Abstract
Homeobox genes are an evolutionarily conserved class of transcription factors that are key regulators during developmental processes such as regional specification, patterning and differentiation. In this review, we summarize the expression pattern, loss-and/or gain-of-function mouse models, and naturally occurring mouse and human mutations of known homeobox genes required for the development of ectodermal appendages.
Keywords: homeobox genes, development, tooth, hair follicle, mammary gland, nail, ectodermal appendages
Introduction
Homeobox genes are characterized by a conserved 180-bp DNA sequence coding for a 60-aminoacid DNA-binding domain called the ‘homeodomain’. They function as transcription factors recognizing specific DNA sequences and regulate target genes (McGinnis and Krumlauf, 1992; Quiring et al., 1994). The first homeobox genes were identified in Drosophila but homologous genes were rapidly found in all animal species as well as in fungi and plants. There are two classes of homeobox genes. Class I genes, called Hox genes, share a high degree of identity in their homeodomain and are organized in clusters. In mammals, 39 Hox genes have been identified and are organized in four clusters located on four different chromosomes and labeled A, B, C and D. Individual genes within each cluster have been classified into 13 paralogous families on the basis of sequence similarity and chromosomal position within each linkage group (Scott, 1993). Interestingly, in both invertebrates and vertebrates, there is a correlation between the position of Hox genes in the cluster and their expression pattern along the anterior-posterior axis of the body, with genes located most 5’ in the cluster expressed most posteriorly while the more 3’ located genes are expressed in more anterior regions (Duboule and Dolle, 1989; Gaunt, 1988; Graham et al., 1989). Class II genes share a lower degree of identity in their homeodomain. They are grouped into subfamilies based on the presence of additional conserved functional domains besides the homeodomain.
Homeobox genes play a crucial roles in specifying cell identity and positioning during embryonic development, and mutations in these genes can cause dramatic developmental defects including loss of specific structures as well as changes in the identity of a body part or segment, which are known as ‘homeotic transformations’ (Lewis et al., 1999a).
In this review, we focus on the role of homeobox genes and summarize what is known on their function in the specification, patterning and differentiation of ectodermal appendages. Ectodermal appendages are structures derived during embryonic development from the interaction between the ectoderm and the underlying mesoderm. In mammals, ectodermal appendages include: tooth, hair, mammary gland, nail, sweat and sebaceous gland. Teeth, hair and mammary gland are among the organs in which a substantial amount of information on developmental regulation by homeobox genes has been accumulated in recent years. While only few studies reported the involvement of homeobox genes in nail development, the role of these transcription factors in sweat and sebaceous gland formation has not been documented. Figure 1 illustrates the principal stages of tooth, hair and mammary gland development and the effect of different knockouts of homeobox genes on these developmental processes. The phenotype resulting from the knockout of these genes is summarized in Table 1. Human diseases associated with mutations in homeobox genes and defects in ectodermal appendage formation are listed in Table 2.
Figure 1. Key steps in the development of three major ectodermal appendages.
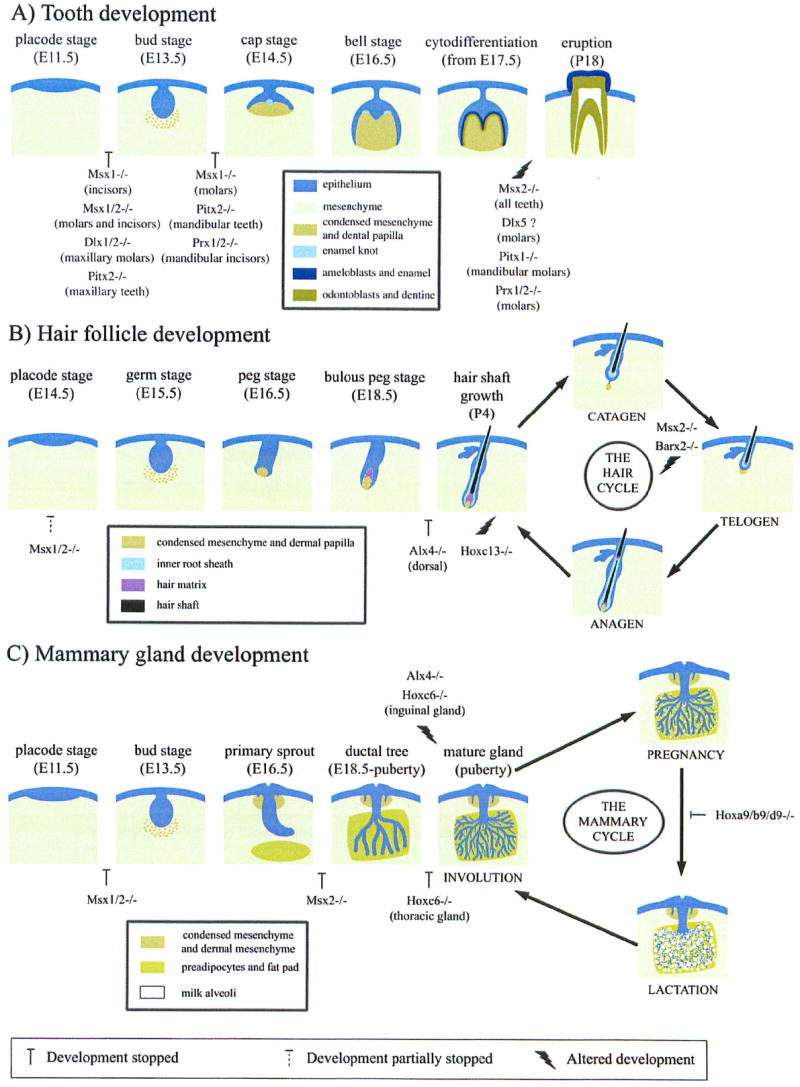
Schematic representation of tooth (A), hair follicle (B) and mammary gland (C) development. Stages are based on mouse embryonic (E) and postnatal (P) development. In A, stages correspond to the development of the first molar. In B, stages correspond to the development of body hair. Homeobox gene knockouts leading to either a halt in or an alteration of development at a specific stage are represented.
Table 1.
Knockout of genes expressed in ectodermal appendages
| Gene | Defects in knockout mice | Ref. | |
|---|---|---|---|
| TOOTH | Msx 1 | Lethal (after birth) - Molar development stops at bud stage while incisors development stops at lamina stage | (Satokata and Maas, 1994) |
| Msx2 | Development of all teeth but malformation of both molars and incisors | (Satokata et al., 2000) | |
| Msx1/Msx2 | Lethal (after birth) - Molar development stops at lamina stage | (Bei and Maas, 1998; Satokata et al., 2000) | |
| Dlx1 | Normal tooth development | (Qiu et al., 1997) | |
| Dlx2 | Normal tooth development | (Qiu et al., 1995) | |
| Dlx1/Dlx2 | Development of maxillary molar teeth stops at lamina stage - Ectopic cartilage | (Qiu et al., 1997; Thomas et al., 1997) | |
| Dlx3 | Embryos die at E9.5 | (Morasso et al, 1999) | |
| Dlx5 | Malformation of mandibular and maxillary molars | (Acampora et al, 1999; Depew et al, 1999) | |
| Pitx1 | Development of all teeth but abnormal morphology of mandibular molars | (Mitsiadis and Drouin, 2008) | |
| Pitx2 | Mandibular teeth development stops at bud stage while maxillary teeth development stops at placode stage | (Lin et al, 1999; Lu et al, 1999b) | |
| Prx1 | Normal tooth development | (Martin et al, 1995) | |
| Prx2 | Normal tooth development | (ten Berge et al, 1998) | |
| Prx1/Prx2 | Single or no mandibular incisor - Malformation of molars | (Lu et al, 1999a; Mitchell et al, 2006; ten Berge et al, 1998) | |
| Alx4 | Craniofacial defects without tooth defects | (Qu et al, 1997) | |
| Lhx6 | No tooth defect described | (Liodis et al, 2007) | |
| Lhx8 | Cleft palate but no tooth defects | (Zhao et al, 1999) | |
| Barx1 | No tooth defect described | (Kim et al, 2005; Kim et al, 2007) | |
| Tlx 1 | No tooth defect described | (Roberts et al, 1994) | |
| Isl1 (Islet1) | Embryos die at E10.5 | (Ahlgren et al, 1997; Cai et al, 2003; Pfaff et al, 1996) | |
|
| |||
| HAIR | Hoxc13 | Brittle hair resulting in alopecia | (Godwin and Capecchi, 1998) |
| Msx1 | No hair defects | (Satokata and Maas, 1994) | |
| Msx2 | Pelage loss at P14 (nude) – Regrowth – Loss of second pelage – Regrowth of a patchy pelage | (Satokata et al, 2000) | |
| Msx1/2 | Number of hair follicles reduced to one-third | (Satokata et al, 2000) | |
| Alx4 | Dorsal alopecia | (Qu et al, 1997) | |
| Barx2 | Extended first catagen resulting in shorter hair | (Olson et al, 2005) | |
|
| |||
| MAMMARY GLAND | Hoxc6 | Normal development until birth - Absence of ductal structure in adult thoracic mammary gland - Malformation of ductal structures in adult inguinal mammary gland | (Garcia-Gasca and Spyropoulos, 2000) |
| Hoxa9/b9/d9 | Hypoplasia of the mammary gland during pregnancy and lactation - Reduction of the amount of milk produced | (Chen and Capecchi, 1999) | |
| Msx1 | No mammary gland defects | (Satokata and Maas, 1994) | |
| Msx2 | Development stopped at the primary sprout stage | (Satokata et al, 2000) | |
| Msx1/2 | Development stopped at the placode stage | (Satokata et al, 2000) | |
| Alx4 | Altered branching morphogenesis of the ductal epithelium | (Joshi et al, 2006) | |
Table 2.
Mutations in human homeobox genes affecting development of ectodermal appendages
| Mutated Gene | Syndrome | Phenotype (teeth defects) | Ref. |
|---|---|---|---|
| MSX1 | Tooth agenesis | Absence of second premolars and third molars | (Vastardis et al., 1996) |
| MSX2 | Craniosynostosis | Skull malformation – no teeth defects | (Jabs et al., 1993; Wilkie et al., 2000) |
| RIEG/Pitx2 | Rieger syndrome | Dental hypoplasia | (Semina et al., 1996) |
| DLX3 | Tricho-dento-osseous | Enamel hypoplesia and taurodontism (enlargement of pulp chamber) | (Price et al., 1998) |
| Hair defects (kinky hair) | |||
| Occasionally brittle nails |
Homeobox genes involved in the patterning of the dentition and in tooth morphogenesis
The dentition is derived from the first branchial arch, where complex interactions between the stomodeal epithelium (derived from the ectoderm) and the underlying mesenchyme derived from cranial neural crest (CNC, migrating from the neurectoderm during early neurulation) drive the development of the appendages (Fig. 1A). The first sign of odontogenesis is the thickening and invagination of the stomodeal epithelium forming the dental placode. The dental placode then further invaginates into the surrounding dental mesenchyme to form the tooth bud. At that stage, the mesenchyme proliferates and condensates around the tooth bud. At the cap stage, the invaginated epithelium expands laterally and covers the condensed mesenchyme that will become the dental papilla. From that stage on, different structures can be distinguished in the dental epithelium, including the enamel knot (ek) which is known as an organizing center of tooth morphogenesis. At the late bell stage, cytodifferentiation starts: the cells of the internal enamel epithelium differentiating into ameloblasts (enamel-producing cells) while adjacent cells in the dental papilla differentiate into odontoblasts (dentine-producing cells). Mice develop two types of teeth (Fig. 2): incisors develop in the distal part of the jaw (one maxillary and one mandibular incisor on each side) while molars develop in the proximal part of the jaw (three maxillary and three mandibular molars on each side).
Figure 2. Expression of homeobox genes in the developing dentition.
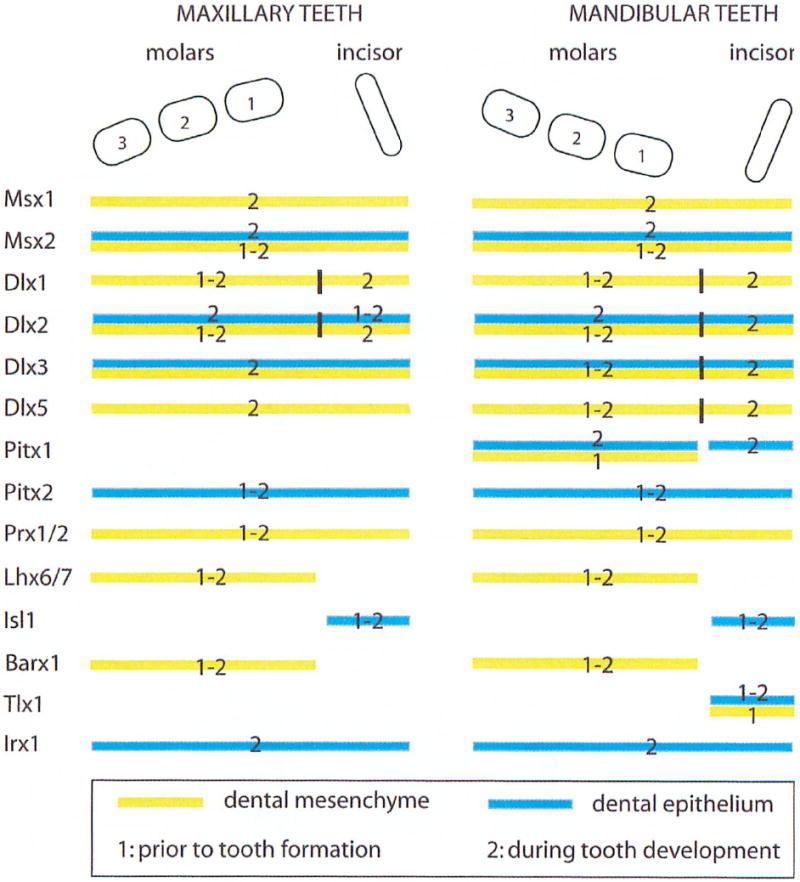
Diagram representing the distribution of homeobox genes during odontogenesis. Areas of expression are defined using three parameters: maxillary versus mandibular teeth, molars versus incisors, and epithelium versus mesenchyme. Since several markers are expressed in stomodeal tissues prior to tooth formation but have a different distribution after the initiation of tooth development, a temporal dimension was added to the diagram: distribution prior to tooth formation (1) and distribution during tooth development (2).
Although Hox genes are not expressed in the first branchial arch derivatives and are thus not involved in tooth development, several genes belonging to the class II homeobox families are expressed during odontogenesis where they are involved in both spatial and temporal control of dentition formation. When approaching the development of the dentition, one must not only consider the different stages of the development of an individual tooth but also what determines the nature (incisor or molar) of the tooth that is forming at a particular place. Moreover, even though teeth of mandibular (lower jaw) and maxillary (upper jaw) origin are histologically and morphologically identical, they involve different developmental pathways. In 1998, Thomas and Sharpe proposed that the patterning of murine dentition was determined by the complex and specific distribution of homeobox genes (“odontogenic homeobox code”) in the first arch mesenchyme prior to the initiation of tooth formation (Thomas and Sharpe, 1998). Several studies using in vitro recombination of dental tissues and culture of dental explants in the presence of purified secreted factor have shown that BMP4 is produced by the distal epithelium while FGF8 is produced by the proximal epithelium, and that the interplay between those two growth factors (inductions and inhibitions) is the main determinant of the expression pattern of homeobox genes in the dental mesenchyme prior to tooth formation (Bei and Maas, 1998; Grigoriou et al., 1998; Mandier and Neubuser, 2001; Mitsiadis et al., 2003; Thomas et al., 2000; Tucker and Sharpe, 2004; Tucker et al., 1998a; Tucker et al., 1998b; Vainio et al., 1993). Figure 1A and Table 1 illustrate and summarize the effects of different homeobox genes knockouts on tooth development. Figure 2 presents a diagram of the mouse dentition showing the expression domains (maxillary/mandibular, incisor/molars and epithelium/mesenchyme) of different homeobox genes.
Role of Msx genes in tooth development
Msx1 is expressed in the mesenchyme of all teeth, from the dental placode stage to the early bell stage (Mackenzie et al., 1991). Msx1-/- mice die after birth of major craniofacial defects. The development of the first and second molars is delayed and stops at the bud stage, while the development of incisors is not even initiated (Satokata and Maas, 1994). Msx1 seems to be critical for proliferation and fate determination of the cranial neural crest (CNC) which exhibit properties consistent with a neuronal fate in Msx1-/- mice (Han et al., 2003). The importance of MSX1 in human tooth development is supported by the discovery of a missense mutation in MSX1 which is responsible for a dominant negative agenesis of the second premolars and third molars (Vastardis et al., 1996). Another member of the Msx family, Msx2, is also expressed in the developing tooth. While Msx1 is exclusively expressed in the dental mesenchyme, Msx2 exhibits a dynamic expression in both the mesenchymal and epithelial compartments of all developing teeth (MacKenzie et al., 1992). Msx2 is first expressed in the mesenchyme in the area where the first molar will develop and is then localized in specific areas of the dental epithelium. At the late bell stage, Msx2 is expressed in both ameloblasts and odontoblasts. Msx2-/- mice develop all their teeth but both incisors and molars exhibit malformations and degenerate, which leads to an inability to chew solid food (Aioub et al., 2007; Satokata et al., 2000). Defects were shown in the structure of the enamel knot that does not express BMP4 in the absence of Msx2, and ameloblasts exhibit defective cell-cell contacts due to their inability to express laminin α3 subunit (Bei et al., 2004). Interestingly, when both Msx1 and Msx2 are knocked out, tooth development stops at the placode stage for both incisors and molars (Bei and Maas, 1998; Satokata et al., 2000). Thus, although Msx2 expression is not required for the initiation of tooth development, its early expression in the proximal mesenchyme probably compensates for the absence of Msx1 and enables the formation of the tooth bud in Msx1-/- mice (Satokata et al., 2000). In human, mutations in MSX2 and MSX2 haploinsufficiency were shown to cause skull malformations (craniosynostosis) but no defects in tooth formation (Jabs et al., 1993; Wilkie et al., 2000).
Role of Dlx genes in tooth development
Members of the Distal-less (Dlx) family of homeobox transcription factor are also involved in tooth morphogenesis. In mammals, there are six Dlx genes organized into three pairs of inverted, convergently transcribed genes, termed Dlx1-2, Dlx3-4 and Dlx5-6. Prior to tooth formation, Dlx1 and Dlx2 are expressed in the proximal mesenchyme of both mandibular and maxillary processes where molars will form, while Dlx2 is also detected in the distal epithelium of the maxillary process where incisors will develop (Thomas et al., 2000; Thomas et al., 1995; Thomas et al., 1997). At the same stage, Dlx5 and Dlx6 are expressed in the proximal mesenchyme of the mandibular process but not in the maxillary process (Qiu et al., 1997). Later, Dlx genes are expressed in all developing teeth, with a very complex expression pattern (Zhao et al., 2000). While Dlx1 is exclusively located in the dental mesenchyme of the developing tooth, Dlx2 follows a very dynamic expression pattern in both the dental mesenchyme and the dental epithelium. Although Dlx1-/- and Dlx2-/- mice have craniofacial defects with no alteration of tooth development (Qiu et al., 1997; Qiu et al., 1995), the deletion of both Dlx1 and Dlx2 results in the absence of maxillary molars whose development stops after epithelial thickening (placode stage), while incisors and mandibular molars have no defects (Qiu et al., 1997; Thomas et al., 1997). It is very interesting that although Dlx1 and Dlx2 are expressed in both mandibular and maxillary teeth, Dlx1/2 deletion affects specifically the teeth of the maxillary jaw. One explanation for the absence of defects in mandibular molars is a compensation by Dlx5 and Dlx6 which are expressed in the mandible but not in the maxillary prior to tooth formation. Interestingly, ectopic pieces of cartilage were detected in place of the missing molars, suggesting a change in the fate of a subpopulation of CNC in the absence of Dlx1 and Dlx2. As it is the case for Msx1, the redundant function of Dlx1 and Dlx2 could be to specify a distinct population of CNC. Although Dlx5 and Dlx6 are restricted to the proximal mesenchyme of the mandibular process before the first signs of tooth formation, they are expressed later in the dental mesenchyme of all teeth (Zhao et al., 2000). The dental phenotype of Dlx5-/- mice, which have multiple defects in craniofacial structures, has not been extensively described (Acampora et al., 1999; Depew et al., 1999). One of the two studies on Dlx5-/- mice reports that, although all teeth were formed, both maxillary and mandibular molars are malformed and have poorly mineralized crowns while both sets of incisors are shortened and misshapen (Depew et al., 1999). Prior to tooth formation, Dlx3 is detected in the distal epithelium of the mandible (Zhao et al., 2000). Subsequently, Dlx3 is detected in the mesenchyme as well as the epithelium of all developing teeth (Robinson and Mahon, 1994; Zhao et al., 2000). At the late bell stage, Dlx3 expression was seen in both ameloblasts and odontoblasts (Robinson and Mahon, 1994). Dlx3-/- mice die of placental defects and thus did not provide any information on the function of Dlx3 in tooth development (Morasso et al., 1999). However, a frameshift mutation in the C-terminal domain of Dlx3 has been associated to an ectodermal dysplasia called Tricho Dento Osseous (TDO) that is characterized by defects in hair, teeth and bone (Price et al., 1998). Teeth defects in TDO patients include enamel hypoplasia and taurodontism (enlargement of the pulp chamber) suggesting that Dlx3 is involved in the development of both epithelial- and mesenchymal-derived structures of the tooth. The roles of Dlx4 and Dlx6 have not been extensively studied.
Role of Pitx genes in tooth development
Pitx2 is a paired-like homeobox gene homologous to the human gene RIEG which is responsible for the Rieger syndrome, an autosomal dominant disorder whose features include dental hypoplasia (Semina et al., 1996). Pitx2 is expressed early in the proximal and distal stomodeal ectoderm of both mandibular and maxillar processes, before any sign of odontogenesis. Pitx2 expression is then localized at the sites of tooth formation and remains in the dental epithelium throughout tooth development (Mitsiadis et al., 1998; Mucchielli et al., 1997). Pitx2-/- mice exhibit severe craniofacial defects including an arrest of all mandibular teeth (incisors and molars) at the bud stage and an arrest of all maxillary teeth development at the placode stage (Lin et al., 1999; Lu et al., 1999b). Another member of the family, Pitx1 is expressed in the mesenchyme and epithelium of developing incisors and molars, exclusively in the mandible. Before the placode stage, Pitx1 is first expressed in the proximal mesenchyme of the mandible. At the placode stage, its expression is then transfered to the epithelium. Pitx1 is then detected in the epithelium of both molars and incisors, and its mesenchymal expression declines (Mitsiadis and Drouin, 2008; St Amand et al, 2000). Initial studies on Pitx1-/- mice reported no obvious defects in tooth morphogenesis at least until E14.5 which corresponds to the cap stage of the first molar (Lanctot et al., 1999; Szeto et al., 1999). A recent study revealed an altered morphology of mandibular teeth in Pitx1-/- mice, where molars form one cusp instead of two and the first and second molars tend to fuse (Mitsiadis and Drouin, 2008).
Role of paired-related homeobox genes in tooth development
The two paired-related homeobox genes, Prx1 and Prx2, are expressed in the mesenchyme of all teeth throughout odontogenesis (Cserjesi et al., 1992; Lu et al., 1999a; Martin et al., 1995; Mitchell et al., 2006; ten Berge et al, 1998). Although Prx1-/- and Prx2-/- mice develop normal teeth (Martin et al., 1995), when both genes are deleted dental abnormalities are observed. The first reports on Prx1/Prx2 knockout mice described defects in the development of mandibular incisors (Lu et al., 1999a; ten Berge et al., 1998). The distal mandible forms a single tooth bud that stops developing or in some cases develops as a single incisor. It is however unclear whether this phenotype reflects a direct function of Prx1/2 in mandibular incisor development or an indirect consequence of a shortening of the mandible. More recently, 3D reconstructions of molars from the same mice revealed that the absence of Prx1 and Prx2 results in defects in the morphology of mandibular and maxillary molars (Mitchell et al, 2006).
Aristaless-like 4 (Alx4), another paired-like homeodomain transcription factor, is expressed in the dental papilla of developing teeth (Hudson et al., 1998). Alx4-/- mice have several abnormalities including polydactyly (Qu et al., 1997). Moreover, a mouse strain with polydactyly known as Strong’s luxoid mutant (Alx41stD/1stD), was shown to harbor a mutation in Alx4 (Qu et al., 1998). Although both Alx4-/- mice and Strong’s luxoid mice have major skeletal defects including abnormal craniofacial structures, neither has been associated with teeth defects.
Role of LIM homeobox genes in tooth development
The two LIM homeobox genes Lhx6 and Lhx8 are expressed in the proximal mesenchyme of both maxillary and mandibular processes and are maintained in the mesenchyme of developing molars (Grigoriou et al., 1998). While Lhx6-/- mice have no craniofacial defects (Liodis et al., 2007), Lhx8-/- mice exhibit a cleft palate with no defects in other craniofacial structures (Zhao et al., 1999).
Isl1 (Islet-1), another member of the LIM homeobox family, is also specifically expressed in developing incisors, in both mandibular and maxillary processes. Isl1 expression is restricted to the dental epithelium throughout tooth development and a strong expression is maintained in differentiated ameloblasts (Mitsiadis et al., 2003). Unfortunately, since Isl1-/- mice die at E10.5, no further role in dentition could be assessed (Ahlgren et al., 1997; Cai et al., 2003; Pfaff et al., 1996).
Other homeobox genes expressed during tooth development
The homeobox gene Barx1 is expressed in the proximal mesenchyme of the mandibular and maxillary processes and is maintained in the dental mesenchyme of all molars throughout their development while incisors develop in a Barx1-negative area (Mitsiadis et al., 1998; Tissier-Seta et al., 1995). Interestingly, when the expression domain of Barx1 is extended to the distal mesenchyme, molars develop in place of incisors (Tucker et al., 1998b). This phenomenon was shown using Noggin beads to inhibit BMP4, which was shown to repress Barx1 expression in the proximal mesenchyme. However, in this experiment, Barx1 is just an indicator of the extension of the molar-inducing mesenchyme in the distal part of the jaw, and only one of the many genes whose expression is repressed by BMP4 in the distal mesenchyme. Moreover, the fact that Barx1-/- mice exhibit stomach and intestine defects but no alteration of tooth development suggest that Barx1 does not play a major role or at least has a redundant role in odontogenesis (Kim et al., 2005; Kim et al., 2007).
The T-cell leukemia homeobox containing gene Tlx1, is first expressed in the distal mesenchyme of the mandibular component of the first branchial arch before appearing in the dental epithelium at the site where lower incisors will form. From the placode stage to the bell stage, Tlx1 expression is restricted to the dental epithelium of mandibular incisors (Raju et al., 1993). Inhibition of BMP4 using Noggin beads was shown to shut down Tlx1 expression in the distal mesenchyme where molars form in place of incisors (Tucker et al., 1998b). However, since the only defect observed on Tlx1-/- mice is the absence of spleen, Tlx1 does not seem to be indispensable for incisor development (Roberts et al., 1994).
Irx1, a member of the Iroquois family of homeobox transcription factors, is expressed in the dental epithelium of all developing teeth from the placode to the cap stage. However, Irx1 is not required for tooth morphogenesis (Ferguson et al., 2001)
Homeobox genes involved in hair follicle development
As for other ectodermal appendages, hair follicle (HF) morphogenesis involves a series of reciprocal and sequential interactions between the ectoderm and underlying mesenchyme. The initial step is the formation of the hair placode, in response to inductive (Wnt) and inhibitory (BMP) signals. After the induction of the dermal condensate, there is subsequent epithelial proliferation and down-growth that progresses to form larger hair germs, and later hair pegs (Fig. 1B). The specialized cells in the mesenchyme (dermal condensate) become the dermal papilla (DP) cells, that remains engulfed and associated with the overlaying epithelial matrix cells, which undergo an upward differentiation process to give rise to the different HF compartments of the hair shaft and the inner root sheath (IRS). The growing hair shaft is surrounded by the keratinocyte-derived outer root sheath (ORS) (Fuchs, 2007; Millar, 2002). The hair matrix cells are derived from epithelial stem cells located in the bulge region of the HF (Cotsarelis et al., 1990; Levy et al., 2005; Oshima et al., 2001; Taylor et al., 2000).
After the initial follicle morphogenesis, the HF continues in growth phase (anagen) generating the first pelage approximately 2 weeks after birth. The length of the hair is determined by the length of the anagen phase. The HF undergoes cyclic transformations, with the anagen phase terminating with the remodeling catagen process (regression) followed by a resting telogen phase. After this, a new anagen phase reinitiates to start a new cycle of hair growth making the HF an ideal system to allow the study of essential stages of proliferation, regression and regeneration that occur throughout mammalian life (Hardy, 1992; Schmidt-Ullrich and Paus, 2005) (Fig 1B). Cyclical postnatal renewal of the hair is thought to recapitulate some of the signaling and regulatory mechanisms found between the DP and overlying epithelial cells during the embryonic onset of hair formation (Hardy, 1992; Oliver and Jahoda, 1988; Schmidt-Ullrich and Paus, 2005). Figure 1B and Table 1 illustrate and summarize the effects of different homeobox gene knockouts on HF development.
Hox genes and hair follicle development
Hox genes from all four clusters are expressed in mammalian skin and present differential spatial and temporal expression in the stratified epidermis, in the developing HF as well as in the sebaceous gland and the sweat gland. Findings show that while H0XA4, H0XA7 and HOXC4 are detected in all epidermal layers of the HF, HOXA5 is restricted to the IRS (Awgulewitsch, 2003; Stelnicki et al., 1997; Stelnicki et al., 1998). HOXC8 localizes to the DP in developing and cycling hair and two HoxD genes, D9 and D11 are expressed in the epidermal peg of the hair germ, the matrix, hair shaft and the ORS of the developing caudal hair. In the mouse, Hoxd13 transcripts are the last to be detected and are limited to the hair matrix cells of the caudal pelage HFs (Kanzier et al., 1994).
Hoxc12 expression is restricted to the epithelial part of the HF excluding the ORS and interfollicular epidermis. During anagen, Hoxc12 overlaps in the upper-most part of the bulb with the more proximally expressed Hoxc13 (Shang et al., 2002).
The spatio-temporal expression of other Hox family members would lead to propose distinct functions during HF development and cycling. As for other structures such as the limb buds, there might exist a complex code of transcriptional regulation that is required for HF patterning and differentiation. Despite the expression patterns of Hox genes in the HF, only the analysis of Hoxc13 loss- and/or gain-of-function mouse models has shown this gene to play an essential role in HF development. As embryonic hair morphogenesis reaches completion (post natal day 9, P9), expression of Hoxc13 is detected in the hair shaft forming compartments of the matrix, cuticle, cortex and medulla (Godwin and Capecchi, 1998; Langbein and Schweizer, 2005; Potter et al., 2006). Hoxc13 directly regulates expression of the hair keratin genes, hair-specific keratin-associated proteins (KAPs) and the transcription factors Foxq1 (Jave-Suarez et al., 2002; Potter et al., 2006; Pruett et al., 2004), and its role in hair development is supported by the findings that Hoxc13-deficient or overexpressing mice show hair growth and differentiation defects that result in a hairless phenotype (Godwin and Capecchi, 1998; Tkatchenko et al., 2001).
Role of Msx genes in hair follicle development
During mouse embryogenesis, Msx1 and Msx2 are expressed in the HF placode ectoderm. When the HF begins the downward growth, both genes are detected in the DP, the IRS and the developing overlying epithelium. Subsequently Msx1 and Msx2 are co-expressed in the epithelial matrix cells suggesting they may play overlapping roles (Noveen et al., 1995; Satokata and Maas, 1994; Stelnicki et al., 1997). Msx2 is known to play a role in maintaining anagen phase and Msx2-/- mice show progressive hair loss, with successive cycles of re-growth and loss, accounting for a cyclic alopecia phenotype (Ma et al., 2003; Satokata et al., 2000). Although Msx1-/- mice present a variety of craniofacial defects, development of the hair occurs normally (Satokata and Maas, 1994). The Msx1/2 double mutant have reduced number of HFs indicating that these genes play important roles in promoting placode fate (Satokata et al., 2000).
Other homeobox genes expressed in the developing hair follicle
No hair phenotype has been reported for Dlx1, Dlx2 or Dlx1/2 mutants. Interestingly, Dlx3 is expressed in the hair matrix cells of the HF (Morasso et al., 1995; Robinson and Mahon, 1994). Although early embryonic lethality of Dlx3-/- mice precluded the analysis of its role in hair development, mutations in human DLX3 lead to Tricho-Dento-Osseous syndrome, an ectodermal dysplasia that is characterized by hair, tooth and bone defects (Price et al., 1998) indicating a role for this homeobox in hair development.
As mentioned earlier, Alx4 is a paired-like homeodomain transcription factor that is expressed in the mesenchymal compartment of several ectodermal appendages, including in the DP of developing HF (Hudson et al., 1998). Contrary to other organs where Alx4 is exclusively expressed in tissues of mesenchymal origin, developing HFs express Alx4, not only in the DP, but also in parts of the ORS. Interestingly, one of the phenotypes of the Strong’s luxoid mutant (Alx41stD/1stD) is dorsal alopecia (Qu et al., 1998), demonstrating that Alx4 plays a crucial role in HF development. Moreover, the fact that only dorsal pelage is affected suggests that Alx4 function is compensated for by other factors in some parts of the body.
The homeodomain Lhx2 is expressed in the postnatal bulge compartment, and studies show that Lhx2 functions to specify and maintain the HF stem cell character but does not have a role in their differentiation (Andl et al., 2002; Rhee et al., 2006).
The Otx genes, Otx1 and Otx2, known to play a crucial role in the development of the brain, show expression in the head epithelia where their domains overlap and persist during the development of the hair vibrissae follicles.
Initial studies on the expression of ovine Barx2 in the ORS of wool follicles suggested a possible role in hair biology (Sander et al., 2000). Barx2 was reported to be absent from all other HF compartments, i.e. IRS and dermal cells associated with the follicle. More recently, the generation of Barx2-/- mice have corroborated its function in hair development, with Barx2-deleted mice presenting defects in initiation and progression of catagen, resulting in an extended first catagen that leads to short hair in mutant mice (Olson et al., 2005).
Another non-Hox homeobox that has been identified in the HF is M0X1. MOX1 expression is confined to cells of the ORS during HF development, however its expression is completely lost in adults. These findings support a role for MOX1 in the initial growth and development events, but indicate lack of function in the maintenance of hair growth in adulthood (Stelnicki et al., 1997).
Homeobox genes involved in the development of the mammary gland
The development of the mammary gland, which is highly controlled by estrogen and testosterone, can be divided into a linear phase starting early during development and continuing after birth until females reach puberty, and a cyclic phase related to the estrous cycle initiated every time females become pregnant (Figure 1C). In mouse, the first sign of MG development appears around embryonic day 11.5 (E11.5) with two lines of thickened epithelium forming ventrally between the limb buds on both sides of the embryo. Then, the underlying mesenchyme locally induces further thickening and invagination of the epithelium forming the mammary bud (E13.5) and further elongates to form the primary sprout (E16.5). There are two distinct mesenchymal compartments in the mouse fetal MG: a dense mesenchyme consisting of layers of fibroblasts of dermal origin, and a less dense mesenchyme composed of preadipocytes. By E18.5, the mammary epithelium has branched into the deeper adipocyte mesenchyme that will later be called the stromal fat pad. At birth and during all the period preceding the onset of puberty, the MG contains only rudimentary ductal network that will develop with the onset of ovarian functions. Indeed, at the onset of puberty, the ductal network proliferates and sends ramifications into the fat pad. This phase of MG development called ductal growth is dependent on the presence of estrogen and is completed by the time females reach sexual maturity. Further changes in MG morphology and physiology occur only with pregnancy and follow a defined cycle that can be divided into three phases: 1) During pregnancy, lobular-alveolar structures develop from the existing ductal system in a process that is driven by pregnancy-induced hormones and estrogen. 2) During lactation, the lobular system grows and differentiates to form alveoli in which milk protein synthesis takes place. 3) During involution, period following the weaning of the pups, the MG undergoes extensive remodeling, leading to a selective loss of alveolar structures, in a process involving large-scale apoptosis. Figure 1C and Table 1 summarize the different stages of MG development and illustrates the effect of different gene knockouts in this process.
Hox genes involved in mammary gland development
Members of all four clusters the Hox family are expressed in the developing MG with some of them playing essential roles (Friedmann et al., 1994; Lewis, 2000). The expression of Hoxc6 has been analyzed most extensively. Hoxc6 was detected in glands from pubescent and mature virgin animals but downregulated during pregnancy and lactation. In response to ovariectomy, Hoxc6 transcript levels were substantially elevated compared to glands from intact virgin mice, suggesting that Hoxc6 expression is downregulated by ovarian secretions (Friedmann et al., 1994). While in intact females Hoxc6 is primarily expressed in the epithelial compartment of the MG, ovariectomized females express Hoxc6 in both the stroma and the ductal epithelium, suggesting that ovarian hormones are involved in the regulation of Hoxc6 expression and distribution (Garcia-Gasca and Spyropoulos, 2000). The phenotype of Hoxc6-/- mice demonstrates the importance of this Hox gene in the spatial and temporal regulation of MG development (Garcia-Gasca and Spyropoulos, 2000). Newborn Hoxc6-/- mice exhibit slightly underdeveloped mammary epithelium, with limited ductal branching. However, these defects are relatively subtle when compared to the phenotype observed in the adult mice. Interestingly, thoracic and inguinal MGs exhibit profoundly distinct phenotypes. Thoracic MGs of adult Hoxc6-/- mice are devoid of mammary epithelium (absence of ductal structures), while inguinal glands develop ductal structures that are dilated and variably orientated, demonstrating a position specific role for Hoxc6 in MG development. The malformed inguinal mammary epithelium does not regress in response to ovariectomy, suggesting that these glands are not responsive to ovarian hormone signal.
The paralogous Hox genes, Hoxa9, Hoxb9 and Hoxd9 give a nice example of genes playing a redundant role in the cyclic phase of MG development and physiology, without affecting the linear phase preceding pregnancy. The three paralogous genes are expressed in the mammary primordia of E12.5 embryos as well as in the MG of adult virgin females, pregnant females and females in lactation (Chen and Capecchi, 1999). Similarly to mice lacking each of the three Hoxa9, Hoxb9 and Hoxd9 genes, and triple knockout mice did not show any defect in MG development and adult virgin mice seem to have normally structured glands. However, during and after pregnancy, triple mutant females exhibit an hypoplasia of the mammary tissue with a reduction of the quantity of milk produced, resulting in the death of the pups (Chen and Capecchi, 1999).
Hoxb7 is expressed in MG of adult virgin mice, while its expression is downregulated during lactation (Srebrow et al., 1998). However, Hoxb7 knockout mice have no defects in MG development and physiology (Chen et al., 1998).
Role and regulation of Msx1 and Msx2 in mammary gland development
At E13.5 of mouse embryonic development, both Msx1 and Msx2 were localized in the epithelium of the mammary bud (Phippard et al., 1996) while in newborn mice Msx1 expression is downregulated and Msx2 expression switches to the mesenchyme (Satokata et al., 2000). Both Msx1 and Msx2 are expressed in the MG of virgin females and during early pregnancy, but their expression is down-regulated during lactation and increases again in the involuting gland (Friedmann and Daniel, 1996; Phippard et al., 1996). Msx2 expression was shown to be up-regulated by estrogen while the expression of Msx1 does not depend on ovarian hormones (Friedmann and Daniel, 1996; Phippard et al., 1996). More recently, progesterone was also shown to induce Msx2 expression (Satoh et al., 2007). However, different groups report diverse descriptions of the expression pattern of Msx1 and Msx2 in the adult MG. On the one hand, Friedmann and Daniel report that Msx2 is expressed in the mesenchymal compartment while Msx1 is localized in the epithelial compartment in the MG of both virgin and pregnant females (Friedmann and Daniel, 1996). On the other hand, Phippard et al. find that Msx2 is expressed in the epithelium while Msx1 is found in both the epithelium and the stroma (Phippard et al., 1996). More recently, a third group found the majority of Msx2 expression is in the epithelium and lesser amounts in the stromal cells near the ductal epithelium from 12-week-old mice (Satoh et al., 2004; Satoh et al., 2007). These discrepancies remain to be clarified.
Msx2-/- mice fail to develop MGs that are arrested at the mammary sprout stage, suggesting that Msx2 is involved in the promotion of branching morphogenesis (Satokata et al., 2000). Msx1 deficiency in mice does not cause any defect in MG development (Satokata and Maas, 1994). However, when both Msx1 and Msx2 are deleted, epithelium invagination fails and MG development stops at the placode stage (Satokata et al., 2000). Thus, although Msx1 is not required for MG development, its early expression in the mammary epithelium probably compensates for the absence of Msx2 and enables the formation of the mammary bud and sprout in Msx2-/- mice. As described earlier, such redundant function of Msx1 and Msx2 is also observed during early tooth development (Bei and Maas, 1998; Satokata et al., 2000). However, in the case of tooth development the roles are inverted: Msx1-/- mice exhibit an arrest at the bud stage while Msx2-/- mice develop teeth that are defective.
Paired-like homeodomain transcription factors and mammary gland development
Alx4 is strongly expressed in a subpopulation of mesenchymal cells adjacent to the ductal epithelium (near the end buds) in the MG of pubescent mice (Hudson et al., 1998). MG morphogenesis in Strong’s luxoid mutant mice (Alx41stD/1stD) has recently been analyzed (Joshi et al., 2006). During puberty, these mice exhibit major defects in mammary epithelial ductal elongation and branching morphogenesis, demonstrating that Alx4 in the mammary stroma is required for normal development of the ductal epithelia. Otex, another member of the paired-like class is specifically expressed in tissues targeted by steroid hormones including the MG (Geserick et al., 2002).
Other homeobox genes expressed the in mammary gland
Other homeobox genes have been shown to be expressed in the MG, but until now, no reports show that they play an essential role in breast morphogenesis. Irx2, a member of the Iroquois family of homeobox genes, is differentially expressed in major mammary epithelial cell lineages and its expression is dynamic thoughout both the linear and cyclic phases of MG development (Lewis et al., 1999b). The homeobox gene Hex is expressed in epithelial cells of the end buds, and its nuclear expression is up-regulated during lactation (Puppin et al, 2006). The expression of Barx2 in the MG has been reported based on northern blot data but no description of its expression pattern in this organ has been provided (Krasner et al, 2000).
Homeobox genes involved in the development of nail and other ectodermal appendages
The mechanism driving nail development is poorly documented when compared to teeth, hair and MG. Only a few studies have shown the involvement of homeobox genes in the nail development.
Hoxc13 is expressed in digit tips starting at E13.5 and is maintained precisely in the area of nail formation throughout development (Godwin and Capecchi, 1998). Hoxc13-/- mice develop malformed nails that have a flatter appearance when compared to wild type nails, and tend to overgrow and form long spirals, supporting a role for Hoxc13 in nail differentiation (Godwin and Capecchi, 1998). Given that hair and nail are both keratinized structures, it is likely that the defects observed in Hoxc13-/- mice for these two organs involve similar mechanisms. However, although several studies have been carried out to investigate further the role of Hoxc13 in HF development (Jave-Suarez et al., 2002; Pruett et al., 2004), none of these studies have included a parallel with nail development.
Msx1 and Msx2 are expressed in the developing digit tips where nails are formed (Reginelli et al., 1995). Msx1 is expressed exclusively in the mesenchyme, while Msx2 is found in both the mesenchyme and the epithelium in a more distal region of the digit tip when compared to Msx1. Digit amputations performed on both fetal and neonatal mice showed that full digit regeneration (including nail) could be obtained only when the amputation plane contained Msx1-positive cells, even in the absence of Msx2-expressing cells (Reginelli et al., 1995). These observations suggest that Msx1-positive cells act like an organizing center of digit tip development. However, Msx1-/- mice do not show any defects in digit formation (Satokata and Maas, 1994). On the other hand, Msx2-/- mice have enlarged nails (Satokata et al., 2000), suggesting a role for Msx2 in nail differentiation. No report has been published on Msx1/Msx2 double knockout regarding nail structure.
Dlx3 was also shown to be expressed in digit tips, at the place where nails form (Morasso et al, 1995). Since Dlx3-/- mice die of placental defects, they did not provide any information on the function of Dlx3 in nail development (Morasso et al., 1999). However, patients with TDO syndrome (mutation in Dlx3) occasionally have brittle nails, suggesting that Dlx3 is involved in nail differentiation (Price et al., 1998).
Lmx1B is a LIM-homeodomain protein expressed in the dorsal mesenchyme of the developing limb bud where it makes a sharp dorsoventral boundary that is maintained throughout development (Riddle et al., 1995; Vogel et al., 1995). Lmx1B has been shown to be essential for the formation of dorsal limb structures. Indeed, Lmx1B-/- mice have abnormalities that are similar to the clinical features seen in the Nail Patella Syndrome (NPS), including hypoplastic nails and absence of patella (Dreyer et al., 1998). Since then, NPS has been associated to different mutations in Lmx1B (Dunston et al., 2005; Sweeney et al., 2003). However, the nail phenotype observed in Lmx1B-/- mice as well as in patients with NPS is not due to a specific role of Lmx1B in nail development but more generally to a role of Lmx1B in dorso-ventral determination of limb development.
Ectodermal appendages also include sebaceous glands that are associated to HFs and sweat glands that are essentially localized in the foot pads in mice. The role of homeobox genes in the development of these two appendages has been barely documented. Although the initial characterization of the expression of different Hox genes in skin development included the expression pattern of theses genes in sebaceous and sweat glands (Stelnicki et al., 1998), their role in the morphogenesis of these structures has not been investigated. In addition to having defects in hair cycling, Barx2-/- mice exhibit an enlarged sebaceous gland (Olson et al., 2005). Hoxc13 is strongly expressed in the foot pads were sweat glands are concentrated (Godwin and Capecchi, 1998). However, the role of Hoxc13 in the development and physiology of the sweat gland remains to be addressed.
Conclusion
The generation of distinct tissues often employs common developmental regulatory factors and pathways. This is particularly evident in the development of organs dependent on the invagination of ectodermally-derived epithelial cells into the underlying mesenchyme. A general function of homeobox genes during embryonic development is to specify cell identity and positioning. Although teeth, HFs and MGs go through similar morphogenetic processes during the early steps of their formation (placode and bud stages), few homeobox genes are expressed in all ectodermal appendages. Homeobox genes expressed in a specific type of ectodermal appendage can also have different involvements in specific subtypes of this appendage (ie: different types of teeth). The dentition, in which homeobox genes can play a role in the specific determination of incisors versus molars while they can also discriminate between maxillary and mandibular teeth, gives a nice example of the role of homeobox genes in deciding where to form a specific structure.
Functional redundancy has been demonstrated for some homeobox genes expressed in ectodermal appendages: Dlx1/2, Prx1/2, Hoxa9/b9/d9, as well as Msx1/2 at early stages of organogenesis. The fact that the number of human diseases currently linked to mutations in homeobox genes is low also reflects a certain degree of functional redundancy between family members. When approaching the role of some specific homeobox gene in any developmental process, one should keep in mind that they act in concert with other homeobox genes. The patterning of the embryo results from a complex network of overlapping expression of homeobox genes. For that reason, it is difficult to address the role of individual homeobox genes in embryonic development, and especially to identify direct target genes that are acting downstream of that same gene.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126(17):3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385(6613):257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Aioub M, Lezot F, Molla M, Castaneda B, Robert B, Goubin G, Nefussi JR, Berdal A. Msx2 -/- transgenic mice develop compound amelogenesis imperfecta, dentinogenesis imperfecta and periodental osteopetrosis. Bone. 2007;41(5):851–859. doi: 10.1016/j.bone.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Developmental Cell. 2002;2(5):643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Awgulewitsch A. Hox in hair growth and development. Naturwissenschaften. 2003;90(5):193–211. doi: 10.1007/s00114-003-0417-4. [DOI] [PubMed] [Google Scholar]
- Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125(21):4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- Bei M, Stowell S, Maas R. Msx2 controls ameloblast terminal differentiation. Developmental Dynamics. 2004;231(4):758–765. doi: 10.1002/dvdy.20182. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(2):541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Greer J, Capecchi MR. Analysis of Hoxa7/Hoxb7 mutants suggests periodicity in the generation of the different sets of vertebrae. Mechanisms of Development. 1998;77(1):49–57. doi: 10.1016/s0925-4773(98)00126-9. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61(7):1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115(4):1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126(17):3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nature Genetics. 1998;19(1):47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO Journal. 1989;8(5):1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunston JA, Lin S, Park JW, Malbroux M, Mclntosh I. Phenotype severity and genetic variation at the disease locus: an investigation of nail dysplasia in the nail patella syndrome. Annals of Human Genetics. 2005;69(Pt 1):1–8. doi: 10.1046/j.1529-8817.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Heikinheimo K, Nomura M, Oh P, Li E, Sharpe PT. The role of effectors of the activin signalling pathway, activin receptors IIA and IIB, and Smad2, in patterning of tooth development. Development. 2001;128(22):4605–4613. doi: 10.1242/dev.128.22.4605. [DOI] [PubMed] [Google Scholar]
- Friedmann Y, Daniel C. Regulated expression of homeobox genes Msx-1 and Msx-2 in mouse mammary gland development suggests a role in hormone action and epithelial-stromal interactions. Dev Biol. 1996;177(1):347–355. doi: 10.1006/dbio.1996.0168. [DOI] [PubMed] [Google Scholar]
- Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Research. 1994;54(22):5981–5985. [PubMed] [Google Scholar]
- Fuchs E. Scratching the surface of skin development. Nature. 2007;445(7130):834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gasca A, Spyropoulos DD. Differential mammary morphogenesis along the anteroposterior axis in Hoxc6 gene targeted mice. Developmental Dynamics. 2000;219(2):261–276. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1048>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ. Mouse homeobox gene transcripts occupy different but overlapping domains in embryonic germ layers and organs: a comparison of Hox-3.1 and Hox-1.5. Development. 1988;103(1):135–144. doi: 10.1242/dev.103.1.135. [DOI] [PubMed] [Google Scholar]
- Geserick C, Weiss B, Schleuning WD, Haendler B. OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochemical Journal. 2002;366(Pt 1):367–375. doi: 10.1042/BJ20020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes & Development. 1998;12(1):11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development. 1998;125(11):2063–2074. doi: 10.1242/dev.125.11.2063. [DOI] [PubMed] [Google Scholar]
- Han J, Ito Y, Yeo JY, Sucov HM, Maas R, Chai Y. Cranial neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19(INK4d) expression during odontogenesis. Developmental Biology. 2003;261(1):183–196. doi: 10.1016/s0012-1606(03)00300-2. [DOI] [PubMed] [Google Scholar]
- Hardy MH. The secret life of the hair follicle. Trends in Genetics. 1992;8(2):55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- Hudson R, Taniguchi-Sidle A, Boras K, Wiggan O, Hamel PA. Alx-4, a transcriptional activator whose expression is restricted to sites of epithelial-mesenchymal interactions. Developmental Dynamics. 1998;213(2):159–169. doi: 10.1002/(SICI)1097-0177(199810)213:2<159::AID-AJA1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75(3):443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- Jave-Suarez LF, Winter H, Langbein L, Rogers MA, Schweizer J. HOXC13 is involved in the regulation of human hair keratin gene expression. Journal of Biological Chemistry. 2002;277(5):3718–3726. doi: 10.1074/jbc.M101616200. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Chang H, Hamel PA. Loss of Alx4, a stromally-restricted homeodomain protein, impairs mammary epithelial morphogenesis. Developmental Biology. 2006;297(1):284–294. doi: 10.1016/j.ydbio.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Kanzier B, Viallet JP, Le Mouellic H, Boncinelli E, Duboule D, Dhouailly D. Differential expression of two different homeobox gene families during mouse tegument morphogenesis. International Journal of Developmental Biology. 1994;38(4):633–640. [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Developmental Cell. 2005;8(4):611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Kim BM, Miletich I, Mao J, McMahon AP, Sharpe PT, Shivdasani RA. Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen. Development. 2007;134(20):3603–3613. doi: 10.1242/dev.009308. Epub 2007 Sep 3612. [DOI] [PubMed] [Google Scholar]
- Krasner A, Wallace L, Thiagalingam A, Jones C, Lengauer C, Minahan L, Ma Y, Kalikin L, Feinberg AP, Jabs EW, Tunnacliffe A, Baylin SB, Ball DW, Nelkin BD. Cloning and chromosomal localization of the human BARX2 homeobox protein gene. Gene. 2000;250(12):171–180. doi: 10.1016/s0378-1119(00)00169-4. [DOI] [PubMed] [Google Scholar]
- Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126(9):1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- Langbein L, Schweizer J. Keratins of the human hair follicle. International Review of Cytology. 2005;243:1–78. doi: 10.1016/S0074-7696(05)43001-6. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Developmental Cell. 2005;9(6):855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Lewis DL, DeCamillis MA, Brunetti CR, Halder G, Kassner VA, Selegue JE, Higgs S, Carroll SB. Ectopic gene expression and homeotic transformations in arthropods using recombinant Sindbis viruses. Current Biology. 1999a;9(22):1279–1287. doi: 10.1016/s0960-9822(00)80049-4. [DOI] [PubMed] [Google Scholar]
- Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Research. 2000;2(3):158–169. doi: 10.1186/bcr49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MT, Ross S, Strickland PA, Snyder CJ, Daniel CW. Regulated expression patterns of IRX-2, an Iroquois-class homeobox gene, in the human breast. Cell & Tissue Research. 1999b;296(3):549–554. doi: 10.1007/s004410051316. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401(6750):279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. Journal of Neuroscience. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MF, Cheng HT, Kern MJ, Potter SS, Tran B, Diekwisch TG, Martin JF. prx-1 functions cooperatively with another paired-related homeobox gene, prx-2, to maintain cell fates within the craniofacial mesenchyme. Development. 1999a;126(3):495–504. doi: 10.1242/dev.126.3.495. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999b;401(6750):276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Ma L, Liu J, Wu T, Plikus M, Jiang TX, Bi Q, Liu YH, Muller-Rover S, Peters H, Sundberg JP, Maxson R, Maas RL, Chuong CM. ‘Cyclic alopecia’ in Msx2 mutants: defects in hair cycling and hair shaft differentiation. Development. 2003;130(2):379–389. doi: 10.1242/dev.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie A, Ferguson MW, Sharpe PT. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115(2):403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- Mackenzie A, Leeming GL, Jowett AK, Ferguson MW, Sharpe PT. The homeobox gene Hox 7.1 has specific regional and temporal expression patterns during early murine craniofacial embryogenesis, especially tooth development in vivo and in vitro. Development. 1991;111(2):269–285. doi: 10.1242/dev.111.2.269. [DOI] [PubMed] [Google Scholar]
- Mandier M, Neubuser A. FGF signaling is necessary for the specification of the odontogenic mesenchyme. Developmental Biology. 2001;240(2):548–559. doi: 10.1006/dbio.2001.0490. [DOI] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes & Development. 1995;9(10):1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Millar SE. Molecular mechanisms regulating hair follicle development. Journal of Investigative Dermatology. 2002;118(2):216–225. doi: 10.1046/j.0022-202x.2001.01670.x. [DOI] [PubMed] [Google Scholar]
- Mitchell JM, Hicklin DM, Doughty PM, Hicklin JH, Dickert JW, Jr, Tolbert SM, Peterkova R, Kern MJ. The Prx1 homeobox gene is critical for molar tooth morphogenesis. Journal of Dental Research. 2006;85(10):888–893. doi: 10.1177/154405910608501003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Angeli I, James C, Lendahl U, Sharpe PT. Role of Islet1 in the patterning of murine dentition. Development. 2003;130(18):4451–4460. doi: 10.1242/dev.00631. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Drouin J. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol. 2008;313(2):887–896. doi: 10.1016/j.ydbio.2007.10.055. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Mucchielli ML, Raffo S, Proust JP, Koopman P, Goridis C. Expression of the transcription factors Otlx2, Barx1 and Sox9 during mouse odontogenesis. European Journal of Oral Sciences. 1998;106(Suppl 1):112–116. doi: 10.1111/j.1600-0722.1998.tb02161.x. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(1):162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso MI, Mahon KA, Sargent TD. A Xenopus distal-less gene in transgenic mice: conserved regulation in distal limb epidermis and other sites of epithelial-mesenchymal interaction. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(9):3968–3972. doi: 10.1073/pnas.92.9.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Developmental Biology. 1997;189(2):275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- Noveen A, Jiang TX, Ting-Berreth SA, Chuong CM. Homeobox genes Msx-1 and Msx-2 are associated with induction and growth of skin appendages. Journal of Investigative Dermatology. 1995;104(5):711–719. doi: 10.1111/1523-1747.ep12606960. [DOI] [PubMed] [Google Scholar]
- Oliver RF, Jahoda CA. Dermal-epidermal interactions. Clinics in Dermatology. 1988;6(4):74–82. doi: 10.1016/0738-081x(88)90069-7. [DOI] [PubMed] [Google Scholar]
- Olson LE, Zhang J, Taylor H, Rose DW, Rosenfeld MG. Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3708–3713. doi: 10.1073/pnas.0500519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104(2):233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Phippard DJ, WeberHall SJ, Sharpe PT, Naylor MS, Jayatalake H, Maas R, Woo I, RobertsClark D, FrancisWest PH, Liu YH, Maxson R, Hill RE, Dale TC. Regulation of Msx-1, Msx-2, Bmp-2 and Bmp-4 during foetal and postnatal mammary gland development. Development. 1996;122(9):2729–2737. doi: 10.1242/dev.122.9.2729. [DOI] [PubMed] [Google Scholar]
- Potter CS, Peterson RL, Barth JL, Pruett ND, Jacobs DF, Kern MJ, Argraves WS, Sundberg JP, Awgulewitsch A. Evidence that the satin hair mutant gene Foxql is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. Journal of Biological Chemistry. 2006;281(39):29245–29255. doi: 10.1074/jbc.M603646200. [DOI] [PubMed] [Google Scholar]
- Price JA, Wright JT, Kula K, Bowden DW, Hart TC. A common DLX3 gene mutation is responsible for tricho-dento-osseous syndrome in Virginia and North Carolina families. Journal of Medical Genetics. 1998;35(10):825–828. doi: 10.1136/jmg.35.10.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett ND, Tkatchenko TV, Jave-Suarez L, Jacobs DF, Potter CS, Tkatchenko AV, Schweizer J, Awgulewitsch A. Krtap16, characterization of a new hair keratin-associated protein (KAP) gene complex on mouse chromosome 16 and evidence for regulation by Hoxc13. Journal of Biological Chemistry. 2004;279(49):51524–51533. doi: 10.1074/jbc.M404331200. [DOI] [PubMed] [Google Scholar]
- Puppin C, Puglisi F, Pellizzari L, Manfioletti G, Pestrin M, Pandolfi M, Piga A, Di Loreto C, Damante G. HEX expression and localization in normal mammary gland and breast carcinoma. BMC Cancer. 2006;6:192. doi: 10.1186/1471-2407-6-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Developmental Biology. 1997;185(2):165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes & Development. 1995;9(20):2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Qu S, Niswender KD, Ji Q, van der Meer R, Keeney D, Magnuson MA, Wisdom R. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997;124(20):3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- Qu S, Tucker SC, Ehrlich JS, Levorse JM, Flaherty LA, Wisdom R, Vogt TF. Mutations in mouse Aristaless-like4 cause Strong’s luxoid polydactyly. Development. 1998;125(14):2711–2721. doi: 10.1242/dev.125.14.2711. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265(5173):785–789. doi: 10.1126/science.7914031. see comment. [DOI] [PubMed] [Google Scholar]
- Raju K, Tang S, Dube ID, Kamel-Reid S, Bryce DM, Breitman ML. Characterization and developmental expression of Tlx-1, the murine homolog of HOX11. Mechanisms of Development. 1993;44(1):51–64. doi: 10.1016/0925-4773(93)90016-q. [DOI] [PubMed] [Google Scholar]
- Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development. 1995;121(4):1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312(5782):1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle RD, Ensini M, Nelson C, Tsuchida T, Jessell TM, Tabin C. Induction of the LIM homeobox gene Lmx1 by WNT7a establishes dorsoventral pattern in the vertebrate limb. Cell. 1995;83(4):631–640. doi: 10.1016/0092-8674(95)90103-5. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368(6473):747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Mahon KA. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mechanisms of Development. 1994;48(3):199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Sander G, Bawden CS, Hynd PI, Nesci A, Rogers G, Powell BC. Expression of the homeobox gene, Barx2, in wool follicle development. Journal of Investigative Dermatology. 2000;115(4):753–756. doi: 10.1046/j.1523-1747.2000.00122.x. [DOI] [PubMed] [Google Scholar]
- Satoh K, Ginsburg E, Vonderhaar BK. Msx-1 and Msx-2 in mammary gland development. Journal of Mammary Gland Biology & Neoplasia. 2004;9(2):195–205. doi: 10.1023/B:JOMG.0000037162.84758.b5. [DOI] [PubMed] [Google Scholar]
- Satoh K, Hovey RC, Malewski T, Warri A, Goldhar AS, Ginsburg E, Saito K, Lydon JP, Vonderhaar BK. Progesterone enhances branching morphogenesis in the mouse mammary gland by increased expression of Msx2. Oncogene. 2007;26(54):7526–7534. doi: 10.1038/sj.onc.1210555. [DOI] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nature Genetics. 2000;24(4):391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nature Genetics. 1994;6(4):348–356. doi: 10.1038/ng0494-348. see comment. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Paus R. Molecular principles of hair follicle induction and morphogenesis. Bioessays. 2005;27(3):247–261. doi: 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- Scott MP. A rational nomenclature for vertebrate homeobox (HOX) genes. Nucleic Acids Research. 1993;21(8):1687–1688. doi: 10.1093/nar/21.8.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nature Genetics. 1996;14(4):392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Shang L, Pruett ND, Awgulewitsch A. Hoxc12 expression pattern in developing and cycling murine hair follicles. Mechanisms of Development. 2002;113(2):207–210. doi: 10.1016/s0925-4773(02)00022-9. [DOI] [PubMed] [Google Scholar]
- Srebrow A, Friedmann Y, Ravanpay A, Daniel CW, Bissell MJ. Expression of Hoxa-1 and Hoxb-7 is regulated by extracellular matrix-dependent signals in mammary epithelial cells. Journal of Cellular Biochemistry. 1998;69(4):377–391. doi: 10.1002/(sici)1097-4644(19980615)69:4<377::aid-jcb1>3.0.co;2-k. erratum appears in J Cell Biochem 1998 Nov 1;71(2):310-2. [DOI] [PubMed] [Google Scholar]
- St Amand TR, Zhang Y, Semina EV, Zhao X, Hu Y, Nguyen L, Murray JC, Chen Y. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Developmental Biology. 2000;217(2):323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Stelnicki EJ, Komuves LG, Holmes D, Clavin W, Harrison MR, Adzick NS, Largman C. The human homeobox genes MSX-1, MSX-2, and MOX-1 are differentially expressed in the dermis and epidermis in fetal and adult skin. Differentiation. 1997;62(1):33–41. doi: 10.1046/j.1432-0436.1997.6210033.x. [DOI] [PubMed] [Google Scholar]
- Stelnicki EJ, Komuves LG, Kwong AO, Holmes D, Klein P, Rozenfeld S, Lawrence HJ, Adzick NS, Harrison M, Largman C. HOX homeobox genes exhibit spatial and temporal changes in expression during human skin development. Journal of Investigative Dermatology. 1998;110(2):110–115. doi: 10.1046/j.1523-1747.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- Sweeney E, Fryer A, Mountford R, Green A, Mclntosh I. Nail patella syndrome: a review of the phenotype aided by developmental biology. Journal of Medical Genetics. 2003;40(3):153–162. doi: 10.1136/jmg.40.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto DP, Rodriguez-Esteban C, Ryan AK, O’Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes & Development. 1999;13(4):484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102(4):451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brouwer A, Korving J, Martin JF, Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125(19):3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Liu JK, Rubenstein JL, Sharpe PT. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127(2):217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Porteus MH, Rubenstein JL, Sharpe PT. The spatial localization of Dlx-2 during tooth development. Connective Tissue Research. 1995;32(14):27–34. doi: 10.3109/03008209509013702. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Sharpe PT. Patterning of the murine dentition by homeobox genes. European Journal of Oral Sciences. 1998;106(Suppl 1):48–54. doi: 10.1111/j.1600-0722.1998.tb02153.x. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124(23):4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- Tissier-Seta JP, Mucchielli ML, Mark M, Mattei MG, Goridis C, Brunet JF. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mechanisms of Development. 1995;51(1):3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- Tkatchenko AV, Visconti RP, Shang L, Papenbrock T, Pruett ND, Ito T, Ogawa M, Awgulewitsch A. Overexpression of Hoxc13 in differentiating keratinocytes results in downregulation of a novel hair keratin gene cluster and alopecia. Development. 2001;128(9):1547–1558. doi: 10.1242/dev.128.9.1547. [DOI] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nature Reviews Genetics. 2004;5(7):499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Developmental Dynamics. 1998a;212(4):533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998b;282(5391):1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75(1):45–58. [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nature Genetics. 1996;13(4):417–421. doi: 10.1038/ng0896-417. see comment. [DOI] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Warnken W, Izpisua Belmonte JC. Dorsal cell fate specified by chick Lmx1 during vertebrate limb development. Nature. 1995;378(6558):716–720. doi: 10.1038/378716a0. erratum appears in Nature 1996 Feb 29;379(6568):848. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Tang Z, Elanko N, Walsh S, Twigg SR, Hurst JA, Wall SA, Chrzanowska KH, Maxson RE., Jr Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nature Genetics. 2000;24(4):387–390. doi: 10.1038/74224. see comment. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, Huang S, Westphal H. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):15002–15006. doi: 10.1073/pnas.96.26.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Stock D, Buchanan A, Weiss K. Expression of Dlx genes during the development of the murine dentition. Development Genes & Evolution. 2000;210(5):270–275. doi: 10.1007/s004270050314. [DOI] [PubMed] [Google Scholar]


