. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends in biochemical sciences. 2002;27:122–6. doi: 10.1016/s0968-0004(01)02038-2. [DOI] [PubMed] [Google Scholar]
- 25.Aricescu AR, Jones EY. Immunoglobulin superfamily cell adhesion molecules: zippers and signals. Current opinion in cell biology. 2007;19:543–50. doi: 10.1016/j.ceb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence S, Love J, Colman D. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu Rev Neurosci. 2007;30:751–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- 27.Wang JH, Smolyar A, Tan K, Liu JH, Kim M, Sun ZY, Wagner G, Reinherz EL. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- 28.Davis SJ, Davies EA, Barclay AN, Daenke S, Bodian DL, Jones EY, Stuart DI, Butters TD, Dwek RA, van der Merwe PA. Ligand binding by the immunoglobulin superfamily recognition molecule CD2 is glycosylation-independent. The Journal of biological chemistry. 1995;270:369–75. doi: 10.1074/jbc.270.1.369. [DOI] [PubMed] [Google Scholar]
- 29.Davis SJ, Ikemizu S, Wild MK, van der Merwe PA. CD2 and the nature of protein interactions mediating cell-cell recognition. Immunological reviews. 1998;163:217–36. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 30.Engel J. Role of oligomerization domains in thrombospondins and other extracellular matrix proteins. The international journal of biochemistry & cell biology. 2004;36:997–1004. doi: 10.1016/j.biocel.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Tan K, Duquette M, Liu JH, Shanmugasundaram K, Joachimiak A, Gallagher JT, Rigby Alan C, Wang JH, Lawler J. Heparin-induced cis- and trans-Dimerization Modes of the Thrombospondin-1 N-terminal Domain. J Biol Chem. 2008;283:3932–3941. doi: 10.1074/jbc.M705203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capila I, Linhardt RJ. Heparin-protein interactions. Angewandte Chemie (International ed in English) 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Ha Y. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Molecular cell. 2004;15:343–53. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 34.Miao WM, Vasile E, Lane WS, Lawler J. CD36 associates with CD9 and integrins on human blood platelets. Blood. 2001;97:1689–96. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- 35.CCP4. The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica Section D, Biological crystallography. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis PJ. MOLSCRIPT: A Program to Produce Both Detailed and Schematic Plots of Protein Structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–96. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]


