Abstract
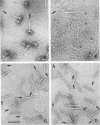
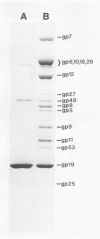
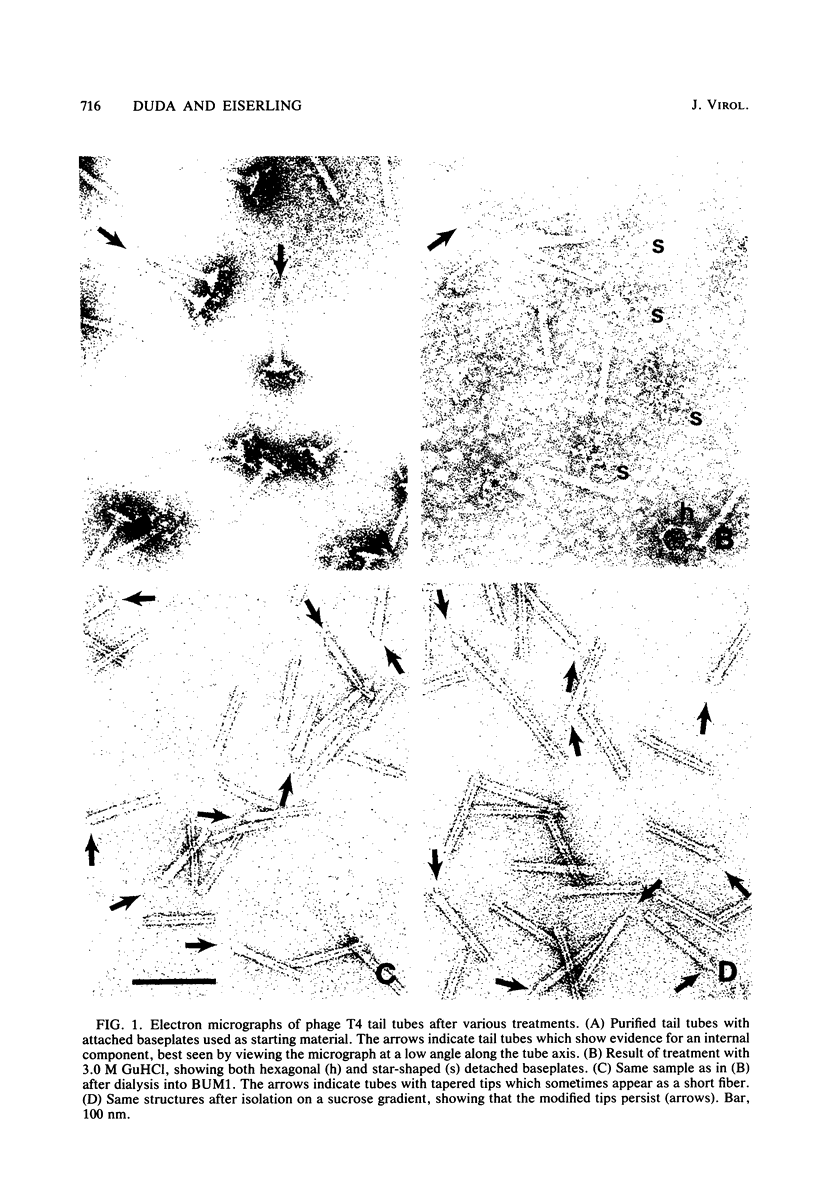
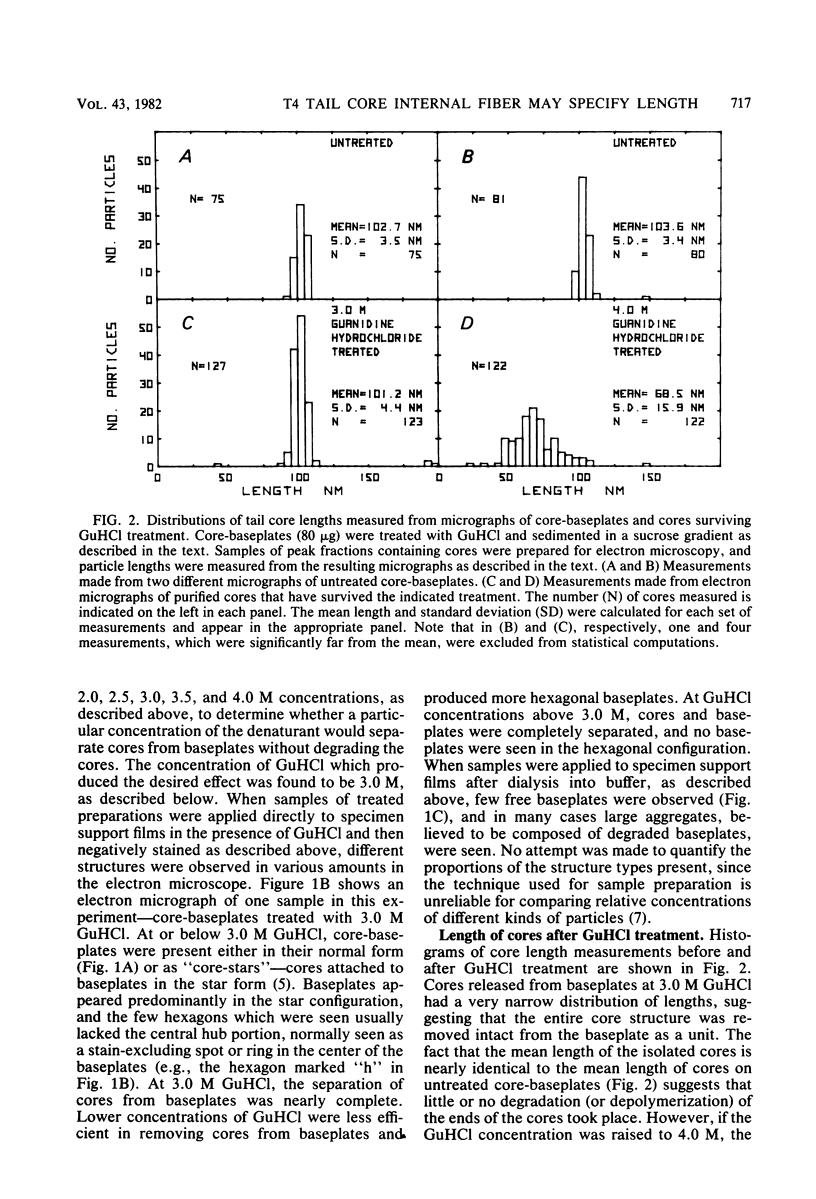
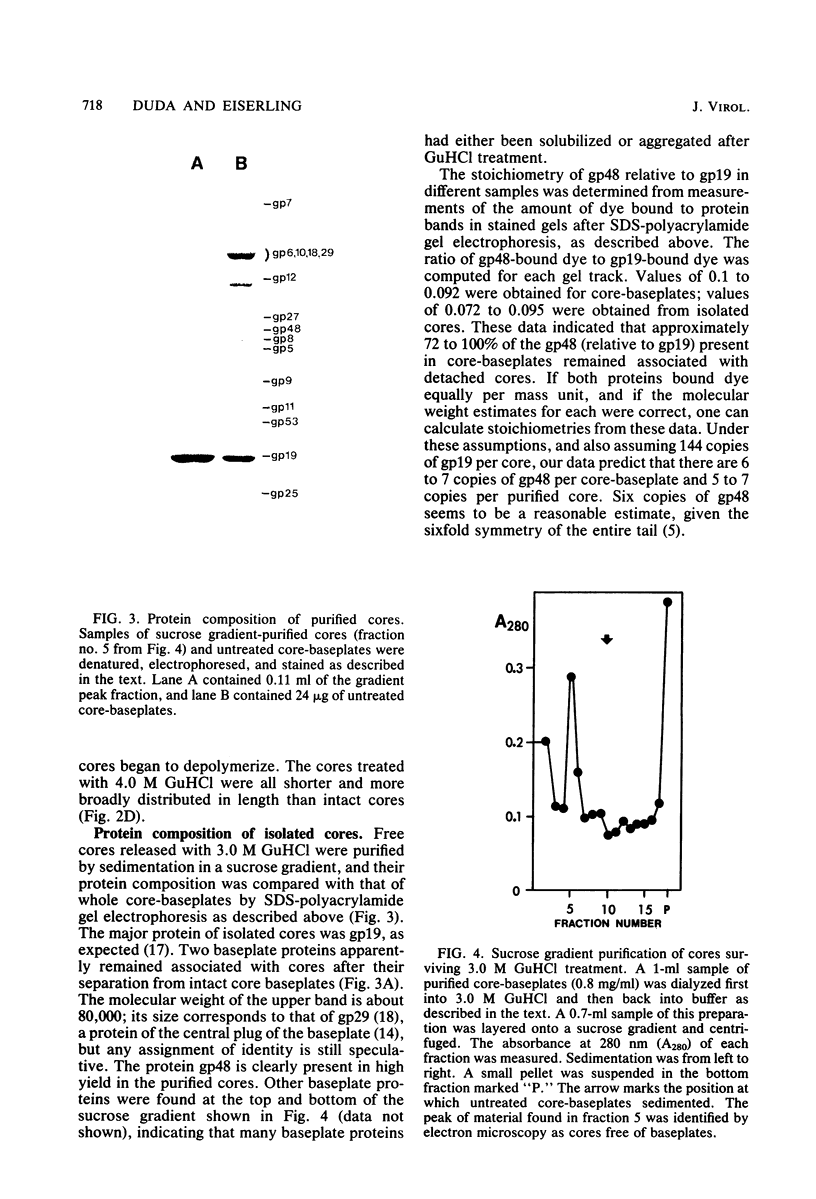
The length of the T4 tail is precisely regulated in vivo at the time of polymerization of the tail core protein onto the baseplate. Since no mutations which alter tail length have been identified, a study of in vivo-assembled tail cores was begun to determine whether the structural properties of assembled cores would reveal the mechanism of length regulation. An assembly intermediate consisting of a core attached to a baseplate (core-baseplate) was purified from cells infected with a T4 mutant in gene 15. When core-base plates were treated with guanidine hydrochloride, cores were released from baseplates. The released cores had the same mean length as cores attached to baseplates. Electron micrographs of these cores showed partial penetration of negative stain into one end, and, at the opposite end, a modified tip which often appeared as a short fiber projecting from the core. When cores were purified and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, two minor proteins and the major core protein were detected. One minor protein, the product of gene 48 (gp48), was present in at least 72% of the amount found in core-baseplates, relative to the amount of the major core protein. These findings suggest that cores contain a fibrous structure, possibly composed of gp48, which may form a "ruler" that specifies the length of the T4 tail.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berget P. B., Warner H. R. Identification of P48 and P54 as components of bacteriophage T4 baseplates. J Virol. 1975 Dec;16(6):1669–1677. doi: 10.1128/jvi.16.6.1669-1677.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. M., Eiserling F. A. T4 gene 40 mutants. II. Phenotypic properties. Virology. 1979 Aug;97(1):77–89. doi: 10.1016/0042-6822(79)90374-x. [DOI] [PubMed] [Google Scholar]
- Caspar D. L. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980 Oct;32(1):103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs D. H. Density gradient fractionation by piston displacement. Anal Biochem. 1975 Sep;68(1):95–101. doi: 10.1016/0003-2697(75)90683-1. [DOI] [PubMed] [Google Scholar]
- Crowther R. A., Lenk E. V., Kikuchi Y., King J. Molecular reorganization in the hexagon to star transition of the baseplate of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):489–523. doi: 10.1016/0022-2836(77)90081-x. [DOI] [PubMed] [Google Scholar]
- Cummings D. J. A remeasurement of the molecular weights of T-even bacteriophage substructural proteins. J Virol. 1972 Mar;9(3):547–550. doi: 10.1128/jvi.9.3.547-550.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J., Kellenberger E. Selective adsorption of particles to the supporting film and its consequences on particle counts in electron microscopy. Microsc Acta. 1972 Jul;72(2):119–130. [PubMed] [Google Scholar]
- Fenner C., Traut R. R., Mason D. T., Wikman-Coffelt J. Quantification of Coomassie Blue stained proteins in polyacrylamide gels based on analyses of eluted dye. Anal Biochem. 1975 Feb;63(2):595–602. doi: 10.1016/0003-2697(75)90386-3. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys J. 1980 Oct;32(1):139–153. doi: 10.1016/S0006-3495(80)84930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KELLENBERGER E., SECHAUD J. Electron microscopical studies of phage multiplication. II. Production of phage-related structures during multiplication of phages T2 and T4. Virology. 1957 Apr;3(2):256–274. doi: 10.1016/0042-6822(57)90092-2. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975 Dec 25;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol. 1975 Dec 25;99(4):673–694. doi: 10.1016/s0022-2836(75)80179-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975 Dec 25;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971 Jun 28;58(3):693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Bacteriophage T4 tail assembly: structural proteins and their genetic identification. J Mol Biol. 1973 Apr 5;75(2):315–337. doi: 10.1016/0022-2836(73)90024-7. [DOI] [PubMed] [Google Scholar]
- King J., Mykolajewycz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J Mol Biol. 1973 Apr 5;75(2):339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- Meezan E., Wood W. B. The sequence of gene product interaction in bacteriophage T4 tail core assembly. J Mol Biol. 1971 Jun 28;58(3):685–692. doi: 10.1016/0022-2836(71)90033-7. [DOI] [PubMed] [Google Scholar]
- Moody M. F., Makowski L. X-ray diffraction study of tail-tubes from bacteriophage T2L. J Mol Biol. 1981 Aug 5;150(2):217–244. doi: 10.1016/0022-2836(81)90450-2. [DOI] [PubMed] [Google Scholar]
- Poglazov B. F., Nikolskaya T. I. Self-assembly of the protein of bacteriophage T2 tail cores. J Mol Biol. 1969 Jul 14;43(1):231–233. doi: 10.1016/0022-2836(69)90094-1. [DOI] [PubMed] [Google Scholar]
- STEINBERG C. M., EDGAR R. S. A critical test of a current theory of genetic recombination in bacteriophage. Genetics. 1962 Feb;47:187–208. doi: 10.1093/genetics/47.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotton D. M., Burke B. E., Branton D. The molecular structure of human erythrocyte spectrin. Biophysical and electron microscopic studies. J Mol Biol. 1979 Jun 25;131(2):303–329. doi: 10.1016/0022-2836(79)90078-0. [DOI] [PubMed] [Google Scholar]
- To C. M., Kellenberger E., Eisenstark A. Disassembly of T-even bacteriophage into structural parts and subunits. J Mol Biol. 1969 Dec 28;46(3):493–511. doi: 10.1016/0022-2836(69)90192-2. [DOI] [PubMed] [Google Scholar]
- Tschopp J., Smith P. R. Extra-long bacteriophage T4 tails produced under in vitro conditions. J Mol Biol. 1977 Aug 5;114(2):281–286. doi: 10.1016/0022-2836(77)90211-x. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T., Bloomfield V. A. Bacteriophage T4 tail length is controlled by its baseplate. Biochem Biophys Res Commun. 1978 Jun 14;82(3):1049–1055. doi: 10.1016/0006-291x(78)90889-6. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T., Bloomfield V. A. In vitro polymerization of bacteriophage T4D tail core subunits. J Mol Biol. 1977 Nov 5;116(3):347–359. doi: 10.1016/0022-2836(77)90074-2. [DOI] [PubMed] [Google Scholar]